Monocytes are required for arteriogenesis after injury and one of their major roles is to produce VEGF; however, the mechanisms behind this have not been identified. Morrison et al. find a link between chemokine and integrin stimulation through Rac2 that involves myosin heavy chain to redistribute the RNA stabilizing protein HuR to augment VEGF expression.
Abstract
Myeloid cells are important contributors to arteriogenesis, but their key molecular triggers and cellular effectors are largely unknown. We report, in inflammatory monocytes, that the combination of chemokine receptor (CCR2) and adhesion receptor (β2 integrin) engagement leads to an interaction between activated Rac2 and Myosin 9 (Myh9), the heavy chain of Myosin IIA, resulting in augmented vascular endothelial growth factor A (VEGF-A) expression and induction of arteriogenesis. In human monocytes, CCL2 stimulation coupled to ICAM-1 adhesion led to rapid nuclear-to-cytosolic translocation of the RNA-binding protein HuR. This activation of HuR and its stabilization of VEGF-A mRNA were Rac2-dependent, and proteomic analysis for Rac2 interactors identified the 226 kD protein Myh9. The level of induced Rac2–Myh9 interaction strongly correlated with the degree of HuR translocation. CCL2-coupled ICAM-1 adhesion-driven HuR translocation and consequent VEGF-A mRNA stabilization were absent in Myh9−/− macrophages. Macrophage VEGF-A production, ischemic tissue VEGF-A levels, and flow recovery to hind limb ischemia were impaired in myeloid-specific Myh9−/− mice, despite preserved macrophage recruitment to the ischemic muscle. Micro-CT arteriography determined the impairment to be defective induced arteriogenesis, whereas developmental vasculogenesis was unaffected. These results place the macrophage at the center of ischemia-induced arteriogenesis, and they establish a novel role for Myosin IIA in signal transduction events modulating VEGF-A expression in tissue.
Multiple cell types, including endothelial cells, smooth muscle cells, and BM-derived cells, contribute to developmental and inducible arteriogenesis (Carmeliet and Jain, 2011). Although some data exist for cells of hematopoietic lineage incorporating into neovascular structures, there is evidence that factors secreted by macrophages may contribute to arteriogenesis. Many of these factors are encoded by mRNAs bearing AU-rich elements (AREs) that confer intrinsically short half-lives, limiting protein production. RNA-binding proteins (RBPs), and in particular HuR, have been shown to bind to and stabilize ARE-bearing transcripts, prolonging mRNA half-life and increasing translation, in part through a complex interplay between RBPs and noncoding RNAs (van Kouwenhove et al., 2011). Engagement of leukocyte β2 integrin (LFA-1 and Mac-1) triggers a rapid signaling molecular switch that results in HuR-dependent stabilization of multiple transcripts, including TNF, IFN-γ, GM-CSF, IL-3, vascular endothelial growth factor A (VEGF-A), and MMP-9 (Wang et al., 2006; Ramgolam et al., 2010; Zhang et al., 2012). In the setting of ischemia, chemokine release and endothelial activation result in recruitment of inflammatory monocytes through β2 integrin engagement, circulatory arrest, and transmigration (Kuziel et al., 1997; Lu et al., 1998; Gerszten et al., 1999). The major monocyte chemokine CCL2 is a well-established β2 integrin activator and vital to flow recovery in animal models of hind limb ischemia (Carr et al., 1996; Ito et al., 1997; van Royen et al., 2003). Thus, we identified that CCL2 stimulation has the potential to promote and couple with integrin adhesion to activate HuR-dependent pathways in primary cells. Disrupting combinatorial signaling of CCL2-CCR2 engagement with β2 integrin adhesion in vivo has resulted in monocyte adhesion, and migratory and recruitment defects, to which vascular phenotypes have largely been attributed (Hoefer et al., 2004; Voskuil et al., 2004; Waeckel et al., 2005; Shireman et al., 2007). The inability to uncouple adhesion, migration, and recruitment from membrane receptor signal transduction influencing gene expression limits a more thorough in vivo assessment of the molecular mechanisms behind macrophage-driven arteriogenesis. We sought to define critical intermediaries that might regulate this integrin-driven, HuR-dependent switch during in vivo neovascular responses that were independent of adhesion and migration.
β2 integrin engagement leads to activation (GTP loading) of the Rho family GTPases Rac1 and Rac2, key modulators of actin cytoskeletal networks and membrane signal transduction events (Etienne-Manneville and Hall, 2002). Constitutively active mutants of Rac promote stabilization of a chimeric, ARE-bearing RNA reporter when expressed in cell lines (Ramgolam et al., 2010). Because Rac2 expression is relatively restricted to cells of hematopoietic lineage (Shirsat et al., 1990; Gu et al., 2003), we sought novel Rac2 interactors that, as a consequence of integrin adhesion, may drive the noted posttranscriptional switch in primary macrophages. Here, we identify myosin 9 (Myh9), the heavy chain of nonmuscle myosin IIA (NMMIIA), as a critical Rac2 interactor in this mRNA stabilizing axis. NMMIIA is an actin-binding protein and extended component of integrin-based adhesion complexes important for cell spreading and motility (Vicente-Manzanares et al., 2009). As a form of inside-out signaling to enhance adhesion avidity, NMMIIA mediates nascent adhesion maturation through membrane integrin clustering (Humphries et al., 2007). Force generated through NMMIIA can result in membrane protein conformational changes that expose cryptic critical adhesion sites (Sawada et al., 2006; del Rio et al., 2009; Friedland et al., 2009). Despite this large body of data regarding NMMIIA and integrins in cytoskeletal dynamics and cell adhesion, little is known about how integrin-based signaling networks affecting gene expression depend on nonmuscle myosins. This study demonstrates that macrophage NMMIIA is critical to integrin signal transduction events that activate HuR and augment expression of VEGF-A during adult ischemia-induced arteriogenesis in a manner independent of developmental vasculogenesis.
RESULTS AND DISCUSSION
Chemokine-coupled β2 integrin engagement induces rapid HuR translocation in human inflammatory monocytes
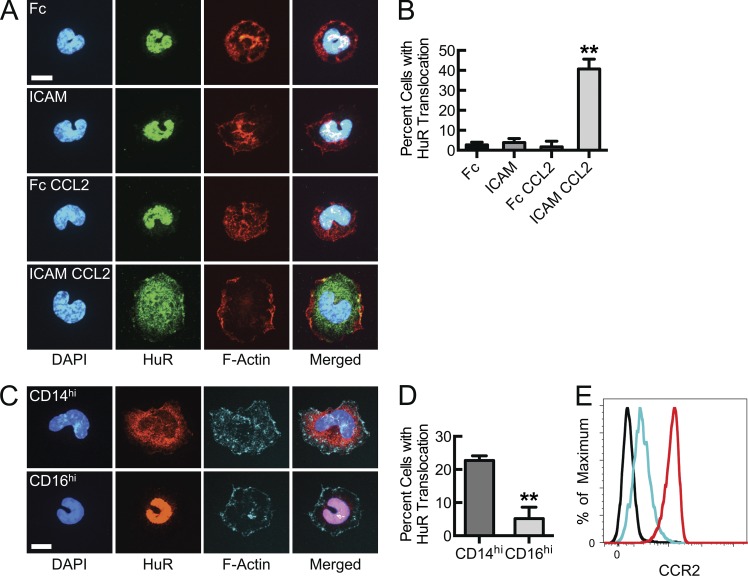
Nuclear-to-cytosolic translocation of HuR is a marker of, and required for, its mRNA stabilizing effect. In our previous studies, either ligand-mimetic anti–LFA-1 antibodies or adhesion to ICAM-1 in the presence of supraphysiologic divalent cation concentrations were used to promote integrin activation and signaling. Although this does lead to integrin affinity modulation and productive ICAM-1 adhesion, stimulation with the monocyte chemokine CCL2 represents a physiological mechanism that during inflammation can promote and couple with β2 integrin engagement to drive signaling and HuR-dependent transcript stabilization. Freshly isolated human peripheral blood monocytes adhered to and spread on recombinant ICAM-1. However, rapid and robust HuR translocation occurred only when a CCL2 stimulus was coupled with β2 integrin engagement on ICAM-1 (Fig. 1, A and B). Peripheral monocytes are heterogeneous and the maximal stimulus (CCL2 plus ICAM-1) resulted in robust HuR translocation in <50% of cells, suggesting that only a subset of monocytes may be capable of responding. Thus, to address whether inflammatory (CD14hi) monocytes (recruited to sites of chemokine release and endothelial activation) are more likely to activate this posttranscriptional expression program upon ICAM-1 adhesion than “atypical” or “patrolling” (CD16hi) monocytes, freshly isolated cells were sorted based on these two markers. Although the overall responsiveness was less after the duration of sorting, the HuR translocation response was almost exclusively found within the CD14hiCD16lo inflammatory monocyte population (Fig. 1, C and D). In part, this may be a consequence of greater CCR2 expression levels on the inflammatory monocytes (Fig. 1 E and Fig. S1; Geissmann et al., 2003; Swirski et al., 2009). This is the first report that chemokine signaling can couple with integrin engagement to activate HuR in primary human monocytes and supports the likely scenario that, in response to chemokine release and endothelial activation, inflammatory monocytes are recruited, adhere, and transmigrate through integrin engagement, after which there is rapid, enhanced production of multiple factors.
Figure 1.
β2 Integrin engagement (adhesion to ICAM-1) of CCL2-stimulated inflammatory monocytes results in HuR nuclear-to-cytosolic translocation. (A) Confocal micrographs of human peripheral blood monocytes adhered to immobilized ICAM-1 or control Fcγ fragment in the presence or absence of 50 ng/ml CCL2 for 30 min, after which HuR was immunostained to detect induced translocation. (B) Quantification of cells in A with nuclear-to-cytosolic HuR translocation, using automated confocal tracking and threshold-based, algorithmic analysis. (**, P < 0.05, n = 3). (C) Confocal micrographs of inflammatory or patrolling human peripheral blood monocytes, sorted based upon CD14hi and CD16hi, respectively. Cells were CCL2-stimulated and ICAM-bound, followed by HuR immunostaining for translocation analysis. (D) Quantification of cells in C with nuclear-to-cytosolic HuR translocation (**, P = 0.009, n = 3). (E) Relative CCR2 surface expression determined for NK cells (black), CD14hi monocytes (red), and CD16hi monocytes (cyan; n = 4). Bars, 7 µm. Data are representative of at least 3 independent experiments. Quantitative data are displayed as mean ± SEM.
Chemokine-coupled β2 integrin engagement triggers Rac2-dependent VEGF-A mRNA stabilization and a Rac2–Myh9 interaction
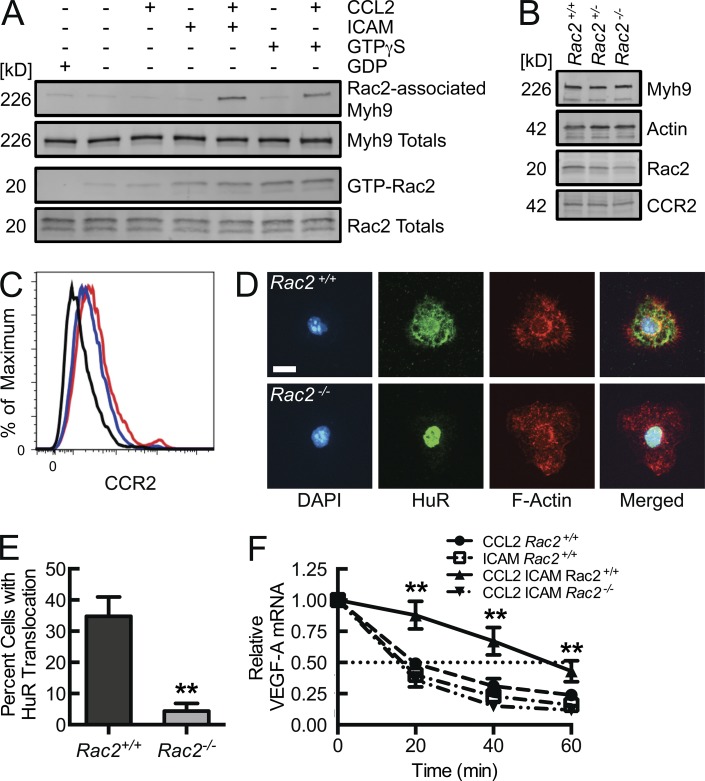
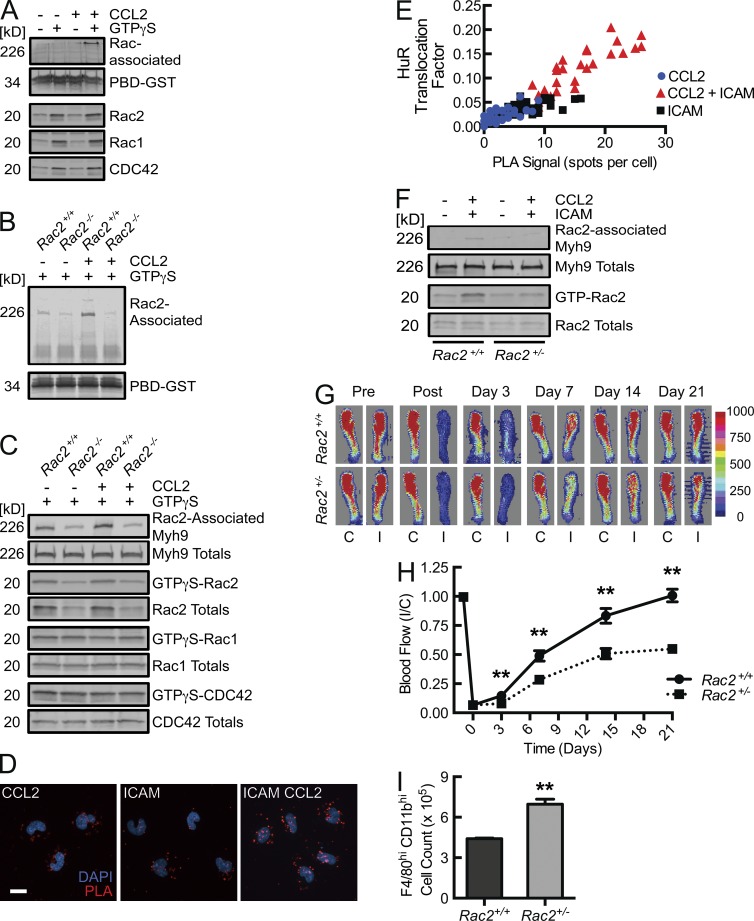
Although β2 integrin engagement does trigger Rac2 activation (GTP-loading), as seen in a p21-activated kinase (PAK) binding domain (PBD) affinity pulldown assay (Fig. 2 A, lane 4), it is apparent that this stimulus alone is insufficient to promote HuR translocation. BM-derived macrophages (BMDMs) isolated from Rac2 gene-deleted mice maintain WT levels of CCR2 (Fig. 2 B). There is also no significant difference in thioglycollate-elicited peritoneal macrophage (CD11bhiF4/80hi) surface expression of CCR2 between Rac2+/+ and Rac2−/− mice (Fig. 2 C). However, BMDMs from Rac2−/− mice were not able to translocate HuR in the setting of CCL2-coupled, ICAM-1 adhesion despite adequate adhesion and cell spreading (Fig. 2, D and E). Consequential to the effects on HuR translocation, the threefold increase in VEGF-A mRNA half-life induced by the combination of CCL2 stimulation and β2 integrin engagement observed in BMDMs from Rac2+/+ mice was lost in BMDMs from Rac2−/− mice (Fig. 2 F). The HuR translocation and VEGF-A mRNA decay analyses support that integrin signal transduction leading to HuR activation depends on Rac2 as an intermediary. Thus, in efforts to define novel Rac2 interactors that trigger this posttranscriptional switch, the screening activation stimulus included both Rac2 activation (through either integrin engagement or, as a surrogate, Rac2 loading with nonhydrolyzable GTPγS) and CCR2 engagement. In the human monocytic cell line THP-1, upon GTPγS loading, Rac2, Rac1, and Cdc42 were detected in the PBD pulldown material (Fig. 3 A). With the additional CCL2 stimulus, a 226 kD protein was detected in the PBD pulldown material by Coomassie staining (Fig. 3 A). This stimulus-driven protein association was Rac2-specific, as it was undetectable in pulldown material from CCL2-activated, GTPγS-loaded Rac2−/− BMDM lysates (Fig. 3, B and C). By liquid chromatography mass spectrometry analysis of in-gel trypsin-digested material, this 226-kD Rac2-associated protein was identified as Myh9, the heavy chain of NMMIIA. 61.5% of the Myh9 protein sequence was mapped by peptides identified from this tryptic digestion (Fig. S2). This proteomic identification was validated in Myh9 immunoblots of PBD pulldown material, where Myh9 was detected in lysates from GTPγS-loaded CCL2-treated Rac2+/+, but not from Rac2−/− BMDMs or from GTPγS-loaded non–CCL2-treated Rac2+/+ BMDMs (Fig. 3 C). This again demonstrates Rac2 specificity, as Rac1-GTPγS and Cdc42-GTPγS were both present in pulldowns of Rac2−/− BMDMs but without associated Myh9. Additional validation was obtained by the presence of Myh9 in PBD pulldown material obtained from WT BMDMs that were CCL2-stimulated and adhered to ICAM-1, but not from cells singly CCL2-stimulated or ICAM-1–bound, despite the latter leading to Rac2 activation (Fig. 2 A).
Figure 2.
Chemokine-coupled β2 integrin engagement drives Rac2 activation (GTP-loading), Rac2-dependent HuR translocation, and consequent VEGF mRNA stabilization. (A) Rac2 and Myh9 immunoblots from lysates of WT BMDMs adhered to immobilized ICAM-1 or Fcγ fragment control for 5 min at 37°C in the presence or absence of 50 ng/ml CCL2, after which GTP-Rac2 was affinity precipitated by PBD pulldown (n = 4). (B) Rac2, Myh9, Actin, and CCR2 immunoblots from lysates of Rac2+/+, Rac2+/−, and Rac2−/− BMDMs (n = 3). (C) Relative CCR2 surface expression on thioglycollate-elicited peritoneal macrophages (CD11bhiF4/80hi) from Rac2+/+ (red) and Rac2−/− (blue) mice (isotype control; black, n = 4). (D) Confocal micrographs of Rac2+/+ and Rac2−/− BMDMs, CCL2-stimulated and ICAM-bound for 30 min, followed by HuR immunostaining for translocation analysis. Bar, 10 µm. (E) Quantification of percentage of cells in D with HuR nuclear-to-cytosolic translocation (**, P < 0.002; n = 3). (F) VEGF-A mRNA decay assays, using the indicated BMDMs, adhered to immobilized ICAM-1 or Fcγ fragment control in the presence or absence of CCL2 for 30 min, after which transcription was arrested with 10 mg/ml actinomycin d at time 0, and RNA collected at the noted time points for VEGF-A mRNA quantification by RT-qPCR (**, P = 0.0001; n = 5). Thioglycollate-elicited peritoneal macrophage data are from two independent experiments, each with two mice per treatment group. Remaining data are representative of at least 3 independent experiments. Quantitative data are displayed as mean ± SEM.
Figure 3.
Chemokine-coupled integrin-driven activation of Rac2 leads to a unique Rac2-Myh9 complex that correlates with HuR translocation, and in vivo neovascular responses depend on efficient integrin activation of Rac2. (A) THP-1 cells in the presence or absence of CCL2 (50 ng/ml × 10 min) were lysed and GTPγS (or vehicle control)-loaded, followed by PBD affinity pulldown and Coomassie staining (top) or immunoblotting for the indicated Rho family GTPases (bottom; n = 3). (B) Chemokine stimulation and GTP loading as in A, in Rac2+/+ or Rac2−/− BMDMs, followed by PBD affinity pulldown and Coomassie staining for the 226 kD protein noted above (n = 3). (C) Immunoblots from lysates of Rac2+/+ and Rac2−/− BMDMs, which were CCL2-treated for 10 min, GTPγS (100 µM)-loaded for 10 min, and affinity precipitated with PBD beads to determine the association of Myh9 with the indicated Rac proteins (n = 4). (D) Confocal micrographs of human peripheral blood monocytes adhered to ICAM-1 or Fcγ fragment control in the presence or absence of 50 ng/ml CCL2 for 10 min, followed by PLA reaction for Rac2–Myh9 interaction (red signal) and DAPI (cyan) staining (n = 3). Bar, 15 µm. (E) PLA-HuR translocation correlation graph. THP-1 HuR-YFP stable transfectants were ICAM-1– or poly-l-lysine control-bound in the presence or absence of CCL2, after which PLA and HuR translocation analysis were performed (r = 0.9, P < 0.0001, n = 3). (F) Rac2 and Myh9 immunoblots from lysates of Rac2+/+ or Rac2+/− BMDMs adhered to immobilized ICAM-1 in the presence of 50 ng/ml CCL2, after which Rac2 was affinity precipitated by PBD pulldown (n = 3). (G and H) Laser Doppler scan images (G) of flow in the ischemic (I) and contralateral control (C) hind limb of Rac2+/+ and Rac2+/− mice at the indicated time points before and after femoral artery ligation with quantitative blood flow analysis (H; **, P < 0.0025; n = 10). (I) Quantification of CD11bhi F4/80hi macrophages isolated from collagenase-treated ischemic muscle tissue at day 3 after femoral artery ligation (**, P < 0.0001; n = 6). Animal data are from 2 independent experiments, each with 3 mice per treatment group. Remaining data are representative of at least 3 independent experiments. Quantitative data are displayed as mean ± SEM.
Strength of Rac2–Myh9 interaction signal correlates with extent of HuR translocation
The proximity ligation assay (PLA) was used to identify the native, intracellular Rac2–Myh9 interaction, and to quantify whether there is a relationship between this interaction and the noted downstream posttranscriptional switch. Fig. 3 D displays an augmented PLA signal in CCL2-stimulated ICAM-1–bound human peripheral blood monocytes above either stimulus alone. When performed in THP-1 cells stably expressing basal, nuclear HuR-YFP, we were able to quantify and correlate the PLA signal with induced HuR translocation. Fig. 3 E displays a strong, direct correlation (r = 0.90, P < 0.0001) between the induced PLA signal and HuR translocation, with the strongest PLA signal and HuR translocation in the combined CCL2-stimulated ICAM-1–bound cells. Although PLA does not definitively determine direct protein–protein interactions, Rac2 and Myh9 must be within 40 nm to generate a PLA signal, consistent with either a direct interaction or a close association within a larger adhesion-based signaling complex (Fredriksson et al., 2002, 2007; Söderberg et al., 2006). The correlation between PLA signal and degree of HuR translocation supports that the Rac2 and Myh9 interaction is required for the signal transduction events leading to the activation of the HuR-dependent posttranscriptional switch.
Flow recovery to hind limb ischemia is dependent on efficient integrin-induced Rac2 activation
Rac2 gene deletion leads to decreased flow recovery after femoral artery ligation in a manner independent of collateralization potential and collateral maturation (De et al., 2009; Distasi et al., 2009). Rac2 expression is gene dose dependent, yet Rac2+/− heterozygote animals have not been demonstrated to have an in vivo phenotype to date (Roberts et al., 1999; Li et al., 2002). Rac2+/− neutrophils are known to have minimal defects in neutrophil superoxide production; however, heterozygote neutrophils have moderate reductions of FMLP-stimulated F-actin formation, chemotaxis, and NADPH oxidase activity. We hypothesized that with a 50% reduction in Rac2 (Fig. 2 B), macrophages would demonstrate a decreased β2 integrin–induced Rac2 activation and consequent Myh9 interaction, as the reduced availability of Rac2 would fall below threshold levels required to promote such signaling. Indeed, BMDMs from Rac2+/− mice demonstrated a deficiency in CCL2-coupled, ICAM adhesion-driven Rac2 activation and consequent Myh9 association (Fig. 3 F). If β2 integrin–induced Rac2 activation were required for macrophage VEGF-A mRNA stabilization and subsequent vasculogenic responses, we would expect a defect in the femoral artery ligation model of hind limb ischemia. Laser Doppler-imaged flow recovery to hind limb ischemia was reduced at day 3 after ligation and reached a 50% reduction by 3 wk (Fig. 3, G and H). This was not a consequence of impaired monocyte/macrophage recruitment and localization, as numbers of CD11bhiF4/80hi cells recovered from day 3 collagenase-treated ischemic muscle tissue were not reduced (Fig. 3 I). This is consistent with intact β2 integrin engagement (required for localization) but impaired integrin-mediated Rac2 activation and Myh9 association (required for maximal VEGF-A production) as seen in vitro, allowing us to segregate the membrane receptor-dependent adhesion from this important integrin-induced signaling cascade in vivo.
Macrophage HuR translocation and consequent VEGF-A mRNA stabilization depend on Myh9
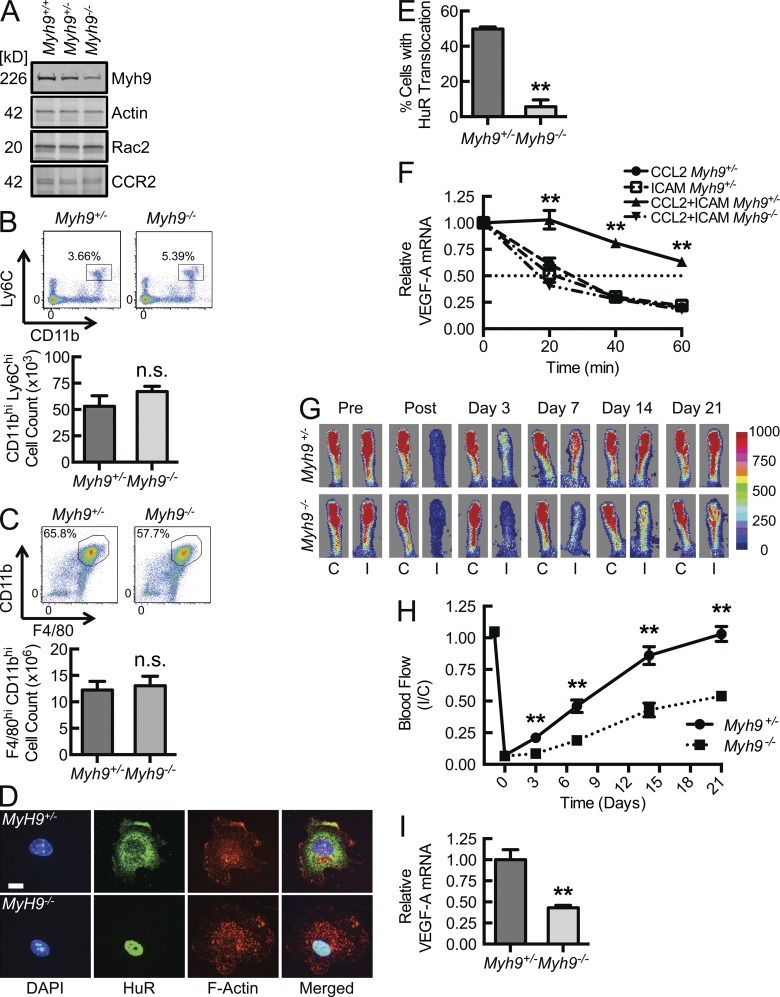
Myh9, in its rapidly induced association with Rac2, appears to be a key component of a chemokine-coupled integrin-induced pathway, resulting in labile mRNA stabilization in macrophages. To address this requirement for Myh9 in vitro and in vivo, myeloid-specific Myh9 gene-deleted mice were generated, using Myh9 gene-targeted and Lysozyme M–Cre (LysM+/Cre) mice. NMMIIA heavy chain levels were 20 and 80% of WT (Myh9+/+) levels in homozygous (LysM+/CreMyh9fl/fl, herein Myh9−/−) and heterozygous (LysM+/CreMyh9+/fl, herein Myh9+/−) gene-deleted mice, respectively (Fig. 4 A). Neither Rac2 nor Myh9 gene deletion affects each other’s or CCR2’s expression in BMDMs (Figs. 2 B and 4 A). Myeloid Myh9−/− animals were of similar size and body weight, and appeared to develop normally, compared with their heterozygous littermates. Myh9−/− mice did not have a deficiency in peripheral, circulating inflammatory monocyte levels, as determined by CD11bhiLy6Chi flow cytometric analysis (Fig. 4 B; Geissmann et al., 2003; Swirski et al., 2009). Most importantly for our in vivo studies, similar numbers of macrophages were recruited to an inflammatory thioglycollate-induced peritonitis (Fig. 4 C). This is consistent with a recent report that Myh9-deficient cells can migrate efficiently despite diminished traction forces at the cell periphery (Jorrisch et al., 2013). Thus, a myeloid-specific Myh9−/− mouse phenotype has the potential to reflect a deficient chemokine-coupled integrin-induced signaling cascade rather than just impaired myeloid mobility/migration.
Figure 4.
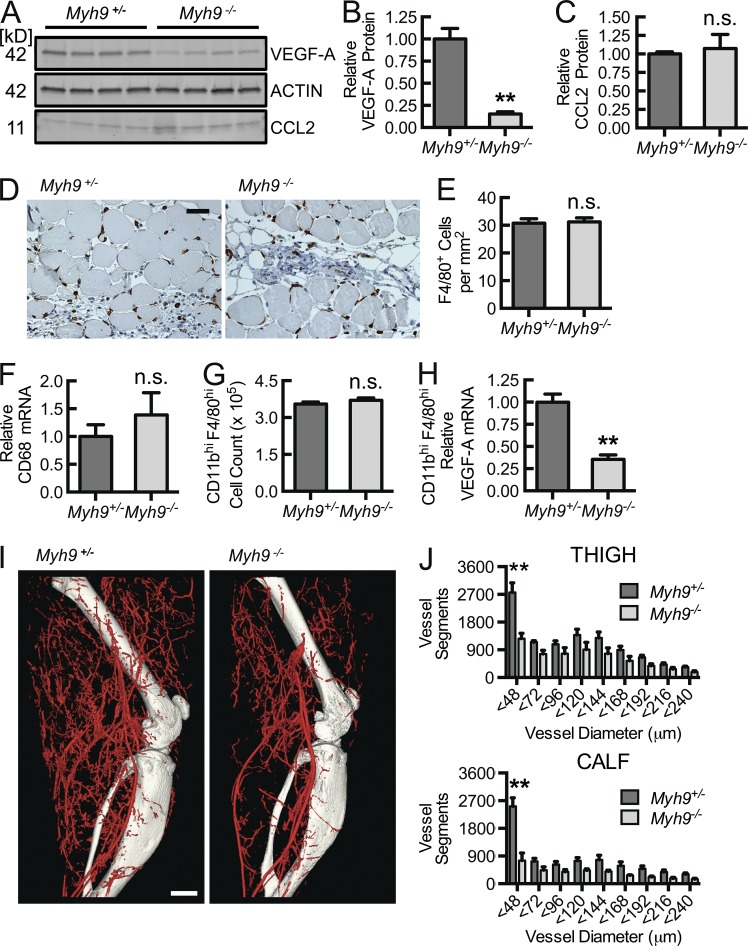
Macrophage HuR translocation with consequent VEGF-A mRNA stabilization, flow recovery during hind limb ischemia, and tissue VEGF-A mRNA levels depend on myeloid Myh9. (A) Myh9, Actin, Rac2, and CCR2 immunoblots of lysates obtained from BMDMs isolated from LysM+/creMyh9+/+(Myh9 +/+), LysM+/creMyh9+/fl (Myh9 +/−), and LysM+/creMyh9 fl/fl(Myh9 −/−) mice (n = 3). (B) Flow cytometric CD11b and Ly6C phenotyping (top) of density gradient–enriched peripheral blood mononuclear cells obtained from myeloid cell–specific Myh9+/− and Myh9−/− mice along with quantification of absolute cell count per ml blood (lower panel, n = 4). (C) Flow cytometric CD11b and F4/80 phenotyping (top) of cells recovered from the peritoneal cavity 3 d after initiation of thioglycollate-induced peritonitis in myeloid cell–specific Myh9+/− and Myh9−/− mice along with quantification of absolute cell count per ml blood (bottom, n = 5). (D) Confocal micrographs of Myh9+/− and Myh9−/− BMDMs, CCL2-stimulated and ICAM-bound, followed by HuR immunostaining for translocation analysis. Bar, 10 µm. (E) Quantification of percentage of cells in D with HuR nuclear-to-cytosolic translocation (**, P < 0.002; n = 3). (F) VEGF-A mRNA decay assays, using the indicated BMDMs, adhered to immobilized ICAM-1 or Fcγ fragment control in the presence or absence of CCL2 for 30 min, after which transcription was arrested with 10 mg/ml actinomycin (time 0), and RNA collected at the noted time points for VEGF-A mRNA quantification by RT-qPCR (**, P = 0.0001; n = 5). (G and H) Laser Doppler scan images (G) of flow in the ischemic (I) and contralateral control (C) hind limb of Myh9+/− and Myh9−/− mice at the indicated time points before and after femoral artery ligation with quantitative blood flow analysis (H; **, P < 0.0006; n = 7). (I) Real-time PCR quantification of VEGF-A mRNA in thigh (adductor) muscle tissue obtained 3 d after femoral artery ligation (**, P = 0.003; n = 4). Animal data are from 3 independent experiments, each with 2–3 mice per treatment group. Remaining data are representative of at least 3 independent experiments. Quantitative data are displayed as mean ± SEM.
As observed with Rac2−/− BMDMs (Fig. 2, C and D), CCL2-coupled β2 integrin engagement failed to induce HuR translocation in BMDMs isolated from myeloid-specific Myh9−/− mice, despite adequate cell spreading (Fig. 4, D and E). This is further reflected in VEGF-A mRNA decay assays, in which the threefold increase in VEGF-A mRNA half-life induced by the combination of CCL2 stimulation and adhesion to ICAM-1in Myh9+/− BMDMs is lost in Myh9−/− BMDMs (Fig. 4 F). The in vitro adhesion data are consistent with a chemokine–integrin signal transduction axis that depends on Myh9, resulting in HuR nuclear-to-cytosolic translocation and labile VEGF mRNA stabilization.
VEGF-A–induced arteriogenesis and flow recovery to hind limb ischemia is myeloid MyH9-dependent
If Myh9 were required for inflammation-based (chemokine plus integrin signaling) VEGF-A mRNA stabilization, we would expect a defect in macrophage-dependent vascular responses that require leukocyte recruitment and tissue localization, and so we returned to the femoral artery ligation model. 3 d after femoral artery ligation, there is a significant reduction in laser Doppler-imaged flow recovery in the myeloid-specific Myh9−/− mice, compared with Myh9+/− littermate controls, and this early difference persists as a 50% decrease in perfusion recovery out to 21 d (Fig. 4, G and H). There was no difference in absolute perfusion at baseline nor in the non-ischemic limbs throughout the analysis, confirming that the relative difference is driven by a macrophage-dependent absolute flow recovery deficiency in the ischemic limbs. At day 3 after ligation, Myh9−/− ischemic thigh muscle tissue VEGF-A mRNA levels were 40% that of control, with a consequent 80% reduction in VEGF-A protein (Fig. 4 I; Fig. 5, A and B). Of note, CCL2 levels were comparable in ischemic tissue homogenates from both Myh9−/− and Myh9+/− mice (Fig. 5, A and C). This difference is not a consequence of reduced macrophage recruitment and localization to the ischemic limb, as quantity of F4/80+ cells by immunohistochemistry and relative CD68 mRNA levels in ischemic tissue homogenates are not decreased in Myh9−/− mice (Fig. 5, D–F). Moreover, CD11bhiF4/80hi cells isolated from collagenase-treated ischemic muscle tissue at day 3 reflected no difference in early recruitment of macrophages between Myh9−/− and Myh9+/− mice. However, there was a 70% reduction in Myh9−/− macrophage VEGF-A mRNA expression, demonstrating the global tissue reduction in VEGF-A protein to be a consequence of the macrophage VEGF-A expression defect (Fig. 5, G and H).
Figure 5.
Despite adequate macrophage recruitment to ischemic muscle tissue, macrophage Myh9 gene deletion results in decreased VEGF-A protein in muscle tissue, reduced macrophage VEGF-A expression, and decreased arteriogenic response during hind limb ischemia. (A–C) VEGF-A and CCL2 immunoblots (A) from corresponding muscle tissue in H with each lane representing a separate animal along with quantification (B and C) of β-actin–normalized protein levels (**, P < 0.0005; n = 4). (D and E) Micrographs of 3 d after ligation thigh muscle tissue sections immunostained for macrophages (D; F4/80) and quantified as F4/80+ cells per mm2 (E; n = 4). Bar, 50 µm. (F) Real-time PCR quantification of GAPDH-normalized CD68 mRNA in thigh muscle tissue obtained 3 d after femoral artery ligation (n = 4). (G) Quantification of CD11bhi F4/80hi macrophages isolated from collagenase-treated ischemic muscle tissue at day 3 after femoral artery ligation (n = 6). (H) Real-time PCR quantification of VEGF-A mRNA expression in CD11bhi F4/80hi macrophages isolated from collagenase-treated ischemic muscle tissue at day 3 after femoral artery ligation (**, P < 0.0001; n = 6). (I and J) Micro-CT arteriograms 14 d after femoral artery ligation (I; bar, 1,000 µm), with segmental quantification (J) as total number of specified diameter vascular structures in the adductor (thigh) and gastrocnemius (calf; **, P < 0.05; n = 5). Animal data are from 2–3 independent experiments, each with 2 mice per treatment group. Quantitative data are displayed as mean ± SEM.
On day 14 after ligation, we performed micro-computed tomography (micro-CT) arteriography on a subset of mice to quantify arteriogenesis as a consequence of early macrophage VEGF-A expression in the ischemic tissue. Fig. 5 I displays micro-CT angiograms at the thigh (adductor) and calf (soleus, gastrocnemius) levels. Quantitative segmentation analysis demonstrates that there was a trend of decreased arteries overall with significantly fewer small arteries (<48 µm) at both the thigh and calf levels in Myh9−/− mice (Fig. 5 J). This defines the macrophage, and its expression of Myh9, as a critical cell responder and molecular effector, respectively, in ischemia-induced arteriogenesis. These results confirm that Myosin IIA, a molecule involved in macrophage integrin-triggered signal transduction leading to VEGF-A mRNA stabilization and enhanced protein production, is critical to tissue ischemia–driven arteriogenesis.
Normal developmental vasculogenesis appears independent of myeloid MyH9 expression
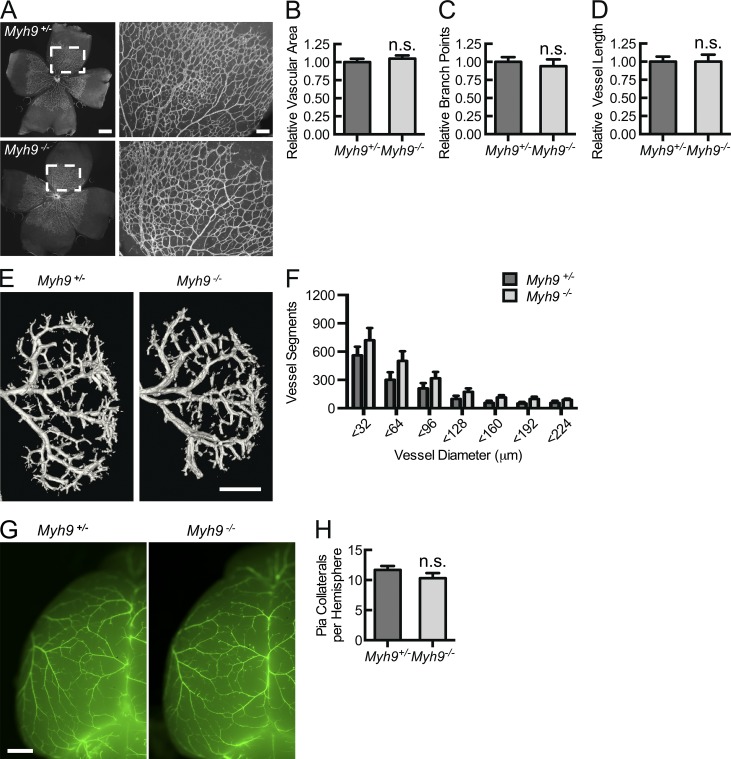
Because the impairment in flow recovery was strongly evident at an early time point (day 3) and there was a trend toward decreased arteries overall in the ischemic limb, we addressed whether macrophage Myh9 deletion confers differences in developmental vasculogenesis by studying baseline angiogenesis, arteriogenesis, and arteriolar collateral formation. Quantitative measures of postnatal retinal angiogenesis, renal arteriogenesis, or pial arterial collateral networks have been established (Jones et al., 2008; del Toro et al., 2010; Pitulescu et al., 2010; Tirziu et al., 2012). In the developmental model of retinal capillary angiogenesis, analyzed at postnatal day 4 (P4), there was no difference in relative vessel area, vessel branch points, and vessel length between Myh9−/− and Myh9+/− pups (Fig. 6, A–D). Micro-CT arteriograms at P7 failed to display baseline differences in development of the kidney arterial system (Fig. 6, E and F). Pial collateral artery quantification using smooth muscle actin–based imaging at P4 demonstrated an equal number of collateral arteries per cerebral hemisphere in Myh9−/− and Myh9+/− pups (Fig. 6, G and H). Thus, there were no detectable deficiencies in developmental capillary angiogenesis, arteriogenesis, and arterial collateral network formation in myeloid Myh9-deleted mice. Although this strengthens our inflammation-based, ischemia-induced arteriogenesis data, we were surprised by these results, as resident-tissue macrophages have been shown to play a role in developmental angiogenesis (Fantin et al., 2010). However, early resident macrophages are derived from the yolk sac, predating liver hematopoiesis at embryonic day 10.5 (E10.5), when monocyte-derived macrophages first appear and Lysozyme M expression begins (Clausen et al., 1999; Lichanska et al., 1999; Fantin et al., 2010). Also, macrophages are involved in capillary development, downstream of VEGF-A–mediated tip cell induction, facilitating tip cell fusion to capillary networks in a VEGF-A–independent manner, leading to increased vessel intersection and capillary network complexity (Fantin et al., 2010). It is likely that both the early timing of the tissue macrophage role (relative to Lysozyme M promoter activity for Cre expression) and VEGF-A–independent effects lead to the absence of an apparent developmental phenotype in these Myh9−/− models.
Figure 6.
Macrophage Myh9 is not required for developmental vasculogenesis by measures of angiogenesis, arteriogenesis, and collateral artery formation. (A–D) Isolectin B4 fluorescent micrographs of whole-mounted retina at P4 (bar, 500 µm), with magnified images (bar, 100 µm) of boxed areas (A), allowing for quantification of relative (to Myh9+/− controls) vascular area (B), relative vessel branch points (C), and relative vessel length (D; n = 6). (E and F) Micro-CT renal arteriograms at P7 (E; bar, 750 µm), quantified (F) as total number of vascular structures of specified diameters (n = 5). (G and H) Cy3-SMA immunostaining of brain whole mount sections (G; bar, 500 µm), displaying pial collateral arteries at P4, quantified (H) as mean number of collaterals per hemisphere (n = 7). Data are representative of at least 3 independent experiments with 2–3 mice in each treatment group. Quantitative data are displayed as mean ± SEM.
In summary, our results add a new mechanistic dimension to the cellular contributors and soluble factors involved in arteriogenesis, defining an inflammatory macrophage Myh9-dependent signaling cascade that drives VEGF-A production in tissue with consequent arteriogenesis. In the setting of ischemia (or other perturbations of tissue homeostasis, such as disturbed shear stress or wounding), chemokine release (CCL2) and endothelial activation (ICAM-1 augmentation) result in recruitment of inflammatory monocytes, which undergo phenotypic modulation of secreted factors through a Rac2–Myh9 interaction-dependent posttranscriptional switch. This switch is certainly required for stabilization of VEGF-A mRNA with consequent enhanced translation and secretion of VEGF-A, a critical mediator of arteriogenesis. It is highly likely that transcripts encoding other molecular mediators of arteriogenesis are also affected. That is, VEGF-A is absolutely required, but we believe this to be a powerful switch, affecting a larger gene expression program. Although our data support a role for this mechanism in adult ischemic (inflammatory) arteriogenic responses but not in developmental vasculogenesis, it remains possible that earlier embryonic deletion of macrophage Myh9 may result in a developmental phenotype. This integrin–Rac2–Myh9 axis highlights the interdependence of signal transduction and cytoskeletal network dynamics, as the major phenotype was reflected by the loss of a signaling cascade that modulates gene expression rather than the loss of macrophage adhesion and cell spreading or migration and recruitment to sites of inflammation. We believe this molecular pathway has the potential to be therapeutically targeted, either for the enhancement (during ischemia) or inhibition (during tumor metastasis) of macrophage-induced neovascular responses.
MATERIALS AND METHODS
Materials.
Anti-Rac2 mouse monoclonal antibody was purchased from Proteintech. Anti–Myosin IIA was purchased from Abcam (mouse monoclonal) and Cell Signaling Technology (rabbit polyclonal). Anti-HuR (clone 3A2, mouse monoclonal) and Anti-Actin (rabbit and goat polyclonals) were purchased from Santa Cruz Biotechnology, Inc. Goat anti–human IgG, Fcγ fragment-specific, was purchased from Jackson ImmunoResearch Laboratories, Inc. Recombinant ICAM-1 chimera with Fcγ fragment of human IgG1, Fcγ of human IgG1 fragment alone, and human and murine CCL2 were purchased from R&D Systems. Where appropriate, murine ICAM-1 and murine CCL2 were used with mouse cells, and human ICAM-1 and human CCL2 were used with human cells. PAK1 PBD agarose beads were purchased from Cell Biolabs. Alexa Fluor–conjugated secondary antibodies and phalloidin, DAPI (4,6 diamidino-2-phenylindole), and RPMI 1640 culture medium were purchased from Invitrogen. Actinomycin d, poly-l-lysine, Ficoll-Histopaque, GTPγS, and endotoxin-free BSA were purchased from Sigma-Aldrich. Duolink II Proximity Ligation kit was purchased from Olink. THP-1 cell line was purchased from American Type Culture Collection. Sequences of primers used in quantitative real-time PCR were all synthesized by the Keck Biotechnology Resource Facility at Yale University School of Medicine as follows: VEGF, 5′-ACTGGACCCTGGCTTTACTGCT-3′ (sense) and 5′-TGATCCGCATGATCTGCATGGTG-3′ (antisense); CD68, 5′-CTTCCCACAGGCAGCACAG-3′ (sense) and 5′-AATGATGAGAGGCAGCAAGAGG-3′ (antisense); and GAPDH, 5′-AAGGCCGGGGCCCACTTGAA-3′ (sense) and 5′-GGACTGTGGTCATGAGCCCTTCCA-3′ (antisense).
Animals.
C57BL/6 mice were obtained from The Jackson Laboratory. Rac2−/− mice were a gift from D. Williams (Harvard Medical School, Boston, MA; Roberts et al., 1999). LysM+/cre were obtained as described and backcrossed onto C57BL/6 for 11 generations by our laboratory (Clausen et al., 1999; Zhang et al., 2012). Myh9fl/fl mice were a gift from R.S. Adelstein (National Institutes of Health, Bethesda, MD; Jacobelli et al., 2010). For BMDMs, animals 6–8 wk of age were used. For hind limb ischemia or thioglycollate-elicited macrophage experiments, animals 12–14 wk of age were used. Animals were excluded from the study if BMDMs demonstrated a <50% reduction in Myh9 levels normalized to Actin, as measured by Western blotting. Animal studies did not involve randomization; however, the investigators were blinded to the genotypes of individual animals in each litter until after the study outcomes were assessed. The Yale University School of Medicine Institutional Animal Care and Use Committee approved all housing protocols and experimental animal procedures.
Human primary monocytes.
Human peripheral blood monocytes were isolated by density centrifugation and, where indicated, fluorescence-activated cell sorting (Geissmann et al., 2003; Repnik et al., 2003; Swirski et al., 2009). Monocytes were isolated from anonymous healthy donors. Informed consent was obtained before drawing blood from all donors. The Yale University School of Medicine Human Investigation Committee approved all protocols and experiments involving human leukocytes.
BMDM isolation and culture.
BMDMs were obtained by in vitro differentiation of primary femur and tibia BM cells using an established protocol (Davies and Gordon, 2005). In brief, femurs and tibiae from Rac2+/+, Rac2−/−, myeloid-specific Myh9+/−, or myeloid-specific Myh9−/− mice were dissected, cleaned, disinfected in 70% ethanol, and washed with fully supplemented RPMI 1640 medium. Red blood cells were removed by ammonium-chloride-potassium lysis buffer and subsequent centrifugation. Remaining BM cells were then cultured at a density of 4 × 106 cells/10 cm Petri dish in fully supplemented RPMI with 30% (vol/vol) L929 cell-conditioned medium. Cells were matured to phenotypic macrophages over 6–8 d. Nonadherent cells were washed away with PBS. Adherent cells were recovered with gentle pipetting in PBS with 1 mM EDTA. Flow cytometric analysis confirmed the recovered cells to be >97% CD11bhiF4/80hi.
Cell adhesion protocol.
Petri dishes or glass coverslips were coated with 10 µg/ml goat anti–human IgG, Fcγ fragment-specific, in 50 mM Tris-HCl, pH 9.5, for 1 h at room temperature. Blocking was performed for 1 h at room temperature with PBS containing 2% (wt/vol) BSA. Dishes or coverslips were then incubated with 100 ng/ml human or mouse rICAM-1-Fcγ fragment chimeric protein or, as a negative control, 100 ng/ml Fcγ alone or 20 mg/ml poly-l-lysine, overnight at 4°C. Where appropriate, cells were resuspended in fully supplemented RPMI 1640 with or without 50 ng/ml human or mouse recombinant CCL2 at the time of adhesion.
HuR translocation assay.
Plated cells were incubated for 30 min at 37°C, washed with PBS, and then fixed with 4% PFA for 15 min at room temperature. Cells were then washed with PBS and permeabilized with 0.1% Triton X-100 in PBS, followed by blocking with 5% goat serum for 60 min at room temperature. Immunostaining with 1/250 dilution of anti-HuR antibody was performed overnight at 4°C. Cells were incubated with Alexa Fluor–conjugated secondary antibodies for 2 h in the dark at room temperature. Counterstaining was performed with phalloidin and 0.005% DAPI as per manufacturer protocols. Confocal microscopy was performed on a five-laser inverted microscope (Ti-E Eclipse; Nikon) equipped with a Perfect Focus, motorized XY stage on a Nano Focusing Piezo Stage. Oil immersion objective lenses (60 and 100×; CFI Plan Apochromat VC Series; Nikon) with a numerical aperture of 1.4 were used. Samples were imaged at 21°C. Flourochromes included Alexa Fluor 488, 568, and 647 (Invitrogen). Cells were mounted in Fluorogel with Tris Buffer (Electron Microscopy Sciences). Images were taken with the UltraVIEW VoX confocal imaging system (Perkin Elmer) using Volocity 6.2.1 (Perkin Elmer) acquisition software. We have defined an algorithm (Matlab-based) to quantify the relative translocation of HuR in each cell. Background subtracted cell images were identified and segmented using the watershed algorithm and then tracked across the z-plane by using the centroid of the nucleus. From this point, the relative translocation of HuR was calculated by multiplying the relative external (to nucleus) intensity with the relative external (to nucleus) volume. Threshold for the quantification of cells with a robust translocation in a 30-min period are based on a minimum intensity-volume product of 0.25.
PLA.
To demonstrate a complex between native Rac2 and Myh9 in adhered primary human monocytes, a PLA kit was used as per manufacturer protocols (Fredriksson et al., 2002, 2007; Söderberg et al., 2006). Primary antibodies to Rac2 and Myosin IIA were used, and secondary antibodies were conjugated to oligonucleotides for ligation and subsequent rolling circle amplification. To correlate Rac2–Myh9 complex formation with HuR translocation, HuR was cloned into a modified pEGFP-C3 vector. The monocytic cell line, THP-1, was transfected with the construct and stable clones expressing YFP-HuR fusion protein were selected by a combination of antibiotic and fluorescence-activated cell sorting. Clones expressing YFP-HuR were used to track HuR translocation without use of an antibody. We have defined an algorithm (Matlab-based) to quantify both the proximity ligation signal and relative HuR translocation simultaneously in the same cell for correlative analysis.
mRNA decay assay.
After cells were adhered for 30 min, transcriptional arrest was imparted by 10 µg/ml actinomycin d (time 0 in assay). RNA was harvested at 20-min intervals for 60 min mRNA expression was analyzed by RT-qPCR as previously described (Wang et al., 2006). VEGF-A mRNA levels were normalized to GAPDH mRNA.
GTP-Rac PBD affinity pulldown assay.
Rac1, Rac2, and CDC42 activity was measured by an affinity precipitation assay with Pak1 PBD agarose beads (del Pozo et al., 2000; Knaus et al., 2007). Cells were stimulated with CCL2 and adhered as described above, after which they were lysed in 500 µl lysis buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 4 mM EDTA, 4 mM EGTA, and 10% glycerol, supplemented with protease inhibitor cocktail [Roche]). 50 µl was set aside for whole lysate Rac analysis. To the remainder of the sample, 10 µg Pak1 PBD beads were added, followed by a 45-min incubation at 4°C. Where GTPγS-loading was required, cell lysates were incubated with 0.1 mM GTPγS for 10 min at 37°C before addition of PBD beads. Beads were washed with lysis buffer and subsequently brought up in Laemmli sample buffer. Activated (GTP- or GTPγS-bound) Rac1, Rac2, or CDC42 was detected by Western blot analysis with whole cell lysates as loading controls.
FACS analysis.
Human peripheral blood monocytes were enriched by density gradient centrifugation in Histopaque (Sigma-Aldrich), followed by Percoll (GE Healthcare) and stained with the following mouse monoclonal antibodies to gate out contaminating NK cells and to separate monocyte subpopulations: PE-conjugated anti-CD56, allophycocyanin (APC)-conjugated anti-CD14, Alexa Fluor 700–conjugated anti-CD16, and PerCP-Cy5.5–conjugated anti-CCR2. Mouse peripheral blood mononuclear cells were obtained from whole blood by density gradient centrifugation and stained with the following rat monoclonal antibodies to identify monocytes: PE-conjugated anti-CD11b and FITC-conjugated anti-Ly6C. Mouse thioglycollate-elicited peritoneal macrophages were stained with rat monoclonal PE-conjugated anti-CD11b and APC-conjugated anti-F4/80 antibodies. Staining of mouse cells was always performed in the presence of FcBlock to eliminate nonspecific binding by Fc receptors. The antibodies were purchased from BD, eBioscience, and BioLegend. LSR II flow cytometer and FACS Diva software (BD) were used to acquire fluorescence signal for live single cells based on gating for forward and side scatter; at least 2 × 105 cells were acquired for each sample. Subsequent analyses of cell populations were performed using FlowJo software (v. 7.6.4; Tree Star). Sorting of monocytes was performed using iCyt Reflection high-speed sorter (Sony) at Yale University School of Medicine Cell Sorter Facility.
Hind limb ischemia model.
Hind limb ischemia surgery was performed by the Microsurgery Core Facility at the Yale Cardiovascular Research Center. In brief, mice were anesthetized by intraperitoneal injection of ketamine/xylazine (100/10 mg/kg) solution. The femoral artery was ligated at two positions spaced 5 mm apart, one just below the inguinal ligament and the second distal to superficial epigastric artery. All branches between the two ligatures were ligated and the femoral artery segment was excised. Flow images of the foot were acquired using a Moor Laser Doppler Imager (Moor Instruments) at 37 ± 0.50°C under ketamine/xylazine anesthesia. The data were analyzed with moorLDI image processing software (V5.3; Moor instruments) and reported as the ratio of flow in the ischemic/non-ischemic contralateral control hind limbs (Tirziu et al., 2005, 2012; Lanahan et al., 2010). Tissue perfusion was assessed before (day −1), after (day 0), and 3, 7, 14, and 21 d after surgery. Early inflammatory assessment was performed on day 3 after ligation, where thigh and calf muscles were dissected, harvested, and segmented for subsequent analysis. Portions of thigh and calf were homogenized for subsequent TRIzol-based fractionation and isolation of DNA, RNA, and protein (Chomczynski, 1993). RNA was treated with a DNase kit (Life Technologies) to remove genomic contaminant and then mRNA was reverse transcribed and analyzed by RT-qPCR for relative VEGF-A and CD68 expression, using GAPDH for normalization. VEGF-A protein was quantified by densitometry of immunoblot analysis with β-actin normalization. In some animals, ischemic muscle tissue was dissected on day 3 after femoral artery ligation and then treated with collagenase digestion to generate a single cell suspension (Swirski et al., 2009). After density gradient centrifugation with Percoll (GE Healthcare), cells were stained for CD11b and F4/80 and fixed in 1% paraformaldehyde for 5 min. Macrophages (CD11bhi F4/80hi cells) were isolated using FACS. Cross-linking was reversed and macrophage mRNA purified for subsequent RT-qPCR using the miRNeasy FFPE kit (QIAGEN). Other segmented portions of thigh and calf were fixed, dehydrated, embedded, and sectioned for histology. Quantitative high-resolution micro-CT was performed on ischemic limbs at day 14 after surgery.
Micro-CT.
Quantitative micro-CT was performed after bismuth-gelatin perfusion of small arteries by the Yale Translational Research Imaging Center. Ischemic hind limbs or P7 kidneys were staged in a high-resolution micro-CT scanner (GE eXplore Locus SP; GE Healthcare) with a cone beam filtered back projection algorithm, set to an 8-µm effective detector pixel size. Micro-CT was operated with 80-kVp x-ray tube voltage, 80-mA tube current, 3,000-millisecond per frame, 1 × 1 detector binning model, 360° angle, and 0.5° increments per view.
Labeling of the retinal vasculature.
Retina labeling was done as previously described with some modification (del Toro et al., 2010; Pitulescu et al., 2010). In brief, the eyeballs were removed from neonates at P4 and prefixed win 4% paraformaldehyde for 15 min at room temperature. The dissected retinas were blocked overnight at 4°C in TNB (0.1 M Tris-HCl, pH 7.4, 150 mM NaCl, and 0.2% Blocking reagent [PerkinElmer]) supplemented with 0.5% Triton X-100. After blocking, the retinas were incubated with IsolectinB4, Alexa Fluor 488 Conjugate (Molecular Probes) in Pblec (1 mM MgCl2, 1 mM CaCl2, 0.1 mM MnCl2, and 1% Triton X-100 in PBS) for 2 h at room temperature, washed 6× in PBS, fixed briefly in 4% PFA, and mounted in fluorescent mounting medium (DAKO). 20–25 images per group were acquired on the above-described 80i Ti-E Eclipse inverted microscope, and Biological CMM Analyzer software was used to quantify vessel branch points, vessel length, and number of segments per 10× magnification fields (Jones et al., 2008).
Whole mount brain staining.
Whole brains were harvested and stained as described previously (Tirziu et al., 2012). In brief, at P4, mouse brains were extracted, fixed in 1:4 dimethyl sulfoxide/methanol overnight at 4°C, washed in methanol, and stored in methanol at −20°C. The samples were then rehydrated, washed in PBS, and permeabilized overnight at 4°C in TNBT (0.1 M Tris-HCl, pH 7.4, 150 mM NaCl, 0.2% Blocking reagent [PerkinElmer], and 0.5% Triton X-100). After incubation with smooth muscle α-actin antibody (1A4-Cy3, 1:200) overnight at 4°C, the brains were washed in PBS and imaged on a microscope (M205 FA; Leica) with Leica Application Suite (LAS) software. Pial arteriolar–arteriolar collateral anastomoses were quantified as mean number of collateral vessels between the anterior cerebral artery (ACA) and the middle cerebral artery (MCA) territories, per cerebral hemisphere.
Statistical analysis.
Based on previous studies (Tirziu et al., 2005, 2012; Lanahan et al., 2010; Zhang et al., 2012), the pooled standard deviation of hind limb ischemia recovery over 21 d is assumed to be 0.13. Alpha is specified as 0.05, using a two-tailed test of statistical significance. The study had a 95% power to detect a between group difference of 25% with a sample size of 7. All data that demonstrated a normal distribution are presented as the mean ±SEM. Differences between multiple groups were assessed with one-way ANOVA, followed by Tukey’s post hoc multiple comparisons test. Comparisons between two independent groups were performed with a two-sample t test. All p-values were calculated with two-tailed statistical tests. Differences were considered significant when P < 0.05. Statistical data were analyzed with the use of Prism 6 (GraphPad Software).
Online supplemental material.
Fig. S1 shows flow cytometric serial selection, identification, and subsequent separation (sorting) of human peripheral blood inflammatory versus patrolling monocytes. Fig. S2 shows MH+ ions detected in trypsin digests by LCMS. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132130/DC1.
Supplementary Material
Acknowledgments
We thank David Williams and Robert Adelstein for their generous gifts of the Rac2−/− and Myh9fl/fl mice, respectively. We thank Gautham Rao for his generation of the HuR-YFP construct.
This work was supported by the following National Institutes of Health grants: F32 HL097422 (A.R. Morrison), T32 HL007950 (B.D. Young), R01 HL043331 (J.R. Bender), R01 HL084619 (M. Simons), and R01 GM47214 (M.A. Schwartz). This work was also supported by a Raymond and Beverly Sackler Foundation Award (J.R. Bender) and an AIRC-IG (R. Pardi).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ARE
- AU-rich element
- BMDM
- BM-derived macrophage
- micro-CT
- micro-computed tomography
- NMMIIA
- nonmuscle myosin IIA
- PAK
- p21-activated kinase
- PBD
- PAK binding domain
- PLA
- proximity ligation assay
- VEGF-A
- vascular endothelial growth factor A
References
- Carmeliet, P., and Jain R.K.. 2011. Molecular mechanisms and clinical applications of angiogenesis. Nature. 473:298–307 10.1038/nature10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, M.W., Alon R., and Springer T.A.. 1996. The C-C chemokine MCP-1 differentially modulates the avidity of β1 and β2 integrins on T lymphocytes. Immunity. 4:179–187 10.1016/S1074-7613(00)80682-2 [DOI] [PubMed] [Google Scholar]
- Chomczynski, P.1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 15:532–534: 536–537 [PubMed] [Google Scholar]
- Clausen, B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Davies, J.Q., and Gordon S.. 2005. Isolation and culture of murine macrophages. Methods Mol. Biol. 290:91–103 [DOI] [PubMed] [Google Scholar]
- De, P., Peng Q., Traktuev D.O., Li W., Yoder M.C., March K.L., and Durden D.L.. 2009. Expression of RAC2 in endothelial cells is required for the postnatal neovascular response. Exp. Cell Res. 315:248–263 10.1016/j.yexcr.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, M.A., Price L.S., Alderson N.B., Ren X.D., and Schwartz M.A.. 2000. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19:2008–2014 10.1093/emboj/19.9.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio, A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J.M., and Sheetz M.P.. 2009. Stretching single talin rod molecules activates vinculin binding. Science. 323:638–641 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro, R., Prahst C., Mathivet T., Siegfried G., Kaminker J.S., Larrivee B., Breant C., Duarte A., Takakura N., Fukamizu A., et al. 2010. Identification and functional analysis of endothelial tip cell-enriched genes. Blood. 116:4025–4033 10.1182/blood-2010-02-270819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distasi, M.R., Case J., Ziegler M.A., Dinauer M.C., Yoder M.C., Haneline L.S., Dalsing M.C., Miller S.J., Labarrere C.A., Murphy M.P., et al. 2009. Suppressed hindlimb perfusion in Rac2−/− and Nox2−/− mice does not result from impaired collateral growth. Am. J. Physiol. Heart Circ. Physiol. 296:H877–H886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall A.. 2002. Rho GTPases in cell biology. Nature. 420:629–635 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Fantin, A., Vieira J.M., Gestri G., Denti L., Schwarz Q., Prykhozhij S., Peri F., Wilson S.W., and Ruhrberg C.. 2010. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 116:829–840 10.1182/blood-2009-12-257832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson, S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S.M., Ostman A., and Landegren U.. 2002. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20:473–477 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- Fredriksson, S., Dixon W., Ji H., Koong A.C., Mindrinos M., and Davis R.W.. 2007. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods. 4:327–329 [DOI] [PubMed] [Google Scholar]
- Friedland, J.C., Lee M.H., and Boettiger D.. 2009. Mechanically activated integrin switch controls α5β1 function. Science. 323:642–644 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- Geissmann, F., Jung S., and Littman D.R.. 2003. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19:71–82 10.1016/S1074-7613(03)00174-2 [DOI] [PubMed] [Google Scholar]
- Gerszten, R.E., Garcia-Zepeda E.A., Lim Y.C., Yoshida M., Ding H.A., Gimbrone M.A. Jr, Luster A.D., Luscinskas F.W., and Rosenzweig A.. 1999. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 398:718–723 10.1038/19546 [DOI] [PubMed] [Google Scholar]
- Gu, Y., Filippi M.D., Cancelas J.A., Siefring J.E., Williams E.P., Jasti A.C., Harris C.E., Lee A.W., Prabhakar R., Atkinson S.J., et al. 2003. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 302:445–449 10.1126/science.1088485 [DOI] [PubMed] [Google Scholar]
- Hoefer, I.E., van Royen N., Rectenwald J.E., Deindl E., Hua J., Jost M., Grundmann S., Voskuil M., Ozaki C.K., Piek J.J., and Buschmann I.R.. 2004. Arteriogenesis proceeds via ICAM-1/Mac-1-mediated mechanisms. Circ. Res. 94:1179–1185 10.1161/01.RES.0000126922.18222.F0 [DOI] [PubMed] [Google Scholar]
- Humphries, J.D., Wang P., Streuli C., Geiger B., Humphries M.J., and Ballestrem C.. 2007. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179:1043–1057 10.1083/jcb.200703036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, W.D., Arras M., Winkler B., Scholz D., Schaper J., and Schaper W.. 1997. Monocyte chemotactic protein-1 increases collateral and peripheral conductance after femoral artery occlusion. Circ. Res. 80:829–837 10.1161/01.RES.80.6.829 [DOI] [PubMed] [Google Scholar]
- Jacobelli, J., Friedman R.S., Conti M.A., Lennon-Dumenil A.M., Piel M., Sorensen C.M., Adelstein R.S., and Krummel M.F.. 2010. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nat. Immunol. 11:953–961 10.1038/ni.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E.A., Yuan L., Breant C., Watts R.J., and Eichmann A.. 2008. Separating genetic and hemodynamic defects in neuropilin 1 knockout embryos. Development. 135:2479–2488 10.1242/dev.014902 [DOI] [PubMed] [Google Scholar]
- Jorrisch, M.H., Shih W., and Yamada S.. 2013. Myosin IIA deficient cells migrate efficiently despite reduced traction forces at cell periphery. Biol. Open. 2:368–372 10.1242/bio.20133707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus, U.G., Bamberg A., and Bokoch G.M.. 2007. Rac and Rap GTPase activation assays. Methods Mol. Biol. 412:59–67 10.1007/978-1-59745-467-4_5 [DOI] [PubMed] [Google Scholar]
- Kuziel, W.A., Morgan S.J., Dawson T.C., Griffin S., Smithies O., Ley K., and Maeda N.. 1997. Severe reduction in leukocyte adhesion and monocyte extravasation in mice deficient in CC chemokine receptor 2. Proc. Natl. Acad. Sci. USA. 94:12053–12058 10.1073/pnas.94.22.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan, A.A., Hermans K., Claes F., Kerley-Hamilton J.S., Zhuang Z.W., Giordano F.J., Carmeliet P., and Simons M.. 2010. VEGF receptor 2 endocytic trafficking regulates arterial morphogenesis. Dev. Cell. 18:713–724 10.1016/j.devcel.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Yamauchi A., Marchal C.C., Molitoris J.K., Quilliam L.A., and Dinauer M.C.. 2002. Chemoattractant-stimulated Rac activation in wild-type and Rac2-deficient murine neutrophils: preferential activation of Rac2 and Rac2 gene dosage effect on neutrophil functions. J. Immunol. 169:5043–5051 10.4049/jimmunol.169.9.5043 [DOI] [PubMed] [Google Scholar]
- Lichanska, A.M., Browne C.M., Henkel G.W., Murphy K.M., Ostrowski M.C., McKercher S.R., Maki R.A., and Hume D.A.. 1999. Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.1. Blood. 94:127–138 [PubMed] [Google Scholar]
- Lu, B., Rutledge B.J., Gu L., Fiorillo J., Lukacs N.W., Kunkel S.L., North R., Gerard C., and Rollins B.J.. 1998. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J. Exp. Med. 187:601–608 10.1084/jem.187.4.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu, M.E., Schmidt I., Benedito R., and Adams R.H.. 2010. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat. Protoc. 5:1518–1534 10.1038/nprot.2010.113 [DOI] [PubMed] [Google Scholar]
- Ramgolam, V.S., DeGregorio S.D., Rao G.K., Collinge M., Subaran S.S., Markovic-Plese S., Pardi R., and Bender J.R.. 2010. T cell LFA-1 engagement induces HuR-dependent cytokine mRNA stabilization through a Vav-1, Rac1/2, p38MAPK and MKK3 signaling cascade. PLoS ONE. 5:e14450 10.1371/journal.pone.0014450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repnik, U., Knezevic M., and Jeras M.. 2003. Simple and cost-effective isolation of monocytes from buffy coats. J. Immunol. Methods. 278:283–292 10.1016/S0022-1759(03)00231-X [DOI] [PubMed] [Google Scholar]
- Roberts, A.W., Kim C., Zhen L., Lowe J.B., Kapur R., Petryniak B., Spaetti A., Pollock J.D., Borneo J.B., Bradford G.B., et al. 1999. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 10:183–196 10.1016/S1074-7613(00)80019-9 [DOI] [PubMed] [Google Scholar]
- Sawada, Y., Tamada M., Dubin-Thaler B.J., Cherniavskaya O., Sakai R., Tanaka S., and Sheetz M.P.. 2006. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 127:1015–1026 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shireman, P.K., Contreras-Shannon V., Ochoa O., Karia B.P., Michalek J.E., and McManus L.M.. 2007. MCP-1 deficiency causes altered inflammation with impaired skeletal muscle regeneration. J. Leukoc. Biol. 81:775–785 10.1189/jlb.0506356 [DOI] [PubMed] [Google Scholar]
- Shirsat, N.V., Pignolo R.J., Kreider B.L., and Rovera G.. 1990. A member of the ras gene superfamily is expressed specifically in T, B and myeloid hemopoietic cells. Oncogene. 5:769–772 [PubMed] [Google Scholar]
- Söderberg, O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Swirski, F.K., Nahrendorf M., Etzrodt M., Wildgruber M., Cortez-Retamozo V., Panizzi P., Figueiredo J.L., Kohler R.H., Chudnovskiy A., Waterman P., et al. 2009. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 325:612–616 10.1126/science.1175202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirziu, D., Moodie K.L., Zhuang Z.W., Singer K., Helisch A., Dunn J.F., Li W., Singh J., and Simons M.. 2005. Delayed arteriogenesis in hypercholesterolemic mice. Circulation. 112:2501–2509 10.1161/CIRCULATIONAHA.105.542829 [DOI] [PubMed] [Google Scholar]
- Tirziu, D., Jaba I.M., Yu P., Larrivée B., Coon B.G., Cristofaro B., Zhuang Z.W., Lanahan A.A., Schwartz M.A., Eichmann A., and Simons M.. 2012. Endothelial nuclear factor-κB-dependent regulation of arteriogenesis and branching. Circulation. 126:2589–2600 10.1161/CIRCULATIONAHA.112.119321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove, M., Kedde M., and Agami R.. 2011. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 11:644–656 10.1038/nrc3107 [DOI] [PubMed] [Google Scholar]
- van Royen, N., Hoefer I., Böttinger M., Hua J., Grundmann S., Voskuil M., Bode C., Schaper W., Buschmann I., and Piek J.J.. 2003. Local monocyte chemoattractant protein-1 therapy increases collateral artery formation in apolipoprotein E-deficient mice but induces systemic monocytic CD11b expression, neointimal formation, and plaque progression. Circ. Res. 92:218–225 10.1161/01.RES.0000052313.23087.3F [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares, M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil, M., Hoefer I.E., van Royen N., Hua J., de Graaf S., Bode C., Buschmann I.R., and Piek J.J.. 2004. Abnormal monocyte recruitment and collateral artery formation in monocyte chemoattractant protein-1 deficient mice. Vasc. Med. 9:287–292 10.1191/1358863x04vm571oa [DOI] [PubMed] [Google Scholar]
- Waeckel, L., Mallat Z., Potteaux S., Combadière C., Clergue M., Duriez M., Bao L., Gerard C., Rollins B.J., Tedgui A., et al. 2005. Impairment in postischemic neovascularization in mice lacking the CXC chemokine receptor 3. Circ. Res. 96:576–582 10.1161/01.RES.0000159389.55544.20 [DOI] [PubMed] [Google Scholar]
- Wang, J.G., Collinge M., Ramgolam V., Ayalon O., Fan X.C., Pardi R., and Bender J.R.. 2006. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J. Immunol. 176:2105–2113 10.4049/jimmunol.176.4.2105 [DOI] [PubMed] [Google Scholar]
- Zhang, J., Modi Y., Yarovinsky T., Yu J., Collinge M., Kyriakides T., Zhu Y., Sessa W.C., Pardi R., and Bender J.R.. 2012. Macrophage β2 integrin-mediated, HuR-dependent stabilization of angiogenic factor-encoding mRNAs in inflammatory angiogenesis. Am. J. Pathol. 180:1751–1760 10.1016/j.ajpath.2011.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.