Heise et al. find that the NF-κB subunits c-REL and RELA in B cells play distinct roles during the germinal center reaction. While RELA stimulates the emergence of plasma cells from the germinal center, c-REL supports maintenance of the reaction over time, possibly by inducing a metabolic gene program connected to cell proliferation.
Abstract
Germinal centers (GCs) are the sites where memory B cells and plasma cells producing high-affinity antibodies are generated during T cell–dependent immune responses. The molecular control of GC B cell maintenance and differentiation remains incompletely understood. Activation of the NF-κB signaling pathway has been implicated; however, the distinct roles of the individual NF-κB transcription factor subunits are unknown. We report that GC B cell–specific deletion of the NF-κB subunits c-REL or RELA, which are both activated by the canonical NF-κB pathway, abolished the generation of high-affinity B cells via different mechanisms acting at distinct stages during the GC reaction. c-REL deficiency led to the collapse of established GCs immediately after the formation of dark and light zones at day 7 of the GC reaction and was associated with the failure to activate a metabolic program that promotes cell growth. Conversely, RELA was dispensable for GC maintenance but essential for the development of GC-derived plasma cells due to impaired up-regulation of BLIMP1. These results indicate that activation of the canonical NF-κB pathway in GC B cells controls GC maintenance and differentiation through distinct transcription factor subunits. Our findings have implications for the role of NF-κB in GC lymphomagenesis.
B cells with high specificity to T cell–dependent antigens are generated in the germinal center (GC) reaction, where their antibody genes are modified by somatic hypermutation. GC B cells with improved antigen affinity are selected and undergo further rounds of hypermutation, or differentiate into plasma cells or memory B cells expressing high-affinity antibodies (MacLennan, 1994; Rajewsky, 1996). The GC microenvironment is largely compartmentalized (Allen et al., 2007; Victora and Nussenzweig, 2012), resulting in effective GC responses (Bannard et al., 2013; Gitlin et al., 2014). Somatic hypermutation primarily occurs in centroblasts which localize in the dark zone of the GC. In the GC light zone, the descendants of centroblasts, the centrocytes, are subjected to selection for improved antigen binding and eventually differentiation. Consequently, centrocytes undergo marked changes in their transcriptional program, including the down-regulation of the transcriptional repressor BCL6, the master regulator of GC formation, and the activation of the transcription factors IRF4 and BLIMP1 (prdm1), which have key functions in class-switch recombination and/or plasma cell differentiation (Klein and Dalla-Favera, 2008). An important role for NF-κB signaling during the GC reaction has been implicated based on its specific activation pattern in GC B cells (see below).
Ligation of a range of cell surface receptors results in activation of the NF-κB signaling pathway, which is commonly associated with cell growth and survival, and occurs via two different routes, the canonical and the alternative pathways, each mediated by specific NF-κB subunits that occur as hetero- or homodimers (Hayden and Ghosh, 2004; Vallabhapurapu and Karin, 2009). RELA (p65), c-REL, and p50 compose the subunits of the canonical NF-κB pathway, and the major heterodimers are RELA/p50 and c-REL/p50. The subunits of the alternative NF-κB pathway are RELB and p52, which occur in heterodimeric form. Only c-REL, RELA, and RELB are able to drive transcription of target genes upon nuclear translocation due to the presence of a transactivation domain.
Studies in humans, somewhat unexpectedly, have shown that the vast majority of GC B cells are not subjected to NF-κB signaling (Shaffer et al., 2001; Basso et al., 2004). A subset of centrocytes, however, does show nuclear translocation of the canonical subunits RELA, c-REL, and p50 (Basso et al., 2004). One possible function of this activation process is suggested by the observation made in human cell lines that CD40-mediated NF-κB activation results in the expression of IRF4, which in turn represses the bcl6 gene, thus extinguishing the GC program (Saito et al., 2007). The analysis of the in vivo function of NF-κB transcription factors in GC B cell development has been hampered by the circumstance that the individual NF-κB subunits have important roles before the GC reaction (Gerondakis and Siebenlist, 2010; Kaileh and Sen, 2012), revealing a biphasic activation pattern of the canonical NF-κB subunits in T-dependent B cell responses. For example, the analysis of rel (c-REL) knockout mice has demonstrated that both B and T cells require c-REL for their activation in vitro (Köntgen et al., 1995; Tumang et al., 1998), suggesting that this subunit is essential for the B cell activation step that precedes GC formation, and RELA (rela) deficiency causes embryonic lethality (Beg et al., 1995). Therefore, the in vivo functions of the canonical NF-κB subunits in GC B cells cannot be investigated with existing knockout mice.
Understanding the functions of NF-κB subunits in GC B cell differentiation is highly relevant because constitutive activation of NF-κB signaling due to genetic mutations has recently been implicated in the pathogenesis of several GC-derived B cell lymphoma subtypes (Lenz et al., 2008; Compagno et al., 2009; Kato et al., 2009; Schmitz et al., 2009), identifying NF-κB as a critical player in GC-lymphomagenesis (Shaffer et al., 2012). Of note, evidence suggests a predominant activation of either c-REL or RELA in particular lymphoma subtypes.
To investigate the specific functions of c-REL and RELA in GC B cell development, we have generated mice in which rel and rela can be conditionally deleted in GC B cells. We show that both c-REL and RELA are required for the completion of the GC B cell reaction, although at distinct developmental stages and via different mechanisms. c-REL is required for the maintenance of the GC reaction, whereas RELA is required during the GC exit.
RESULTS
Conditional deletion of rela and rel in GC B cells
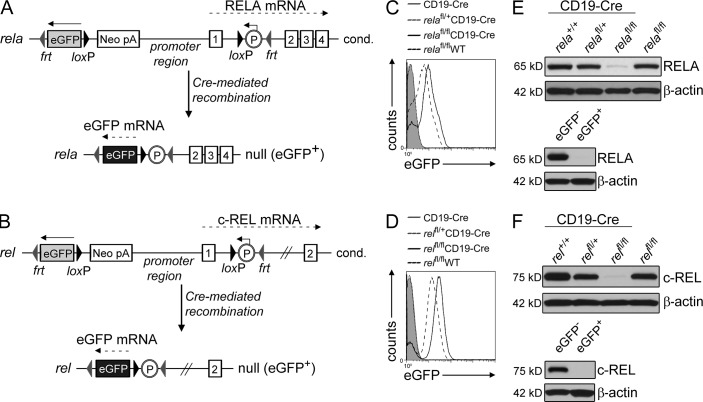
To determine the in vivo role of RELA and c-REL in GC B cell development, we generated transgenic mouse strains carrying loxP-flanked alleles of rela or rel, respectively, and crossed these mice to mice expressing Cre recombinase in B cells activated upon T cell–dependent immunization (Cγ1-Cre; Casola et al., 2006). The promoter regions and the exons encoding the translational start site of rela and rel were flanked by loxP sites to allow the creation of a null allele upon Cre-mediated recombination of loxP sites (Fig. 1, A and B; Fig. S1, A and D). To enable tracking of RELA or c-REL–deficient cells, a gene encoding enhanced GFP (eGFP) was placed in the opposite orientation upstream of the rela and rel promoter region, similar to a strategy previously used for the conditional deletion of the irf4 gene (Klein et al., 2006). Expression of eGFP after Cre-mediated recombination is achieved by juxtaposition of a mouse phosphoglycerate kinase promoter (placed in intron 1 of rela or rel) to the eGFP cassette (Fig. 1, A and B; Fig. S1, A and D). Correctly targeted embryonic stem cell lines were identified by Southern blot analysis, and germline transmission of the conditional rela and rel alleles was confirmed (Fig. S1, A and D). An independently generated rela conditional mouse line has been described previously (p65fl/+ mice; Luedde et al., 2008).
Figure 1.
Generation of mice with conditional deletion of rela or rel in GC B cells and simultaneous expression of eGFP. (A and B) Targeting strategy showing the status of rela and rel before (top) and after (bottom) Cre-mediated recombination. Numbers indicate the respective exons. (C and D) Flow cytometry of eGFP expression by splenic B cells of the indicated genotypes (n = 4 per group, one representative experiment shown). (E and F) Western blot analysis of RELA and c-REL protein levels in purified B cells of the genotypes shown in C and D, respectively, and of flow-sorted eGFP+ B cells from relafl/flCD19-Cre (E) or relfl/flCD19-Cre (F) mice and the corresponding eGFP− CD19-Cre control mice. One representative of 2 independent experiments is shown for each.
The functionality of the newly generated floxed rela and rel alleles was confirmed by crossing the alleles to mice carrying a Cre-recombinase specifically expressed in B cells (CD19-Cre). Deletion of the loxP-flanked alleles together with simultaneous activation of eGFP expression upon Cre-mediated recombination was observed via PCR and flow cytometric analysis for eGFP expression in the majority of B cells of mice homo- or heterozygous for the floxed allele (Fig. 1, C and D; and Fig. S1, B, C, and E). Of note, bi- and monoallelically deleted cells could be distinguished by their eGFP mean fluorescence. Western blot analysis confirmed that biallelically deleted B cells from rela and rel conditional mice had strongly reduced amounts of RELA or c-REL protein (Fig. 1, E and F, top), with the remaining protein likely to be derived from nondeleted (eGFP−) B cells as a result of incomplete Cre-mediated deletion (Fig. 1, C and D). This was confirmed by Western analysis for RELA and c-REL protein expression on purified eGFP+ B cells, demonstrating that eGFP+ B cells from relfl/flCD19-Cre and relafl/flCD19-Cre mice do not produce c-REL or RELA protein, respectively (Fig. 1, E and F, bottom). In the un-rearranged configuration, the conditional rela and rel alleles produced physiological amounts of RELA and c-REL protein, respectively (Fig. 1, E and F; and Fig. S1 F).
Rela is dispensable for GC formation and affinity maturation
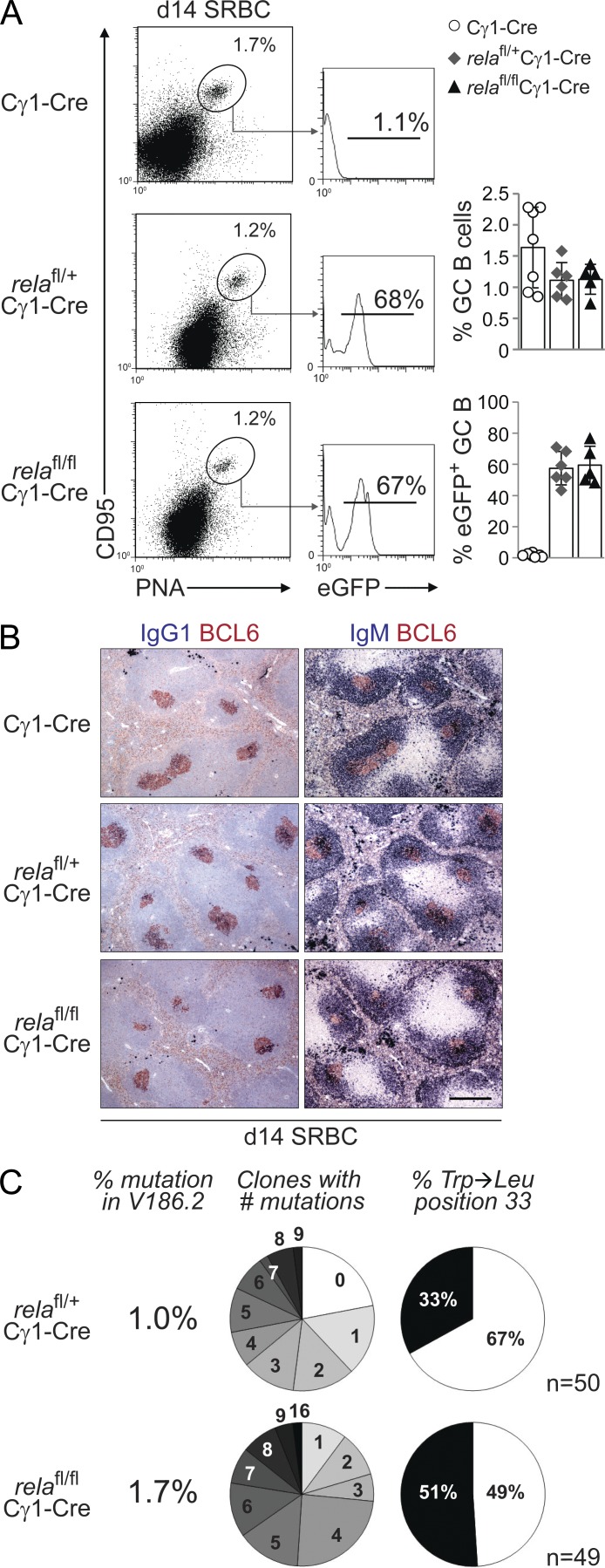
To determine how ablation of the canonical NF-κB subunit RELA in GC B cells affects GC B cell development, relafl/flCγ1-Cre, relafl/+Cγ1-Cre, and rela+/+Cγ1-Cre (henceforth referred to as Cγ1-Cre) control mice were immunized with the T cell–dependent antigen sheep RBCs (SRBCs) which elicits a robust GC response. 14 d after immunization, the fraction of splenic CD95hiPNAhi GC B cells in relafl/flCγ1-Cre mice did not significantly differ from Cγ1-Cre and relafl/+Cγ1-Cre mice (Fig. 2 A). Accordingly, histological analysis of spleen sections demonstrated robust formation of BCL6+ GCs (Fig. 2 B). Moreover, the fractions of eGFP+ GC B cells in relafl/flCγ1-Cre and relafl/+Cγ1-Cre mice were similar (Fig. 2 A) and were equally distributed among the centroblast and centrocyte compartments of the GC as defined by Victora et al. (2010; not depicted).
Figure 2.
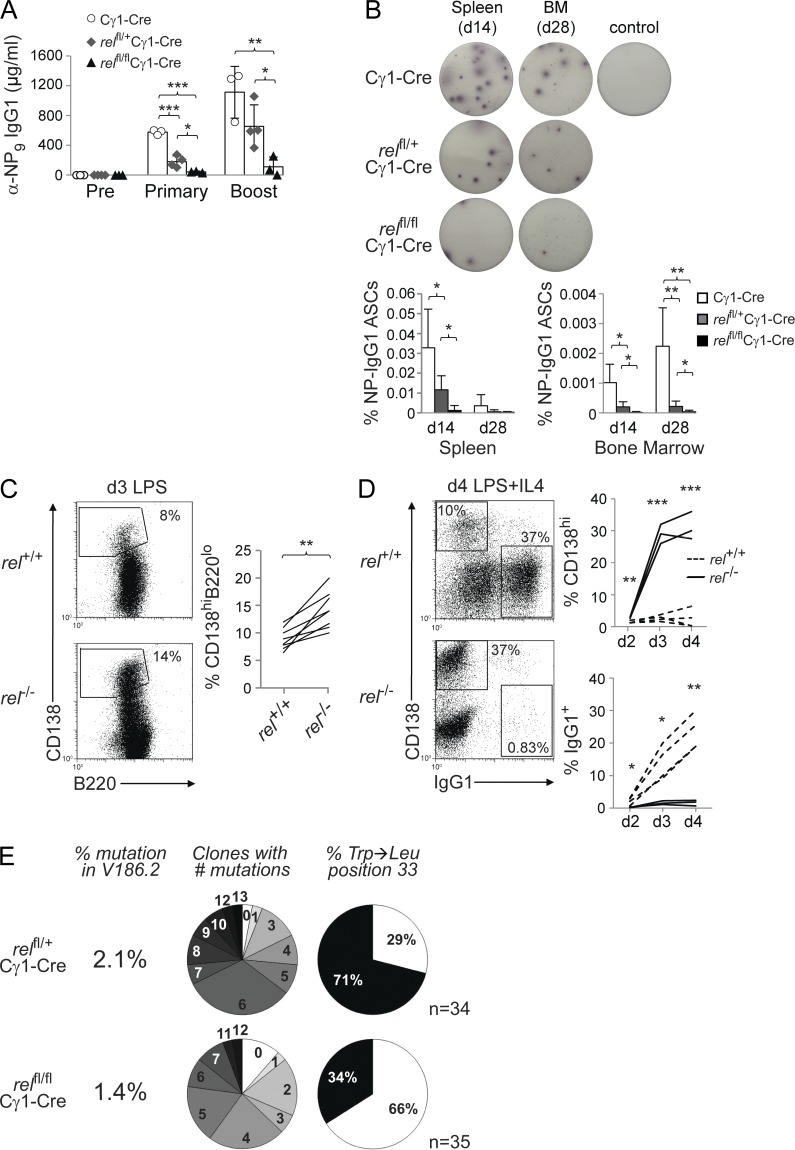
Rela is dispensable for GC formation and affinity maturation. relafl/flCγ1-Cre, relafl/+Cγ1-Cre, and Cγ1-Cre mice were immunized with SRBC (A and B) or NP-KLH (C) and analyzed 14 d later. (A) CD95, PNA, and eGFP expression by splenic B cells from mice of the corresponding genotypes were analyzed by flow cytometry. Numbers above gates indicate the percentage of CD95hiPNAhi (dot plots) or eGFP+CD95hiPNAhi (histograms) GC B cells. (Right) Data are cumulative from two experiments with 5–7 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± SD. Statistical significance was determined by Student’s t test. (B) Spleen sections from mice of the corresponding genotypes were analyzed for the expression of BCL6 and IgG1 or IgM; IgG1 stainings were counterstained with hematoxylin. One representative mouse out of three per group is shown. Bar, 300 µm. (C) Summary of sequence analysis of V186.2 γ1 transcripts amplified from eGFP+ B cells purified from three relafl/flCγ1-Cre and four relafl/+Cγ1-Cre mice 14 d after immunization with NP-KLH. n indicates number of clones analyzed. Pie charts: fraction of clones with the number of mutations indicated and percentage of clones with (black) or without (white) Trp-Leu amino acid exchange at position 33.
We then investigated the generation of high-affinity B cells during the GC reaction in relafl/flCγ1-Cre mice by immunization with the hapten NP (4-hydroxy-3-nitrophenyl-acetyl) coupled to a carrier protein (keyhole limpet hemocyanin [KLH]), which allows the analysis of antigen-specific immune responses at the level of somatic hypermutation, as described previously (Klein et al., 2006). Sequence analysis of VH186.2-Cγ1 transcripts amplified from eGFP+ splenic B cells isolated by flow cytometry from relafl/flCγ1-Cre and relafl/+Cγ1-Cre mice 14 d after NP-KLH immunization showed that there was no significant difference in the percentage of rearrangements carrying a tryptophan-to-leucine substitution at position 33 (P = 0.07, Fisher’s exact probability test; P = 0.09, Chi-Square test of association), which leads to a 10-fold increase in antibody affinity for NP, among the genotypes (Fig. 2 C). Together, the findings demonstrate that RELA is not required for GC formation, and that RELA-deficient GC B cells undergo normal affinity maturation.
Rela is required for the generation of GC-derived plasma cells
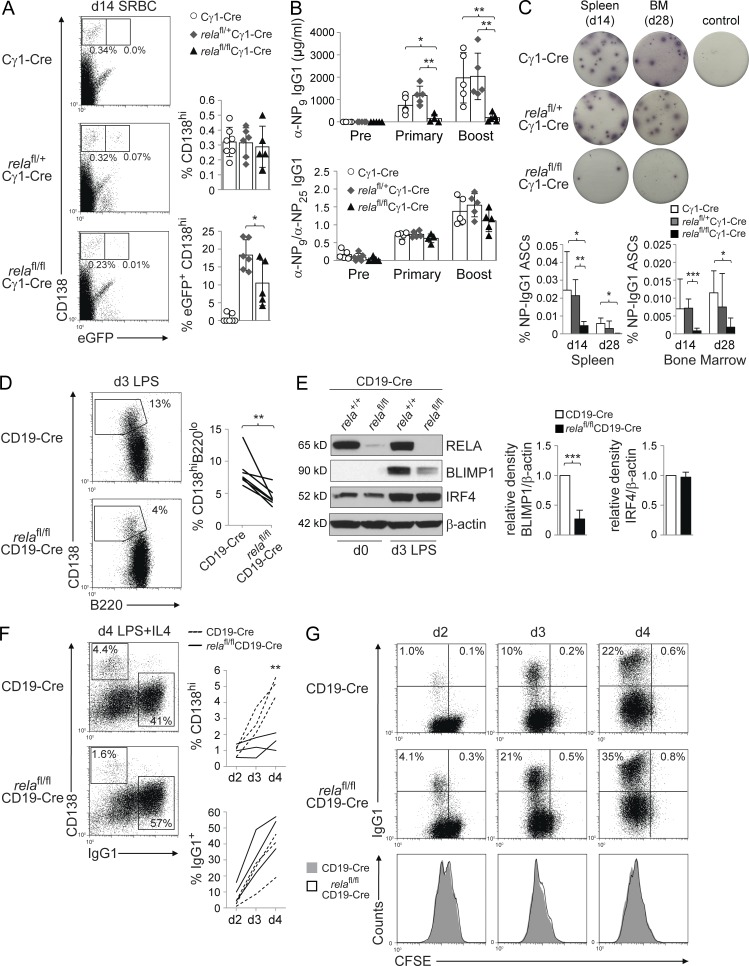
Despite normal GC formation, eGFP+CD138hi plasma cells were significantly reduced in the spleen of SRBC-immunized relafl/flCγ1-Cre mice compared with relafl/+Cγ1-Cre control mice (Fig. 3 A). In accordance, total IgG1 serum titers (not depicted) and NP-specific IgG1 serum titers after primary and secondary challenge with NP-KLH were strongly reduced in relafl/flCγ1-Cre compared with Cγ1-Cre and relafl/+Cγ1-Cre mice (Fig. 3 B). These results were further supported by ELISPOT analysis for NP-specific IgG1 antibody-secreting cells (ASCs) that revealed a strong reduction of these cells in spleen and BM of relafl/flCγ1-Cre mice 14 and 28 d after immunization (Fig. 3 C). Our observation that GC B cells in relafl/flCγ1-Cre mice undergo normal antigen selection (Fig. 2 C) is further supported by the finding that the affinity of the remaining plasma cell compartment in relafl/flCγ1-Cre mice did not differ from the controls, as indicated by the ratio of anti-NP9 to anti-NP25 IgG1 antibodies (Fig. 3 B, bottom), which serves as a measure for affinity maturation of a humoral immune response. Thus, our observations suggest a role for RELA in GC B cells after affinity maturation and during the differentiation of GC B cells into plasmablasts, reminiscent of mice that are deficient for BLIMP1 or IRF4 in GC B cells which show normal GC development but impaired generation of GC-exiting plasma cell precursors (Shapiro-Shelef et al., 2003; Klein et al., 2006).
Figure 3.
Rela is required for the generation of GC-derived plasma cells. (A–C) relafl/flCγ1-Cre, relafl/+Cγ1-Cre, and Cγ1-Cre mice were immunized with SRBC (A) or NP-KLH (B and C) and analyzed 14 or 28 d later. (A) CD138 and eGFP expression by splenic mononuclear cells from mice of the corresponding genotypes were analyzed by flow cytometry. Numbers below gates indicate the percentage of CD138hieGFP+ or CD138hieGFP− cells. (Right) Data are cumulative from two experiments with 5–7 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± SD. Statistical significance was determined by Student’s t test (*, P < 0.05). (B) Serum response to NP-KLH immunization before (Pre), 12 d after primary (Primary), and 7 d after secondary (Boost) immunization with NP-KLH (top). Ratio of anti-NP9 to anti-NP25 IgG1 antibodies (bottom). Data are cumulative from two experiments with 5 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student’s t test (*, P < 0.05; **, P < 0.01). (C) Elispot analysis for NP-specific IgG1 ASCs in spleen and BM on days 14 and 28 after NP-KLH immunization. One representative Elispot experiment per genotype is shown for day 14 spleen and day 28 BM. Control, unimmunized mouse. (Bottom) Data from 3–5 independent experiments/genotype/tissue/time point are summarized. Data are shown as mean ± standard deviation. Statistical significance was determined by Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (D) Purified B cells from relafl/flCD19-Cre and CD19-Cre mice were stimulated in vitro with LPS and analyzed 3 d later for CD138 and B220 expression by flow cytometry. Numbers besides gates indicate the percentage of CD138hiB220low cells. Representative example (left) and pairwise representation of individual experiments (right) for CD138hiB220lo expression of LPS-stimulated B cells, with each line representing an independent experiment (n = 6). Statistical significance was determined by Student’s t test (**, P < 0.01). (E) Representative Western blot analysis (left) of purified B cells ex vivo (day 0) and stimulated for 3 d with LPS for expression of BLIMP1 and IRF4 protein, and mean relative density of 3 experiments (right) compared with CD19-Cre B cells. Data are shown as mean ± standard deviation. Statistical significance was determined by Student’s t test (***, P < 0.001). (F) Purified B cells from relafl/flCD19-Cre and CD19-Cre mice were stimulated in vitro with LPS+IL-4 and analyzed 4 d later for CD138 and IgG1 expression by flow cytometry. Numbers within gates indicate the percentage of CD138hiIgG1− and CD138−IgG1+ cells. Representative example (left) and pairwise representation of individual experiments (right), with each line representing an independent experiment (n = 3). Statistical significance was determined by Student’s t test (**, P < 0.01). Experiments shown in C–E were performed with two different RELA-conditional mouse lines (see experimental procedures) that gave similar results. (G) Purified B cells from relafl/flCD19-Cre and CD19-Cre mice (p65fl/+ cohort) were stained with CFSE on day 0, stimulated with LPS+IL-4 and analyzed by flow cytometry on days 2–4 for IgG1 expression to determine the fraction of class-switched cells in relation to cell cycle as determined by CFSE dilution. Numbers indicate the percentage of cells in the corresponding quadrants. One out of three independent experiments is shown.
In accordance with the impaired generation of plasma cells in vivo, RELA-deficient B cells purified from relafl/flCD19-Cre mice developed into fewer CD138hiB220lo plasmablasts compared with control B cells upon stimulation with LPS, an inducer of plasmablastic differentiation in vitro (Fig. 3 D). We then investigated the corresponding cultures via Western blot for the induction of the transcription factors BLIMP1 and IRF4, which are jointly required for plasma cell differentiation, and found that RELA-deficient B cells showed dramatically impaired up-regulation of BLIMP1 compared with the control B cells (Fig. 3 E), which was also observed at the mRNA level (not depicted). Conversely, IRF4, which in addition to plasma cell differentiation also regulates class switch recombination (Klein et al., 2006; Sciammas et al., 2006), was normally up-regulated in RELA-deficient B cells (Fig. 3 E). Consistent with the requirement of IRF4 for class-switch recombination, cultures of RELA-deficient B cells generated similar numbers of IgG1+ switched B cells under conditions that induce both plasmablastic differentiation and class-switch recombination (LPS and IL-4) compared with the control cultures, despite the reduction in CD138hiB220lo plasmablasts (Fig. 3 F) and similar proliferation properties of both RELA-deficient and wild-type B cells (Fig. 3 G). These results suggest that the impaired plasma cell generation in relafl/flCγ1-Cre mice in vivo is due to a requirement for RELA in the induction of prdm1 expression. In support of this notion, RELA has been shown to bind to the prdm1 locus and activate prdm1 transcription in a mouse lymphoma cell line (Morgan et al., 2009). Collectively, these findings implicate RELA as a regulator of plasma cell differentiation during the GC reaction and show that c-REL does not compensate for RELA in this process.
GC B cell–specific deletion of rel causes disruption of GC B cell development
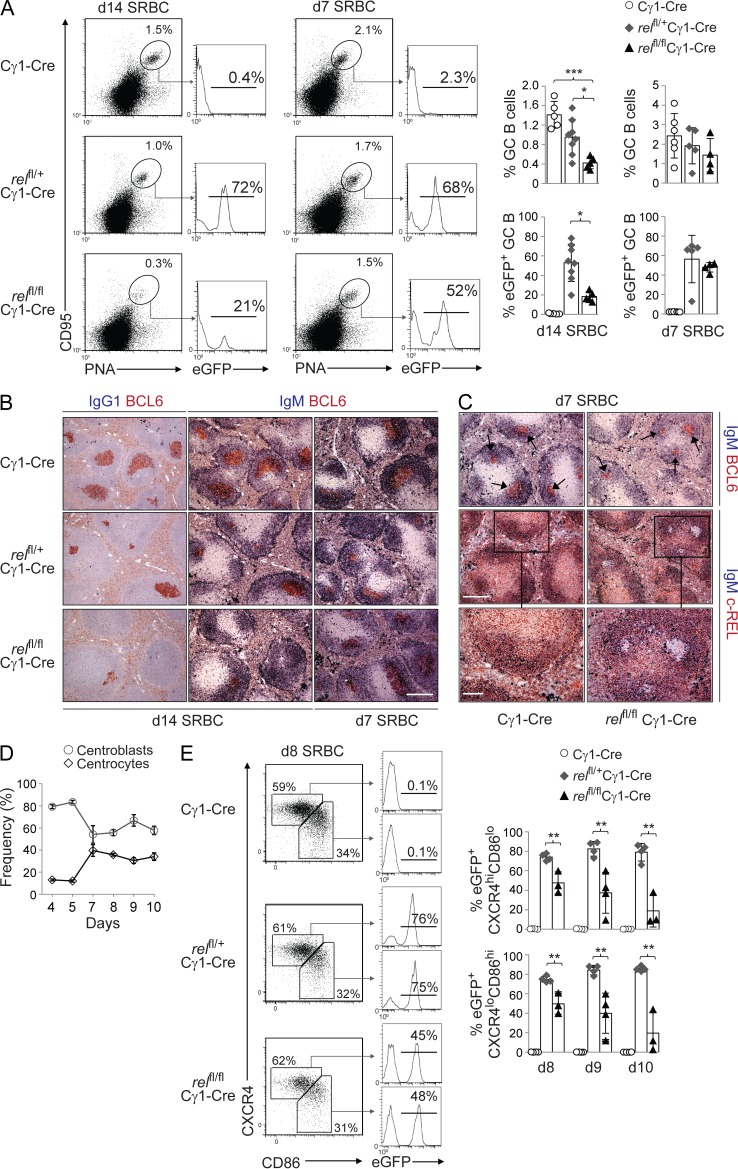
c-REL and RELA function within the same pathway of NF-κB activation. However, in striking contrast to relafl/flCγ1-Cre mice, the fraction of CD95hiPNAhi GC B cells on day 14 after SRBC immunization was strongly reduced in relfl/flCγ1-Cre mice compared with relfl/+Cγ1-Cre and Cγ1-Cre mice (Fig. 4 A). The marked reduction of c-REL–deficient GC B cells was particularly evident from the analysis for eGFP expression that demonstrated the preferential loss of rel-homozygous– over rel-heterozygous–deleted GC B cells (Fig. 4 A). (Of note, heterozygous deletion of the rel allele resulted in a partial reduction of GC B cells, consistent with the reported haploinsufficiency of rel deletion in B cells [Köntgen et al., 1995].) In agreement with the flow cytometric data, histological analysis revealed fewer and smaller BCL6+ GCs in the spleen upon homozygous deletion of rel (Fig. 4 B). Interestingly, however, GC formation was not affected in relfl/flCγ1-Cre mice 7 d after SRBC immunization (Fig. 4, A and B) despite the lack of c-REL protein expression in GC B cells at this time point (Fig. 4 C). Day 7 is the time at which GCs featuring the characteristic ∼2:1 centroblast/centrocyte ratio (Victora et al., 2010) have formed (Fig. 4 D). Thus, our data suggest that GCs in relfl/flCγ1-Cre mice begin to involute only after dark and light zones have been fully established, which is the time point when somatically mutated B cells start to appear and when the selection of antigen-specific B cells is thought to begin (Jacob et al., 1993). Because gene deletion occurs in Cγ1-Cre mice as early as day 4 after immunization (Casola et al., 2006), our findings indicate that c-REL is dispensable during the establishment phase of the GC until day 7, but that it is required at a later stage.
Figure 4.
GC-specific deletion of rel impairs GC development and the generation of IgG1-producing plasma cells. relfl/flCγ1-Cre, relfl/+Cγ1-Cre, and Cγ1-Cre mice were immunized with SRBC (A–C) and analyzed 7 or 14 d later. (A) CD95, PNA, and eGFP expression by splenic B cells from mice of the corresponding genotypes were analyzed by flow cytometry. Numbers above gates indicate the percentage of CD95hiPNAhi (dot plots) or eGFP+CD95hiPNAhi (histograms) GC B cells. (Right) Data are cumulative from two experiments with 5–8 mice per genotype, with each symbol representing a mouse. Data shown are as mean ± SD. Statistical significance was determined by Student’s t test (*, P < 0.05; ***, P < 0.001). (B) Spleen sections from mice of the corresponding genotypes were analyzed for the expression of BCL6 and IgG1 or IgM; IgG1 stainings were counterstained with hematoxylin. One representative mouse out of three per group is shown. Bar, 300 µm. (C) Spleen sections of mice were stained for IgM and BCL6 (top) and IgM and c-REL (middle, bottom) 7 d after immunization. One representative mouse out of 2 per group is shown. Bars: (top and middle) 300 µm; (bottom) 100 µm. (D) Percentage of splenic B220+CD38loCD95hiCXCR4hiCD86lo centroblasts and B220+CD38loCD95hiCXCR4loCD86hi centrocytes in Cγ1-Cre mice on the indicated time points after SRBC immunization. GCs consisting of the characteristic centroblast/centrocyte ratio of ∼2:1 have not formed until day 7. Data from 3–4 mice per time point are shown. Data are shown as mean ± SD. (E) Mice of the corresponding genotypes were immunized with SRBC and splenic B cells were analyzed for B220, CD38, CD95, CXCR4, CD86, and eGFP expression by flow cytometry at the indicated time points. Numbers besides gates indicate the percentage of B220+CD38loCD95hiCXCR4hiCD86lo centroblasts and B220+CD38loCD95hiCXCR4loCD86hi centrocytes (dot plots) or the eGFP+ cells of the corresponding fractions (histograms). (Bottom) Data are cumulative from two experiments with 3–4 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student’s t test (**, P < 0.01).
The loss of rel-deleted GC B cells in vivo could be due to either a role of c-REL in the development of the centroblast or centrocyte subpopulations or in the maintenance of the GC B cell reaction. Centroblasts and centrocytes can be distinguished by surface expression of the activation markers CD83 or CD86 and the chemokine receptor CXCR4 (Victora et al., 2010). We observed that whereas the percentage of total splenic GC B cells (B220+CD38loCD95hi) gradually decreased in relfl/flCγ1-Cre mice from days 8 to 10 after SRBC immunization (not depicted), the ratio of CXCR4hiCD86lo centroblasts to CXCR4loCD86hi centrocytes of ∼2:1 was maintained over time at a level comparable to that seen in relfl/+Cγ1-Cre and Cγ1-Cre mice (not depicted). Of note, rel-deleted, eGFP+ GC B cells were lost equally from the centroblast and centrocyte compartments in relfl/flCγ1-Cre mice (Fig. 4 E). Collectively, our findings suggest a role for c-REL in the maintenance of the GC B cell reaction as opposed to a role in the development of a particular GC B cell subset (see Discussion).
In accordance with the disrupted GC B cell development in immunized relfl/flCγ1-Cre mice, we observed a strong reduction in serum IgG1 titers (not depicted) and the secretion of antigen-specific IgG1 antibodies after primary and secondary immunization with NP-KLH in relfl/flCγ1-Cre mice compared with both Cγ1-Cre and relfl/+Cγ1-Cre mice (Fig. 5 A). In agreement, ELISPOT analysis for NP-specific IgG1 ASCs revealed a strong reduction of these cells in spleen and BM of relfl/flCγ1-Cre mice 14 and 28 d after immunization (Fig. 5 B). The strong reduction of GC-derived plasma cells is unlikely to be due to a c-REL–dependent defect in plasma cell differentiation because in contrast to RELA deficiency (Fig. 3 D), ablation of c-REL function did not impair the development of CD138hiB220lo cells upon induction of plasmablastic differentiation by LPS (Fig. 5 C). Rather, c-REL–deficient B cells had a higher propensity to differentiate into plasmablasts compared with control B cells (P < 0.01). Under conditions that induce both plasmablastic differentiation and class-switch recombination (LPS and IL-4), cultures of c-REL–deficient B cells generated elevated numbers of plasmablasts compared with wild-type B cells (Fig. 5 D), whereas a strong impairment in class switch recombination was observed, as previously described (Köntgen et al., 1995; Tumang et al., 1998). Together, these findings indicate that unlike RELA, c-REL is dispensable for plasmablastic differentiation, further supporting the notion that c-REL exerts its specific function in GC B cells before differentiation. Of note, V168.2 gene sequence analysis of the minute fraction of rel-deleted eGFP+ B cells that is detectable in the spleen of relfl/flCγ1-Cre mice at day 14 after immunization identified a reduced somatic hypermutation frequency and percentage of V168.2 rearrangements with Trp→Leu replacements compared with the control, which was statistically significant as determined by Fisher’s exact probability test (P = 0.004) or chi-squared test of association (P = 0.005; Fig. 5 E). These results suggest that the few rel-deleted cells that were detectable in relfl/flCγ1-Cre mice represent NP-specific cells that have escaped counterselection in the GC, most probably because they were generated early in the GC response before or during the time the Cre-mediated deletion of rel has occurred, when c-REL protein amounts were still sufficient to allow completion of the selection process.
Figure 5.
The strong reduction of plasma cells in relfl/flCγ1-Cre mice is unlikely to be due to an impairment in the generation of plasmablasts. (A) relfl/flCγ1-Cre, relfl/+Cγ1-Cre, and Cγ1-Cre mice were immunized with NP-KLH and analyzed for the serum response to NP-KLH immunization before (pre), 12 d after primary (primary), and 7 d after secondary (boost) immunization with NP-KLH. Data are cumulative from two experiments with 3–4 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± SD. Statistical significance was determined by Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Elispot analysis for NP-specific IgG1 ASCs in spleen and BM on days 14 and 28 after NP-KLH immunization. One representative Elispot experiment per genotype is shown for day 14 spleen and day 28 BM. (Bottom) Data from 3–4 (day 14) and 5–6 (day 28) independent experiments/genotype/tissue/time point are summarized. Data are shown as mean ± standard deviation. Statistical significance was determined by Student’s t test (*, P < 0.05; **, P < 0.01). Control, unimmunized mouse. (C) Purified B cells from rel−/− and rel+/+ mice were stimulated in vitro with LPS and analyzed 3 d later for CD138 and B220 expression by flow cytometry. Numbers besides gates indicate the percentage of CD138hiB220low cells. Representative example (left) and pairwise representation of individual experiments (right) for CD138hiB220lo expression of LPS-stimulated B cells, with each line representing an independent experiment (n = 8). Statistical significance was determined by Student’s t test (**, P < 0.01). (D) Purified B cells from rel−/− and rel+/+ mice were stimulated in vitro with LPS+IL-4 and analyzed 4 d later for CD138 and IgG1 expression by flow cytometry. Numbers within gates indicate the percentage of CD138hiIgG1− and CD138−IgG1+ cells. Representative example (left) and pairwise representation of individual experiments (right), with each line representing an independent experiment (n = 3–4). Statistical significance was determined by Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (E) Summary of sequence analysis of V186.2 γ1 transcripts amplified from eGFP+ B cells purified from each two pooled relfl/flCγ1-Cre and relfl/+Cγ1-Cre mice 14 d after immunization with NP-KLH. n indicates number of clones analyzed. Pie charts: fraction of clones with the number of mutations indicated and percentage of clones with (black) or without (white) Trp-Leu amino acid exchange at position 33.
Expression of a bcl2 transgene does not rescue the loss of c-REL–deficient GC B cells in vivo
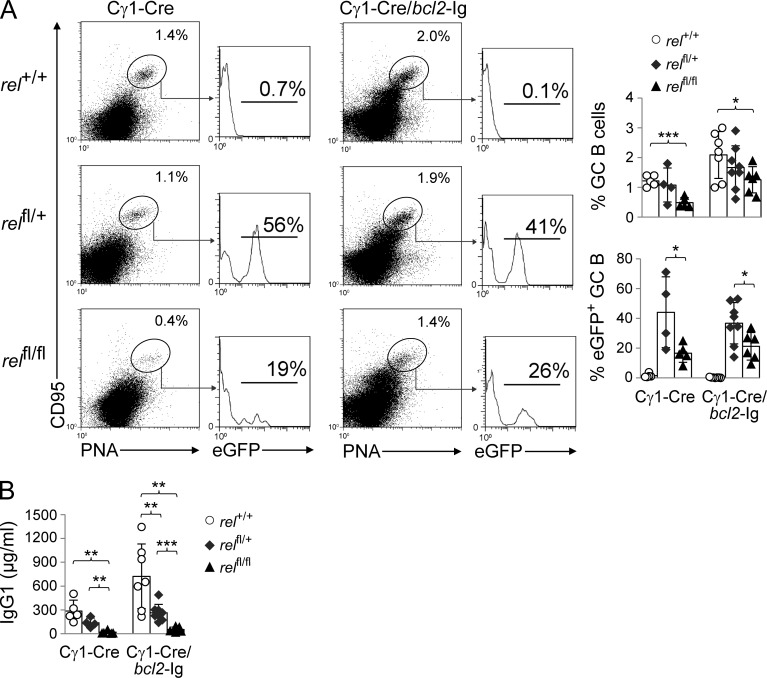
NF-κB transcription factors are known regulators of B cell survival (Gerondakis and Strasser, 2003; Kaileh and Sen, 2012), raising the possibility that the gradual disappearance of rel-deleted GC B cells may be due to an inability of these cells to escape apoptosis, the default pathway of GC B cells unless they are positively selected (MacLennan, 1994). To investigate this matter, we crossed the conditional rel allele into mice carrying a constitutively expressed bcl2 gene (bcl2–Ig; McDonnell et al., 1989). Expression of a bcl2 transgene has been shown to inhibit the elimination of counterselected GC B cells by apoptosis (Smith et al., 2000). We observed that, whereas the percentage of GC B cells was generally higher in the bcl2-transgenic mice irrespective of the genotype due to enhanced B cell survival (McDonnell et al., 1989), the fraction of GC B cells in relfl/flCγ1-Cre/bcl-2–Ig mice remained significantly lower compared with the Cγ1-Cre/bcl-2–Ig control mice 14 d after SRBC immunization (1.3 vs. 2.1%; P = 0.04; Fig. 6 A). Importantly, the expression of the transgene was unable to rescue the loss of eGFP+ GC B cells upon homozygous rel deletion in relfl/flCγ1-Cre/bcl-2–Ig versus relfl/flCγ1-Cre littermates (21 vs. 17%), whereas they were significantly reduced compared with the corresponding heterozygously deleted mice (Fig. 6 A). In agreement with these findings, bcl2–Ig expression in c-REL–deficient B cells did not rescue the strong reduction in IgG1 serum titers observed upon immunization in relfl/flCγ1-Cre mice versus the c-REL–proficient and heterozygous rel-deleted mice (Fig. 6 B). These findings indicate that the observed loss of rel-deleted GC B cells is not due to the inability of c-REL–deficient GC B cells to transmit prosurvival signals.
Figure 6.
Bcl2 transgene expression fails to rescue the loss of c-REL–deficient GC B cells in vivo. (A) relfl/flCγ1-Cre, relfl/+Cγ1-Cre, and Cγ1-Cre mice and the corresponding genotypes with a bcl2-Ig transgene were immunized with SRBC and analyzed 14 d later. CD95, PNA, and eGFP expression by splenic B cells from mice of the corresponding genotypes were analyzed by flow cytometry. Numbers above gates indicate the percentage of CD95hiPNAhi (dot plots) or eGFP+CD95hiPNAhi (histograms) GC B cells. (Right) Data are cumulative from two experiments with 4–8 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± SD. Statistical significance was determined by Student’s t test (*, P < 0.05; ***, P < 0.001). (B) relfl/flCγ1-Cre, relfl/+Cγ1-Cre, and Cγ1-Cre mice and the corresponding genotypes with a bcl2-Ig transgene were immunized with SRBC and analyzed for serum IgG1 antibodies 14 d later. Data are cumulative from two experiments with 4–7 mice per genotype, with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student’s t test (**, P < 0.01; ***, P < 0.001).
c-REL–deficient GC B cells fail to up-regulate a metabolic program that directs cell growth
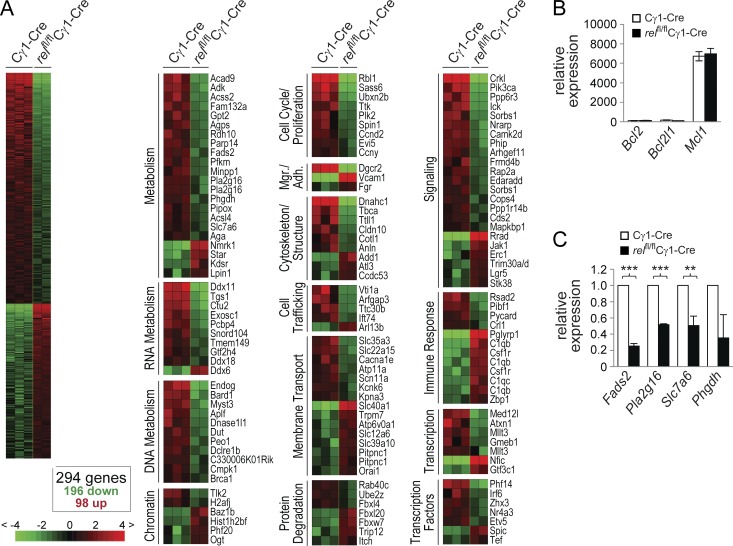
To gain insight into the biological programs controlled by c-REL to facilitate the maintenance of the GC B cell reaction, we performed a gene expression profile (GEP) analysis to identify genes differentially expressed between rel-deleted and wild-type GC B cells. Specifically, eGFP+ and eGFP− GC B cells (B220+CD95hiPNAhi) were sorted from the spleens of relfl/flCγ1-Cre and Cγ1-Cre mice, respectively, 7 d after SRBC immunization, when the GCs of the former mice were indistinguishable in size from the controls (Fig. 4, A and B). We anticipated that alterations in transcriptional programs that ultimately lead to the loss of rel-deleted GC B cells past day 8 would already be evident at day 7 after immunization. Supervised analysis identified a signature of 294 genes that were differentially expressed between rel-deleted and wild-type GC B cells (Fig. 7 A; Table S1), and that were assigned to putative functional categories. No differences were observed in the expression of regulators of cell survival (Fig. 7, A and B), consistent with the observation that expression of a bcl2–Ig transgene failed to rescue rel-deleted GC B cells. Also, genes associated with the proliferation of GC B cells were expressed at similar levels in rel-deleted and wild-type GC B cells, with the exception of the cell cycle regulator cyclin D2 (ccnd2), which is repressed in GC B cells by BCL6 (Shaffer et al., 2000) and is thus not thought to play a role in the proliferation of GC B cells (not depicted). These findings are in agreement with the observation that the initial burst of proliferation required for the formation of fully established GCs by day 7 seems to occur normally in relfl/flCγ1-Cre mice. In accordance, GC B cell–associated genes (Green et al., 2011), including bcl6, aicda, and ezh2, were equally expressed among the rel-deleted and wild-type GC B cell fractions (not depicted), demonstrating that c-REL does not modulate the expression of key GC developmental regulators.
Figure 7.
c-REL–deficient GC B cells fail to up-regulate a set of genes involved in metabolic functions. (A) RNA was isolated from eGFP+ or eGFP− B220+CD95+PNA+ GC B cells of, respectively, relfl/flCγ1-Cre and Cγ1-Cre mice purified by flow cytometry and processed for hybridization on microarrays. B cell fractions were isolated separately from 2 or 3 mice per genotype and independently processed for microarray hybridization. Gene expression differences between GC B cells from relfl/flCγ1-Cre and Cγ1-Cre mice were determined by supervised analysis (n = 2 or 3 each group). Color changes within a row indicate expression levels relative to the mean of the sample population. Values are quantified by the scale bar that visualizes the difference in the zge score (expression difference/standard deviation) relative to the mean (0). Genes are ranked according to their zg score (mean expression difference of the respective gene between phenotype and control group/standard deviation). Shown are only those gene segments that differ twofold or more in their zg score. TFs, transcription factors; Mgr/Adh., migration/adhesion; Trf., trafficking. (B and C) Total RNA from relfl/flCγ1-Cre and Cγ1-Cre GC B cells was analyzed for the expression of Bcl2, Bcl2l1, and Mcl1 (B), and Fads2, Pla2d16, Slc7a6, and Phgdh (C) by qRT-PCR. The expression was corrected for β-actin levels and then normalized to wild-type B cells. Data are shown as mean ± SD (n = 3, 2 independent experiments). Statistical significance was analyzed by Student’s t test (**, P < 0.01; ***, P < 0.001); p-values for Bcl2l1 and Phgdh were P = 0.088 and P = 0.085, respectively.
Strikingly, several genes differentially expressed between c-REL–deficient and wild-type GC B cells encode known regulators of cellular metabolism that are instrumental in meeting the heightened demands of proliferating cells for both energy and building blocks for anabolic processes (Fig. 7 A). For representative genes, the differential expression among the GC fractions was confirmed by qRT-PCR (Fig. 7 C). The genes identified as down-regulated in rel-deleted versus wild-type GC B cells include phosphofructokinase (PFKM), the enzyme which catalyzes the rate-limiting step in glycolysis, phosphoglycerate dehydrogenase (PHGDH), which directs glycolytic carbon into serine and glycine metabolism, and the amino acid transporter solute carrier family 7, member 6 (SLC7A6), which transports cationic amino acids—including glutamine, a major provider of carbon blocks for anabolic reactions (Le et al., 2012)—across the cell membrane (Fig. S2). In addition, several enzymes involved in the oxidation of fatty acids (Fig. S2) were significantly down-regulated in rel-deleted GC B cells. Together, these results suggest that c-REL establishes a metabolic program that allows the cell to meet the energy demands required by the rapidly proliferating GC B cells.
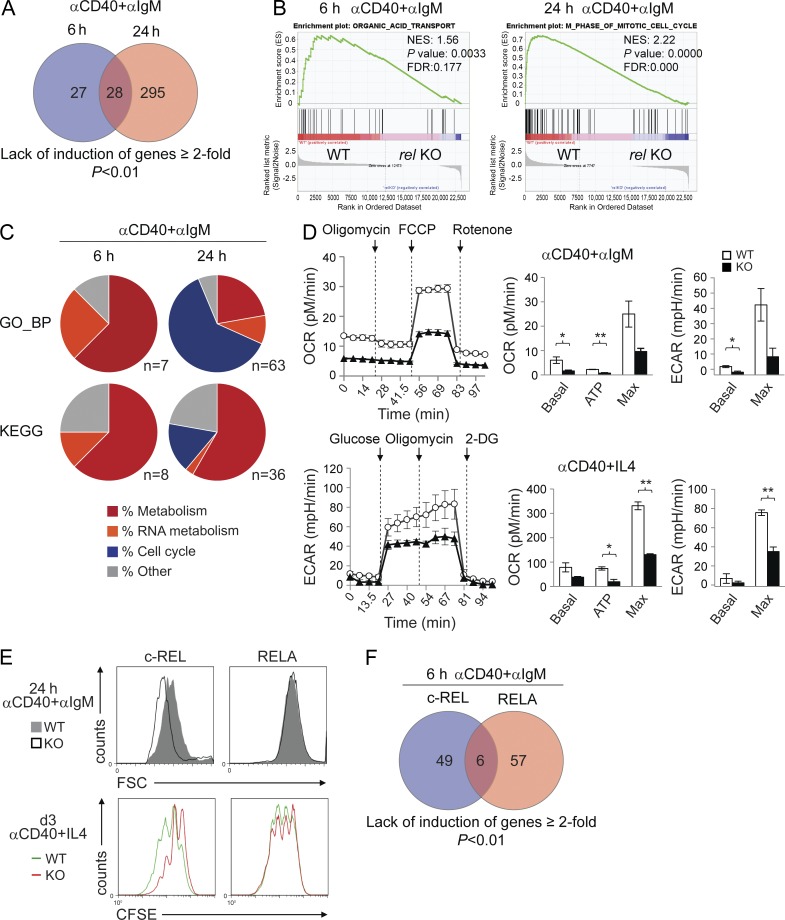
Because GC B cells showing active NF-κB signaling presumably represent only a small fraction among the eGFP+ GC B cells isolated from relfl/flCγ1-Cre mice, we used an ex vivo activation system that allowed us to more comprehensively investigate the genes that are directly or indirectly regulated by c-REL. To this end, we mimicked the stimuli a B cell is likely to encounter during the T cell–dependent process of antigen selection within the GC by activating splenic B cells isolated from relfl/flCD19-Cre and CD19-Cre control mice with anti-CD40 and anti-IgM. RNA from three samples per genotype was harvested after 6 and 24 h of stimulation and subjected to RNA sequencing. Upon identifying genes expressed at ≥2-fold lower levels in the c-REL-deficient cultures compared with wild-type B cells at each of these two time points, three categories emerged. These categories comprised genes whose expression was significantly reduced only at 6 h (27/55 genes) or 24 h (295/323 genes), or at both of these time points (28/55 and 28/323 genes; Fig. 8 A; for the identity of genes, see Table S2). These results indicate that, upon in vitro stimulation, the gene expression changes in rel-deleted B cells progress over time.
Figure 8.
c-REL–deficient in vitro–activated B cells are impaired in establishing a metabolic program. (A) Venn diagram depicting the overlap of genes that failed to be up-regulated ≥2-fold upon CD40+IgM stimulation at both 6 and 24 h in relfl/flCD19-Cre versus CD19-Cre mice as determined by RNA-seq analysis. (B) Representative examples of a metabolic signature and a proliferation signature identified at 6 and 24 h of CD40+IgM stimulation, respectively, by GSEA using the gene set category GO_BP. Normalized enrichment scores (NES), p-values, and false discovery rates (FDRs) are indicated. (C) GSEA was used to identify gene sets of the categories GO_BP and KEGG that were enriched in B cells from CD19-Cre versus relfl/flCD19-Cre mice after 6 and 24 h of CD40+IgM stimulation. Gene sets showing significant enrichment (FDR <25%; P ≤ 0.05) were grouped into functional categories. For the identity of the gene sets, see Table S3. (D) Measurement of OCR and ECAR of c-REL–deficient and wild-type B cells stimulated with αCD40+αIgM or αCD40+IL-4 for 24 h by extracellular flux analysis. OCR was determined upon sequential administration of the indicated drugs to assess basal respiration, the respiration needed to sustain ATP consumption (oligomycin), and the maximal respiration that reflects how the cells react to an increased ATP demand (FCCP), followed by administration of the mitochondrial NADH dehydrogenase complex 1 inhibitor rotenone. ECAR was determined upon administration of glucose, the ATP synthase inhibitor oligomycin, and the glucose analogue 2-DG. (Left) Representative experiments. (Right) Statistical analysis of one representative out of 3 (OCR) or 2 (ECAR) independently performed measurements, each with 2 mice per genotype. Data are shown as mean ± SD (n = 2). Statistical significance was analyzed by Student’s t test (*, P < 0.05; **, P < 0.01); p-values for the OCR Max and ECAR Max in the αCD40+αIgM stimulation were P = 0.058 and P = 0.057, respectively. (E) Forward scatter analysis (top) at 24 h of CD40+IgM stimulation and CFSE dilution (bottom) in CD40+IL-4–stimulated B cells (day 3). (F) Venn diagram depicting the overlap of genes that failed to be up-regulated ≥2-fold upon CD40+IgM stimulation at 6 h in both relfl/flCD19-Cre or relafl/flCD19-Cre versus CD19-Cre as determined by RNA-sequencing analysis.
We next used gene set enrichment analysis (GSEA; Subramanian et al., 2005) to determine the nature of the gene expression changes. Whereas several signatures associated with cellular metabolism were up-regulated in the control B cells at 6 h of stimulation, these signatures were absent in the rel-deleted B cells (Fig. 8 C; a representative gene set is shown in Fig. 8 B; and Table S3). At 24 h, in addition to a further numerical enrichment for cellular and RNA metabolism gene sets in the control B cells (Table S3), the enriched signatures were dominated by those relating to cell cycle entry and progression and DNA metabolism (Fig. 8 C; a representative gene set is shown in Fig. 8 B, right; see Table S3 for the identity of the remaining signatures). Our results suggest that in the early stages of B cell activation, c-REL controls the expression of genes that may be necessary for an initial burst of metabolism to enable biosynthesis of building blocks and the generation of energy. In agreement with this notion, measurement of the oxygen consumption rate (OCR) by extracellular flux assay revealed that CD40+IgM and CD40+IL-4–stimulated c-REL–deficient B cells showed reduced baseline and maximal oxygen consumption compared with control B cells at 24 h (Fig. 8 D). Also, ATP production measured upon administration of the ATP-synthase inhibitor oligomycin was significantly lower in the c-REL–deficient B cells. In addition, we observed that the glycolytic capacity of c-REL–deficient B cells under these stimulation conditions, as measured by the extracellular acidification rate (ECAR), was reduced compared with that of the control B cells (Fig. 8 D). These results support our observations from the RNA-seq analysis by demonstrating at the functional level that activated c-REL–deficient B cells have impaired metabolic functions. Due to their metabolic deficiency, rel-deleted B cells may be unable to complete the growth necessary for eventual cell division at later time points (starting at 36–48 h); this, in turn, could explain the marked absence of cell cycle entry and progression signatures in the rel-deleted B cells at 24 h compared with the wild-type B cells. In agreement with this notion is the observation that c-REL–deficient (but not RELA-deficient) B cells had a smaller cell size compared with wild-type B cells 24 h after stimulation (Fig. 8 E). In addition, a defect in the proliferation of c-REL–deficient B cells compared with wild-type and RELA-deficient B cells is evident 3 d after stimulation via CFSE staining in the presence of strong proliferation signals (Fig. 8 E), which correlates with the reduced expression of gene sets involved in cell cycle entry and progression observed in the rel-deleted B cells after 24 h of stimulation. Integrating the results obtained from the in vivo GEP and ex vivo RNA-seq analyses, we propose that c-REL is required for the maintenance of the GC reaction through the establishment of a growth program that is necessary for the proliferation of GC B cells after the formation of dark and light zones has been completed by day 7.
To determine whether the observed gene expression changes were specifically associated with activation of the c-REL subunit, we analyzed the transcriptome of B cells from relafl/flCD19-Cre and control mice stimulated for 6 h with CD40+IgM. The results revealed that 63 genes were expressed at ≥2-fold lower levels in the RELA-deficient cultures compared with wild-type B cells, compared with the 55 genes in the corresponding c-REL–deficient cultures (Fig. 8 F and Table S4). However, only 6 genes demonstrated ≥2-fold reduced expression in the absence of both c-REL and RELA compared with wild-type, demonstrating that c-REL and RELA control largely distinct sets of genes under these activation conditions.
DISCUSSION
The biphasic activation pattern of the canonical NF-κB subunits RELA and c-REL in T cell–dependent B cell responses, i.e., immediately upon antigen activation and later during the GC reaction in centrocytes, made it impossible to study their functions in GC B cell development by using constitutional rela and rel knockout mice. We took advantage of newly generated conditional mouse models to show that RELA and c-REL play essential, nonredundant roles in distinct stages of the GC reaction, ultimately impairing the generation of high-affinity B cells. Whereas c-REL was found to be required for GC maintenance and consequently plasma cell formation, RELA was dispensable for the GC reaction but essential for the development of GC-derived plasma cells. Our results have implications for the role of NF-κB in GC physiology and GC lymphomagenesis.
Because constitutional rela deletion causes embryonic lethality (Beg et al., 1995), previous studies on the role of RELA in T cell–dependent immunity were performed by reconstituting irradiated SCID mice with rela−/− fetal liver cells (Doi et al., 1997) and could thus not address B cell autonomy of rela deletion. Using mice with a conditional rela allele, we demonstrate here a B cell–intrinsic requirement of RELA for the generation of antigen-specific plasma cells in the GC reaction in vivo. The impeded plasma cell generation is most likely due to an impairment of RELA-deficient B cells to up-regulate BLIMP1 expression upon induction of plasmablastic differentiation, in the presence of normal IRF4 up-regulation. The normal up-regulation of IRF4 in RELA-deficient B cells is consistent with the ability of these cells to undergo Ig class switching, a process which is also dependent on IRF4. Moreover, it is not inconsistent with the impaired generation of plasmablasts because prdm1 can be expressed in an IRF4-independent fashion, and because BLIMP1 and IRF4 are jointly required for plasmablastic differentiation (De Silva et al., 2012). Collectively, our results provide evidence for a major role of RELA in T cell–dependent antibody responses in facilitating plasma cell generation by regulating BLIMP1 expression.
Mice with constitutional rel deletion (Carrasco et al., 1998; Pohl et al., 2002) and relfl/flCD19-Cre mice (unpublished data) show severely impaired formation of GCs upon immunization, consistent with a critical role of c-REL in the pre-GC B cell activation step (Köntgen et al., 1995; Tumang et al., 1998). The NF-κB pathway is, however, not activated in centroblasts, the descendants of antigen-activated B cells which clonally expand to form the GC (Shaffer et al., 2001; Basso et al., 2004). Accordingly, conditional deletion of rel in GC B cells did not affect GC development in the early phase that is characterized by the rapid proliferation of centroblasts. However, a dramatic change occurred on day 8 when GCs began to collapse in the absence of c-REL. Of note, centroblasts and centrocytes were lost equally, demonstrating that c-REL is required for the maintenance of the GC reaction and suggesting a role in licensing the cyclic reentry of centrocytes from the light into the dark zone (discussed further below).
We propose that the deletion of rel in GC B cells affects a subset of centrocytes in the light zone that is subjected to positive selection, rather than the GC B cell population as a whole, based on two observations. First, CD40 stimulation is known to induce nuclear translocation of NF-κB subunits including c-REL, and evidence suggests that this activation indeed occurs in centrocytes in the light zone, presumably due to B cell–T cell interaction (Basso et al., 2004). Importantly, antibody-mediated inhibition of the CD40–CD40L interaction leads to a rapid involution of the GC, indicating that CD40 signaling has a critical role in the maintenance of the GC reaction (Han et al., 1995). Thus, our observations raise the possibility that c-REL is an essential mediator of CD40 signaling in the GC. Second, c-REL is known to be strongly activated by BCR stimulation in vitro (Damdinsuren et al., 2010). The recent demonstration that BCR signaling is short circuited and thus inactivated in the vast majority of GC B cells (Khalil et al., 2012) may contribute to the observed lack of NF-κB activation in most GC B cells, and it is consistent with the possibility that NF-κB, and thus c-REL activation, may be restricted to the fraction of cells that undergoes BCR-mediated selection of the hypermutated antigen receptor for improved antigen affinity in the light zone.
Our results suggest that the loss of c-REL–deficient GC B cells is not due to an impaired transmission of prosurvival signals in rel-deleted B cells because constitutive expression of BCL2 in GC B cells was unable to rescue their disappearance in relfl/flCγ1-Cre mice. Consistent with this notion, mRNA encoding mcl1, which is required for the formation and persistence of GCs (Vikstrom et al., 2010), was expressed at similar levels in rel-deleted and control B cells. Surprisingly, the GEP analysis of rel-deleted and wild-type GC B cells performed before GC B cells are lost revealed the significantly reduced expression of genes involved in metabolism—including glucose, amino acid, and lipid metabolism—in the c-REL-deficient versus wild-type GC B cells. These findings were confirmed in in vitro–activated c-REL–deficient versus control B cells by RNA-seq analysis and at the biological level by extracellular flux analysis. Together, these results indicate that c-REL controls a metabolic program that mediates cell growth by providing energy and building blocks for the biosynthesis of protein, DNA, and phospholipids. The failure to activate a metabolic program may render the activated B cell unable to undergo cell growth before cell division. This observation is reminiscent of recent reports that identified metabolic reprogramming upon lymphocyte activation as a prerequisite for differentiation in various cellular contexts (Wang et al., 2011; van der Windt and Pearce, 2012; Man et al., 2013; Sinclair et al., 2013), and it places c-REL at a critical position in the control of cellular metabolism after B cell activation. Thus, the findings provide additional evidence for the regulation of metabolism by NF-κB that is emerging in different cellular contexts and that is only beginning to be explored (Grumont et al., 2002; Mauro et al., 2011). With regard to the GC B cell reaction, we propose that around day 7, at the time the GC has been fully established into dark and light zones and when somatically mutated B cells begin to appear (Jacob et al., 1993), positively selected B cells receive signals that ultimately activate c-REL. Because cell growth is a prerequisite for cell division, we hypothesize that the c-REL–dependent metabolic program may sustain the growth of these cells and would thus license the B cells for cyclic reentry to undergo consecutive rounds of hypermutation and selection. The need to reestablish a growth program in the GC B cell each time it reaches the point of selection would effectively eliminate from the GC reaction B cells that do not receive such signals as they cannot continue to clonally expand. Future work is needed to test this hypothesis.
Our observations may be interesting in light of recent findings that c-MYC expression is up-regulated in centrocytes and required for the maintenance of the GC reaction (Calado et al., 2012; Dominguez-Sola et al., 2012). In these studies, functional inactivation of c-MYC in GC B cells led to a rapid disappearance of both centrocytes and centroblasts in a fashion strikingly similar to what we observed in rel-deleted GC B cells beyond day 7. In this regard, it is interesting to note that the percentage of c-MYC–positive cells, after an initial spike immediately after immunization (∼40% of B cells at days 1 and 2), decreased dramatically in the early stages of the GC reaction before increasing to ∼25% of GC B cells (mostly comprising centrocytes) on day 8 (Dominguez-Sola et al., 2012). Thus, the expression pattern of c-MYC and the functional consequences of c-REL deficiency in the GC temporally coincide, consistent with the possibility that both c-MYC and c-REL exert critical functions in mediating the cyclic reentry of antigen-selected B cells into the dark zone, and thus in sustaining the GC response. Because NF-κB–dependent control of c-MYC expression has been observed during B cell activation in vitro (Grumont et al., 2002), it will be interesting to determine the relation between c-REL and c-MYC in GC B cell development.
Rel has been identified as a viral oncogene (v-rel) in birds that is causally involved in the pathogenesis of reticuloendotheliosis (Gilmore, 1999). The rel locus is amplified in certain human B cell lymphomas. Moreover, the recent identification of genetic mutations in NF-κB pathway components in B lymphoma subtypes (Lenz et al., 2008; Compagno et al., 2009; Kato et al., 2009; Schmitz et al., 2009) that lead to constitutive activation of the canonical pathway implicates a potential oncogenic role for c-REL in humans. Our results raise the possibility that unrestrained c-REL activity may promote lymphomagenesis by dysregulating cellular metabolism and thereby promoting cell survival and proliferation. How RELA—whose activation has been associated with certain non-Hodgkin lymphomas and multiple myeloma (Annunziata et al., 2007)—could promote tumorigenesis may be an interesting area of investigation. Our findings that c-REL and RELA exert entirely different functions during GC development highlights the importance of dissecting the roles of the individual NF-κB subunits in cancers with constitutive NF-κB activation, a circumstance which may be exploited for developing targeted therapies. Finally, our findings have general implications for NF-κB biology as they may help to improve our understanding of the selectivity of the NF-κB response (Sen and Smale, 2010).
MATERIALS AND METHODS
Generation of mice carrying a floxed rel or rela allele.
The vector to target rela and rel has been described previously (Klein et al., 2006). The vector was constructed such that upon Cre-mediated deletion, the promoter regions and the exons comprising the first ATG of rela or rel were deleted with simultaneous activation of eGFP expression, and Flp-mediated recombination produced deletion of the targeted region plus the eGFP gene (performed to obtain rel germ-line knockouts; Fig. S1). See the Fig. S1 legend for details on cloning of the targeting vectors and the generation of the mice. The conditional rel and rela and the knockout rel allele were backcrossed to C57BL/6 mice (n > 10). Cγ1-Cre, CD19-Cre, and Bcl2-Ig mice have been previously described (McDonnell et al., 1989; Rickert et al., 1997; Casola et al., 2006). In some experiments, conditional rela mice (referred to as p65fl/+; Luedde et al., 2008)—in which exons 2–4 of rela were flanked with loxP sites—were used that do not express eGFP upon gene deletion. Mice were housed and treated in compliance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and according to the guidelines of the Institute of Comparative Medicine at Columbia University. The animal protocol was approved by Columbia University’s IACUC.
Immunization.
Mice were immunized i.p. with 109 SRBCs (Cocalico Biologicals) in PBS, or NP28-KLH (100 µg per mouse; Biosearch Technologies) in complete Freund’s adjuvant (Sigma-Aldrich). For secondary antibody responses, the immunized mice were rechallenged on day 21 with NP-KLH (20 µg per mouse) in incomplete Freund’s adjuvant (Sigma-Aldrich). Peripheral blood and spleens were removed at the indicated time points for analysis.
B cell isolation.
Single cell suspensions of mouse spleen were subjected to hypotonic lysis and B cells were purified by depletion of magnetically labeled non–B cells using the MACS B cell isolation kit (Miltenyi Biotec). NP-binding B cells were enriched by MACS-based depletion of T cells, macrophages, granulocytes, and unswitched B cells, using biotinylated antibodies (from BD if not stated otherwise) to CD90.2 (clone 53–2.1), F4/80 (clone A3-1; Serotec), Ly-6G and Ly-6C (clone RB6-8C5), and IgM (clone R6-60.2) and IgD (clone 11–26; Southern Biotech), respectively, followed by incubation with streptavidin microbeads (Miltenyi Biotec) and magnetic depletion of the labeled cell fractions. eGFP+ and eGFP− B220+ B cells, or B220+CD95hiPNAhi eGFP+ or eGFP− GC B cell subpopulations were isolated by flow cytometry on a FACSAria (BD). Cells were sorted into 50% FBS in PBS and cell pellets were either lysed with TRIzol reagent (Life Technologies) for RNA isolation or washed once with PBS and subjected to NP-40–based lysis for immunoblot analysis.
B cell culture.
Purified B cells from the indicated genotypes were cultured in the presence of: 1 µg/ml anti–mouse CD40 (clone HM40-3; BD), 20 µg/ml LPS (Sigma-Aldrich), 20 ng/ml IL-4 (R&D Systems), and 16.25 µg/ml anti–mouse IgM (Jackson ImmunoResearch Laboratories, Inc.). Cell density in CD40+IgM stimulation experiments was 1.5 × 106 cells/ml. In all other experiments, cells were cultured at a density of 106 cells/ml.
Extracellular flux analysis.
In vitro–stimulated B cells were harvested and seeded in triplicates at 3 × 105 (CD40+IgM stimulation) or 2 × 105 cells per well on XF96 microplates (Seahorse Bioscience) coated with Cell-TAK (Corning). The plates were then centrifuged to attach the cells to the bottom of the plate. The ECAR was assessed in glucose- and bicarbonate-free DMEM (Sigma-Aldrich) supplemented with 2 mM L-Ala-Gln (Glutamax; Gibco) in response to 10 mM glucose, 1 µM oligomycin (Sigma-Aldrich), and 100 mM 2-deoxyglucose (2-DG; Sigma-Aldrich). The OCR was measured in DMEM containing 1 mM sodium pyruvate, 11 mM glucose, and 2 mM L-Ala-Gln upon sequential addition of 1 µM oligomycin, 1 µM fluorocarbonyl cyanide phenylhydrazone (FCCP; Sigma-Aldrich), and 1 µM rotenone (Sigma-Aldrich). All measurements were performed on an XF96 Extracellular Flux Analyzer (Seahorse Bioscience). The cells of each well were subsequently detached, and cell number and viability were assessed by trypan blue exclusion. All OCR and ECAR measurements were normalized to the yielded live cell counts to adjust for unequal cell death during the assay.
Flow cytometry.
Spleen cell suspensions or cultured B cells were stained on ice in PBS/0.5% BSA with specific antibodies listed in Table S5. Anti–mouse CD16-CD32 (BD) was used to block FcγIII/II receptors before staining for NP-binding cells. The cells were washed and analyzed on a FACSCalibur or an LSRII (BD). Data were analyzed using FlowJo software.
CFSE analysis.
The proliferation capacity of purified B cells was assessed by staining using the CellTrace CFSE Cell Proliferation kit (Life Technologies) according to the manufacturer’s protocol. Successful labeling was confirmed by flow cytometry, and cells were cultured with CD40 plus IL-4 for 3 d.
ELISA.
For SRBC immunization experiments, 96-well immune plates (Thermo Fisher Scientific) were coated with anti–mouse Ig(H+L) (Southern Biotech). For NP immunization experiments, plates were coated with NP9-BSA or NP25-BSA (Biosearch Technologies). Mouse serum samples were incubated for 2 h at room temperature. Standard curves were generated using mouse IgM and mouse IgG1 (Southern Biotech). Bound antibodies were detected by AP-conjugated anti–mouse IgM and anti–mouse IgG1 antibodies (Table S5). Plates were developed with p-nitrophenylphosphate (Southern Biotech) dissolved in substrate buffer. For ELISPOT analysis, the spleens and BM of NP-KLH–immunized mice were removed at the indicated time points, and single cell suspensions were subjected to hypotonic lysis. The samples were plated overnight on 96-well filtration plates (Millipore) coated with NP25-BSA (Biosearch Technologies). The cells were plated in 1:2 serial dilutions, starting at 8 × 105 cells for BM-derived samples, and at 105 cells for spleen-derived fractions. NP-specific IgG1-secreting cells were detected by AP-conjugated anti–mouse IgG1 antibody (Table S5). Plates were developed with nitro blue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Roche).
Immunohistochemistry.
Sections 2–3 µm in thickness were cut from spleen tissue that was fixed overnight in 10% formalin and embedded in paraffin. Unlabeled antibodies (Table S5) were incubated over night at 4°C and counterstained with either anti–rabbit HRP-labeled polymer (Dako) developed in aminoethylcarbazole (AEC; Sigma-Aldrich) or alkaline peroxidase-conjugated streptavidin (Dako) developed in nitro blue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Roche). Sections stained for BCL6/IgG1 were counterstained with hematoxylin.
Immunoblot analysis.
Purified B cells were subjected to NP-40–based lysis, separated by SDS-PAGE, and blotted on nitrocellulose membranes (GE Healthcare). Samples were incubated with primary antibodies (Table S5) overnight at 4°C. Horseradish peroxidase–conjugated secondary antibodies (Table S5) and ECL Western Blotting Substrate or West Dura Extended Duration Substrate (Thermo Scientific) were used for detection.
Somatic hypermutation analysis.
Specific amplification of VH186.2 transcripts from sorted eGFP+ and eGFP− B cell fractions from the spleen of NP-KLH–immunized mice was performed as previously described (Klein et al., 2006). In brief, cDNA was generated using a Cγ1-specific primer. Semi-nested PCR for amplification of cDNA fractions was performed using a Cγ1 and a VH186.2 leader primer (first round PCR, 20 cycles), followed by amplification of 3 µl of the first-round reaction product for 30 cycles with the Cγ1 primer and a VH186.2 nested primer. PCR products were cloned into pGEM-T Easy Vector (Promega). Sequencing of PCR products corresponding to single colonies was performed as previously described (Klein et al., 2006), using the VBASE2 database for sequence analysis.
GEP analysis.
Purified B cells were sorted from SRBC-immunized mice (day 7) into GFP+ and GFP− GC (CD95hiPNAhi) B cell fractions. Total RNA of ∼3 × 104 cells was isolated using the Nucleospin RNA XS isolation kit (MACHEREY-NAGEL) according to the manufacturer’s protocol, and RNA integrity >8 was determined on a BioAnalyzer 2100 (Agilent Technologies). Approximately 10 ng of total RNA was reverse transcribed into cDNA with the Ovation RNA Amplification System (NuGEN) according to the manufacturer’s instructions, followed by purification of the amplified cDNA using the QIAquick PCR purification kit (QIAGEN). 3 µg of total cDNA was labeled and fragmented using the Encore Biotin Module (NuGEN) and hybridized onto GeneChip Mouse Genome 430 2.0 arrays (Affymetrix). Data were analyzed using Affymetrix MAS5. Supervised analysis was performed with the GENES@WORK software platform as previously described (Klein et al., 2001). GEP data are available through the GEO database (GSE58839).
Quantitative RT-PCR.
qRT-PCR was performed using gene-specific primers spanning at least one intron in the target gene and Power SYBR Green PCR Master Mix (Applied Biosystems; Table S6). All reactions contained 10 ng of amplified cDNA and were performed in triplicate with the 7500 Fast Real Time PCR System (Applied Biosystems). Results were analyzed using the 2−ΔΔCT method with Hprt1 as reference gene.
RNA-seq.
RNA was harvested from three culture replicates/genotype of CD40+IgM-stimulated B cells and submitted for RNA sequencing at the Columbia Genome Center. 30 million single-end 100 bp reads were sequenced per sample on the HiSeq2000/2500 V3 instrument (Illumina). DESeq analysis was used to identify differentially expressed genes and determine fold expression changes between genotypes. RNA-seq data are available through the GEO database (GSE58983).
GSEA analysis.
The GSEA tool (Subramanian et al., 2005) available from the website of the Broad Institute was used to identify gene signatures enriched in wild-type control versus rel-deleted B cells. We screened the collection of signatures under the category GO_BP and the curated category KEGG to determine significant enrichment (FDR < 25%, P ≤ 0.05).
Online supplemental material.
Fig. S1 shows generation and functional analysis of the conditional rela and rel alleles. Fig. S2 shows a chart depicting metabolic pathways that seem to be affected by the failed up-regulation of genes (indicated by green color) in c-REL–deficient GC B cells. Table S1 shows genes differentially expressed in c-REL–deficient versus wild-type GC B cells. Table S2 shows genes not induced in c-REL–deficient versus wild-type B cells by ≥2-fold. Table S3 shows a c-REL–dependent metabolic program precedes a proliferation program in activated B cells. Table S4 shows genes not induced in RELA-deficient versus wild-type B cells by ≥2-fold. Table S5 shows antibodies used for flow cytometry, immunohistochemistry, ELISA, and Western blot analysis. Table S6 shows primer sequences used for quantitative RT-PCR analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132613/DC1.
Supplementary Material
Acknowledgments
We thank S. Casola and K. Rajewsky for Cγ1-Cre mice; L. Pasqualucci and R. Dalla-Favera for critical reading of this manuscript; D. Dominguez-Sola for insights and discussion; S. Reiner for discussion; G. Victora and K. Gordon for advice on flow cytometric assays; C. Liu and S. Tetteh for cell sorting; Q. Shen for help in the generation of the transgenic mouse lines; T. Ludwig for advice on the targeting vectors; and H. Tang for advice on staining procedures. We also thank Kevin Man and Axel Kallies for advice on metabolism experiments and Xue-Liang Du at the Metabolomics Core at Albert Einstein Medical College for help with the Seahorse experiments. We are grateful to R. Dalla-Favera for his support in the generation of the transgenic mouse lines.
This work was supported by National Cancer Institute (NCI)/National Institutes of Health (NIH) grant R01-CA157660 to U. Klein, a grant from the Stewart Trust Foundation (USA), the HICCC, and through fellowships of the German Research Council (DFG) to N. Heise, a Cancer Biology Training Program fellowship (NCI/NIH grant 5T32-CA009503-26) to N.S. De Silva, and through a fellowship of the Fondazione Cariplo (Italy) to G. Simonetti. The Metabolomics Core of the Diabetes Training and Research Center of AECOM is supported by NIH grant P60DK20541.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ASC
- antibody-secreting cell
- ECAR
- extracellular acidification rate
- eGFP
- enhanced GFP
- GC
- germinal center
- GEP
- gene expression profiling
- GSEA
- gene set enrichment analysis
- KLH
- keyhole limpet hemocyanin
- OCR
- oxygen consumption rate
- SRBC
- sheep RBC
References
- Allen, C.D., Okada T., and Cyster J.G.. 2007. Germinal-center organization and cellular dynamics. Immunity. 27:190–202 10.1016/j.immuni.2007.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata, C.M., Davis R.E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., et al. 2007. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 12:115–130 10.1016/j.ccr.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannard, O., Horton R.M., Allen C.D., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso, K., Klein U., Niu H., Stolovitzky G.A., Tu Y., Califano A., Cattoretti G., and Dalla-Favera R.. 2004. Tracking CD40 signaling during germinal center development. Blood. 104:4088–4096 10.1182/blood-2003-12-4291 [DOI] [PubMed] [Google Scholar]
- Beg, A.A., Sha W.C., Bronson R.T., Ghosh S., and Baltimore D.. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 376:167–170 10.1038/376167a0 [DOI] [PubMed] [Google Scholar]
- Calado, D.P., Sasaki Y., Godinho S.A., Pellerin A., Köchert K., Sleckman B.P., de Alborán I.M., Janz M., Rodig S., and Rajewsky K.. 2012. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immunol. 13:1092–1100 10.1038/ni.2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, D., Cheng J., Lewin A., Warr G., Yang H., Rizzo C., Rosas F., Snapper C., and Bravo R.. 1998. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J. Exp. Med. 187:973–984 10.1084/jem.187.7.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola, S., Cattoretti G., Uyttersprot N., Koralov S.B., Seagal J., Hao Z., Waisman A., Egert A., Ghitza D., and Rajewsky K.. 2006. Tracking germinal center B cells expressing germ-line immunoglobulin γ1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA. 103:7396–7401 10.1073/pnas.0602353103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno, M., Lim W.K., Grunn A., Nandula S.V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A., et al. 2009. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 459:717–721 10.1038/nature07968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damdinsuren, B., Zhang Y., Khalil A., Wood W.H. III, Becker K.G., Shlomchik M.J., and Sen R.. 2010. Single round of antigen receptor signaling programs naive B cells to receive T cell help. Immunity. 32:355–366 10.1016/j.immuni.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva, N.S., Simonetti G., Heise N., and Klein U.. 2012. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol. Rev. 247:73–92 10.1111/j.1600-065X.2012.01113.x [DOI] [PubMed] [Google Scholar]
- Doi, T.S., Takahashi T., Taguchi O., Azuma T., and Obata Y.. 1997. NF-κB RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med. 185:953–962 10.1084/jem.185.5.953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola, D., Victora G.D., Ying C.Y., Phan R.T., Saito M., Nussenzweig M.C., and Dalla-Favera R.. 2012. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 13:1083–1091 10.1038/ni.2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis, S., and Strasser A.. 2003. The role of Rel/NF-κB transcription factors in B lymphocyte survival. Semin. Immunol. 15:159–166 10.1016/S1044-5323(03)00036-8 [DOI] [PubMed] [Google Scholar]
- Gerondakis, S., and Siebenlist U.. 2010. Roles of the NF-κB pathway in lymphocyte development and function. Cold Spring Harb. Perspect. Biol. 2:a000182 10.1101/cshperspect.a000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, T.D.1999. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 18:6925–6937 10.1038/sj.onc.1203222 [DOI] [PubMed] [Google Scholar]
- Gitlin, A.D., Shulman Z., and Nussenzweig M.C.. 2014. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 509:637–640 10.1038/nature13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M.R., Monti S., Dalla-Favera R., Pasqualucci L., Walsh N.C., Schmidt-Supprian M., Kutok J.L., Rodig S.J., Neuberg D.S., Rajewsky K., et al. 2011. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl. Acad. Sci. USA. 108:2873–2878 10.1073/pnas.1019537108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont, R.J., Strasser A., and Gerondakis S.. 2002. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-κB regulated c-myc transcription. Mol. Cell. 10:1283–1294 10.1016/S1097-2765(02)00779-7 [DOI] [PubMed] [Google Scholar]
- Han, S., Hathcock K., Zheng B., Kepler T.B., Hodes R., and Kelsoe G.. 1995. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J. Immunol. 155:556–567 [PubMed] [Google Scholar]
- Hayden, M.S., and Ghosh S.. 2004. Signaling to NF-κB. Genes Dev. 18:2195–2224 10.1101/gad.1228704 [DOI] [PubMed] [Google Scholar]
- Jacob, J., Przylepa J., Miller C., and Kelsoe G.. 1993. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med. 178:1293–1307 10.1084/jem.178.4.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaileh, M., and Sen R.. 2012. NF-κB function in B lymphocytes. Immunol. Rev. 246:254–271 10.1111/j.1600-065X.2012.01106.x [DOI] [PubMed] [Google Scholar]
- Kato, M., Sanada M., Kato I., Sato Y., Takita J., Takeuchi K., Niwa A., Chen Y., Nakazaki K., Nomoto J., et al. 2009. Frequent inactivation of A20 in B-cell lymphomas. Nature. 459:712–716 10.1038/nature07969 [DOI] [PubMed] [Google Scholar]
- Khalil, A.M., Cambier J.C., and Shlomchik M.J.. 2012. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 336:1178–1181 10.1126/science.1213368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, U., and Dalla-Favera R.. 2008. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 8:22–33 [DOI] [PubMed] [Google Scholar]
- Klein, U., Tu Y., Stolovitzky G.A., Mattioli M., Cattoretti G., Husson H., Freedman A., Inghirami G., Cro L., Baldini L., et al. 2001. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194:1625–1638 10.1084/jem.194.11.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Köntgen, F., Grumont R.J., Strasser A., Metcalf D., Li R., Tarlinton D., and Gerondakis S.. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965–1977 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- Le, A., Lane A.N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C.J., Slusher B.S., Zhang H., et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15:110–121 10.1016/j.cmet.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz, G., Davis R.E., Ngo V.N., Lam L., George T.C., Wright G.W., Dave S.S., Zhao H., Xu W., Rosenwald A., et al. 2008. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 319:1676–1679 10.1126/science.1153629 [DOI] [PubMed] [Google Scholar]
- Luedde, T., Heinrichsdorff J., de Lorenzi R., De Vos R., Roskams T., and Pasparakis M.. 2008. IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc. Natl. Acad. Sci. USA. 105:9733–9738 10.1073/pnas.0800198105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan, I.C.1994. Germinal centers. Annu. Rev. Immunol. 12:117–139 10.1146/annurev.iy.12.040194.001001 [DOI] [PubMed] [Google Scholar]
- Man, K., Miasari M., Shi W., Xin A., Henstridge D.C., Preston S., Pellegrini M., Belz G.T., Smyth G.K., Febbraio M.A., et al. 2013. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14:1155–1165 10.1038/ni.2710 [DOI] [PubMed] [Google Scholar]
- Mauro, C., Leow S.C., Anso E., Rocha S., Thotakura A.K., Tornatore L., Moretti M., De Smaele E., Beg A.A., Tergaonkar V., et al. 2011. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 13:1272–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell, T.J., Deane N., Platt F.M., Nunez G., Jaeger U., McKearn J.P., and Korsmeyer S.J.. 1989. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 57:79–88 10.1016/0092-8674(89)90174-8 [DOI] [PubMed] [Google Scholar]
- Morgan, M.A., Magnusdottir E., Kuo T.C., Tunyaplin C., Harper J., Arnold S.J., Calame K., Robertson E.J., and Bikoff E.K.. 2009. Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation. Mol. Cell. Biol. 29:5813–5827 10.1128/MCB.00670-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl, T., Gugasyan R., Grumont R.J., Strasser A., Metcalf D., Tarlinton D., Sha W., Baltimore D., and Gerondakis S.. 2002. The combined absence of NF-κB1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc. Natl. Acad. Sci. USA. 99:4514–4519 10.1073/pnas.072071599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky, K.1996. Clonal selection and learning in the antibody system. Nature. 381:751–758 10.1038/381751a0 [DOI] [PubMed] [Google Scholar]
- Rickert, R.C., Roes J., and Rajewsky K.. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, M., Gao J., Basso K., Kitagawa Y., Smith P.M., Bhagat G., Pernis A., Pasqualucci L., and Dalla-Favera R.. 2007. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 12:280–292 10.1016/j.ccr.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Schmitz, R., Hansmann M.L., Bohle V., Martin-Subero J.I., Hartmann S., Mechtersheimer G., Klapper W., Vater I., Giefing M., Gesk S., et al. 2009. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 206:981–989 10.1084/jem.20090528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas, R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Sen, R., and Smale S.T.. 2010. Selectivity of the NF-κB response. Cold Spring Harb. Perspect. Biol. 2:a000257 10.1101/cshperspect.a000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, A.L., Yu X., He Y., Boldrick J., Chan E.P., and Staudt L.M.. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212 10.1016/S1074-7613(00)00020-0 [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., Rosenwald A., Hurt E.M., Giltnane J.M., Lam L.T., Pickeral O.K., and Staudt L.M.. 2001. Signatures of the immune response. Immunity. 15:375–385 10.1016/S1074-7613(01)00194-7 [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L. III, Young R.M., and Staudt L.M.. 2012. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 30:565–610 10.1146/annurev-immunol-020711-075027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro-Shelef, M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., and Calame K.. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Sinclair, L.V., Rolf J., Emslie E., Shi Y.B., Taylor P.M., and Cantrell D.A.. 2013. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14:500–508 10.1038/ni.2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K.G., Light A., O’Reilly L.A., Ang S.M., Strasser A., and Tarlinton D.. 2000. bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J. Exp. Med. 191:475–484 10.1084/jem.191.3.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumang, J.R., Owyang A., Andjelic S., Jin Z., Hardy R.R., Liou M.L., and Liou H.C.. 1998. c-Rel is essential for B lymphocyte survival and cell cycle progression. Eur. J. Immunol. 28:4299–4312 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu, S., and Karin M.. 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- van der Windt, G.J., and Pearce E.L.. 2012. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol. Rev. 249:27–42 10.1111/j.1600-065X.2012.01150.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora, G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Victora, G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom, I., Carotta S., Lüthje K., Peperzak V., Jost P.J., Glaser S., Busslinger M., Bouillet P., Strasser A., Nutt S.L., and Tarlinton D.M.. 2010. Mcl-1 is essential for germinal center formation and B cell memory. Science. 330:1095–1099 10.1126/science.1191793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., and Green D.R.. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 35:871–882 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.