Jacque et al. investigate the functions of NF-κB1 p105 and its associated NF-κB–binding partners in B cells, using a mutant mouse strain that carries a form of the NF-κB1 precursor that is resistant to IKK-induced proteolysis. They identify a critical B cell–intrinsic role for this IKK signaling pathway in the antigen-induced survival and differentiation of follicular mature B cells.
Abstract
The importance of IκB kinase (IKK)–induced proteolysis of NF-κB1 p105 in B cells was investigated using Nfkb1SSAA/SSAA mice, in which this NF-κB signaling pathway is blocked. Nfkb1SSAA mutation had no effect on the development and homeostasis of follicular mature (FM) B cells. However, analysis of mixed bone marrow chimeras revealed that Nfkb1SSAA/SSAA FM B cells were completely unable to mediate T cell–dependent antibody responses. Nfkb1SSAA mutation decreased B cell antigen receptor (BCR) activation of NF-κB in FM B cells, which selectively blocked BCR stimulation of cell survival and antigen-induced differentiation into plasmablasts and germinal center B cells due to reduced expression of Bcl-2 family proteins and IRF4, respectively. In contrast, the antigen-presenting function of FM B cells and their BCR-induced migration to the follicle T cell zone border, as well as their growth and proliferation after BCR stimulation, were not affected. All of the inhibitory effects of Nfkb1SSAA mutation on B cell functions were rescued by normalizing NF-κB activation genetically. Our study identifies critical B cell-intrinsic functions for IKK-induced NF-κB1 p105 proteolysis in the antigen-induced survival and differentiation of FM B cells, which are essential for T-dependent antibody responses.
NF-κB transcription factors, which are composed of dimers of Rel polypeptides, regulate gene expression by binding to κB elements in the promoters and enhancers of target genes (Ghosh et al., 1998). Inactive NF-κB dimers are sequestered in the cytoplasm of unstimulated cells by interaction with proteins of the inhibitor of NF-κB (IκB) family, which includes IκBα, IκBβ, IκBε, and NF-κB2 p100. After appropriate agonist stimulation, the canonical NF-κB signaling pathway stimulates the IκB kinase (IKK) complex, which is composed of IKK1 (IKKα) and IKK2 (IKKβ) kinases and the regulatory ubiquitin-binding protein NEMO (IKKγ), to phosphorylate IκBα (Karin and Ben-Neriah, 2000). This promotes K48-linked ubiquitination of IκBα and subsequent degradation by the proteasome, releasing associated NF-κB1 p50-RelA and NF-κB1 p50-c-Rel dimers to translocate into the nucleus and modulate gene expression. The proteolysis of both IκBβ and IκBε is controlled by the IKK complex in a similar fashion. A subset of NF-κB agonists activates an alternative NF-κB signaling pathway, which induces IKK1 to phosphorylate NF-κB2 p100 promoting its partial proteolysis by the proteasome to produce p52, which is principally associated with RelB (Beinke and Ley, 2004).
Most of our knowledge about the specific functions of NF-κB activation in mature B cells is based on in vitro experiments with purified splenic B cells from mice deficient in specific Rel proteins (Kaileh and Sen, 2012). These studies have suggested important roles for canonical NF-κB activation in B cell growth, proliferation, and survival after B cell antigen receptor (BCR) stimulation (Grumont et al., 1999; Grumont et al., 1998, 2002). Whole animal studies have also demonstrated a requirement for NF-κB family members in the B cell response to antigen. For example, NF-κB1 or c-Rel deficiency diminishes the antibody response, whereas compound NF-κB1 and c-Rel deficiency results in a complete block (Pohl et al., 2002). However, because both NF-κB1 and c-Rel have essential roles in dendritic cells and T cells (Gerondakis and Siebenlist, 2010a), it has remained unclear whether NF-κB activation in B cells is required for optimal antibody responses.
The cell-intrinsic functions of canonical NF-κB activation in B cell physiology in vivo have been investigated genetically by conditional deletion of components of the IKK complex in the B cell lineage, using a CD19-Cre driver mouse strain. Although ablation of either IKK2 or NEMO does not affect B cell development in the BM, it does lead to the disappearance of mature B lymphocytes (Pasparakis et al., 2002; Li et al., 2003). In line with this, mature B cells fail to accumulate in the periphery in the combined absence of c-Rel and RelA (Grossmann et al., 2000). Similarly, mice with mutations in components of the alternative NF-κB signaling pathway, which regulates NF-κB2 p100 proteolysis to p52, are also deficient in mature B cells, whereas B cell development in the BM is largely unaffected (Gerondakis and Siebenlist, 2010b; Kaileh and Sen, 2012). The alternative pathway is activated downstream of the receptor for B cell activation factor (BAFF), which promotes peripheral B cells survival and determines the size of the B cell compartment (Mackay et al., 2010), and CD40 (Kaileh and Sen, 2012). Together these genetic studies have established that NF-κB activation has a critical role for the development and/or homeostasis of mature B cells. However, the requirement for NF-κB activation to maintain normal mature B cell numbers has precluded the use of conditional knockout strains lacking IKK subunits in B cells to determine the B cell–intrinsic function of NF-κB activation in humoral immunity (Pasparakis et al., 2002; Li et al., 2003; Derudder et al., 2009).
NF-κB1 p105 functions as a cytoplasmic IκB through binding to preformed NF-κB dimers via its C-terminal ankyrin repeat region, and to Rel monomers via its N-terminal Rel homology domain (Savinova et al., 2009). NF-κB1 p105 is also the precursor of p50, which is produced constitutively by partial proteolysis (processing) of p105 by the proteasome after its monoubiquitination on multiple lysine residues by an unknown E3 ligase (Kravtsova-Ivantsiv et al., 2009). Activation of the canonical NF-κB pathway induces IKK2 to phosphorylate two serines in the C-terminal PEST region of p105 (Lang et al., 2003; Yang et al., 2012), creating a binding site for SCFβ-TrCP E3 ligase, which catalyzes the subsequent K48-linked polyubiquitination of p105 and proteolysis by the proteasome. This can result either in the complete degradation of p105, releasing associated Rel subunits, or the processing of p105 to p50, both of which can potentially modulate NF-κB–dependent transcription (Sriskantharajah et al., 2009; Yang et al., 2012). In addition, IKK-induced p105 proteolysis is an essential step in the activation of the p105-associated MEK1/2 kinase TPL-2, which regulates ERK-1/2 MAP kinase activation in TLR-stimulated macrophages (Beinke et al., 2004; Waterfield et al., 2004; Yang et al., 2012). The role of IKK-induced proteolysis of NF-κB1 p105 in regulating the development and function of B cells, via NF-κB and/or TPL-2, remain unknown.
In the present study, the effect of blocking IKK phosphorylation of NF-κB1 p105 in B cells was investigated using Nfkb1SSAA/SSAA knock-in mice, which express mutant p105SSAA that is resistant to signal-induced proteolysis due to mutation of the IKK2-target serines to alanines (Sriskantharajah et al., 2009). We found that Nfkb1SSAA mutation had no effect on B cell development in the BM, or on the number of follicular mature (FM) B cells in the spleen or LNs. However, analysis of mixed BM chimeras in which the Nfkb1SSAA mutation was restricted to B cells revealed that Nfkb1SSAA FM B cells were completely unable to mount an immune response to a T cell–dependent (TD) antigen, failing to develop into antigen-specific plasmablasts or germinal center (GC) B cells. In contrast, TPL-2 deficiency in FM B cells did not affect a TD antibody response. The Nfkb1SSAA mutation was found to decrease BCR activation of NF-κB in FM B cells, which prevented BCR signaling promoting cell survival, but had minimal inhibitory effects on BCR-induced growth and proliferation. Significantly, we also discovered that Nfkb1SSAA mutation reduced NF-κB–dependent induction of the IRF4 transcription factor after BCR ligation, which prevented antigen-induced differentiation of FM B cell to plasmablasts and GC B cells. All of the inhibitory effects of Nfkb1SSAA mutation on FM B cell function were overcome by normalizing NF-κB activation genetically. Our results demonstrate that the regulation of NF-κB activation in B cells by IKK-induced proteolysis of NF-κB1 p105 is essential for antibody responses to TD antigens.
RESULTS
Nfkb1SSAA mutation does not affect steady-state numbers of mature follicular B cells
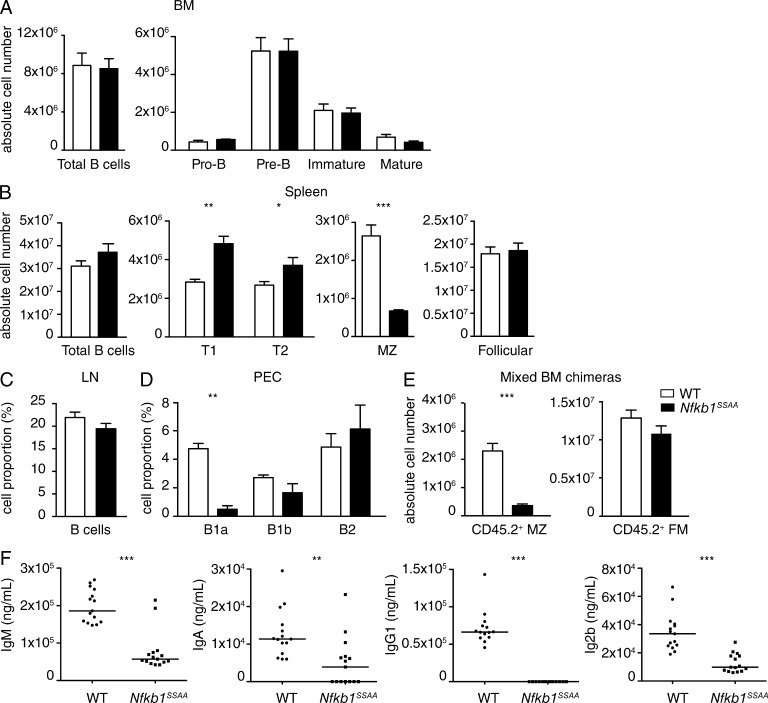
To investigate the role of IKK-induced p105 proteolysis in B cell development and homeostasis, we analyzed B cell subpopulations in the BM, secondary lymphoid organs (spleen and lymph nodes), and peritoneal cavity of Nfkb1SSAA/SSAA (Nfkb1SSAA) and WT mice by flow cytometry. B cell development in the BM was similar between both genotypes, with similar proportions and absolute numbers of pro–B cells, pre–B cells, and immature B cells (Fig. 1 A and Fig. S1 A). The numbers of mature recirculating B cells in the BM were also unaffected by Nfkb1SSAA mutation. Total numbers and proportions of B cells in spleen and peripheral lymph nodes, respectively, were equivalent in Nfkb1SSAA/SSAA and WT mice (Fig. 1, B and C; and Fig. S1, B and C). Numbers of splenic FM B cells were also similar. In contrast, marginal zone (MZ) B cells number in the spleen were substantially reduced by Nfkb1SSAA mutation, whereas the numbers of transitional type 1 (T1) and type 2 (T2) B cells were fractionally increased (Fig. 1 B). The proportion of B1a cells in the peritoneal cavity was also significantly reduced in Nfkb1SSAA/SSAA mice compared with WT (Fig. 1 D and Fig. S1 D). However, the fractions of peritoneal B1b and B2 cells were similar between genotypes.
Figure 1.
Nfkb1SSAA/SSAA mice have normal numbers of FM B cells. (A–D) Flow cytometric analysis of B cell populations in WT and Nfkb1SSAA/SSAA mice from the indicated organs. The gating strategies used are shown in Fig. S1. (A) Absolute cell numbers of pro–B (B220+CD19+IgD−IgM−CD2−), pre–B (B220+CD19+IgD−IgM−CD2+), immature mature B (B220+CD19+IgD−IgM+CD2+), and mature B cells (B220+CD19+IgD+IgM+CD2+) in the BM (mean ± SEM; n = 10–11 mice/genotype). (B) Absolute cell numbers (mean ± SEM; n = 10–11 mice/genotype) of total B cells (IgM+IgD+), immature B cells (IgDloB220+AA4.1+) separated into transitional T1 B cells (IgMhiCD23−), and T2 B cells (IgMhiCD23+), and mature B cells (IgDhiB220+AA4.1−) separated into FM B cells (IgMhiCD23+) and MZ B cells (IgMhiCD23−) in the spleen. (C) Percentages of B cells (IgM+CD19+) in peripheral lymph nodes (pools of single cervical, axillary and inguinal nodes; mean ± SEM; n = 10–11 mice/genotype). (D) Proportion of B1a (B220+CD19+CD5+CD23−), B1b (B220+CD19+CD5−CD23−), and B2 (B220+CD19+CD5−CD23+) cells in the peritoneal cavity (mean ± SEM; n = 5–7 mice/genotype). In A–D, results are representative of at least three independent experiments. (E) WT or Nfkb1SSAA/SSAA CD45.2+ BM cells were mixed with WT CD45.1+ BM cells at a 1:1 ratio, and transferred into sublethally irradiated Rag1−/− mice. After 8-wk reconstitution, the absolute number of CD45.2+ MZ B cells was assessed by flow cytometry. FM B cells were used as the control population. Graphs show absolute cell numbers (mean ± SEM; n = 14 mice/genotype), results are representative of two independent experiments. (F) Graphs show ELISAs for IgM, IgA, IgG1, and IgG2b, assaying sera from naive WT and Nfkb1SSAA/SSAA mice. n = 18 mice per genotype. PEC, peritoneal cavity. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In summary, mutation of the IKK target serines on NF-κB1 p105 did not alter the overall number of peripheral mature B cells, in marked contrast to deletion of IKK2 or NEMO in the B cell lineage that results in substantially reduced numbers of such cells (Pasparakis et al., 2002). However, MZ B cell numbers were significantly reduced by Nfkb1SSAA mutation, in line with the known importance of NF-κB for the production of this subset of B cells in the spleen (Pillai and Cariappa, 2009). Mixed BM chimeras demonstrated that the reduction in MZ B cells resulted from a B cell–autonomous effect of the Nfkb1SSAA mutation and confirmed that Nfkb1SSAA mutation did not affect FM B cell numbers (Fig. 1 E).
Nfkb1SSAA/SSAA B cells cannot mount an antibody response to a TD antigen
Measurement of steady-state levels of Ig in serum revealed significantly reduced IgM (Fig. 1 F), consistent with the reduction in MZ B cells in the spleen and B1a cells in the peritoneum (Zhang, 2013). Serum titers of IgA and both IgG1 and IgG2b were also significantly decreased in unimmunized mice, raising the possibility that one or several aspects of antibody responses might also be impaired by Nfkb1SSAA mutation. Because we were interested in investigating the role of NF-κB1 p105 proteolysis in B cell function in immunity, we focused our subsequent experiments on the effects of Nfkb1SSAA mutation on TD antibody responses. It has previously been shown that RBP-J–deficient mice, which lack MZ B cells, are still able to generate normal antibody responses to chicken gammaglobulin (CGG; Tanigaki et al., 2002). The reduction in MZ B cells in Nfkb1SSAA/SSAA mice, therefore, should not contribute significantly to TD antibody responses, although this might decrease antibody responses to T cell–independent antigens. Consequently, any detected effects of Nfkb1SSAA mutation on antibody responses to a TD antigen were likely to result from alterations in the function of FM B cells (Oracki et al., 2010), as their numbers were normal in Nfkb1SSAA/SSAA mice (Fig. 1 B).
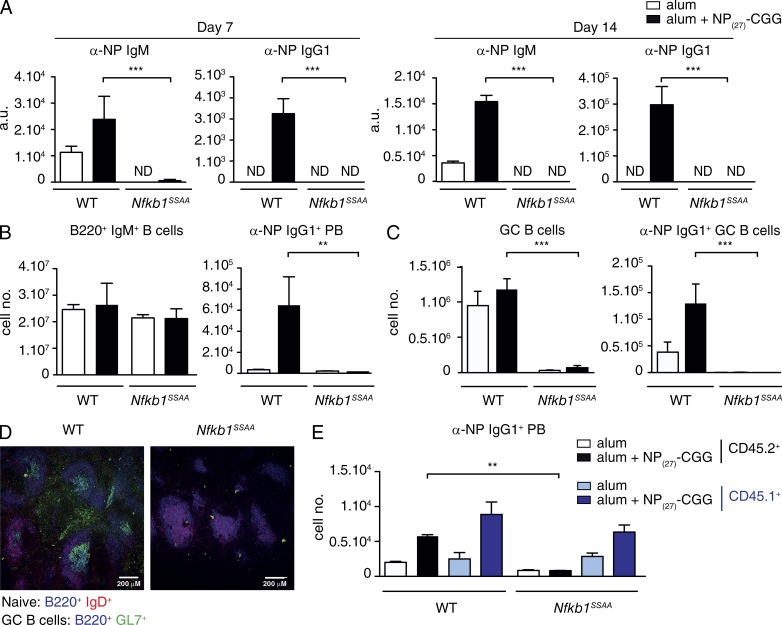
To investigate the B cell–intrinsic effects of Nfkb1SSAA mutation on FM B cell function, we generated mixed BM chimeras. BM cells from μMT−/− mice, lacking B cells (Kitamura et al., 1991), were mixed at 4:1 ratio with BM cells from WT or Nfkb1SSAA/SSAA mice. These were then transferred into irradiated Rag1−/− mice, producing chimeric mice in which all B cells had a WT (Nfkb1+/+) or Nfkb1SSAA/SSAA genotype, whereas 80% of the other hematopoietic cells had a WT genotype. After reconstitution for 8 wk, chimeric mice were then injected with NP27-CGG precipitated in alum or alum-only control. Nfkb1SSAA mutation totally blocked the production of serum anti-NP IgM and IgG1 at 7 and 14 d (Fig. 2 A), although the numbers of splenic B cells were equivalent between WT and Nfkb1SSAA/SSAA chimeric mice (Fig. 2 B, left). Nfkb1SSAA mutation also prevented the generation of anti–NP-specific plasmablasts (Fig. 2 B, right) and anti–NP-specific GC B cells (Fig. 2 C). Furthermore, immunofluorescence analysis of spleen sections demonstrated the absence of GC structures in Nfkb1SSAA/SSAA chimeras after NP27-CGG-alum immunization (Fig. 2 D), which is consistent with a reduction in total GC B cell numbers (Fig. 2 C, left). Analysis of allotype-marked mixed BM chimeras confirmed that the effect of Nfkb1SSAA mutation on generation of anti-NP–specific plasmablasts was B cell intrinsic (Fig. 2 E).
Figure 2.
Nfkb1SSAA/SSAA mutation blocks the B cell antibody response to a TD antigen. WT or Nfkb1SSAA/SSAA BM cells were mixed with μMT−/− BM cells in a 1:4 ratio, and transferred into sublethally irradiated Rag1−/− mice. After 8 wk, chimeras were immunized with NP27-CGG alum or PBS alum controls. (A) Serum antibody response to NP27-CGG immunization of WT or Nfkb1SSAA/SSAA mixed BM chimeras assessed 7 and 14 d after challenge. Data show mean NP-specific serum IgM and IgG1 levels (±SEM) measured by ELISA. (B) Flow cytometric analysis of total naive B cells (IgM+B220+; left) and αNP-IgG1+ plasmablasts (PB; B220loIgMloIgD−CD138+; right) in the spleens of chimeras 7 d after NP27-CGG immunization (mean absolute number ± SEM). (C) Flow cytometric analysis of total GC B cells (IgMloB220+IgD−PNAhiGL7+; left) and αNP-IgG1+ GC B cells (right) in chimeras 14 d after NP27-CGG immunization (mean absolute number ± SEM). (D) Confocal microscopy of spleen sections from WT or Nfkb1SSAA/SSAA chimeras immunized with NP27-CGG (14d). GC formation was measured by staining with anti-IgD (red), anti-B220 (blue) and GL7 (green). In A–D, results are representative of at least three independent experiments with n = 5 mice per genotype. (E) WT or Nfkb1SSAA/SSAA CD45.2+ BM cells were mixed with WT CD45.1+ and μMT−/− BM cells in a 1:1:3 ratio, and transferred into sublethally irradiated Rag1−/− mice. After an 8-wk reconstitution, chimeras were immunized with NP27-CGG alum or alum alone. Graph shows the flow cytometric analysis of αNP+ IgG1+ plasmablasts (PB), 7 and 14 d after NP27-CGG immunization. Results are representative of at least two independent experiments with n = 5–6 mice per genotype. **, P < 0.01; ***, P < 0.001.
The results in this section demonstrated an essential role for IKK-induced proteolysis of NF-κB1 p105 within FM B cells for their participation in TD antibody responses.
Nfkb1SSAA mutation reduces BCR activation of NF-κB
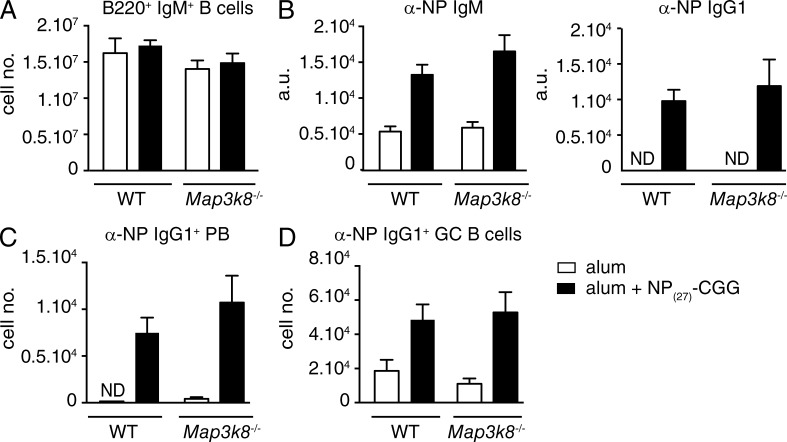
Our earlier work demonstrated that blockade of IKK-induced p105 proteolysis by Nfkb1SSAA mutation can impair the activation of both TPL-2 and NF-κB in macrophages (Yang et al., 2012). Therefore, the inhibitory effects of Nfkb1SSAA mutation on TD antibody responses could have resulted from reduced TPL-2 and/or NF-κB activation in B cells. To investigate whether TPL-2 had important B cell–intrinsic functions in TD antibody responses, mixed BM chimeras were generated in which B cells lacked expression of TPL-2 (Dumitru et al., 2000). Immunization of these chimeric mice with NP27-CGG revealed that TPL-2–deficient B cells produced a TD antibody response equivalent to that of WT controls (Fig. 3). This suggested that the failure of Nfkb1SSAA/SSAA FM B cells to mount an antibody response to NP27-CGG did not result from defective TPL-2 signaling, and was likely due to altered NF-κB activation.
Figure 3.
TPL-2 is not required for a B cell antibody response to TD antigen. WT or Map3k8−/− BM cells were mixed with μMT−/− BM cells in a 1:4 ratio, and transferred into sublethally irradiated Rag1−/− mice. After 8 wk, chimeras were immunized with NP27-CGG alum or PBS alum controls. (A) Flow cytometric analysis of total naive B cells (IgM+B220+). (B) α-NP IgM and IgG1 serum antibody levels were quantified by ELISA (mean ± SEM), 14 d after NP27-CGG immunization. (C) Splenic α-NP IgG1+ plasmablasts (PB; B220loIgMloIgD−CD138+) 7d after NP27-CGG immunization (mean absolute number ± SEM). (D) Splenic α-NP IgG1+ GC B cells 14 d after NP27-CGG immunization (mean absolute number ± SEM). All results are representative of two independent experiments (4–5 mice per genotype each).
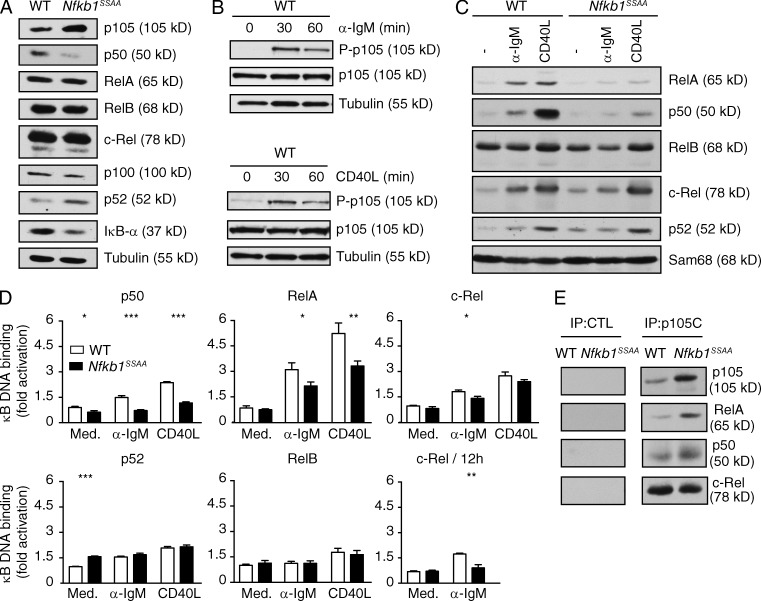
To investigate this possibility, we determined the effect of Nfkb1SSAA mutation on NF-κB activation in purified splenic FM B cells in vitro. Steady-state protein levels of RelA, RelB, and c-Rel were comparable between Nfkb1SSAA/SSAA and WT FM B cells (Fig. 4 A). However, similar to our previous findings with macrophages (Yang et al., 2012), Nfkb1SSAA mutation increased the steady-state concentration of NF-κB1 p105, while NF-κB1 p50 was reduced. Nfkb1 mRNA levels were comparable between Nfkb1SSAA/SSAA and WT FM B cells (unpublished data), implying that these changes in p105 and p50 protein levels resulted from Nfkb1SSAA mutation blocking constitutive p105 phosphorylation by IKK. Consistent with that hypothesis, unstimulated WT FM B cells had small amounts of phosphorylated p105, that increased after stimulation with anti-IgM, CD40L and LPS (Fig. 4 B and not depicted). In addition, steady-state levels of NF-κB2 p52 were increased in Nfkb1SSAA/SSAA B cells compared with WT, whereas levels of p100 were fractionally reduced (Fig. 4 A), suggesting that Nfkb1SSAA mutation increased constitutive activation of the NF-κB alternative pathway.
Figure 4.
Effects of Nfkb1SSAA mutation on NF-κB activation in FM B cells. (A) Lysates of purified splenic FM B cells from WT or Nfkb1SSAA/SSAA mice were immunoblotted for the indicated proteins. (B) Purified splenic FM B cells from WT mice were stimulated with α-IgM or CD40L, and whole-cell extracts immunoblotted. (C) Purified splenic FM B cells were stimulated with α-IgM (1 h) or CD40L (4 h). Nuclear fractions were immunoblotted. (D) Nuclear fractions were prepared as in C, and NF-κB binding of the indicated Rel subunits was determined by ELISA (mean ± SEM). To assay late activation of c-Rel, FM B cells were stimulated with anti-IgM for 12 h. (E) p105 was immunoprecipitated (IP) from lysates of purified splenic FM B cells. Associated Rel proteins were detected by immunoblotting. Pre-immune serum was used for control immunoprecipitations (CTL). All results are representative of at least three separate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Nuclear translocations of RelA and p50, after stimulation with anti-IgM for 1 h, were clearly decreased in Nfkb1SSAA/SSAA FM B cells compared with WT cells (Fig. 4 C). Accordingly, NF-κB ELISAs demonstrated that Nfkb1SSAA mutation impaired BCR activation of p50 and RelA binding activity at 1 h (Fig. 4 D). Although nuclear translocation of c-Rel was not obviously affected after 1h BCR stimulation, c-Rel binding activity was fractionally reduced by Nfkb1SSAA mutation (Fig. 4, C and D). However, Nfkb1SSAA mutation had a more pronounced effect on the second wave of c-Rel activation that results from prolonged BCR stimulation (Damdinsuren et al., 2010), detected at 12 h (Fig. 4 D). Both p50 and RelA nuclear translocations and binding activities were reduced by Nfkb1SSAA mutation after 4-h CD40L stimulation. In contrast, CD40L-induced nuclear translocations and NF-κB binding activities of c-Rel, RelB, and NF-κB2 p52 were similar between Nfkb1SSAA/SSAA and WT FM B cells. Thus, the major effects of Nfkb1SSAA mutation in FM B cells were to reduce the early activation of canonical NF-κB subunits p50 and RelA, after stimulation of either BCR or CD40, and the late activation of c-Rel after prolonged BCR cross-linking. p50, RelA and c-Rel each co-immunoprecipitated with p105, suggesting that Nfkb1SSAA mutation reduced activation of these Rel subunits, in part, by p105SSAA retention in the cytoplasm (Fig. 4 E).
Nfkb1SSAA mutation prevents BCR augmentation of FM B cell survival
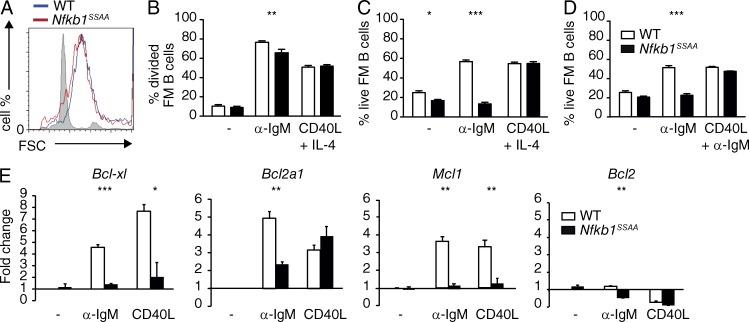
Optimal TD antibody responses depend on the clonal expansion and survival of antigen-stimulated B cells (Oracki et al., 2010). Furthermore, in vitro studies with purified splenic B cells from Rel-deficient mice have suggested that NF-κB promotes the cell growth, proliferation and survival of mature B cells after BCR stimulation (Gerondakis and Siebenlist, 2010a; Kaileh and Sen, 2012). As NF-κB activation was impaired by Nfkb1SSAA mutation, we next compared the growth, proliferation and survival of Nfkb1SSAA/SSAA FM B cells with WT cells. Nfkb1SSAA mutation did not affect FM B cell growth (Fig. 5 A), and only modestly decreased the fraction of FM B cells that divided following anti-IgM stimulation (Fig. 5 B). However, Nfkb1SSAA mutation prevented BCR ligation increasing FM B cell survival (Fig. 5 C). CD40-induced proliferation and survival were similar between Nfkb1SSAA/SSAA and WT FM B cells (Fig. 5, B and C). The survival defect of Nfkb1SSAA/SSAA FM B cells after BCR cross-linking was rescued by co-stimulation with CD40L (Fig. 5 D).
Figure 5.
Effects of Nfkb1SSAA mutation on FM B cell survival and proliferation. (A) Flow cytometric analysis of cell growth (forward scatter) on purified FM B cells cultured in presence of α-IgM, for 24 h. Unstimulated WT FM B cells are represented in gray. (B) Purified splenic FM B cells plus α-IgM, CD40L, and IL-4 or control medium (−) were cultured for 68 h. The fraction of FM B cells that had divided was determined by flow cytometric analysis of CFSE dilution (mean ± SEM; triplicates). (C and D) Purified splenic FM B cells were cultured for 48 h with α-IgM, CD40L plus IL-4, α-IgM plus CD40L and IL-4, or control medium (-). The fraction of live FM B cells was determined by flow cytometric analysis of 7AAD staining (mean ± SEM; triplicates). (E) Triplicate cultures of purified splenic FM B cells were stimulated with α-IgM for 1 h or with CD40L for 4 h. Levels of specific mRNAs were determined by quantitative RT-PCR. Data are normalized to Hprt1 mRNA levels and represent mean fold change (±SEM) relative to unstimulated WT cells. All results are representative of at least three independent experiments (3 mice/genotype each). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
NF-κB was previously shown to maintain the viability of BCR-activated B cells by inducing the expression of Bcl-xl and A1 (Bfl-1) genes (Grumont et al., 1999; Köntgen et al., 1995). Expression of Mcl-1, which is essential for survival of mature B cells (Hettmann et al., 2003) and plasma cells (Peperzak et al., 2013), may also be regulated by NF-κB (Buggins et al., 2010). To gain further insight into how IKK-induced p105 proteolysis regulates B cell survival, we determined the expression of these genes in purified FM B cells from Nfkb1SSAA/SSAA and WT FM mice. Nfkb1SSAA mutation significantly reduced BCR up-regulation of Bcl-xl, A1, and Mcl-1 mRNAs (Fig. 5 E). CD40 induction of Bcl-xl and Mcl-1 mRNAs was also reduced in Nfkb1SSAA/SSAA FM B cells compared with WT. In contrast, CD40 up-regulation of A1 mRNA was similar between genotypes, which may explain why CD40-induced FM B cell survival was not altered by Nfkb1SSAA mutation.
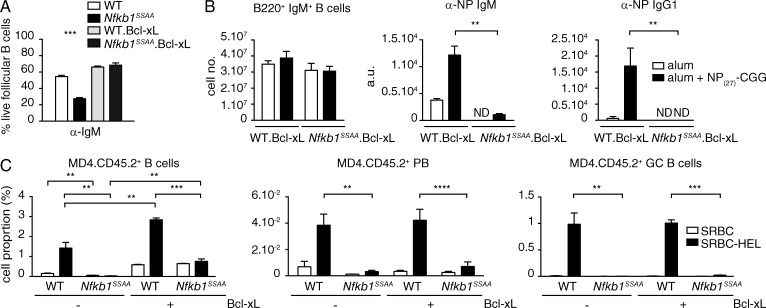
Together these results suggested that failure of the BCR to up-regulate Bcl-2 family proteins in Nfkb1SSAA/SSAA FM B cells increased apoptosis, and might explain the block in TD antibody responses caused by Nfkb1SSAA mutation. To investigate this hypothesis, we crossed Nfkb1SSAA/SSAA mice with a transgenic mouse strain that overexpresses Bcl-xl in the B cell lineage (Takahashi et al., 1999). In vitro experiments demonstrated that Bcl-xl normalized Nfkb1SSAA/SSAA FM B cell survival after BCR stimulation (Fig. 6 A). Nevertheless, chimeric mice in which the B cell compartment was generated from the BM of Bcl-xl Nfkb1SSAA/SSAA mice were still unable to mount an IgG1 antibody response to NP27-CGG, while the IgM anti-NP response was substantially reduced compared with WT Bcl-xl chimera controls (Fig. 6 B). The recovery of transplanted FM B cells from MD4 Nfkb1SSAA/SSAA mice, which express a transgenic BCR specific for HEL (hen egg lysozyme; Goodnow et al., 1988), was increased by Bcl-xl expression (Fig. 6 C). However, MD4 Bcl-xl Nfkb1SSAA/SSAA B cells did not differentiate into plasmablasts or GC B cells after immunization with HEL-SRBC, in contrast to MD4 Bcl-xl Nfkb1+/+ control B cells under the same conditions (Fig. 6 C and Fig. S2). Thus, Nfkb1SSAA mutation did not block TD antibody responses solely by reducing BCR-induced FM B cell survival.
Figure 6.
Bcl-xl overexpression does not rescue the TD antibody response of Nfkb1SSAA/SSAA FM B cells. (A) The fraction of live FM B cells purified from mice of the indicated genotypes was determined as in Fig. 5 C (mean ± SEM). (B) Mixed radiation chimeras were made as in Fig. 2. LH graph show flow cytometric analysis of splenic B220+ IgM+ B cells. Central and RH graphs show anti-NP ELISAs of sera (mean ± SEM) 14 d after NP27-CGG immunization. Results are representative of three independent experiments (4–5 mice per genotype). (C) Mean percentages (±SEM) of transferred CD45.2+ cells (left) differentiating to plasmablasts (B220loIgMloIgD−CD138+CD45.2+; middle) and GC B cells (B220+PNAhiGL7+ CD45.2+; right) in the spleens of CD45.1+ WT hosts after 6d immunization with SRBC-HEL or unconjugated SRBC. Results are representative of two independent experiments (n = 6 mice per condition). Gating strategies used are shown in Fig. S2 (A and B). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Nfkb1SSAA mutation does not block B cell migration to the follicle T cell zone or antigen presenting function
The antigen-specific interaction between B cells and helper T cells in secondary lymphoid organs is an essential step in TD humoral immune responses (Cyster, 2005). T cells provide a second signal via CD40 ligand, which promotes B cell survival and proliferation, and prevents B cell paralysis following BCR stimulation (Goodnow et al., 2010). Our observation that Nfkb1SSAA mutation did not affect CD40L induction of FM B cell proliferation or survival (Fig. 5, B and C), and that CD40 co-stimulation rescued the BCR-induced survival defect of Nfkb1SSAA/SSAA FM B cells in vitro (Fig. 5 D) raised the possibility that these cells failed to receive a signal from CD4+ T cells after NP27-CGG immunization.
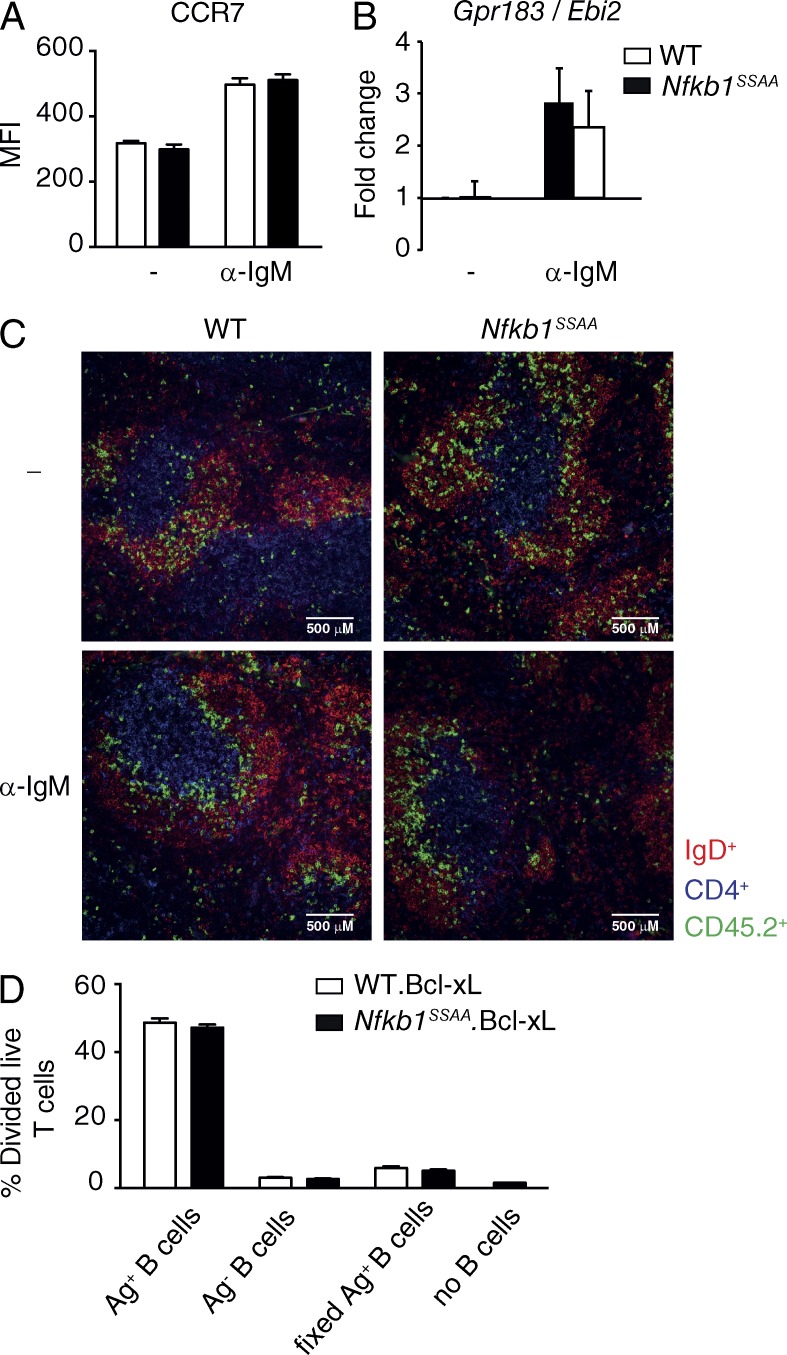
After binding antigen, efficient FM B cell interaction with CD4+ T cells is facilitated by BCR up-regulation of CCR7, which induces FM B cell migration to the B cell zone-T cell zone boundary of the follicle in response to the T zone-expressed CCR7 ligand, CCL21 (Pereira et al., 2010). CCR7 is an NF-κB–regulated gene (Krappmann et al., 2004), but Nfkb1SSAA mutation did not impair the up-regulation of CCR7 expression on FM B cells following BCR ligation (Fig. 7 A). Furthermore, anti-IgM-induced mRNA expression of Epstein-Barr virus-induced molecule 2 (EBI-2), another NF-κB–regulated gene (Glynne et al., 2000) encoding a cytokine which facilitates the initial uniform distribution of B cells along the B/T border (Gatto et al., 2009), was also unaffected by Nfkb1SSAA mutation (Fig. 7 B). Consistent with these data, anti-IgM-stimulated CD45.2+ Nfkb1SSAA/SSAA FM B cells migrated to the follicle-T cell zone border after transfer into wild-type CD45.1+ recipients, similar to their WT counterparts (Fig. 7 C). These results suggest that a failure to co-localize with T cells did not explain the defective function of Nfkb1SSAA/SSAA B cells in TD antibody responses. However, we cannot exclude some contribution of the NF-κB1 signaling pathway in this process, as in vitro stimulation before cell transfer may not completely replicate the in vivo environment.
Figure 7.
Nfkb1SSAA/SSAA B cells migrate normally to follicle T cell zone. (A) Flow cytometric analysis of CCR7 surface expression on purified FM B cells, cultured ± α-IgM for 6 h. Graphs represent CCR7 mean fluorescent intensity (±SEM) of live B cells (7AAD−B220+). (B) Triplicate cultures of purified splenic FM B cells cultured ± α-IgM for 1 h. Expression of Ebi-2 mRNAs was determined by quantitative RT-PCR. Data were normalized to Hprt1 mRNA levels and represent mean fold change (±SEM) relative to unstimulated WT cells. All results are representative of at least two independent experiments done, with 3 mice/genotype each. (C) Splenic distribution of WT and Nfkb1SSAA/SSAA CD45.2+ B cells, prestimulated in vitro for 1 h with α-IgM or control buffer (−), 5.5 h after transfer in to CD45.1+ WT hosts. Transferred B cells were detected by staining for CD45.2 (green), whereas host B cells and T cells were stained with antibodies to IgD (red) and CD4 (blue), respectively. Views are representative of at least two mice of each type. (D) Purified antigen-pulsed (Ag+) or not pulsed (Ag−) B cells were co-culture with purified CTV-labeled OTII CD4+ T cells for 72 h. The fraction of OTII CD4+ that had divided was determined by flow cytometric analysis of CTV dilution (mean ± SEM; triplicates). All results are representative of at least two independent experiments done, with 3 mice/genotype each.
For productive T cell-B cell cooperation in TD antibody responses, B cells must capture external antigens and present them as peptide fragments on MHC class II molecules to CD4+ T cells (Yuseff et al., 2013). It was therefore possible that while Nfkb1SSAA mutation did not inhibit T cell–B cell co-localization, the antigen presenting function of FM B cells was impaired. A model system was used to assay this (Huntington et al., 2006), in which OVA was targeted to the BCR by treating Nfkb1SSAA/SSAA and WT FM B cells sequentially with biotinylated anti-IgM and antibiotin OVA-FITC. Antigen-pulsed B cells were then co-cultured with CTV-labeled OTII CD4+ T cells, which recognize OVA-derived peptides presented on I-Ab class II molecules. Flow cytometric analysis of OTII CD4+ T cell proliferation indicated that Nfkb1SSAA/SSAA FM B cells were able to process and present OVA peptides in an active process requiring viable cells as efficiently as WT cells (Fig. 7 D). In accordance with these data, BCR up-regulation of CD86 and MHC class II was equivalent between Nfkb1SSAA/SSAA and WT FM B cells (unpublished data). The absence of a TD antibody response in Nfkb1SSAA/SSAA chimeric mice, therefore, did not result from a block in the ability of FM B cells to present antigen.
Nfkb1SSAA mutation blocks the differentiation of FM B cells to plasma cells
After antigen exposure and T cell help, proliferating B cells differentiate into either short-lived extrafollicular plasma cells or GC B cells, which subsequently generate plasma cells that secrete high-affinity antibody and persist for a lifetime. Although GC did not develop after NP27-CGG immunization of Nfkb1SSAA/SSAA chimeric mice (Fig. 2 C), the lack of anti-NP IgM and IgG1 at 7d suggested that the inhibitory effect of Nfkb1SSAA mutation might precede GC formation. The complete lack of NP27-specific IgM plasma cells in Nfkb1SSAA/SSAA chimeric mice raised the possibility that Nfkb1SSAA mutation might affect plasmablast differentiation of FM B cells.
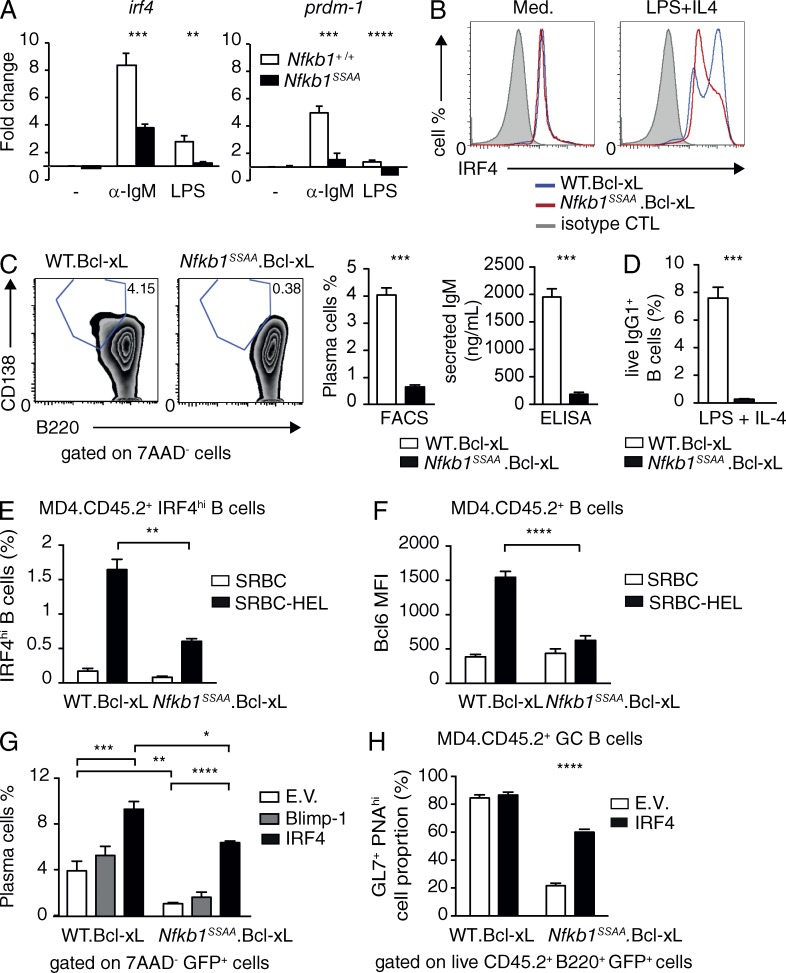
Plasma cell differentiation depends on high expression of the transcription factors BLIMP-1 (B lymphocyte-induced maturation protein 1) and IRF4 (interferon regulatory factor 4). The transcription of Prdm1 (encoding BLIMP-1) and Irf4 genes is regulated by NF-κB (Grumont and Gerondakis, 2000; Saito et al., 2007; Morgan et al., 2009), although the role of NF-κB activation in plasma cell differentiation has not been investigated. The failure to generate antigen-specific plasmablasts in Nfkb1SSAA/SSAA chimeric mice suggested that Nfkb1SSAA mutation might prevent antigen-stimulated FM B cells differentiating into plasmablasts by affecting the expression of these transcription factors. To initially investigate this, qRT-PCR was used to determine Prdm1 and Irf4 mRNA levels in cultured FM B cells. We found that Nfkb1SSAA mutation significantly reduced the mRNA levels of both genes induced by stimulation with anti-IgM (Fig. 8 A). LPS induction of Irf4 mRNA was also decreased in Nfkb1SSAA/SSAA FM B cells compared with WT controls, which correlated with a defect in LPS activation of NF-κB (unpublished data). In accordance with these latter data, flow cytometric analyses demonstrated that Nfkb1SSAA mutation prevented FM B cells (from mice transgenic for Bcl-xl to maintain cell survival) from up-regulating IRF4 protein and differentiating into B220loCD138+ plasmablasts secreting IgM after stimulation with LPS plus IL-4 (Fig. 8, B and C). Nfkb1SSAA mutation also prevented LPS plus IL-4 induced switching to IgG1 (Fig. 8 D), consistent with the critical role of IRF4 in class switch recombination (Sciammas et al., 2006). Importantly, the inhibitory effect of Nfkb1SSAA/SSAA mutation on plasmablast differentiation was confirmed in vivo. Purified FM B cells from MD4 Bcl-xl Nfkb1SSAA/SSAA mice did not differentiate into plasmablasts after transfer to WT recipient mice and concomitant immunization with HEL-SRBC, which contrasted markedly with the MD4 Bcl-xl Nfkb1+/+ control B cells under the same conditions (Fig. 6 C and Fig. S2). This defect in MD4 Bcl-xl Nfkb1SSAA/SSAA B cells correlated with impaired up-regulation of IRF4 (Fig. 8 E and Fig. S3 A) and down-regulation of IgD (Fig. S3 A). Transient expression of Irf4 is essential for up-regulation of Bcl6 and generation of GC B cells (Ochiai et al., 2013). In line with this, Nfkb1SSAA mutation blocked antigen-induced up-regulation of BCL6 (Fig. 8 F and Fig. S3 B) and the formation of GC B cells (Fig. 6 C and Fig. S2 B).
Figure 8.
Nfkb1SSAA mutation impairs plasma cell differentiation. (A) Purified splenic FM B cells were cultured (triplicates) with α-IgM for 1 h or with LPS for 3 h. Expression of Irf4 and Prdm-1 mRNAs was determined by quantitative RT-PCR. Data were normalized to Hprt1 mRNA levels and represent mean fold change (±SEM) relative to unstimulated WT cells. All results are representative of at least three independent experiments (3 mice/genotype each). (B) Histograms show intracellular IRF4 staining in live purified FM B cells cultured ± LPS and IL-4 (40 h). (C) FM B cells were stimulated with LPS and IL-4 for 4 d. CD138+B220lo plasma cells among the live (7AAD−) lymphocytes were quantified by flow cytometry. (middle) Mean percentages of plasma cells (±SEM). (right) IgM secretion as quantified by ELISA (mean ± SEM). (D) Purified FM B cells were stimulated with LPS and IL-4 for 3 d. Fractions of IgG1+ live B cells (B220+7AAD−) were determined by flow cytometry (±SEM; E) Graphs show the mean percentages (±SEM) of transferred CD45.2+ cells up-regulating intracellular IRF4 expression in the spleens of CD45.1+ WT hosts after 6d immunization with SRBC-HEL or unconjugated SRBC. These results are representative of four independent experiments (n = 6 mice per condition). The gating strategies used are shown in Fig. S3 A. (F) Mean fluorescent intensities (MFI) of intracellular Bcl6 in transferred CD45.2+ B cells after 6-d immunization with SRBC-HEL or unconjugated SRBC. Results are representative of three independent experiments (n = 6 mice per condition). Gating strategies used are shown in Fig. S4 B. (G) FM B cells were transduced with recombinant retroviruses. The fraction of GFP+ cells differentiating to plasma cells (B220loIgD−CD138+) after stimulation with LPS and IL-4 was quantified by flow cytometry (±SEM). Results are representative of three independent experiments (3 mice/genotype each). (H) FM B cells were transduced with the indicated retroviruses (E.V., empty vector). The fraction of transferred GFP+ CD45.2+ cells differentiating to GC B cells (B220+PNAhiGL7+ CD45.2+) after 6-d immunization with SRBC-HEL or unconjugated SRBC are shown. Results are representative of two independent experiments (n = 4–6 mice per condition). The gating strategies used are shown in Fig. S5. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To determine whether the reduction of Irf4 and/or Prdm1 mRNA levels caused by Nfkb1SSAA mutation was responsible for the block in plasma cell differentiation, WT and Nfkb1SSAA/SSAA FM B cells were transduced with recombinant retroviruses encoding BLIMP-1 or IRF4. Restoring IRF4 expression increased the fraction of Nfkb1SSAA/SSAA FM B cells that differentiated into plasmablasts sixfold after restimulation with LPS and IL-4 in vitro, whereas BLIMP-1 expression had little effect (Fig. 8 G). Furthermore, retroviral expression of IRF4 in purified FM B cells from MD4 Bcl-xl Nfkb1SSAA/SSAA mice rescued antigen-induced generation of GC B cells after transfer to WT recipient mice (Fig. 8 H and Fig. S4). Insufficient plasmablasts were generated in chimeric mice for quantitation. Together, these results suggested that the inhibitory effect of Nfkb1SSAA mutation on TD antibody responses resulted partly from reduced BCR-mediated up-regulation of Irf4 mRNA, which blocked differentiation of FM B cells to plasmablasts and GC B cells.
Increasing p50 levels overcomes the inhibitory effect of Nfkb1SSAA mutation on TD B cell antibody responses
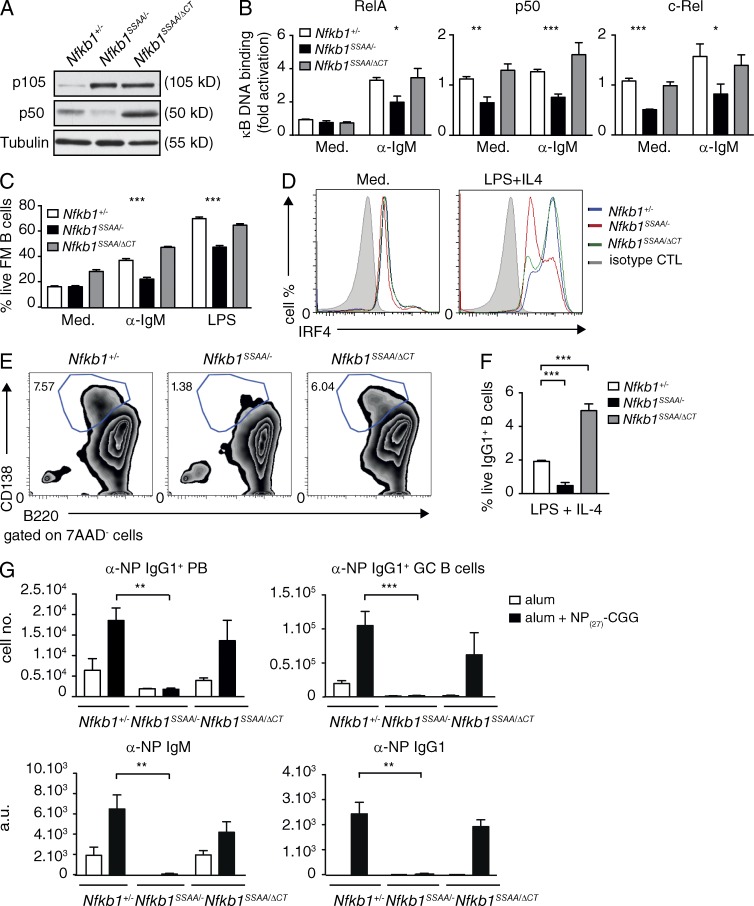
Our in vitro experiments indicated that a major effect of Nfkb1SSAA mutation with respect to NF-κB was to reduce the steady-state levels and activation of NF-κB1 p50 in FM B cells. To investigate the role of reduced p50 levels in the function of Nfkb1SSAA/SSAA FM B cells, Nfkb1SSAA/SSAA mice were crossed with Nfkb1ΔCT/ΔCT mice, which express elevated amounts of p50 compared with WT cells and no p105 (Ishikawa et al., 1998), as described previously (Yang et al., 2012). For comparison with these Nfkb1SSAA/ΔCT mice, Nfbk1+/− and Nfkb1SSAA/− strains were also generated. Nfkb1SSAA/ΔCT FM B cells expressed higher amounts of p50 compared with Nfkb1+/− cells, whereas p50 levels were very low in Nfkb1SSAA/− FM B cells (Fig. 9 A). The steady-state concentrations of p105SSAA were similar between Nfkb1SSAA/− and Nfkb1SSAA/ΔCT FM B cells. NF-κB ELISAs revealed reduced BCR activation of p50, RelA, and c-Rel in Nfkb1SSAA/− FM B cells compared with Nfkb1+/− cells (Fig. 9 B). However, increasing p50 levels with the Nfkb1DCT allele normalized BCR activation of all three Rel subunits.
Figure 9.
Increasing p50 levels overcomes the inhibitory effect of Nfkb1SSAA mutation on FM B cell function. (A) Lysates of purified splenic FM B cells were immunoblotted. (B) Binding of the indicated Rel subunits to an NF-κB oligonucleotide was determined by ELISA (mean ± SEM). (C) The fractions of live FM B cells were determined as described in Fig. 5 C (mean ± SEM). (D) Intracellular IRF4 in live purified FM B cells ± LPS and IL-4 (40 h) was quantified by flow cytometry. (E) Plasma cell differentiation was monitored as in Fig. 8 C. (F) Purified FM B cells were stimulated with LPS and IL-4 for 3 d. The fractions of IgG1+ live B cells (B220+7AAD−) were determined by flow cytometry (±SEM). (G) Mixed radiation chimeras were made as in Fig. 2. Naive B (B220+, IgM+) cells and αNP+ IgG1+ plasmablasts (PB) and GC B cells were quantified by flow cytometry (mean absolute number ± SEM) 7 and 14 d after NP27-CGG immunization, respectively. α-NP IgM and IgG1 serum antibody levels were quantified by ELISA (mean ± SEM) 14 d after NP27-CGG immunization. All results are representative of three independent experiments (4–5 mice per genotype each). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
BCR-induced survival was reduced in Nfkb1SSAA/− FM B cells in comparison with Nfbk1+/− controls (Fig. 9 C), as expected. In contrast, survival of Nfkb1SSAA/ΔCT FM B cells after BCR cross-linking was similar to Nfbk1+/− controls. BCR-induced proliferation was similar between the three genotypes (unpublished data), in line with the small inhibitory effect of homozygous Nfkb1SSAA mutation on anti-IgM–induced cell division (Fig. 5 B). Nfkb1SSAA mutation reduced both survival and proliferation of FM B cells after TLR4 stimulation, and these inhibitory effects were rescued by the Nfkb1ΔCT allele (Fig. 9 C and not depicted). The impairment by Nfkb1SSAA mutation of IRF4 up-regulation, differentiation to CD138+ B220lo plasmablasts and switching to IgG1 after LPS and IL-4 stimulation of FM B cells were also overcome by the Nfkb1ΔCT allele (Fig. 9, D–F). These results showed that the reduced survival, plasmablast differentiation and class switch recombination of FM B cells in vitro caused by Nfkb1SSAA mutation were rescued by increasing p50 levels with the Nfkb1ΔCT allele, which normalized NF-κB activation.
Finally, to investigate the importance of p50 in the inhibitory effect of Nfkb1SSAA mutation on TD antibody responses, we generated mixed BM chimeras and then immunized with NP27-CGG. The titers of anti-NP IgM and IgG1 in the serum, as well as the number of anti-NP IgG1 plasma cells and GC B cells in the spleens of chimeras reconstituted with Nfkb1SSAA/− BM, were significantly reduced compared with Nfbk1+/− control chimeras (Fig. 9 G and not depicted). In contrast, no significant differences were detected between chimeras generated with Nfbk1+/− and Nfkb1SSAA/ΔCT BM cells. Therefore, rescuing NF-κB activation by increasing p50 levels with the Nfkb1DCT allele overcame the inhibitory effect of Nfkb1SSAA mutation on the FM B cell antibody response to a TD antigen.
DISCUSSION
Analyses of conditional knockout mouse strains lacking IKK2 or NEMO expression in the B cell lineage have demonstrated that canonical NF-κB activation is critical for mature B cell generation or homeostasis (Pasparakis et al., 2002; Li et al., 2003). However, this has prevented using such mouse strains for investigation of the potential role for canonical NF-κB activation in B cells for antibody production during an immune response. In the present study, we used Nfkb1SSAA/SSAA mice, in which the development and homeostasis of peripheral FM B cells were normal, to investigate the role of NF-κB activation in FM B cells in humoral immunity. Nfkb1SSAA mutation selectively inhibits the IKK signaling pathway that regulates NF-κB activation by inducing NF-κB1 p105 proteolysis (Sriskantharajah et al., 2009). Our results demonstrate for the first time that IKK-induced p105 proteolysis is required for optimal BCR activation of NF-κB in B cells, and that this is essential for generation of a TD antibody response, specifically regulating the survival of FM B cells and their differentiation to plasmablasts and GC B cells after BCR stimulation.
Nfkb1SSAA mutation reduced activation of p50 and RelA after both BCR and CD40 stimulation. Activation of c-Rel was also significantly reduced in Nfkb1SSAA/SSAA FM B cells compared with WT, particularly after prolonged BCR stimulation. Immunoprecipitation and immunoblotting demonstrated that p50, RelA, and c-Rel were able to interact with p105. The inhibitory effect of Nfkb1SSAA mutation on the activation of these Rel subunits, therefore, probably resulted from their retention in the cytoplasm by p105SSAA. In addition, steady-state levels of p50 were reduced in Nfkb1SSAA/SSAA FM B cells compared with WT controls, suggesting that Nfkb1SSAA mutation reduced p105 processing to p50, similar to the situation we previously described in macrophages (Yang et al., 2012). The Nfkb1ΔCT allele overcame the inhibitory effect of Nfkb1SSAA mutation on BCR activation of p50, RelA, and c-Rel, presumably by increasing p50 expression to a degree that the concentration of p50 exceeded p105 (p105:p50 [Nfkb1SSAA/ΔCT] = 0.63 ± 0.03; p105:p50 [Nfkb1SSAA/−] = 4.04 ± 0.3). Consequently, p105SSAA became saturated with p50 monomers and homodimers, with which it preferentially binds (Moorthy and Ghosh, 2003), and no longer able to block BCR activation of RelA and c-Rel monomers and p50/RelA and p50/c-Rel heterodimers. Remarkably, the Nfkb1ΔCT allele also normalized FM B cell survival and plasmablast differentiation in vitro, and TD antibody responses in vivo, demonstrating that the inhibitory effects of Nfkb1SSAA mutation on FM B cell function resulted from impaired NF-κB activation.
Nfkb1SSAA mutation did not affect RelB and p52 NF-κB–binding activity in CD40L-stimulated cells, presumably because these subunits are predominantly associated with NF-κB2 p100 (Solan et al., 2002). Inability to inhibit activation of the alternative NF-κB pathway probably explains why Nfkb1SSAA mutation did not affect A1 mRNA up-regulation and the survival of FM B cells after CD40 stimulation, which activates both canonical and alternative NF-κB signaling pathways (Rickert et al., 2011). CD40 co-stimulation overcame the BCR-induced survival defect of Nfkb1SSAA/SSAA FM B cells in vitro. However, although these cells could co-localize and presumably interact with CD40 on T cells in the spleen after BCR cross-linking, they still failed to mount an antibody response to NP27-CGG. This suggests that defective survival alone did not block the TD antibody response of Nfkb1SSAA/SSAA FM B cells. In line with this, transgenic overexpression of Bcl-xl, which restored in vitro BCR-induced survival of Nfkb1SSAA/SSAA FM B cells to normal, did not rescue antibody responses to a TD antigen.
Nfkb1SSAA mutation blocked the late activation of c-Rel after BCR cross-linking with anti-IgM, and impaired BCR induction of Irf4 mRNA, a c-Rel–regulated gene (Grumont and Gerondakis, 2000). The up-regulation of IRF4 protein expression in MD4 B cells by HEL antigen in vivo was also prevented by Nfkb1SSAA mutation, confirming the physiological significance of these findings. IRF4 is a critical transcription factor for the induction of plasma cell differentiation and Ig class switch recombination (Sciammas et al., 2006; De Silva et al., 2012). Consistent with this, Nfkb1SSAA mutation reduced the ability of LPS plus IL-4 to induce IRF4 expression in FM B cells, impairing differentiation to IgM-secreting plasmablasts and switching to IgG1. The induction of plasmablast differentiation of MD4 B cells in vivo by HEL was also prevented by Nfkb1SSAA mutation. In addition, retroviral expression of IRF4 rescued plasma cell differentiation of Nfkb1SSAA/SSAAv FM B cells stimulated with LPS and IL-4, confirming a functional effect of reduced IRF4 in these cells. These experiments establish Irf4 as a physiologically important transcriptional target of the NF-κB1 signaling pathway in B cell antibody responses, and demonstrate for the first time the critical role of NF-κB activation in plasmablast differentiation.
CD19-Cre Irf4fl/fl mice have a substantially reduced GC response, and residual GCs form only with B cells in which the Irf4 allele has not been deleted (Ochiai et al., 2013). IRF4, therefore, plays an essential and B cell autonomous role in GC formation, regulating the expression of the key GC transcription factor Bcl6 (Goodnow et al., 2010; Ochiai et al., 2013). Antigen-stimulated Nfkb1SSAA/SSAA FM B cells did not up-regulate Bcl6 or differentiate into GC B cells during a TD immune response. NF-κB signaling activity is very low in GC B cell centroblasts (Shaffer et al., 2001; Basso et al., 2004). However, IRF4 is required for the initiation, but not maintenance of the GC state (Klein et al., 2006; Ochiai et al., 2013). This suggests that Nfkb1SSAA mutation may have prevented the initiation of GC formation by inhibition of NF-κB–dependent transcriptional up-regulation of Irf4. Consistent with this hypothesis, retroviral expression of IRF4 significantly increased antigen-induced differentiation of MD4 Bcl-xl Nfkb1SSAA/SSAA B cells. The failure to induce IRF4 in Nfkb1SSAA/SSAA B cells was a feature of agonist of the BCR and TLR4, but not CD40 (unpublished data). As TLR signaling is not required for antibody responses to antigens delivered in alum (Gavin et al., 2006), this suggests that the immune response defects detected were primarily caused by impaired BCR-mediated signaling.
In this study, we investigated the role of IKK-induced NF-κB1 p105 proteolysis in mature B cells, using Nfkb1SSAA/SSAA mice in which this IKK signaling pathway is blocked (Sriskantharajah et al., 2009). Nfkb1SSAA mutation prevented FM B cells from mounting an antibody response to a TD antigen by selectively blocking BCR-induction of BCL-2 family proteins and IRF4, which impaired FM B cell survival and antigen-induced differentiation of FM B cells to plasmablasts and GC B cells, respectively. Our study demonstrates that the regulation of NF-κB activation in mature B cells by IKK-induced NF-κB1 p105 proteolysis is essential for TD humoral immune responses.
MATERIALS AND METHODS
Mouse strains.
Mouse strains were bred in the specific pathogen–free animal facility of the National Institute for Medical Research (NIMR; London, UK). 7–12-wk-old mice were used for all experiments. Rag1−/− (Mombaerts et al., 1992), μMT−/− (Kitamura et al., 1991), and HEL-specific BCR transgenic mice (MD4; Mason et al., 1992) and Nfkb1SSAA/SSAA (Sriskantharajah et al., 2009), Nfkb1−/− (Sha et al., 1995), and Map3k8−/− (Dumitru et al., 2000) mouse strains have been previously described and were all fully backcrossed on to a C57BL/6 background. Nfkb1+/−, Nfkb1SSAA/- and Nfkb1SSAA/ΔCT (Ishikawa et al., 1998) mice were generated by intercrossing Nfkb1SSAA/SSAA, Nfkb1ΔCT/ ΔCT, and Nfkb1−/− strains. Nfkb1SSAA/SSAA.Bcl-xL and Nfkb1+/+.Bcl-xL mice experimental mice were generated by intercrossing Bcl-xL transgenic mice (Takahashi et al., 1999) with Nfkb1SSAA/SSAA and C57BL/6 mouse strains, respectively. These were subsequently intercrossed with MD4.Rag1−/− mice to generate MD4.Rag1+/−.Nfkb1SSAA/SSAA.Bcl-xL and MD4.Rag1+/−.Nfkb1+/+.Bcl-xL mice.
This study was conducted following authorization by the UK Home Office, under relevant Project Licence authority.
BM chimeras and B cell adoptive transfer.
BM cells were harvested from femora and tibiae of the indicated donor mice, and resuspended in ACK lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 100 µM EDTA) to lyse red blood cells. Cells (4–5 × 106 per mouse) were then transferred by intravenous injection into sublethally irradiated Rag1−/− mice (500 rads; Cs source). For competition experiments, BM cell suspensions from CD45.1+ C57BL/6 were mixed with either Nfkb1SSAA/SSAA or wild-type Nfkb1+/+ CD45.2+ BM cells, at a 1:1 ratio. For mixed BM chimeras, sublethally irradiated Rag1−/− mice were reconstituted with Nfkb1SSAA/SSAA or wild-type Nfkb1+/+ BM cells mixed at 1:4 ratio with μMT−/− BM cells. Chimeric mice were immunized 8 wk after reconstitution.
To analyze B cell positioning in the spleen after BCR stimulation, 8–10 × 106 splenic purified B cells from either Nfkb1SSAA/SSAA or wild-type mice were cultured in vitro with or without anti-IgM (10 µg/ml; Jackson ImmunoResearch Laboratories) for 1 h (Pereira et al., 2009). Cells were then transferred into CD45.1+ C57BL/6 hosts by intravenous injection, and spleens removed 5.5 h later for immunofluorescence analysis.
To monitor plasma cell differentiation and GC B cell formation in vivo, 8–10 × 106 splenic purified B cells from MD4.Rag1+/−.Nfkb1SSAA/SSAA.BcL-xL and MD4.Rag1+/−.Nfkb1+/+.Bcl-xL were cotransferred with 0.5–1.109 HEL-conjugated or control sheep red blood cells into CD45.1+ C57BL/6 hosts by intravenous injection (Reif et al., 2002; Kelly et al., 2011). 6 d after transfer, recipient mice were culled and spleens were removed for flow cytometric analysis.
Assay of antibody responses.
For TD antibody responses, mixed BM chimeras were immunized i.p. with 50 µg NP27-CGG in PBS (Biosearch Technologies) precipitated in alum (Imject; Thermo Fisher Scientific; 3:1 ratio). Alum mixed with PBS was used as a control. The production of NP-specific and total immunoglobulins (IgM and IgG1) was monitored by ELISA, using 96-well Maxisorp ELISA plates pretreated with 50 µl/well of 5 µg/ml NP14.-BSA (Biosearch Technologies). Antibody isotypes and subclasses were detected with horseradish peroxidase–conjugated mouse IgM and subclass IgG-specific antibodies (Southern Biotech).
Histological analyses.
Cryosections (7–10 µM) of spleen were prepared, dried, and fixed with acetone before immunofluorescent analysis (Turner et al., 1997). GC B cells were stained using GL7-FITC mAb (Ly77, BD), revealed with rabbit anti-FITC (Alexa Fluor 488 Signal-Amplification kit; Invitrogen), IgD-PE (11-26c; eBioscience), and B220-Alexa Fluor 647 (RA3-6B2, BD). For B cell migration assays, CD45.2-FITC (104, eBioscience), CD4-Alexa Fluor 647 (ABD Serotec), and IgD-PE (11-26c; eBioscience) were used to identify transferred B cells. Spleen sections were analyzed by immunofluorescence microscopy using either a Leica confocal microscope TSP-SP2 (Leica) or a DeltaVision RT Imaging System (Applied Precision Inc.).
Flow cytometry.
Single-cell suspensions were generated from peripheral lymph nodes, spleen, BM, or peritoneal wash via gentle homogenization through nylon mesh filters (70 µM; BD). Erythrocytes in BMs and spleens were lysed with ACK lysis buffer before staining, and cell concentrations were determined using a Casy Counter (Scharfe Instrument Systems). CD1d-PE and CD1d-bio (clone 1B1); CD21-FITC (eBio8D9); CD23-PE (B3B4); B220-FITC, B220-PE, B220-APC, and B220-eFluor450 (RA3-6B2); CD93-APC (AA4.1); IgM-PECy7 (II/41); IgD-FITC, IgD-eFluor450, IgD-PE, and IgD-bio (11–26); Ly5.1-FITC, and Ly5.1-APC (A20); Ly5.2-FITC, and Ly5.2-APC-eFluor780 (104), CD43-bio (eBioR2/60), IgG1-APC (X56), Gr1-bio (RB6-8C5), CD11b-Bio (M1/70); CD11c-Bio (N418); CD5-PE (53–7.3); CD19-FITC (MB19-1); TCRβ-FITC (H57-597); and CCL19-Fc fusion protein (plus anti–human Fc-PE secondary to stain CCR7) were obtained from eBioscience. CD4-PerCP (RM4-5), B220-PcP (RA3-6B2), and CD8-PerCP (53–6.7) were purchased from BioLegend; CD2-PE (RM2-5), CD9-bio (KMC8), CD21-PE (7G6), Streptavidin-PerCP (554064), CD138-PE (281–2), and GL7-FITC (Ly77) were obtained from BD; PNA-Bio (FL-1075) was purchased from Vector Laboratories; and CD19-APC (RM705) and CD23-APC (MCD2305) were purchased from Life Technologies. For thymus-dependent (TD) antibody response assays, NP27-BSA (Biosearch Technologies) was labeled with the lynx rapid rpe or percp antibody conjugation kits (AbDserotec) according to the manufacturer’s protocol. Intracellular IRF4 was detected using IRF4-PE (eBRG1), after cell permeabilization (Foxp3 Fixation/Permeabilization kit; eBioscience). Live cells (Live/dead Fixable Neat-IR Dead Cell Stain kit; Life Technologies) and extracellular markers were identified by prior staining of intact cells.
Stained cells were analyzed by flow cytometry on a CyAn ADP Analyzer (Beckman Coulter) or FACSCanto II Analyzer (BD). Data analyses were performed with FlowJo 887 software (Tree Star).
B cell isolation and in vitro culture.
Splenic follicular B cells were isolated by sequential hypotonic lysis of RBC and magnetic bead separation using a mixture of biotinylated anti-CD43 and anti-CD1d antibodies and Streptavidin-coupled Dynabeads (Life Technologies). Purity of the resulting FM B cell population (B220+CD23hiCD21lo) was >96% as assessed by flow cytometry. Cells were cultured at a density of 2 × 106 cells/ml in complete medium (DMEM plus 10% heat inactivated FCS [Lonza], 100 U/ml Penicillin, 100 µg/ml Streptomycin, 100 µM nonessential amino acids, 100 mM Na-Pyruvate, and 50 µM 2-mercaptoethanol), and stimulated with anti-IgM (10 µg/ml; Jackson ImmunoResearch Laboratories), CD40L (0.5 µg/ml; R&D Systems), or LPS (1–10 µg/ml; Salmonella minnesota R595; Alexis Biochemicals) ± IL-4 (20 ng/ml; PeproTech).
For in vitro survival assays, 3 × 105 B cells were cultured in a 96-well tissue-culture plate in complete medium (200 µl) with or without stimulation (see above). Cells were harvested 2 d later, stained for a B cell marker (B220 or CD19) resuspended and 7-amino-actinomycin D (7-AAD; Sigma-Aldrich). Cell division was monitored by flow cytometry after labeling of FM B cells with 2 µM CFSE (CellTrace; Invitrogen) in Dulbecco’s PBS (Invitrogen). The frequency of the precursor population triggered into cell division was determined using FlowJo V8.5 proliferation analysis software (Tree Star). To monitor the antigen-presenting function of FM B cells, OVA was targeted to the BCR by treating purified B cells sequentially with biotinylated anti-IgM (0.5 µg/ml; Jackson ImmunoResearch Laboratories) and anti-biotin OVA-FITC (Miltenyi Biotec). Antigen-pulsed B cells (2 × 106/ml) were then co-cultured with Cell Trace Violet-labeled OTII CD4+ T cells (5 µM; Life Technologies; 1 × 106/ml) for 72 h. T cell activation was determined by estimating the ability of T cells to divide. To demonstrate that OVA presentation was actively mediated, antigen-pulsed B cells were fixed in 3% paraformaldehyde before co-culture with T cells.
To monitor plasma cell differentiation, FM B cells (1.5 × 106/ml) were cultured with LPS (40 µg/ml) plus IL-4 (20 ng/ml) for 4 d. Plasma cells (B220loCD138+) were identified by flow cytometry. In vitro IgM production was analyzed by harvesting cells after 4-d culture with LPS and IL-4, and replating washed live cells at 106 cells/ml in complete medium. Supernatants were collected after an additional 1-d culture, and analyzed by ELISA. The yields of Nfkb1SSAA/SSAA and WT FM B cells were similar after reculture.
Retroviral infection.
pMSCV-IRES-GFP, pMSCV-IRF4-IRES-GFP, and pMSCV-Blimp-IRES-GFP plasmids (Kallies et al., 2007) were a gift from A. Kallies (The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia). Retroviruses were prepared as previously described using Plat-E packaging cells (Robinson et al., 2007). FM B cells were cultured for 24 h with CD40L and IL-4 before retroviral infection. To analyze the effect of ectopic IRF4 expression on GC B cell formation, purified MD4.Rag1+/−.Nfkb1SSAA/SSAA.Bcl-xL and MD4.Rag1+/−.Nfkb1+/+.Bcl-xL FM B cells were transduced twice with retrovirus, prior to cotransfer with 0.5 to 1.109 HEL-conjugated or control sheep red blood cells (SRBC) into CD45.1+ C57BL/6 hosts by intravenous injection. Splenic B cells were analyzed by flow cytometry after 6 d.
Protein analyses.
For immunoblots of total lysates, FM B cells (3 × 106 cells per point) were lysed RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 2 mM Na4P2O7, 5 mM dithiothreitol, 1 mM Na3VO4, protease inhibitors [Roche], 0.5% deoxycholate, 1% Triton X-100, and 0.1% SDS). After centrifugation, lysates (20–25 µg) were mixed with an equal volume of 2x SDS-PAGE sample buffer. For immunoprecipitation, cells (5 × 107 per point) were lysed in IP buffer (10 mM Hepes, pH 7.6, 1 mM EGTA, 10 mM KCl, 1 mM dithiothreitol, 20 mM NaF, 1 mM Na4P2O7, protease inhibitors, and 0.6% Nonidet-P40). Immunoprecipitations (150 µg of protein extract) were performed as previously described (Kabouridis et al., 1997).
To analyze nuclear translocation of Rel subunits, washed cells were lysed with buffer N (10 mM Hepes, pH 7.6, 0.1 mM EGTA, 10 mM KCl 1.5 mM MgCl2, 1 mM DTT, 20 mM NaF, 1 mM Na-pyrophosphate, protease inhibitors, and 0.2% NP-40). After 2 min, nuclei were pelleted by centrifugation, washed twice in lysis buffer, and nuclear proteins extracted with RIPA buffer. Rel proteins in nuclear extracts (20 µg) were quantified by immunoblotting and laser densitometry, normalizing to SAM68 content.
The p105C antibody to the p105 C terminus, used for p105 immunoprecipitation, has been described previously (Salmerón et al., 2001). p50 antibody was obtained from (Delta Biolabs) and phospho S933 p105 antibody from Cell Signaling Technology. Antibodies to other Rel family proteins, SAM68 and IκBα, were purchased from Santa Cruz Biotechnology, Inc. Tubulin was detected with TAT-1 α-tubulin mAb (provided by K. Gull, University of Oxford, Oxford, England, UK) and used as loading control protein on immunoblots of total cell lysates.
NF-κB activation assays.
Nuclear extracts were prepared as above, and activation of NF-κB subunits assayed (2–4 µg per point) using a commercial ELISA kit (TransAm NF-κB family kit; Active Motif). To increase sensitivity, c-Rel binding was assayed with anti–c-Rel (1 µg per well; sc-71X; Santa Cruz Biotechnology, Inc.). Data were normalized against SAM68 content of nuclear extracts.
Real-time quantitative PCR.
RNA from stimulated and unstimulated FM B cells (5 × 106/ml) was isolated using the RNeasy kit, and contaminating DNA removed using RNase-free DNase set (QIAGEN) according to manufacturer’s instructions and DNase treated (Roche). cDNA was produced using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Life Technologies) and standard protocols. Expression of target genes was determined by real time PCR using a Perkin Elmer ABI Prism 7000 Sequence Detection System, and commercial FAM labeled probes (Applied Biosystems) with the TaqMan Gene Expression Master Mix (Applied Biosystems). Target gene mRNA levels were normalized against Hprt mRNA levels.
Statistical analysis.
All data analyses were performed using GraphPad software (GraphPad Software Inc.). In vitro data were compared using Student’s t test (two tailed and unpaired test). For in vivo experiments, all statistical comparisons were performed using the nonparametric two-tailed Mann-Whitney test. Statistically significant differences are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material.
Fig. S1 shows gating strategies for Fig. 1. Fig. S2 shows gating strategies for Fig. 6. Fig. S3 shows gating strategies for Fig. 8 (E and F). Fig. S4 shows gating strategies for Fig. 8 H. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132019/DC1.
Supplementary Material
Acknowledgments
We thank the NIMR Photographics department, NIMR Confocal Microscopy, NIMR Biological Services, NIMR flow cytometry service, Harald Hartweger (NIMR), Abduelhakem Ben-Addi, and other members of the Ley laboratory for help during the course of this work. We are also grateful to Anne O’Garra (NIMR), Ben Seddon (NIMR), and Steve Gerondakis (Monash University, Australia) for their helpful comments on the manuscript.
This work was funded by the UK Medical Research Council, a Leukaemia and Lymphoma Research Project Grant (LRF:06050), and an award from the Bettencourt-Schueller Foundation to E. Jacque.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAFF
- B cell activation factor
- BCR
- B cell antigen receptor
- FM
- follicular mature
- GC
- germinal center
- HEL
- hen egg lysozyme
- IκB
- inhibitor of NF-κB
- IKK
- IκB kinase
- NF-κB
- nuclear factor κB
- PC
- plasma cell
- MZ
- marginal zone
- NP-CGG
- 4-Hydroxy-3-nitrophenylacetyl hapten conjugated to chicken gammaglobulin
- SRBC
- sheep red blood cells
- TD
- T cell–dependent
References
- Basso, K., Klein U., Niu H., Stolovitzky G.A., Tu Y., Califano A., Cattoretti G., and Dalla-Favera R.. 2004. Tracking CD40 signaling during germinal center development. Blood. 104:4088–4096 10.1182/blood-2003-12-4291 [DOI] [PubMed] [Google Scholar]
- Beinke, S., and Ley S.C.. 2004. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem. J. 382:393–409 10.1042/BJ20040544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinke, S., Robinson M.J., Hugunin M., Ley S.C., Allen H., and Ley S.C.. 2004. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IkappaB kinase-induced proteolysis of NF-kappaB1 p105. Mol. Cell. Biol. 24:9658–9667 10.1128/MCB.24.21.9658-9667.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggins, A.G.S., Pepper C., Patten P.E.M., Hewamana S., Gohil S., Moorhead J., Folarin N., Yallop D., Thomas N.S.B., Mufti G.J., et al. 2010. Interaction with vascular endothelium enhances survival in primary chronic lymphocytic leukemia cells via NF-kappaB activation and de novo gene transcription. Cancer Res. 70:7523–7533 10.1158/0008-5472.CAN-10-1634 [DOI] [PubMed] [Google Scholar]
- Cyster, J.G.2005. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu. Rev. Immunol. 23:127–159 10.1146/annurev.immunol.23.021704.115628 [DOI] [PubMed] [Google Scholar]
- Damdinsuren, B., Zhang Y., Khalil A., Wood W.H. III, Becker K.G., Shlomchik M.J., and Sen R.. 2010. Single round of antigen receptor signaling programs naive B cells to receive T cell help. Immunity. 32:355–366 10.1016/j.immuni.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva, N.S., Simonetti G., Heise N., and Klein U.. 2012. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol. Rev. 247:73–92 10.1111/j.1600-065X.2012.01113.x [DOI] [PubMed] [Google Scholar]
- Derudder, E., Cadera E.J., Vahl J.C., Wang J., Fox C.J., Zha S., van Loo G., Pasparakis M., Schlissel M.S., Schmidt-Supprian M., and Rajewsky K.. 2009. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat. Immunol. 10:647–654 10.1038/ni.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru, C.D., Ceci J.D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J.-H., Patriotis C., Jenkins N.A., Copeland N.G., Kollias G., and Tsichlis P.N.. 2000. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell. 103:1071–1083 10.1016/S0092-8674(00)00210-5 [DOI] [PubMed] [Google Scholar]
- Gatto, D., Paus D., Basten A., Mackay C.R., and Brink R.. 2009. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 31:259–269 10.1016/j.immuni.2009.06.016 [DOI] [PubMed] [Google Scholar]
- Gavin, A.L., Hoebe K., Duong B., Ota T., Martin C., Beutler B., and Nemazee D.. 2006. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 314:1936–1938 10.1126/science.1135299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis, S., and Siebenlist U.. 2010a. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb. Perspect. Biol. 2:a000182 10.1101/cshperspect.a000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis, S., and Siebenlist U.. 2010b. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb. Perspect. Biol. 2:a000182 10.1101/cshperspect.a000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., May M.J., and Kopp E.B.. 1998. NF-κ B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225–260 10.1146/annurev.immunol.16.1.225 [DOI] [PubMed] [Google Scholar]
- Glynne, R., Akkaraju S., Healy J.I., Rayner J., Goodnow C.C., and Mack D.H.. 2000. How self-tolerance and the immunosuppressive drug FK506 prevent B-cell mitogenesis. Nature. 403:672–676 10.1038/35001102 [DOI] [PubMed] [Google Scholar]
- Goodnow, C.C., Crosbie J., Adelstein S., Lavoie T.B., Smith-Gill S.J., Brink R.A., Pritchard-Briscoe H., Wotherspoon J.S., Loblay R.H., Raphael K., et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682 10.1038/334676a0 [DOI] [PubMed] [Google Scholar]
- Goodnow, C.C., Vinuesa C.G., Randall K.L., Mackay F., and Brink R.. 2010. Control systems and decision making for antibody production. Nat. Immunol. 11:681–688 10.1038/ni.1900 [DOI] [PubMed] [Google Scholar]
- Grossmann, M., O’Reilly L.A., Gugasyan R., Strasser A., Adams J.M., and Gerondakis S.. 2000. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J. 19:6351–6360 10.1093/emboj/19.23.6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont, R.J., and Gerondakis S.. 2000. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor κB. J. Exp. Med. 191:1281–1292 10.1084/jem.191.8.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont, R.J., Rourke I.J., O’Reilly L.A., Strasser A., Miyake K., Sha W., and Gerondakis S.. 1998. B lymphocytes differentially use the Rel and nuclear factor κB1 (NF-κB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J. Exp. Med. 187:663–674 10.1084/jem.187.5.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont, R.J., Rourke I.J., and Gerondakis S.. 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 13:400–411 10.1101/gad.13.4.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont, R.J., Strasser A., and Gerondakis S.. 2002. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-kappaB regulated c-myc transcription. Mol. Cell. 10:1283–1294 10.1016/S1097-2765(02)00779-7 [DOI] [PubMed] [Google Scholar]
- Hettmann, T., Opferman J.T., Leiden J.M., and Ashton-Rickardt P.G.. 2003. A critical role for NF-kappaB transcription factors in the development of CD8+ memory-phenotype T cells. Immunol. Lett. 85:297–300 10.1016/S0165-2478(02)00260-2 [DOI] [PubMed] [Google Scholar]
- Huntington, N.D., Xu Y., Puthalakath H., Light A., Willis S.N., Strasser A., and Tarlinton D.M.. 2006. CD45 links the B cell receptor with cell survival and is required for the persistence of germinal centers. Nat. Immunol. 7:190–198 10.1038/ni1292 [DOI] [PubMed] [Google Scholar]
- Ishikawa, H., Claudio E., Dambach D., Raventós-Suárez C., Ryan C., and Bravo R.. 1998. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p)105 precursor (NF-kappaB1) but expressing p50. J. Exp. Med. 187:985–996 10.1084/jem.187.7.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabouridis, P.S., Magee A.I., and Ley S.C.. 1997. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 16:4983–4998 10.1093/emboj/16.16.4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaileh, M., and Sen R.. 2012. NF-κB function in B lymphocytes. Immunol. Rev. 246:254–271 10.1111/j.1600-065X.2012.01106.x [DOI] [PubMed] [Google Scholar]
- Kallies, A., Hasbold J., Fairfax K., Pridans C., Emslie D., McKenzie B.S., Lew A.M., Corcoran L.M., Hodgkin P.D., Tarlinton D.M., and Nutt S.L.. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 26:555–566 10.1016/j.immuni.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Karin, M., and Ben-Neriah Y.. 2000. Phosphorylation meets ubiquitination: the control of NF-[κ]B activity. Annu. Rev. Immunol. 18:621–663 10.1146/annurev.immunol.18.1.621 [DOI] [PubMed] [Google Scholar]
- Kelly, L.M., Pereira J.P., Yi T., Xu Y., and Cyster J.G.. 2011. EBI2 guides serial movements of activated B cells and ligand activity is detectable in lymphoid and nonlymphoid tissues. J. Immunol. 187:3026–3032 10.4049/jimmunol.1101262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, D., Roes J., Kühn R., and Rajewsky K.. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 350:423–426 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- Klein, U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Köntgen, F., Grumont R.J., Strasser A., Metcalf D., Li R., Tarlinton D., and Gerondakis S.. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965–1977 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- Krappmann, D., Wegener E., Sunami Y., Esen M., Thiel A., Mordmuller B., and Scheidereit C.. 2004. The IkappaB kinase complex and NF-kappaB act as master regulators of lipopolysaccharide-induced gene expression and control subordinate activation of AP-1. Mol. Cell. Biol. 24:6488–6500 10.1128/MCB.24.14.6488-6500.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv, Y., Cohen S., and Ciechanover A.. 2009. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol. Cell. 33:496–504 10.1016/j.molcel.2009.01.023 [DOI] [PubMed] [Google Scholar]
- Lang, V., Janzen J., Fischer G.Z., Soneji Y., Beinke S., Salmeron A., Allen H., Hay R.T., Ben-Neriah Y., and Ley S.C.. 2003. betaTrCP-mediated proteolysis of NF-kappaB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol. Cell. Biol. 23:402–413 10.1128/MCB.23.1.402-413.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.-W., Omori S.A., Labuda T., Karin M., and Rickert R.C.. 2003. IKK β is required for peripheral B cell survival and proliferation. J. Immunol. 170:4630–4637 10.4049/jimmunol.170.9.4630 [DOI] [PubMed] [Google Scholar]
- Mackay, F., Figgett W.A., Saulep D., Lepage M., and Hibbs M.L.. 2010. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol. Rev. 237:205–225 10.1111/j.1600-065X.2010.00944.x [DOI] [PubMed] [Google Scholar]
- Mason, D.Y., Jones M., and Goodnow C.C.. 1992. Development and follicular localization of tolerant B lymphocytes in lysozyme/anti-lysozyme IgM/IgD transgenic mice. Int. Immunol. 4:163–175 10.1093/intimm/4.2.163 [DOI] [PubMed] [Google Scholar]
- Mombaerts, P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., and Papaioannou V.E.. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- Moorthy, A.K., and Ghosh G.. 2003. p105.Ikappa Bgamma and prototypical Ikappa Bs use a similar mechanism to bind but a different mechanism to regulate the subcellular localization of NF-κ B. J. Biol. Chem. 278:556–566 10.1074/jbc.M207515200 [DOI] [PubMed] [Google Scholar]
- Morgan, M.A.J., Magnusdottir E., Kuo T.C., Tunyaplin C., Harper J., Arnold S.J., Calame K., Robertson E.J., and Bikoff E.K.. 2009. Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation. Mol. Cell. Biol. 29:5813–5827 10.1128/MCB.00670-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai, K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., and Sciammas R.. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 38:918–929 10.1016/j.immuni.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracki, S.A., Walker J.A., Hibbs M.L., Corcoran L.M., and Tarlinton D.M.. 2010. Plasma cell development and survival. Immunol. Rev. 237:140–159 10.1111/j.1600-065X.2010.00940.x [DOI] [PubMed] [Google Scholar]
- Pasparakis, M., Schmidt-Supprian M., and Rajewsky K.. 2002. IkappaB kinase signaling is essential for maintenance of mature B cells. J. Exp. Med. 196:743–752 10.1084/jem.20020907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peperzak, V., Vikström I., Walker J., Glaser S.P., LePage M., Coquery C.M., Erickson L.D., Fairfax K., Mackay F., Strasser A., et al. 2013. Mcl-1 is essential for the survival of plasma cells. Nat. Immunol. 14:290–297 10.1038/ni.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, J.P., Kelly L.M., Xu Y., and Cyster J.G.. 2009. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 460:1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, J.P., Kelly L.M., and Cyster J.G.. 2010. Finding the right niche: B-cell migration in the early phases of T-dependent antibody responses. Int. Immunol. 22:413–419 10.1093/intimm/dxq047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, S., and Cariappa A.. 2009. The follicular versus marginal zone B lymphocyte cell fate decision. Nat. Rev. Immunol. 9:767–777 10.1038/nri2656 [DOI] [PubMed] [Google Scholar]
- Pohl, T., Gugasyan R., Grumont R.J., Strasser A., Metcalf D., Tarlinton D., Sha W., Baltimore D., and Gerondakis S.. 2002. The combined absence of NF-κ B1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc. Natl. Acad. Sci. USA. 99:4514–4519 10.1073/pnas.072071599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif, K., Ekland E.H., Ohl L., Nakano H., Lipp M., Förster R., and Cyster J.G.. 2002. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 416:94–99 10.1038/416094a [DOI] [PubMed] [Google Scholar]
- Rickert, R.C., Jellusova J., and Miletic A.V.. 2011. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol. Rev. 244:115–133 10.1111/j.1600-065X.2011.01067.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.J., Beinke S., Kouroumalis A., Tsichlis P.N., and Ley S.C.. 2007. Phosphorylation of TPL-2 on serine 400 is essential for lipopolysaccharide activation of extracellular signal-regulated kinase in macrophages. Mol. Cell. Biol. 27:7355–7364 10.1128/MCB.00301-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, M., Gao J., Basso K., Kitagawa Y., Smith P.M., Bhagat G., Pernis A., Pasqualucci L., and Dalla-Favera R.. 2007. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 12:280–292 10.1016/j.ccr.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Salmerón, A., Janzen J., Soneji Y., Bump N., Kamens J., Allen H., and Ley S.C.. 2001. Direct phosphorylation of NF-kappaB1 p105 by the IkappaB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276:22215–22222 10.1074/jbc.M101754200 [DOI] [PubMed] [Google Scholar]
- Savinova, O.V., Hoffmann A., and Ghosh G.. 2009. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol. Cell. 34:591–602 10.1016/j.molcel.2009.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas, R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Sha, W.C., Liou H.C., Tuomanen E.I., and Baltimore D.. 1995. Targeted disruption of the p50 subunit of NF-κ B leads to multifocal defects in immune responses. Cell. 80:321–330 10.1016/0092-8674(95)90415-8 [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., Rosenwald A., Hurt E.M., Giltnane J.M., Lam L.T., Pickeral O.K., and Staudt L.M.. 2001. Signatures of the immune response. Immunity. 15:375–385 10.1016/S1074-7613(01)00194-7 [DOI] [PubMed] [Google Scholar]
- Solan, N.J., Miyoshi H., Carmona E.M., Bren G.D., and Paya C.V.. 2002. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 277:1405–1418 10.1074/jbc.M109619200 [DOI] [PubMed] [Google Scholar]
- Sriskantharajah, S., Belich M.P., Papoutsopoulou S., Janzen J., Tybulewicz V., Seddon B., and Ley S.C.. 2009. Proteolysis of NF-kappaB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat. Immunol. 10:38–47 10.1038/ni.1685 [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Cerasoli D.M., Dal Porto J.M., Shimoda M., Freund R., Fang W., Telander D.G., Malvey E.N., Mueller D.L., Behrens T.W., and Kelsoe G.. 1999. Relaxed negative selection in germinal centers and impaired affinity maturation in bcl-xL transgenic mice. J. Exp. Med. 190:399–410 10.1084/jem.190.3.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki, K., Han H., Yamamoto N., Tashiro K., Ikegawa M., Kuroda K., Suzuki A., Nakano T., and Honjo T.. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443–450 10.1038/ni793 [DOI] [PubMed] [Google Scholar]
- Turner, M., Gulbranson-Judge A., Quinn M.E., Walters A.E., MacLennan I.C., and Tybulewicz V.L.. 1997. Syk tyrosine kinase is required for the positive selection of immature B cells into the recirculating B cell pool. J. Exp. Med. 186:2013–2021 10.1084/jem.186.12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield, M., Jin W., Reiley W., Zhang M., and Sun S.-C.. 2004. IkappaB kinase is an essential component of the Tpl2 signaling pathway. Mol. Cell. Biol. 24:6040–6048 10.1128/MCB.24.13.6040-6048.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.T., Papoutsopoulou S., Belich M., Brender C., Janzen J., Gantke T., Handley M., and Ley S.C.. 2012. Coordinate regulation of TPL-2 and NF-κB signaling in macrophages by NF-κB1 p105. Mol. Cell. Biol. 32:3438–3451 10.1128/MCB.00564-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuseff, M.-I., Pierobon P., Reversat A., and Lennon-Duménil A.-M.. 2013. How B cells capture, process and present antigens: a crucial role for cell polarity. Nat. Rev. Immunol. 13:475–486 10.1038/nri3469 [DOI] [PubMed] [Google Scholar]
- Zhang, X.2013. Regulatory functions of innate-like B cells. Cell. Mol. Immunol. 10:113–121 10.1038/cmi.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.