SUMMARY
The Cullin 9 (CUL9) gene encodes a putative E3 ligase that localizes in the cytoplasm. Cul9 null mice develop spontaneous tumors in multiple organs, however either the cellular or molecular mechanisms of CUL9 in tumor suppression are currently not known. We show here that deletion of Cul9 leads to abnormal nuclear morphology, increased DNA damage and aneuploidy. CUL9 knockdown rescues the microtubule and mitosis defects in cells depleted for CUL7 or OBSL1, two genes that are mutated in a mutually exclusive manner in 3M growth retardation syndrome and function in microtubule dynamics. CUL9 promotes the ubiquitylation and degradation of survivin and is inhibited by CUL7. Depletion of CUL7 decreases survivin level and overexpression of survivin rescues the defects caused by CUL7 depletion. We propose a 3M–CUL9-survivin pathway in maintaining microtubule and genome integrity, normal development and tumor suppression.
Keywords: cullins, genome integrity, 3M disease, survivin
INTRODUCTION
CUL7 and CUL9 (formerly PARC) belong to the cullin family of proteins which function as scaffold proteins in the assembly of E3 ubiquitin ligases by binding to the small RING finger protein ROC1 (RBX1) and substrates or substrate recruiting factors. CUL7 (1698 amino acids in humans) and CUL9 (2,517 amino acids) are the largest members of the cullin family and contain multiple functional domains. CUL9 and CUL7 have a high sequence identity with each other, but not with other cullins except at the ROC1-binding region. Both CUL7 and CUL9 are relatively newly evolved genes as they originated after the emergence of vertebrates (Marin, 2009), localize predominantly in the cytoplasm and bind to p53 (Andrews et al., 2006; Nikolaev et al., 2003). Deletion of Cul7 in mice leads to intrauterine growth retardation and perinatal death (Arai et al., 2003). Mutations in the human CUL7 gene are associated with 3M syndrome (Miller et al., 1975), a severe autosomal recessive disorder characterized by short stature, unusual triangle-shaped facial features with a broad forehead, and widespread skeletal abnormalities in the neck, chest, shoulder, upper and lower back, fingers and legs. Besides CUL7, two additional genes, OBSL1 and CCDC8, have been found to be mutated in 3M syndrome. These three genes are mutated in a mutually exclusive manner, with CUL7 being the most frequently mutated (~65%), followed by OBSL1 (~30%) and CCDC8 (~5%) (Huber et al., 2011). In the accompanying paper, we demonstrate that these three 3M proteins form a complex and function to maintain microtubule integrity.
Survivin (BIRC5) is a member of the inhibitor of apoptosis protein (IAP) family and plays two critical and yet to be fully reconciled roles in cell proliferation. Survivin is highly expressed in different types of human tumors and promotes cell survival by inhibiting caspase and procaspase. Survivin is also a component of the chromosomal passenger complex (CPC) and recruits the CPC to mitotic chromosomes to control multiple steps of mitosis and maintain genome stability (Watanabe, 2010). In addition to these two extensively investigated roles, survivin also plays an important but less known function in the regulation of microtubule dynamics. Loss of survivin, by knockout, knockdown or injection of survivin antibody, reduces microtubule fiber density, increases EB1 foci in interphase cells, increases microtubule recovery after nocodazole treatment and, conversely, overexpression of survivin stabilizes microtubules (Giodini et al., 2002; Li et al., 1998; Rosa et al., 2006). The level of survivin is regulated both transcriptionally, including repression by p53 (Hoffman et al., 2002; Mirza et al., 2002), and posttranslationally by the ubiquitin-proteasome pathway (Zhao et al., 2000). The identity of the survivin E3 ligase is not known.
Deletion of Cul9 in mice resulted in spontaneous tumor development in multiple organ tissues, including lymphoma, sarcoma and tumors in pituitary, lung, liver and ovary, accelerated Eµ-Myc-induced lymphomagenesis and rendered mice susceptible to carcinogenesis (Pei et al., 2011). The cellular and molecular basis for CUL9 function in tumor suppression is unclear. Prompted by the function of the 3M complex in maintaining microtubule integrity (Yan et al. accompanying paper) and the previous report that CUL7 forms a heterodimer with CUL9 (Skaar et al., 2007), we investigated the function of CUL9 in maintaining genome stability and its functional relationship with CUL7 and the 3M complex. These studies led to the discovery that CUL9 is a critical downstream effector of the 3M complex in the maintenance of microtubules and genome integrity and that survivin is a substrate of CUL9.
RESULTS
Deletion of the Cul9 gene resulted in polyploidy in vivo
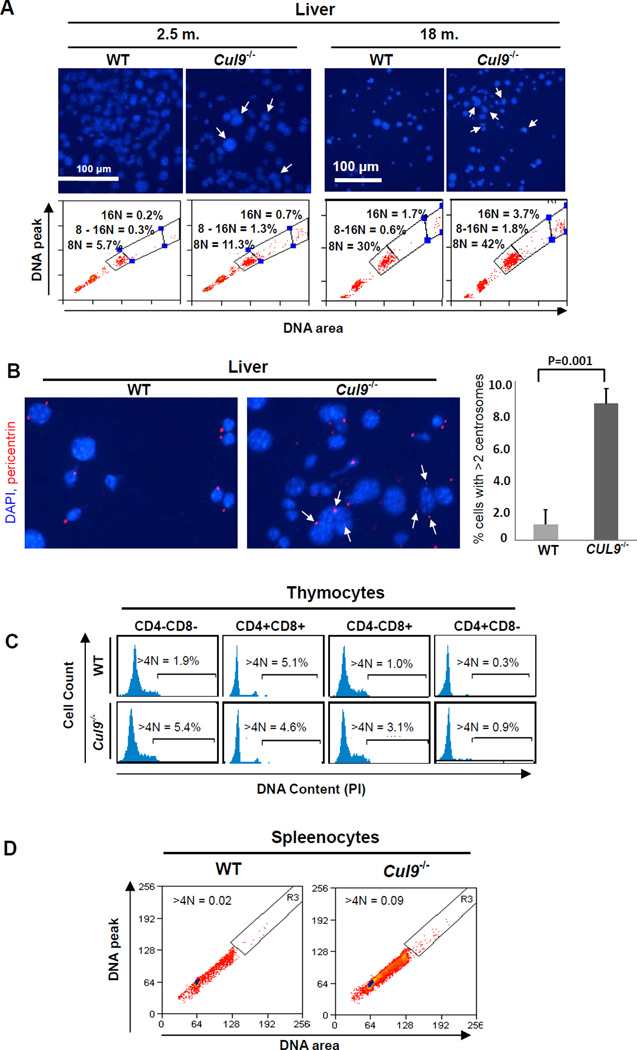
To elucidate the mechanism underlying the tumor suppressor function of CUL9, we carried out extensive flow cytometry analyses of wild-type and Cul9 mutant cells from multiple organs and tissues. These analyses revealed that the loss of Cul9 resulted in widespread polyploidy and aneuploidy. Hepatocytes are one of the few cell lineages where polyploid cells are found in normal adult liver, increasing with age. When compared with wild-type livers, Cul9−/− mice livers, both young and old, contain an increased number of cells with either multiple nuclei or a single nucleus with enlarged sizes or irregular shapes (Figure 1A, upper panel, Figure 1B). FACS analysis of liver cells from littermate mice showed that at a young age (2.5 mon.), 8N and 16N polyploidy increased from 5.7% to 11.3% (98% increase) and from 0.2% to 0.7% (3.5 fold increase), respectively, by the Cul9 deletion. In old mice (18 mon.), 8N and 16N polyploid hepatocytes increased by 40% from 30% to 42% and by 2.2 fold from 1.7% to 3.7%, respectively. In addition, Cul9 loss also increased the percentage of aneuploid hepatocytes with DNA content between 8N and 16N by 4.3 fold from 0.3% to 1.3% in young and by 3 fold from 0.6% to 1.8% in old wild-type mice (Figure 1A lower panel). To confirm the increase of polyploidy in Cul9−/− liver, we stained liver sections from littermate mice for pericentrin, a centrosomal protein, and found that Cul9 deletion increased the number of hepatocytes with more than two centrosomes (Figure 1B). Quantification of three independent sections, each examining 200 hepatocytes, showed that hepatocytes with more than 2 centrosomes increased from 1.0% in wild-type liver to 8.6% in Cul9−/− liver (p = 0.001).
Figure 1. Cul9deletion causes aneupoidy in vivo.
(A) Liver cells isolated from 2.5- and 18-month old mice with indicated genotypes were stained with DAPI and examined microscopically and by FACS. Cells with irregular or enlarged nuclei are indicated by arrows. Polyploid cells with 8N and 16N DNA contents and aneuploid cells with DNA content between 8N and 16N are indicated. Data are representative of separate experiments for two age-matched mice of each genotype.
(B) Liver cells from littermate mice (18 mon.) were stained with pericentrin and DAPI. Cells with 3 or more centrosomes are indicated (left image) and quantification is shown on the right panel (mean ± SD, three independent experiments; >200 cells were measured in each group).
(C) Thymocytes from littermate mice (10 weeks) were isolated and stained with CD4, CD8 and DAPI. DNA contents of each fraction based on CD4 and CD8 expression are shown. The percentages of polyploid and aneuploid (>4N) cells are indicated. Data are representative of separate experiments for two pairs of littermates.
(D) Splenocytes from littermate mice (18 mon.) were stained with PI, analyzed by FACS and the percentages of polyploid (>4N) cells are indicated
In the thymus cells, when compared with wild-type littermate controls, Cul9 deletion resulted in a 2.8-, 3.1- and 3.0 fold increase in polyploid thymocytes (>4N DNA content) in CD4−CD8−, CD4−CD8+ and CD4+CD8− populations, respectively (Figure 1C). A separate ploidy assay which measures the ratio between the peak (width) and area of the DNA fluorescence signal, gating out cell doublets and clumps, confirmed the increase in polyploid spleenocytes in Cul9−/−mice (Figure 1D). Together, these results indicate that Cul9 deletion leads to the development of polyploidy and aneuploidy in vivo, suggesting a potential function of CUL9 in maintaining genome integrity.
Cul9 plays a direct role in preventing polyploidy and aneuploidy in cultured MEFs
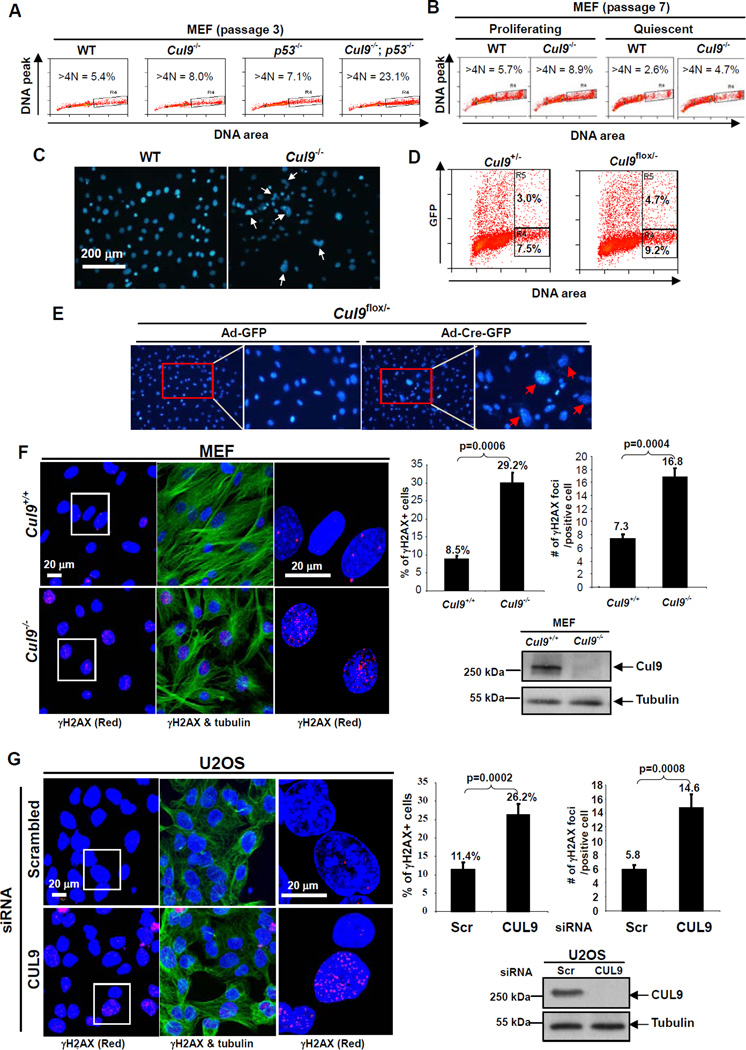
We then investigated the function of Cul9 in suppressing polyploidy and aneuploidy in littermate-matched cultured MEFs. FACS analysis revealed that polyploid and aneuploid cells increased from 5.4% and 5.7% in wild-type to 8.0% and 8.9% in Cul9−/− at both early (passage 3) and late (passage 7) passages, respectively (Figures 2A and 2B), indicating a passage-independent acquisition of polyploidy in Cul9−/− MEFs. Deletion of p53 also increased polyploidy and aneuploidy in cultured MEFs (from 5.4% to 7.1%), similar in extent to that caused by Cul9 deletion. Co-deletion of Cul9 and p53 resulted in a dramatic 23.1% polyploidy and aneuploidy in Cul9−/−; p53−/− MEFs (Figure 2A), suggesting that in maintaining genome integrity, Cul9 and p53 function synergistically, as opposed to in a linear pathway.
Figure 2. Loss of CUL9 function causes spontaneous DNA damage and aneuploidy.
(A) Littermate-matched MEFs were stained with PI and analyzed by FACS. The percentages of aneuploid and polyploid (>4N) populations are indicated.
(B) Logarithmically growing and serum-starved (0.1% FBS for 3 days) littermate-matched MEFs were stained with PI and analyzed by FACS. The percentages of aneuploid and polyploid (>4N) populations are indicated.
(C) MEFs at passage 7 from littermate mice were cultured and stained with DAPI. Representative cells with irregular nuclei, multiple, enlarged or micronuclei are indicated.
(D, E) Cul9flox/− and Cul9+/− MEF cells (passage 2) were infected with Ad-Cre-GFP and analyzed by FACS two days later. The percentages of polyploid cells (>4N) in GFP+ and GFP- populations are indicated. Nuclear morphology was examined by DAPI staining.
(F) Wild-type and Cul9−/− MEFs were stained with antibodies either to γ-H2AX (Red), α-tubulin (Green) and DAPI (Blue). The histogram on the left shows the percentage of γ-H2AX-positive cells (cells with >5 γ-H2AX foci) and the number of cells counted for each genotype. The number of γ-H2AX foci in individual γ-H2AX-positive cells were counted from WT (n = 417) and Cul9−/− MEFs (n = 616), and the average γ-H2AX foci per cell are presented. The error bar indicates ± SD of three independent experiments.
(G) U2OS cells were transfected with scrambled siRNA or siRNA against CUL9. 72 hours after transfection, cells were fixed and stained. The number of γ-H2AX foci in individual γ-H2AX-positive cells were counted from scrambled siRNA (n = 838) and siCUL9 (n = 817) cells. The procedure for quantification was the same as in (F). See also Figure S1.
To determine whether the development of polyploidy and aneuploidy is linked to cell proliferation, we examined proliferating and quiescent littermate-matched wild-type and Cul9−/−MEFs from the same passage and found a reduced population of polyploid cells in quiescent culture (Figure 2B). This reduction appears to be proportional for both wild-type (54% decrease) and Cul9−/− (47%) MEFs, indicating that the development of polyploidy is linked to, if not dependent on, active cell division and is enhanced by Cul9 loss. Microscopic examination of littermate-matched MEFs after DAPI staining revealed a substantial increase in Cul9−/− MEFs with two or three nuclei, and enlarged or irregularly shaped nuclei (Figure 2C). Acute deletion of Cul9 following adenovirus-mediated expression of Cre recombinase in Cul9flox/− MEFs resulted in a 56% increase in polyploid cells (4.7%) after 2 days of culturing when compared with Ad-Cre-GFP infected control Cul9+/− MEFs (3.0%, Figure 2D). This increased polyploidy in acutely deleted Cul9 cells is also accompanied by an evident increase in cells with irregular shapes and with enlarged nuclei within two days (Figure 2E). This result supports the conclusion that loss of CUL9 function results in polyploidy immediately, as opposed to over a period of multiple rounds of cell divisions, thus supporting a direct role of Cul9 in preventing the development of spontaneous polyploidy and aneuploidy.
To further explore the function of CUL9 in maintaining genomic stability, we stained littermate-matched wild-type and Cul9−/− MEFs with antibody recognizing γ-H2AX which is essential for the efficient recognition and repair of DNA double strand breaks (DSBs). Deletion of Cul9 resulted in a substantial increase in γ-H2AX-positive cells (defined as ≥5 foci per cell), from 8.5% (n = 139) in wild-type MEFs to 29.2% (n = 205) in Cul9−/− MEFs (Figure 2F). Moreover, the number of γ-H2AX foci in the γ-H2AX-positive cells was also considerably increased by Cul9 deletion, from an average of 7.3 foci per cell in wild-type MEFs to 16.8 in Cul9−/− MEFs. We confirmed this finding in cultured human cells by knocking down CUL9 using siRNA oligonucleotides (Figure 2G, Figure S1). We found that CUL9 depletion in U2OS cells significantly increased both the percentage of γ-H2AX-positive cells, from 11.4% in control scrambled siRNA cells (n = 279) to 26.2% in CUL9 depleted cells (n = 272), and the number of γ-H2AX foci in γ-H2AX-positive cells, from 5.8 foci per control cell to 14.6 foci per CUL9 depleted cell. Together, these results support the conclusion that CUL9 functions to maintain genome integrity.
Knocking down CUL9 rescues the defects caused by CUL7 depletion
CUL9 shares extensive sequence homology and can form a heterodimer with CUL7 (Skaar et al., 2007). In the accompanying paper, we demonstrate that CUL7 plays an important role in maintaining the integrity of microtubules and regulating mitosis (Yan et al.). Considering the critical role of microtubules in genome stability maintenance, we investigated the possibility that CUL9 and CUL7 function in the same genetic pathway to maintain genome integrity.
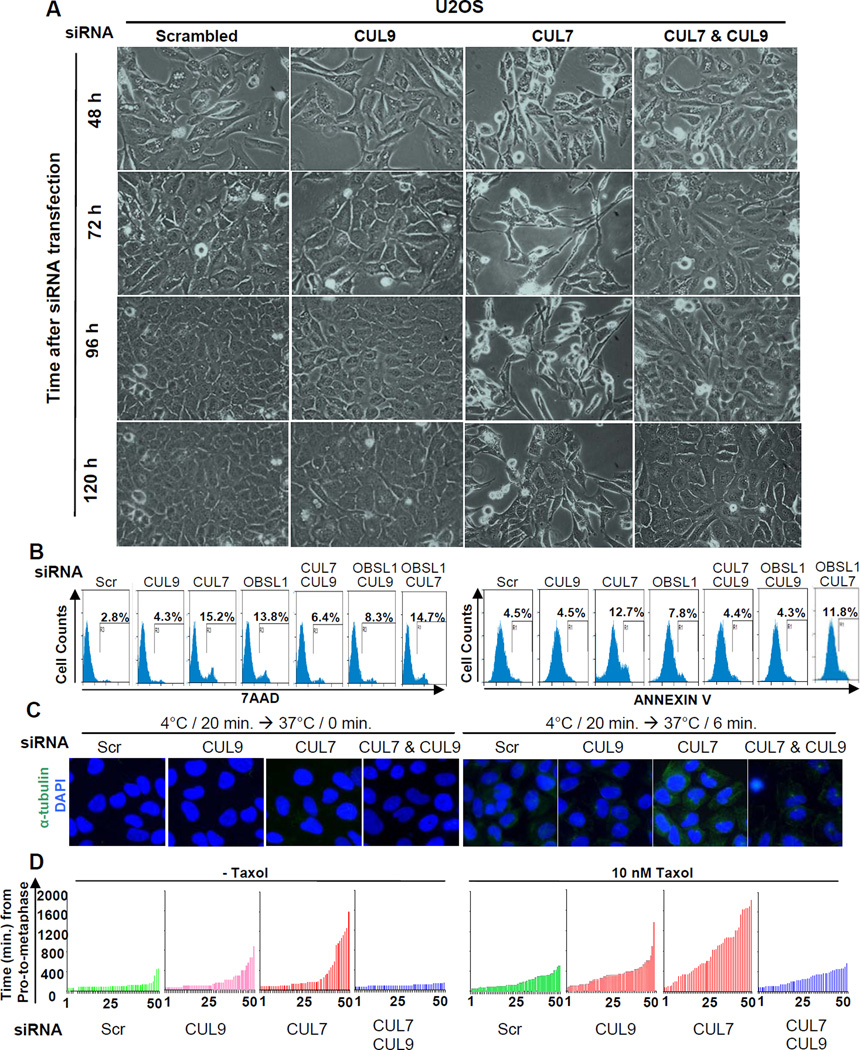
We first examined the effect of individual and combined depletion of CUL7 and CUL9 on cell growth. We found that while depletion of CUL9 did not cause major defects in U2OS cells, co-depletion with CUL7 significantly rescued the growth defects caused by CUL7 depletion, reducing cell death to near background levels (Figure 3A, Figure S2A). To substantiate this finding, we carried out FACS analysis (Figure 3B, Figure S2B). CUL9 depletion did not cause an increase in Annexin V positive (early apoptotic) cells (4.5% in both control and siCUL9 cells), but marginally increased 7-AAD-positive (late apoptotic and non-apoptotic cell death) cells (from 2.8% in control to 4.3% in CUL9-depleted cells). Knocking down CUL7 caused a significant increase in both Annexin V-positive (from 4.5% to 12.7%) and 7AAD-positive (from 2.8% to 15.2%) cells. Notably, co-depletion of CUL7 and CUL9 nearly completely rescued the cell death defects caused by CUL7 depletion, reducing Annexin V-positive and 7AAD-positive cells from 12.7% and 15.2% to 4.4% and 6.4%, respectively.
Figure 3. CUL9 knockdown rescues cells depleted for 3M genes.
(A) U2OS cells were treated with indicated siRNA oligos, and a representative phase contrast view of each sample at different time points after siRNA transfection is shown. See Figure S2A.
(B) U2OS cells were transfected with indicated siRNA oligos, stained with 7AAD and Annexin V and analyzed by FACS. See also Figure S2B and S2E.
(C) U2OS cells were transfected with indicated siRNA oligos for 3 days, switched to 4 °C-cold media for 20 minutes to induce microtubule disassembly and then switched back to 37 °C-warm media to allow microtubule recovery. Representative phase contrast pictures were taken at indicated time of recovery. Cells were stained with DAPI and antibody to tubulin. See also Figure S2C.
(D) NCI-H1155 cells stably expressing EGFP-Histone H2B were transfected with indicated siRNA oligos for 48 hours. The cells were then either untreated or treated with 10 nM paclitaxol for 24 hours, followed by imaging EGFP-Histone H2B every 14.5 minutes for 46 hours to trace mitosis. Duration from nuclear envelope break to anaphase was measured for 50 cells from each sample, and time intervals plotted. See also Figure S2D.
We then determined the effect of CUL9 depletion on the microtubule defects in CUL7-depleted cells by measuring the microtubule recovery time following cold (4 °C)-induced disassembly. CUL7-depleted cells had a significantly faster rate of recovery after switching to warm (37 °C) media, confirming the result from the nocodazole-induced disassembly-recovery experiment that suggested CUL7 may negatively regulate microtubule assembly (Figure 3C, Figure S2C). Depleting CUL9 did not noticeably change the rate of microtubule recovery, but almost completely restored microtubule recovery from cold-induced disassembly to the normal rate in CUL7-depleted cells (Figure 3C).
Finally, we used live cell time-lapse imaging video microscopy to examine the effect of knocking down CUL9 on the chromosome alignment defects caused by CUL7 depletion. As expected, CUL9 depletion alone caused a slight prometaphase delay in NCI-H1155 cells while CUL7 depletion alone caused dramatically prolonged progression from prometaphase to metaphase in the absence or presence of low doses of taxol (Figure 3D, Figure S2D). Co-depletion of CUL9 and CUL7 almost completely restored the rate of prometaphase-to-metaphase progression in both untreated and taxol-treated cells. Together, these results demonstrate that CUL9 acts as a downstream effector of CUL7.
Knocking down CUL9 rescues mitotic and cell death defects caused by OBSL1 depletion
Three genes, CUL7, OBSL1 and CCDC8, are mutated in 3M syndrome patients in a mutually exclusive manner. In the accompanying paper, we demonstrate that CUL7 forms the 3M complex with OBSL1 and CCDC8, and that the 3M complex plays an important role in maintaining microtubule integrity and regulating mitosis (Yan et al.). To determine whether CUL9 is just a downstream effector of CUL7 or mediates the function of the 3M complex, we determined the effects of depleting CUL9 on the cell death caused by OBSL1 depletion. Like CUL7 depletion, OBSL1 depletion also caused a significant increase in both Annexin V-positive (from 4.5% to 7.8%) and 7AAD-positive (from 2.8% to 13.8%) cells. Co-depletion of CUL7 and OBSL1 did not cause a synergistic increase in either Annexin V-positive (11.8%) or 7AAD-positive (14.7%) cells, supporting the notion that the function of OBSL1 in mitotic control is mediated mostly by CUL7. Notably, co-depletion of CUL9 and OBSL1 nearly completely rescued the cell death defects caused by OBSL1 knockdown, reducing Annexin V-positive and 7AAD-positive cells from 7.8% and 13.8% to 4.3% and 8.3%, respectively (Figure 3B and Figure S2E). We conclude that CUL9 is a critical downstream effector of 3M complex.
Loss of CUL9 function sensitizes cells to microtubule damage
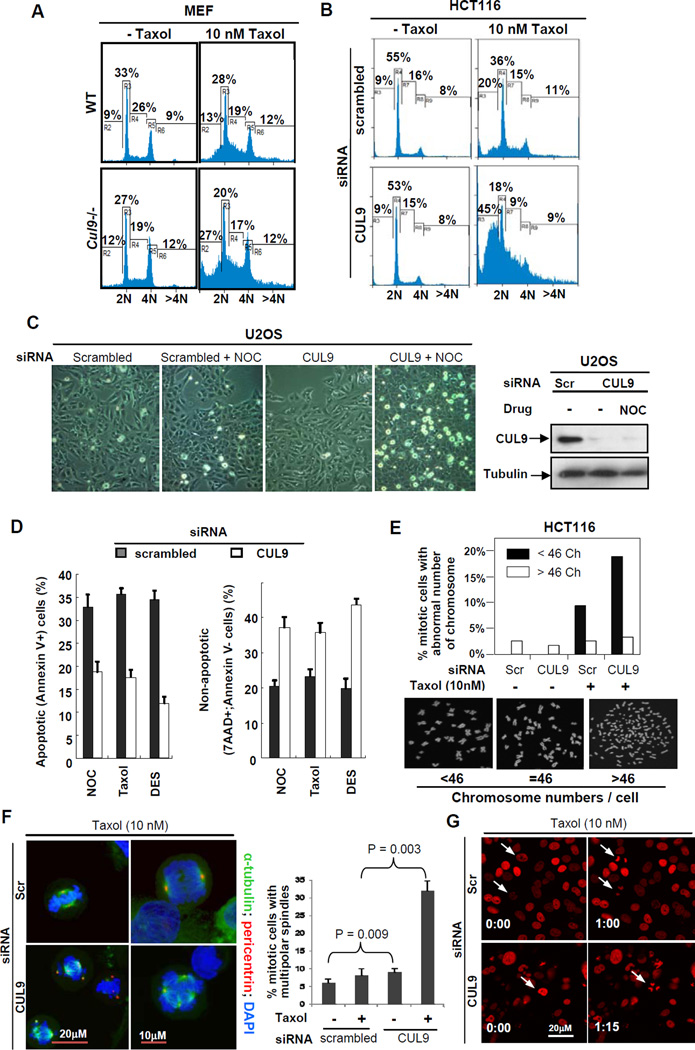
To further demonstrate that microtubule regulation is the molecular basis of CUL9 function in maintaining genome stability and tumor suppression, we examined the effect of CUL9 loss on the cellular response to microtubule damaging agents. In littermate-matched MEFs, while loss of CUL9 function had little effect on the cell cycle phase distribution in the absence of exogenous microtubule damage, it had a profound effect on cells after a low dose (10 nM) taxol treatment, resulting in a 2.08 fold increase in sub-G1 cell population in Cul9−/− MEFs compared to wild-type MEFs (27% vs. 13%, Figure 4A, Figure S3A). Likewise, CUL9 depletion in human colorectal carcinoma derived HCT116 cells had little effect on the cell cycle phase distribution in undamaged cells, but resulted in a significant accumulation (2.25 fold, 45% vs. 20%) of sub-G1 population when cells were treated with a low dose of taxol (Figure 4B, Figure S3B). Similarly, CUL9 depletion did not significantly affect the growth and morphology of U2OS cells, but caused evident cell death and morphological changes in U2OS cells treated with a low dose (50 ng/ml) of nocodazole (Figure 4C). These defects include many rounded, bright and floating cells, indicative of cell death. Collectively, these results suggest a function of CUL9 in protecting cells from microtubule damage.
Figure 4. CUL9 protects cells from microtubule damages.
(A) Wild-type and Cul9−/− MEFs were either untreated or treated with taxol for 24 hours and stained with propidium iodide, followed by FACS analysis. The genotypes were confirmed by immunoblotting (Figure S3A).
(B) HCT116 cells were transfected with either scrambled siRNA or siRNA against CUL9. Transfected cells were either treated with solvent or taxol for 24 hr, then stained with propidium iodide followed by FACS analysis. The knockdown efficiencies were verified by immunoblotting (Figure S3B).
(C) U2OS cells were transfected indicated siRNA oligos for 48 hours. Transfected cells were either treated with solvent or nocodazole (NOC, 5 ng/ml) for an additional 20 hours. Bright-field images are shown; note the presence of increased bright, rounded, and floating cells in CUL9 depleted, nocodazole treated image.
(D) U2OS cells were transfected with indicated siRNA oligos for 48 hours, and then treated with DES (25 µM), nocodazole (50 ng/ml), and taxol (10 nM) for an additional 20 hours. Cells were collected and stained with 7AAD and Annexin V, and analyzed by FACS. The percentages of apoptotic (Annexin V+) and total (7AAD+;Annexin V-) cell death are indicated. Data are representative of three separate experiments; mean ± SD.
(E) HCT116 cells were transfected with indicated siRNA oligos and then treated with either solvent or taxol (10 nM), followed by karyotype analysis. The chromosome number was counted for ≥50 mitotic cells for each sample. See also Figure S3D.
(F) U2OS cells were transfected with indicated siRNA oligos for 48 hours before treated with either vehicle or taxol. Cells were then fixed and stained with DAPI and antibodies to α-tubulin and pericentrin. Fifty mitotic cells were examined microscopically and the number of mitotic cells with multipolar spindle was scored. See also Figure S3E.
(G) HCT116 cells stably expressing mCherry tagged histone H2B were analyzed by live cell imaging and imaged by confocal laser scanning microscopy. Chromosomes are visualized by H2B–mCherry (in red); time is in hour:min. The white arrows indicate cells that have gone through mitosis. See also Figure S3F.
Loss of Cul9 sensitizes cells to non-apoptotic cell death after microtubule damage
To probe the nature of cell death caused by microtubule damage in CUL9-depleted cells, we stained U2OS cells with 7AAD and annexin V to examine total (7AAD+; annexin V+), apoptotic (annexin V+) and non-apoptotic (7AAD+) cell death. CUL9 depletion did not cause a significant change in cell death in untreated U2OS cells (Figure S3C), but it caused a dramatic decrease in apoptotic (annexin V+) cell death (from 37.7% to 19.1%) and concomitantly, a significant (82%) increase in non-apoptotic (7AAD+; annexin V-) cell death (from 18% to 32.7%) in nocodazole-treated cells (Figure 4D). We further extended these observations to two additional microtubule damaging agents, taxol (10 nM) and diethylstilbestrol (DES; 25 µM). We found that similar to nocodazole treatment, CUL9 depletion reduced apoptotic cell death (from 36.5% to 17.9% for taxol and from 35.4% to 12.1% for DES), but increased non-apoptotic cell death (from 20.4% to 31.4% for taxol and from 17.4% to 38.5% for DES) in response to both taxol and DES (Figure 4D). These results suggest that loss of CUL9 function sensitizes cells to microtubule damage, at least in part, by permitting non-apoptotic cell death.
To further examine the nature of the sub-G1 cells accumulated after microtubule damage and loss of CUL9 function, we examined the chromosome number in HCT116 cells that have a stable and near diploid karyotype during in vitro culturing (Roschke et al., 2002). In the absence of exogenous microtubule damage, CUL9 depletion had little effect on the percentage of mitotic cells with abnormal karyotype (Figure 4E, Figure S3D). Although we noted a slight but not significant decrease in cells with >46 chromosomes from 2.9% in control vs. 2.2% in CUL9-depleted populations, while cells with <46 chromosomes in both control and CUL9-depleted populations were negligible. Taxol treatment did not significantly alter the percentage of cells with >46 chromosomes (2.9% vs. 2.1%), but substantially increased the percentage of cells with <46 chromosomes (2.9% to 11.0%). Notably, CUL9 depletion had a significant impact on the karyotype of taxol-treated HCT116 cells with over a two-fold increase in cells with <46 chromosomes (from 11% to 24%), but little effect on the percentage of cells with >46 chromosomes. This result suggests a role for CUL9 in protecting genome integrity in response to microtubule damages.
To determine the basis underlying the chromosome loss in taxol-treated, CUL9-depleted cells, we triply stained cells for α-tubulin, pericentrin and DAPI, and microscopically examined cells. This analysis revealed that taxol-treatment in CUL9-depleted cells increased the percentage of cells with multipolar spindles by nearly 4 fold (8.5% vs. 31.8% between scrambled siRNA and siCUL9 cells; Figure 4F, Figure S3E). Using HCT116 cells stably expressing mCherry fluorescent protein fused to human histone H2B, we monitored cell division in real-time with a time-lapse live cell imaging system. We found that low dose taxol treatment of CUL9-depleted cells caused a slight delay in mitotic progression and increased aberrant chromosome segregation (Figure 4G and Figure S3F). We conclude that CUL9 protects microtubule-damaged cells by preventing the continuation of their mitosis and cytokinesis. Loss of this protective function of CUL9 renders cells with microtubule damages and abnormal centrosomes to incorrectly segregate and consequently lose their chromosomes, leading to subsequent cell death by a non-apoptotic mechanism.
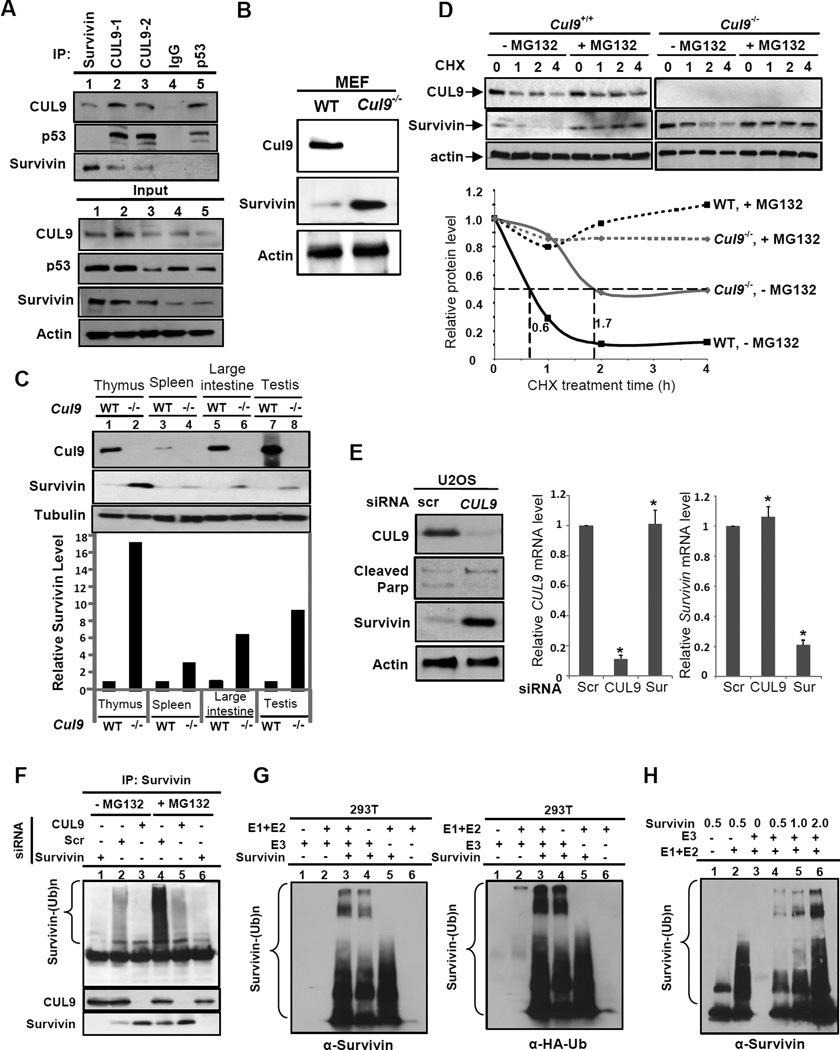
Survivin is a substrate of CUL9
In an effort to understand the mechanistic role of CUL9, we set out to search for CUL9 substrates by both proteomic and candidate approaches. While the former has yet to identify a probable substrate, the candidate approach revealed that among more than a dozen mitotic proteins that we tested, survivin interacts with CUL9 as determined by reciprocal IP-Western analysis (Figure 5A). Notably, Cul9−/− MEFs accumulated a significantly higher level of survivin protein than wild-type MEFs (Figure 5B). Furthermore, we found that Cul9 deletion markedly increased the steady state levels of survivin in multiple organs in vivo (Figure 5C). We then performed cycloheximide chase experiments and found that survivin protein half-life increased from 0.6 hrs in the wild-type MEFs to 1.7 hrs in Cul9−/− MEFs, while the addition of the proteasome inhibitor MG132 stabilized survivin levels in both cell types (Figure 5D). Likewise, CUL9 depletion in U2OS cells significantly stabilized survivin, increasing its half-life from 0.6 hrs in scrambled siRNA cells to 1.8 hrs in CUL9-depleted cells (Figure S4A). The accumulation of survivin proteins in siCUL9 cells did not accompany an increase in survivin mRNA expression (Figure 5E), indicating the posttranscriptional regulation of survivin by CUL9.
Figure 5. Survivin is a substrate of CUL9 E3 ligase.
(A) Endogenous CUL9-survivin interaction was determined in U2OS cells by reciprocal IP-Western analysis using indicated antibodies.
(B, C) The steady state levels of survivin in MEFs and different mouse organs were determined by immunoblotting.
(D) Survivin protein half-life in wild-type and Cul9−/− MEF was determined by cycloheximide chase experiment, normalized to α-actin, and plotted relative to 0 hour (bottom). See also Figure S4A.
(E) U2OS cells were transfected with indicated siRNA oligos. The steady state protein levels and mRNA levels were determined by immunoblotting with indicated antibodies and RT-qPCR analyses, respectively. mRNA levels were normalized to GAPDH (mean ± SD; n = 3; significance was determined by Student's t-test, *p < 0.01).
(F) In vivo ubiquitylation assay was carried out in U2OS cells treated with siRNA or proteasome inhibitor MG132 as indicated. Survivin was immunoprecipitated and its ubiquitylation was determined by immunoblotting with anti-ubiquitin.
(G, H) In vitro ubiquitylation assays were performed using recombinant E1, E2 and survivin. E3 (CUL9) was immunopurified from 293T cells (see Figure S4B).
To test the hypothesis that survivin is a direct substrate of CUL9, we first performed in vivo ubiquitylation assays in U2OS cells and found that CUL9 depletion greatly diminished survivin ubiquitylation (Figure 5F). We then carried out in vitro ubiquitylation assays using recombinant E1, E2, survivin and immunopurified HA-CUL9 as the source of E3 (Figure S4B). We found that CUL9 addition greatly enhanced survivin ubiquitylation with high molecular weight forms of ubiquitylated survivin detected by both survivin and ubiquitin antibodies, and the level of survivin ubiquitylation correlated with the amount of recombinant survivin added to the reaction (Figure 5G and 5H). Together, these results demonstrate the specificity of survivin ubiquitylation by CUL9 E3 ligase complex.
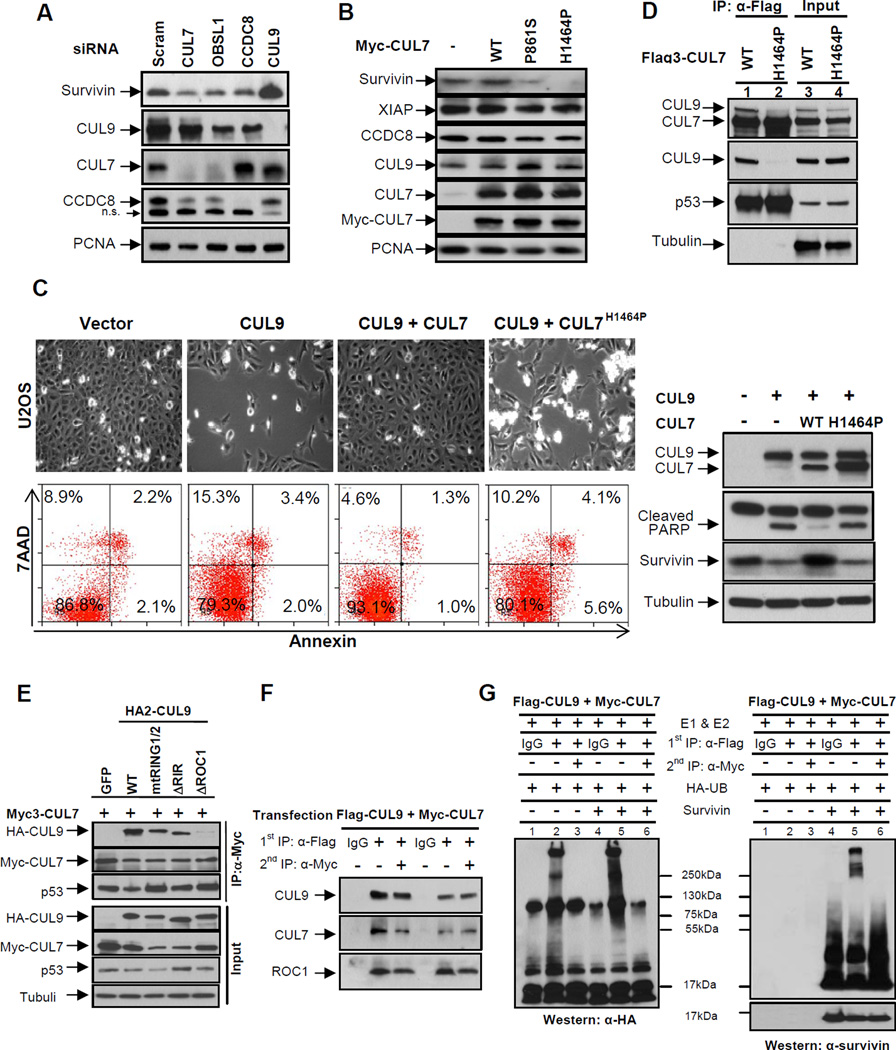
CUL7 inhibits CUL9 by forming a heterodimer
Besides its well-established functions in suppressing apoptosis and regulating mitosis, survivin also has a long observed, but poorly understood, activity in suppressing microtubule dynamics (Giodini et al., 2002; Li et al., 1998; Rosa et al., 2006). Our finding that CUL9 is a key downstream effector of the 3M complex prompted us to test the regulation of survivin by the 3M complex. We first examined the steady state levels of survivin following CUL7, OBSL1, or CCDC8 depletion in U2OS cells. We found that silencing any of these three 3M genes reduced survivin levels (Figure 6A), supporting the notion that the 3M complex negatively regulates CUL9 activity in targeting survivin for ubiquitylation-mediated degradation. We then transduced U2OS cells with retroviruses encoding wild-type or 3M patient-derived CUL7 mutants and examined survivin levels. We found that while wild-type CUL7 overexpression did not change survivin levels, both P861S and H1464P mutants significantly reduced the steady state levels of survivin, but not a related IAP family member such as XIAP (Figure 6B). To confirm that the H1464P mutation disrupts the ability of CUL7 to inhibit CUL9, we overexpressed CUL9, either alone or with CUL7, and examined the function of CUL9 in promoting cell death and the degradation of survivin. As we reported previously (Pei et al., 2011), total cell death was increased from 13.2% in vector transfected cells to 20.7% after CUL9 overexpression. CUL9-promoted cell death was reduced to 6.9% by the co-expression of wild-type CUL7, but remained at 19.9% when CUL9 was co-expressed with H1464P mutant CUL7 (Figure 6C). Confirming this result, overexpression of CUL9 promoted activation of apoptosis as assayed by cleaved poly (ADP-ribose) polymerase (PARP) and downregulation of survivin, both of which were inhibited by co-expression of the wild-type, but not the H1464P mutant CUL7.
Figure 6. CUL7 binds to and inhibits CUL9.
(A) U2OS cells were transfected with indicated siRNA oligos, and 48 hours following transfection, the steady state levels of indicated proteins were determined by immunoblotting.
(B) U2OS cells were transduced with retroviruses expressing indicated wild-type or mutant CUL7 proteins. The steady state levels of proteins were determined by immunoblotting.
(C) U2OS cells were transfected with indicated plasmids for 48 hours, a representative phase contrast view of each sample is shown. The cells were then collected, stained with Annexin V and 7AAD, and then analyzed by FACS. Protein expression was determined by immunoblotting.
(D) Lysates prepared from U2OS cells stably expressing wild-type or mutant CUL7 were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with indicated antibodies to assess CUL7-CUL9 interaction; α-tubulin served as a loading control.
(E) Lysates from transfected U2OS cells were immunoprecipitated with anti-Myc3 antibody, followed by immunoblotting with antibodies against CUL9, CUL7 and p53. See also Figure S5.
(F) Lysates from U2OS cells co-transfected with Flag3-CUL9 and Myc-CUL7 were subjected to sequential IP to assess the direct binding requirement for CUL9 inhibition by CUL7. Flag and Myc antibodies were used for the first IP (lanes 2 and 5) and second IP (lanes 3 and 6), respectively.
(G) Sequential immunoprecipitates from (F) were used for assaying survivin ubiquitylation in vitro. Reactions were resolved by SDS-PAGE and probed with either antibody to HA-ubiquitin or survivin.
P861S and H1464P mutations target the DOC1/APC11 and ROC1-binding domain, respectively (see Figure 3A of accompanying paper). A simple model would be that CUL7 inhibits CUL9 by promoting its ubiquitylation and degradation. In multiple experiments with different cell lines where CUL7 was depleted, however, we did not observe any significant increase in the steady state level of CUL9 protein (see e.g. Figures 6A, Figure S2A, S2C and S2D), nor did we observe a significant decrease in CUL9 protein in cells overexpressing CUL7 (e.g. Figure 6B and 6C). To investigate how these 3M patient-derived mutations impaired the ability of CUL7 to regulate CUL9 and survivin, we examined their interaction with various proteins known to interact with CUL7 and CUL9. Surprisingly, we found that the H1464P mutation not only disrupted CUL7’s binding with ROC1, but also its binding with CUL9 (Figure 6D), suggesting the possibility that ROC1 mediates the heterodimerization between CUL7 and CUL9. To test this prediction, we generated a ROC1 binding deficient CUL9 mutant by deleting six residues (residues 1778–1783, referred to as CUL9DROC1, Figures S5A and S5B) that are highly conserved in cullins and involved in ROC1 binding. We determined its binding with CUL7 and found that disruption of ROC1 binding in CUL9, but not two other mutations disrupting either the two RING fingers (mtRING1/2) or deleting the C-terminal 200 residues comprising the RING-in-between-RING domain (DRIR), abolished CUL9’s binding with CUL7 (Figure 6E). Together, these results indicate that ROC1 mediates CUL7-CUL9 heterodimerization.
To provide direct evidence that the heterodimerization inhibits the E3 ligase activity of CUL9, we co-transfected cells with differentially tagged CUL7 (Myc-CUL7) and CUL9 (Flag-CUL9) and performed sequential immunoprecipitations, first Flag IP, eluted off with Flag peptide and then the second IP using the antibody to Myc. Analysis of protein interaction showed that ROC1, as well as p53, were present in the CUL7-CUL9 heterodimer (Figure 6F). This result is consistent with the notion that ROC1 mediates CUL7-CUL9 heterodimerization and excludes the possibility that CUL7 inhibits CUL9 by dissociating its binding with ROC1. We then examined the in vitro ligase activity of CUL9 after adjusting the amount of CUL9 to a similar level. We found that while the CUL9 complex from the first Flag immunoprecipitation, which contains all CUL9 complexes, exhibited readily detected ubiquitin ligase activity toward survivin, the second immunoprecipitation, enriched for CUL7-CUL9 heterodimer, contained little ligase activity (Figure 6G). Furthermore, we note that the CUL7-CUL9 heterodimer also lacks substrate-independent ligase activity (known as auto-ubiquitylation, lane 3). This result suggests that the CUL7-ROC1-CUL9 complex, while containing a RING finger protein, is catalytically inactive. Together, these results demonstrate that CUL7 forms a heterodimeric complex with CUL9 to inhibit CUL9 E3 ligase activity toward survivin.
Survivin is a target of the 3M complex
To provide functional evidence for the 3M–CUL9-survivin pathway, we determined whether ectopic expression of survivin could rescue the cell growth and microtubule defects caused by the loss of function of 3M genes. We first ectopically expressed different amounts of survivin in U2OS cells depleted of CUL7 and then examined cell viability (Figure 7A). The expression level of exogenous survivin was approximately three times that of endogenous survivin, which did not significantly alter cell morphology, but slightly increased the number of viable cells at lower doses (0.2 and 0.5 µg plasmid). 72 hours after transfection with siRNA against CUL7, there was a substantial loss of endogenous survivin (compare lane 5 and lane 1) and a significant reduction in cell viability (50%). Transfection of 0.2 and 0.5 µg survivin expressing plasmid in CUL7-depleted cells resulted in 68% and 88% rescue of cell viability. Interestingly, at a higher dose of survivin transfection (1 µg), the efficiency of rescue was reduced (43%). These results provide functional support for the notion that the loss of survivin is a major contributing factor to cell death caused by CUL7 depletion.
Figure 7. Overexpression of survivin rescues the defects in CUL7-depleted cells.
(A) U2OS cells were transfected with empty vector or different amounts of plasmids expressing survivin. 24 hours after transfection, cells were transfected again with scrambled or targeting CUL7 targeting siRNA oligos. 48 hours later, the cells were photographed and a representative phase contrast view of each sample is shown (Top); the expression of individual proteins was confirmed by immunoblotting (left). The cells were then collected by trypsinization, stained with trypan blue and viable cells were counted and plotted (right).
(B) U2OS cells were transfected with empty vector or Flag-survivin for 24 hours and then transfected again with indicated siRNA oligos. 48 hours later, the cells were lysed and analyzed by immunoblotting with indicated antibodies, and densitometry analysis with ImageJ software was performed to obtain acetyl-α-tubulin band intensity relative to α-actin (right; *p < 0.001).
(C) U2OS cells stably expressing pEGFP-α-tubulin were transiently transfected as indicated and cultured in nutrient medium for 48 hours before recording images. A representative of confocal Z-stack image (6 slides) is shown. The scale bar is 10 µm.
(D) A schematic model illustrating the 3M–CUL9-survivin pathway.
We then determined whether overexpression of survivin could rescue the microtubule defects in CUL7-depleted cells. We examined the levels of Lys-40 (K40) acetylated α-tubulin, which is more stable than unmodified tubulin, in cells treated with a microtubule-depolymerizing drug (LeDizet and Piperno, 1986; Piperno et al., 1987), although the mechanistic basis and physiological significance of K40 acetylation remains elusive at present. We found that depletion of CUL7 is associated with a decrease in K40-acetylated α-tubulin of over 40% (Figure 7B). Importantly, ectopic expression of survivin in CUL7-depleted cells almost completely restored K40-acetylated α-tubulin levels (Figure 7B). Finally, we examined the microtubule network density by confocal microscopy and found that while CUL7 depletion substantially reduced the microtubule fiber density, this reduction was effectively restored by the ectopic expression of survivin (Figure 7C). Together, these results demonstrate that the loss of survivin is a major contributing factor to the microtubule defects caused by CUL7 depletion.
DISCUSSION
A cellular mechanism for the tumor suppressor function of CUL9
The current study provides a mechanism—maintaining genome integrity–for the function of CUL9 in tumor suppression. This conclusion is supported by three lines of evidence. First, we found that in organs where Cul9 deletion results in tumorigenesis such as the liver, spleen and thymus, there were significant increases in polyploidy and aneuploidy. In vitro, cells deficient for CUL9 accumulated significantly higher amount of DNA damage. Second, we showed in the accompanying paper that the products of three 3M genes, CUL7, OBSL1 and CCDC8 form a complex, referred to as the 3M complex, which maintains microtubule integrity, normal mitosis and cytokinesis. We demonstrate in this paper that CUL9 is a critical downstream effector that mediates the function of 3M in maintaining normal microtubule dynamics. Furthermore, we showed that CUL9 protects cells from microtubule damage. Defects in microtubule dynamics and control in mitosis and cytokinesis are established causes of genomic instability. Third, we discovered that survivin is a substrate of CUL9 E3 ligase. We showed that deletion of Cul9 in mice resulted in elevated survivin levels in multiple organs. Survivin has long been associated with cancer development as its increased expression is found in multiple types of human cancers (Ambrosini et al., 1997; Velculescu et al., 1999). Elevated survivin in Cul9-deficient cells could conceivably promote the survival of microtubule damaged and genetically unstable cells.
The 3M pathway
Collectively, four lines of evidence reported in this and the accompanying paper led us to propose a 3M–CUL9-survivin pathway, or 3M pathway, in maintaining microtubule and genome integrity (Figure 7D). First, three 3M genes form an integral complex to regulate microtubule dynamics and maintain microtubule integrity. Disruption of any of these 3M genes results in severe microtubule damage, abnormal prometaphase and cytokinesis, and severe cell death. Second, depletion of CUL9 substantially rescues cell death, microtubule overgrowth and mitotic defects caused by the depletion of 3M genes. Third, CUL9 binds to and promotes the ubiquitylation and degradation of survivin, a multifunctional protein and a component of CPC that binds to microtubules and histones to regulate multiple steps of mitosis and cell division. Lastly, we showed that CUL7 directly inhibits CUL9 and that depletion of a 3M gene results in down regulation of survivin, indicating that the 3M complex negatively regulates the function of CUL9. We further showed that survivin overexpression, like CUL9 depletion, rescues both cell death and microtubule defects in cells depleted of 3M genes, supporting a critical function of survivin in the CUL7-CUL9 pathway.
The 3M pathway is separate from the well-established spindle assembly checkpoint (SAC) complex in three aspects. First, they are genetically different. Thus far, none of the SAC component, such as MAD, BUB, CDC20 proteins and APC/C E3 ligase, has been linked to 3M syndrome or been found in the 3M complex and vice versa. Second, the 3M complex evolved much more recently than the SAC pathway. While the SAC is evolutionarily conserved from yeast to man, the 3M genes CUL7 and OBSL1 are present only in vertebrates and CCDC8 presents only in placental mammals. This would suggest that the 3M complex evolved to meet newly emerged needs unique to vertebrates or mammals, rather than being an intrinsic component of microtubule biosynthesis or mitotic and cytokinesis apparatus. Third, their targets are different. While the SAC controls the ubiquitylation of multiple mitotic proteins through the regulation of APC E3 ligase, none of the APC substrates we have tested is affected by the depletion of CUL7 or CUL9. Instead, the 3M pathway controls the ubiquitylation of survivin and likely the function of CPC, which is presently not known to be as a major target of APC E3 ligase. It should be pointed out that the 3M pathway does not function as a SAC-independent checkpoint. Instead, as we showed in the accompanying paper, depletion of MAD2 completely rescued prometaphase delay in cells depleted of either CUL7 or OBSL1. It is more likely that the 3M pathway evolved to meet newly emerged needs unique to vertebrates and mammals by acting on the preexisting SAC pathway.
Tumor suppression by the 3M pathway
Survivin, although small (137 residues in human), is a multifunctional protein and has been linked to two separate aspects of biology, cell cycle regulation and cancer development, that are yet to be mechanistically reconciled. A notable feature of survivin is that it is expressed during normal fetal development and is generally no longer present in most adult tissues, but is selectively upregulated in many human tumors, where its overexpression correlates with poor outcome and treatment resistance (Adida et al., 1998; Ambrosini et al., 1997; Velculescu et al., 1999). It remains unclear at present whether the benefits of survivin overexpression to tumor cells come from its ability to promote cell survival by antagonizing caspases or its ability to facilitate cells with spindle or kinetochore defects to continue through cell division. Further complicating the mechanism of survivin is the finding that, in addition to its well-established, CPC-dependent function in the regulation of mitosis (Ruchaud et al., 2007), survivin also has a CPC-independent function in the regulation of microtubule dynamics during interphase (Giodini et al., 2002; Li et al., 1998; Rosa et al., 2006).
Our findings that survivin is directly degraded by a tumor suppressor protein, CUL9, and is indirectly stabilized by the 3M complex which functions to maintain microtubule integrity shed new light to connect the functions of survivin as a regulator of both microtubules and mitosis to the benefit of its overexpression to tumor cells. Abnormally high levels or persistent presence of survivin, such as in Cul9 deficient cells with sustained microtubule damages, would drive cells with mitotic or cytokinesis defects to continue through cell division and result in mitotic death for most cells. A few escaped cells would acquire genetic instability and increased opportunities for transformation. We suggest that the function of survivin in maintaining microtubule and genome integrity, whose impairment commonly leads to aneuploidy and tumorigenesis, is the main contributing factor for the cancer function of survivin.
Because of its broad overexpression in many different types of human tumors and its functions in cytoprotection, survivin has been extensively exploited as a target for anticancer interventions (Kelly et al., 2011; Ryan et al., 2009). Several novel experimental therapeutic strategies have been developed to target survivin. Our study suggests two new opportunities for developing anti-survivin therapy by targeting the 3M pathway. The downregulation of survivin by the depletion of CUL7 suggests that the inhibition of the 3M complex could increase the activity of CUL9 and lead to increased degradation of survivin. In addition, we also showed that the disruption of the 3M complex, through depletion of either OBSL1 or CUL7, substantially increased mitotic abnormalities and cell death caused by sublethal concentrations of the antitumor drug paclitaxel. These findings, together with the fact that survivin inhibitors have shown only modest activity as single agents, suggest that targeting the 3M complex or CUL9 in combination with an anti-survivin agent or cytotoxic chemotherapy merits exploration as a therapy for tumors with increased survivin expression.
Finally, our study raises a number of new and critical questions. What is the source that causes DNA damage in CUL9-deficient cells? When during the cell cycle and where inside the cell does CUL9 ubiquitylate survivin? What is the role of p53, which binds to both CUL7 and CUL9, but is not degraded by either protein, in the 3M pathway? The establishment of the 3M–CUL9-survivin pathway and characterization of cellular defects and mice phenotypes associated with the disruption of the pathway should facilitate the understanding of these and other important questions.
EXPERIMENTAL PROCEDURES
Mice and MEFs
Generation of Cul9 mutant mice has been described previously (Pei et al., 2011). Cul9 heterozygotes were backcrossed for 10 generations with BL/6 mice. p53 null (p53tm1Tyj) and Eµ-Myc transgenic (B6.Cg-Tg(IghMyc)22Bri/J) mice in BL/6 background were obtained from the Jackson Laboratory and bred with the Cul9 mutant mice to create double mutant mice. Total extracts were made from different tissues of most organs by directly lysing in a SDS-lysis buffer. Early passage MEFs (< passage 4) from individual embryos were plated in 100-mm plates and incubated in DMEM plus 10% FBS.
Tetraploidy and polyploidy analyses of thymus, spleen, and liver cells
Cells from thymus, spleen and bone marrow were prepared as previously described (Pei et al., 2011). Hepatocytes were isolated from adult liver tissue by a nonenzymatic procedure as described with modifications (Higgins et al., 1985). Briefly, liver tissue was minced and incubated in 30 mM EDTA in Hank’s solution for 30 min. After dispersion, filtration and sedimentation, cells in the heavy fraction were collected, stained with PI and analyzed by FACS. For hepatocyte staining, fresh liver was gently pressed onto a glass coverslip, and the attached hepatocytes were fixed and stained with DAPI (Nunez et al., 2000). To determine and calculate the ploidy (aneuploidy and polyploidy, in particular) of cells (such as hepatocytes, spleenocytes and MEFs), cells were stained with PI and analyzed by FACS. To eliminate false positive polyploidy resulting from cell clumps, only those cells displaying the ratio between the peak (width) and area of the DNA fluorescence signal were calculated. The cells with DNA contents that do not fall into the 2N, 4N, 8N or 16N peaks were calculated as aneuploidy. For determination of aneuploidy in hepatocytes, only cells with DNA contents that fall between 8N and 16N peaks were calculated as aneuploid to avoid false positive or noise caused by other liver cell populations.
Live-cell microscopy
U2OS cells stably expressing GFP-α-tubulin and GFP-ubiquitin were cultured in 35 mm glass bottom dishes (MatTaK), and transfected with scrambled or indicated siRNA. After 48 hours, live-cell microscopy was carried out on the FV1000 Confocal system (Olympus). Images were obtained every 5 minutes for 16 hours. Human lung cancer NCI-H1155 cells stably expressing EGFP-Histone H2B were generated through infecting cells with pCLNCX-EGFP-Histone H2B retrovirus. For imaging, the NCI-H1155-EGFP-Histone H2B cell line was transfected with indicated siRNA and plated in a 96-well format followed by carrier or paclitaxel treatment 48 hours post transfection. At 72 hours post-transfection, imaging began on a BD Pathway 855 bio-imager using a 20X objective. Images were collected at 14.5 minutes intervals for the next 42 hours. Image sequences were generated using ImageJ. Image sequences were then evaluated for the indicated parameters manually. For statistical analysis, all p values were calculated with Student’s t-test and p < 0.05 was considered as significant.
Karyotype analysis
HCT116 cells were transfected with either scrambled or siCUL9 oligos and were then either untreated or treated with low dose (10 nM) taxol for 24 hours and then treated with colcemid (0.1 µg/ml) for 1.5 hour before mitotic spreading, then the cells are placed in a hypotonic solution (0.075 M KCl) and fixed on a microscope slide. The slides were stained with DAPI and the chromosome number was counted and analyzed for at least 50 mitotic cells.
Additional details and other experimental procedures used in this paper, including plasmids, siRNA and primer sequences, antibodies and immunological procedures, cell culture, transfection procedures, flow cytometry, immunofluorescence, in vitro and in vivo ubiquitylation assays, are available in the Supplemental Information.
Supplementary Material
HIGHLIGHTS.
CUL9 functions in maintaining genomic integrity
CUL9 is a downstream effector of the 3M complex
CUL9 is the first E3 ubiquitin ligase for survivin
A novel 3M–CUL9-survivin pathway
Acknowledgements
We are very grateful to Ted Salmon, Jim Bear, Steve Rogers and Dale Ramsden for insightful discussions and assistance throughout this study, Sarah Jackson, Beezly Groh and Jordon Kardos for critically reading the manuscript. This study was supported by NIH grants CA154699 to A.W.W. and CA068377 to Y.X.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43–49. [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Andrews P, He YJ, Xiong Y. Cytoplasmic localized ubiquitin ligase cullin 7 binds to p53 and promotes cell growth by antagonizing p53 function. Oncogene. 2006;25:4534–4548. doi: 10.1038/sj.onc.1209490. [DOI] [PubMed] [Google Scholar]
- Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci U S A. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giodini A, Kallio MJ, Wall NR, Gorbsky GJ, Tognin S, Marchisio PC, Symons M, Altieri DC. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002;62:2462–2467. [PubMed] [Google Scholar]
- Higgins PJ, Kessler GK, Nisselbaum JS, Melamed MR. Characterization and cell cycle kinetics of hepatocyte populations isolated from adult liver tissue by a nonenzymatic procedure. J Histochem Cytochem. 1985;33:672–676. doi: 10.1177/33.7.2409127. [DOI] [PubMed] [Google Scholar]
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- Huber C, Munnich A, Cormier-Daire V. The 3M syndrome. Best Pract Res Clin Endocrinol Metab. 2011;25:143–151. doi: 10.1016/j.beem.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Kelly RJ, Lopez-Chavez A, Citrin D, Janik JE, Morris JC. Impacting tumor cell-fate by targeting the inhibitor of apoptosis protein survivin. Mol Cancer. 2011;10:35. doi: 10.1186/1476-4598-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Cytoplasmic microtubules containing acetylated alpha-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J Cell Biol. 1986;103:13–22. doi: 10.1083/jcb.103.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Marin I. Diversification of the cullin family. BMC Evol Biol. 2009;9:267. doi: 10.1186/1471-2148-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, McKusick VA, Malvaux P, Temtamy S, Salinas C. The 3-M syndrome: a heritable low birthweight dwarfism. Birth Defects Orig Artic Ser. 1975;11:39–47. [PubMed] [Google Scholar]
- Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- Nunez F, Chipchase MD, Clarke AR, Melton DW. Nucleotide excision repair gene (ERCC1) deficiency causes G(2) arrest in hepatocytes and a reduction in liver binucleation: the role of p53 and p21. Faseb J. 2000;14:1073–1082. doi: 10.1096/fasebj.14.9.1073. [DOI] [PubMed] [Google Scholar]
- Pei XH, Bai F, Li Z, Smith MD, Whitewolf G, Jin R, Xiong Y. Cytoplasmic CUL9/PARC ubiquitin ligase is a tumor suppressor and promotes p53-dependent apoptosis. Cancer Res. 2011;71:2969–2977. doi: 10.1158/0008-5472.CAN-10-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa J, Canovas P, Islam A, Altieri DC, Doxsey SJ. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol Biol Cell. 2006;17:1483–1493. doi: 10.1091/mbc.E05-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Stover K, Tonon G, Schaffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31. doi: 10.1038/sj.neo.7900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Ryan BM, O'Donovan N, Duffy MJ. Survivin: a new target for anti-cancer therapy. Cancer Treat Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Skaar JR, Florens L, Tsutsumi T, Arai T, Tron A, Swanson SK, Washburn MP, DeCaprio JA. PARC and CUL7 form atypical cullin RING ligase complexes. Cancer Res. 2007;67:2006–2014. doi: 10.1158/0008-5472.CAN-06-3241. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, et al. Analysis of human transcriptomes. Nat Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Temporal and spatial regulation of targeting aurora B to the inner centromere. Cold Spring Harb Symp Quant Biol. 2010;75:419–423. doi: 10.1101/sqb.2010.75.035. [DOI] [PubMed] [Google Scholar]
- Zhao J, Tenev T, Martins LM, Downward J, Lemoine NR. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci. 2000;23(113 Pt):4363–4371. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.