Abstract
Odorant molecules bind to their target receptors in a precise and coordinated manner. Each receptor recognizes a specific signal and relays this information to the brain. As such, determining how olfactory information is transferred to the brain, modifying both perception and behavior, merits investigation. Interestingly, there is emerging evidence that cellular transduction and transcriptional factors are involved in the diversification of olfactory receptor neuron. Here we provide a robust whole mount immunological labeling method to assay in vivo olfactory receptor neuron organization. Using this method, we identified all olfactory receptor neurons with anti-ELAV antibody, a known pan-neural marker and Or49a-mCD8::GFP, an olfactory receptor neuron specifically expressed in Nba neuron using anti-GFP antibody.
Keywords: Neuroscience, Issue 87, Developmental biology, Drosophila, Whole mount immunolabeling, olfactory receptor neurons, antennae, sensory organ
Introduction
The olfactory system is used to distinguish between an immense variety of odor molecules and subsequently to send the resulting information to the higher brain centers. This input is used to precisely control fundamental animal behaviors, such as feeding and mating1-6. As each olfactory neuron type is associated with a specific set of odors, the diversification of olfactory receptor neurons (ORN)s is essential for proper olfactory system function7.
Drosophila genetics enables us to perform single cell level investigation involving molecular mechanisms associated with ORN development and physiological function 8-16. Whole mount immunostaining of Drosophila antennae has enabled us to understand in greater detail the molecular mechanisms involved in the diversification of olfactory receptor neurons (ORN)s7. Herein we provide a comprehensive description of a simple method to achieve this.
Protocol
1. Prepare apple plate
Mix 12.5 g agar, 125 ml 100% commercially available apple juice, 12.5 g glucose, and 375 ml H2O. Microwave the mixture for 1 to 2 minutes and pour to the 3 cm cell culture dish. Store at 4 °C.
2. Genetic cross
Use the following representative genetic cross:
Or49a-mCD8::GFP/CyO x w1118
3. Dissection and staining protocol
Anesthetize the fly and then cut the fly head vertically by holding it using a forceps.
Carefully place the antennae containing portion on an apple plate.
Cut the third segment of the antenna using fine dissection scissors.
Place 90 µl of the fixation solution (4% paraformaldehyde in 0.1% PBST (PBS with 0.1% Triton X-100)) to the middle of a glass bottom culture dish.
Gently transfer the dissected antennae with a sharp needle directly to the fixation solution. If necessary, physically submerge the antennae into the solution using needle.
Incubate for 40 minutes at room temperature (RT). Wash the antennae in 0.4% PBST (PBS with 0.4% Triton X-100), 3x 10 minutes in each, keeping them in the same dish. Use yellow tips to remove and add PBST solution. Use 90 µl of the wash solution each time. NOTE: Do not place the dish into a shaker during immunohistochemistry. Before removing or adding the solution into the dish, bring all the antennae into the center of the dish using the needle and carefully add or remove the solution from the edge of the dish.
Block the antennae with 90 µl of 5% normal horse serum in 0.1% PBST for 20 minutes at RT.
After removing the blocking solution, incubate the antennae with 90µl of primary antibodies in 0.1% PBST containing 5% horse serum for 48 hr at 4 °C in a moistened container as described previously8.
Wash the antennae 6x 10 minutes in 0.4% PBST.
Incubate the antennae with 90 µl of secondary antibodies in 0.1% PBST containing 5% horse serum for 48 hr at 4 °C. Wash 6x 10 minutes using 0.4% PBST.
To mount the antennae, remove PBST from the culture dish as much as possible and gradually introduce two different concentrations of glycerol to the antennae. First add 40% glycerol to the dish for 1 to 2 minutes; then remove this and add 80% glycerol.
Carefully extract the antennae (including 80% glycerol) from the culture dish using yellow tip and place them onto a slide. Gently place a coverslip on top and seal the coverslip edges with nail polish. The antennae are now ready to be imaged by fluorescence microscopy.
Representative Results
Ensuring that both the dissection and fixation are performed quickly is a key factor in achieving success with this protocol. Using fine scissors and forceps is also crucial. After immunostaining, fluorescent labeled antennae were examined under a confocal microscope. We normally take 1µm sections using a 20x lens. We labeled Nba7 ORNs using Or49a-mCD8::GFP and counted the number of Nba ORNs in wild-type antenna. The mCD8-GFP reporter is cell membrane localized and so the expression seen in Figure 2 exhibits the cell membrane of OR49a ORNs expressing GFP. In Figure 2 shows Nba ORNs expression using anti GFP antibody and anti ELAV was used as a pan-neural marker. The average number of Nba ORNs per antenna is 20 (n=8).
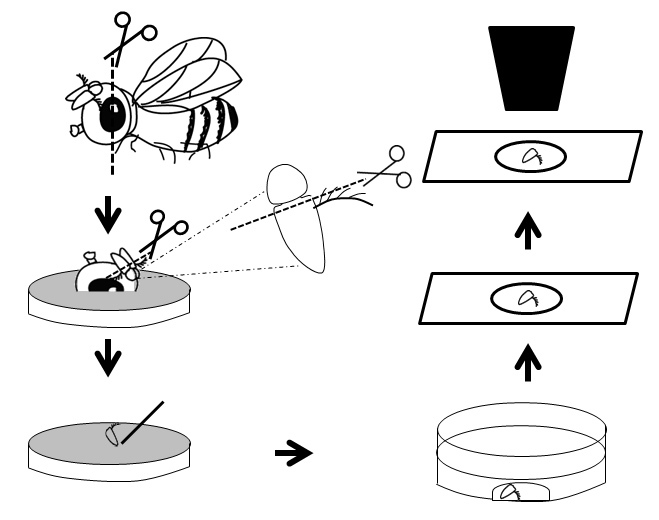 Figure 1: General overview of the dissection procedure.
Figure 1: General overview of the dissection procedure.
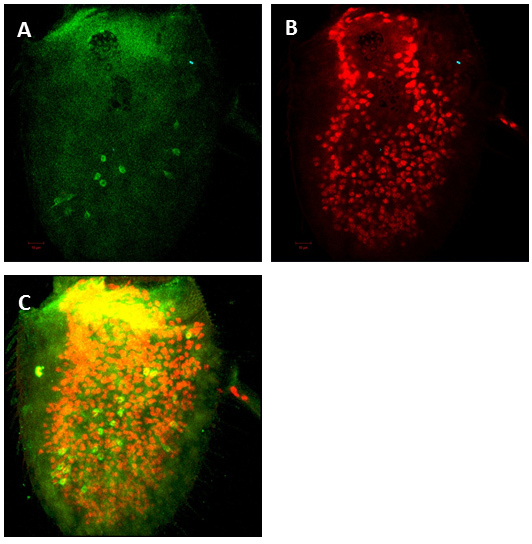 Figure 2: Projection of confocal Z-series of a successfully dissected antenna. To detect neurons anti-ELAV antibody was used as a pan-neural marker (A) and anti-GFP antibody to detect specific odorant receptor expression (B) and merged picture as shown in (C).
Figure 2: Projection of confocal Z-series of a successfully dissected antenna. To detect neurons anti-ELAV antibody was used as a pan-neural marker (A) and anti-GFP antibody to detect specific odorant receptor expression (B) and merged picture as shown in (C).
Discussion
The dissection of the Drosophila antenna we describe is simple and easy to perform in a laboratory setting. To ensure a successful dissection, it is essential to utilize fine-edged scissors. While immunostaining the dissected antenna, it is important to incubate them in a moisture-filled container to avoid evaporation of the antibody solution. The dissected antenna has a tendency to float in the solution. Using 0.1% Triton in PBS during the fixation and blocking steps will facilitate submersion of the antenna in the solution and to ensure better staining. Using "glass bottom culture dish" could reduce the loss of antennae during immunostaining and ensure the small amounts (90 µl) of antibody solution used in each experiment.
Neuronal-class diversification is a central feature of neurogenesis. This physiological process is exemplified in the olfactory system, which utilizes a large array of olfactory receptor neuron (ORN) classes. Generation of a wide variety of ORNs with odorant receptor expression and axonal targeting is crucial to generating the neuronal diversity required for transmitting information from odor molecules to higher brain centers. Our whole mount antenna immunostaining protocol aids in furthering our understanding of the molecular mechanisms underlying ORN diversification.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This study was supported by MEXT-Supported Program for the Strategic Research Foundation at Private Universities and JSPS Young Scientist B grant for H.T. We would like to thank Ohtake Norihito to edit the video clips.
References
- Christensen TA, White J. Representation of olfactory information in the brain In The Neurobiology of Taste and Smell. New York: 2000. pp. 201–232. [Google Scholar]
- Ache BW. Towards a common strategy for transducing olfactory information. Sem. Cell Biol. 1994;5:55–63. doi: 10.1006/scel.1994.1008. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction. C. elegans. Cell. 1993;13:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Barth AL, Justice NJ, Ngai J. Asynchronous onset of odorant receptor expression in the developing zebrafish olfactory system. Neuron. 1996;16:23–34. doi: 10.1016/s0896-6273(00)80020-3. [DOI] [PubMed] [Google Scholar]
- Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- Stockinger P, et al. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Endo K, et al. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat Neurosci. 2011;15:224–233. doi: 10.1038/nn.2998. [DOI] [PubMed] [Google Scholar]
- Karim MR, Moore AW. Morphological analysis of Drosophila larval peripheral sensory neuron dendrites and axons using genetic mosaics. J Vis Exp. 2011. [DOI] [PMC free article] [PubMed]
- Suh GS, et al. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Sachse S, Galizia CG. Role of inhibition for temporal and spatial odor representation in olfactory output neurons: A calcium imaging study. J Neurophysiol. 2002;87:1106–1117. doi: 10.1152/jn.00325.2001. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Clyne P, et al. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, et al. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wang JW, et al. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]


