Abstract
Intrinsic termination signals for multisubunit bacterial RNA polymerases (RNAPs) encode a GC-rich stem-loop structure followed by a polyuridine (poly(U)) tract, and it has been proposed that steric clash of the stem-loop with the exit pore of the RNAP imposes a shearing force on the RNA in the downstream RNA:DNA hybrid, resulting in misalignment of the active site. The structurally unrelated T7 RNAP terminates at a similar type of signal (TΦ), suggesting a common mechanism for termination. In the absence of a hairpin (passive conditions) T7 RNAP slips efficiently in both homopolymeric A and U tracts, and we have found that replacement of the U tract in TΦ with a slippage-prone A tract still allows efficient termination. Under passive conditions, incorporation of a single G residue following a poly(U) tract (which is the situation during termination at TΦ) results in a “locked” complex that is unable to extend the transcript. Our results support a model in which transmission of the shearing force generated by steric clash of the hairpin with the exit pore is promoted by the presence of a slippery tracts downstream, resulting in alterations in the active site and the formation of a locked complex that represents an early step in the termination pathway.
Keywords: RNA pullout, RNA:DNA hybrid, stem loop, mitochondrial RNA polymerase, hybrid shearing
INTRODUCTION
When transcribing through homopolymeric tracts, RNA polymerases (RNAPs) may perform pseudo-templated transcription that results in the synthesis of RNAs whose sequences do not correspond to the encoding template. The mechanism of this phenomenon is unclear and has been attributed to “transcript slippage” (for review see1). Aside from its significance with regard to the fidelity of gene expression, transcript slippage has also been implicated in termination of transcription at certain signals, as well as other regulatory roles1,2.
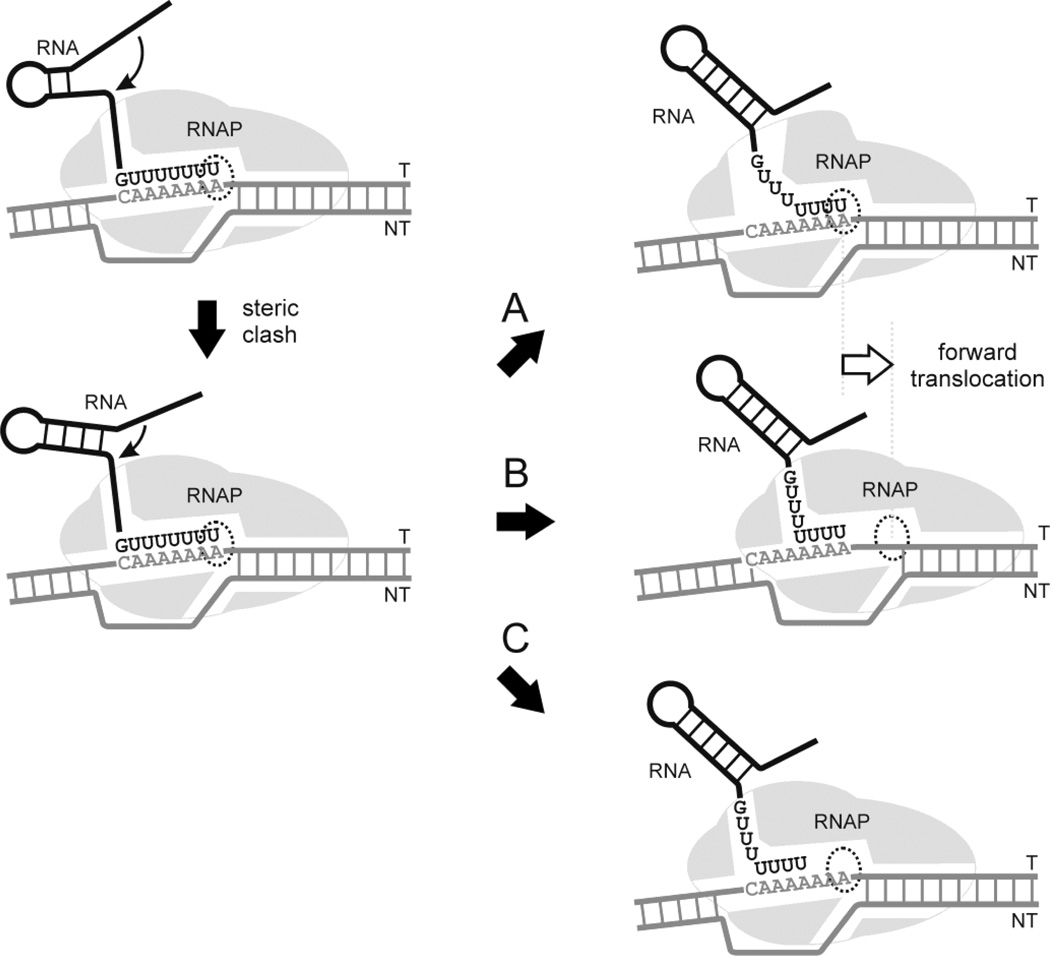
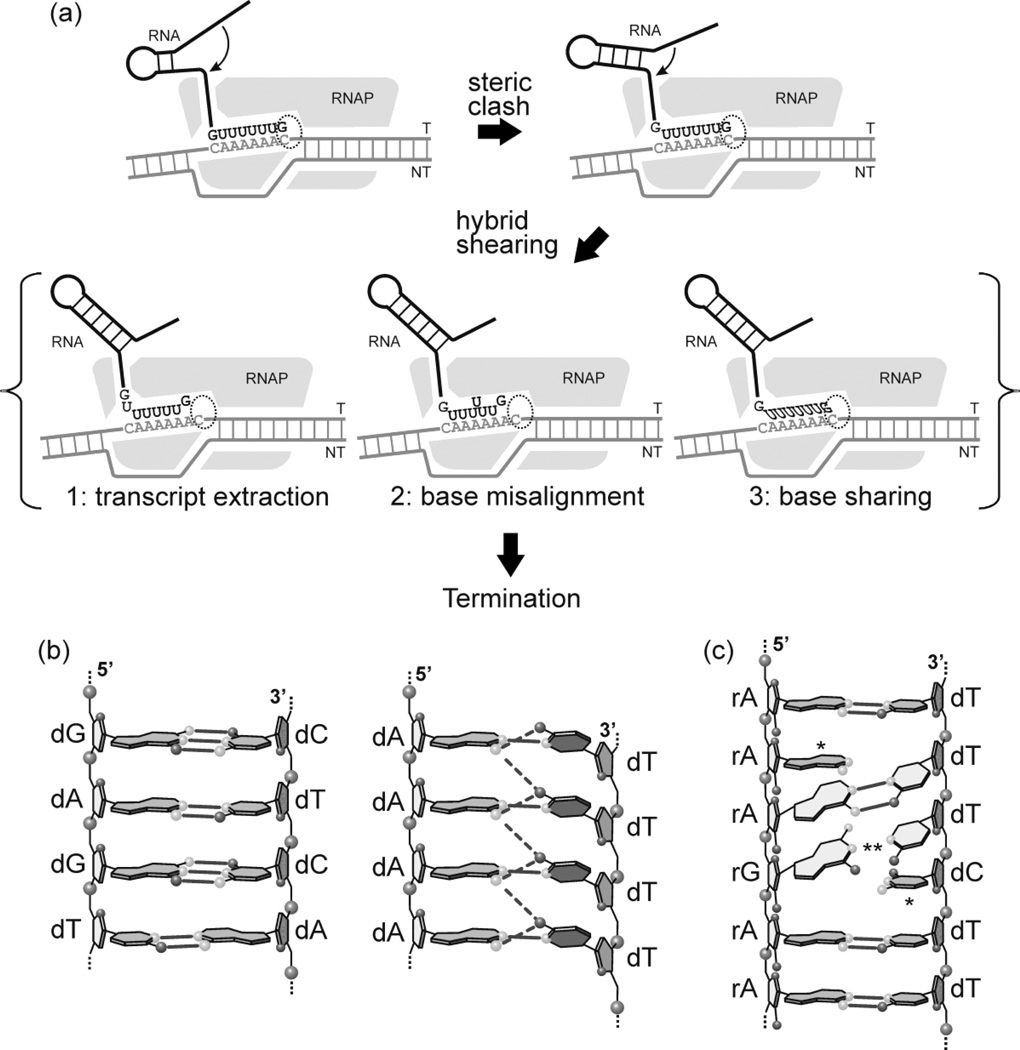
Intrinsic termination signals for bacterial RNAPs encode an RNA that forms a stable G:C-rich stem-loop (hairpin) structure followed by a U-rich tract, and it has been suggested that the particularly low thermodynamic stability of an rU:dA hybrid3–6 may play a role in pausing and termination (for review see7,8). In general, models of termination at such signals fall into three categories (Fig. 1). In the thermodynamic/allosteric model, steric clash of the stem-loop structure with the RNA exit pore of the RNAP disrupts interactions of the nascent transcript with the exit pore, which leads to melting of the upstream region of the RNA:DNA hybrid and changes in protein conformation9–11. In the forward translocation model, steric clash causes forward displacement of the RNAP, shortening of the hybrid due to disruption of the upstream region, and movement of the active site away from the 3’ end of the transcript12–13. In the RNA pullout model, steric clash results in shearing of the RNA in the hybrid, altering the position of the 3’ end of the transcript in the active site14–16. A distinguishing feature of the latter model is that it is expected to be enhanced by the presence of “slippery” tracts that lower the thermodynamic barrier to transcript shearing16.
Figure 1. Models of intrinsic termination.
As the EC approaches the site of termination, complementary sequences emerge from the RNA exit pore and commence the formation of the stem-loop structure (top left panel). Continued folding of the hairpin results in steric clash with the exit pore (lower left). In the thermodynamic/allosteric model (A), steric clash disrupts interactions of the nascent transcript with the exit pore, melting of the upstream region of the RNA:DNA hybrid, and changes in protein conformation. In the forward translocation model (B) steric clash causes forward displacement of the RNAP, shortening of the hybrid due to displacement at the upstream end of the hybrid, and movement of the active site away from the 3’ end of the transcript. In the RNA pullout model (C) steric clash results in shearing of the RNA in the hybrid, altering the position of the 3’ end of the transcript in the active site.
Although the single subunit T7 and multisubunit RNAPs are structurally unrelated, both classes of enzyme terminate at stem-loop signals of the type described above, suggesting a common mechanism of termination that relies primarily upon the properties of the nucleic acid components rather than conserved structural elements in the RNAPs. In this work, we explored the relationship between transcript slippage and termination in the T7 system. Our data support a model of termination in which steric clash of the stem-loop with the exit pore of the RNAP generates a shearing force that is transmitted downstream through an RNA:DNA hybrid that is prone to slippage, resulting in misalignment of the 3’ end of the transcript at the active site and the formation of a blocked complex that represents an initial step in the termination pathway.
RESULTS
Efficient slippage during elongation occurs in both poly(U) and poly(A) tracts
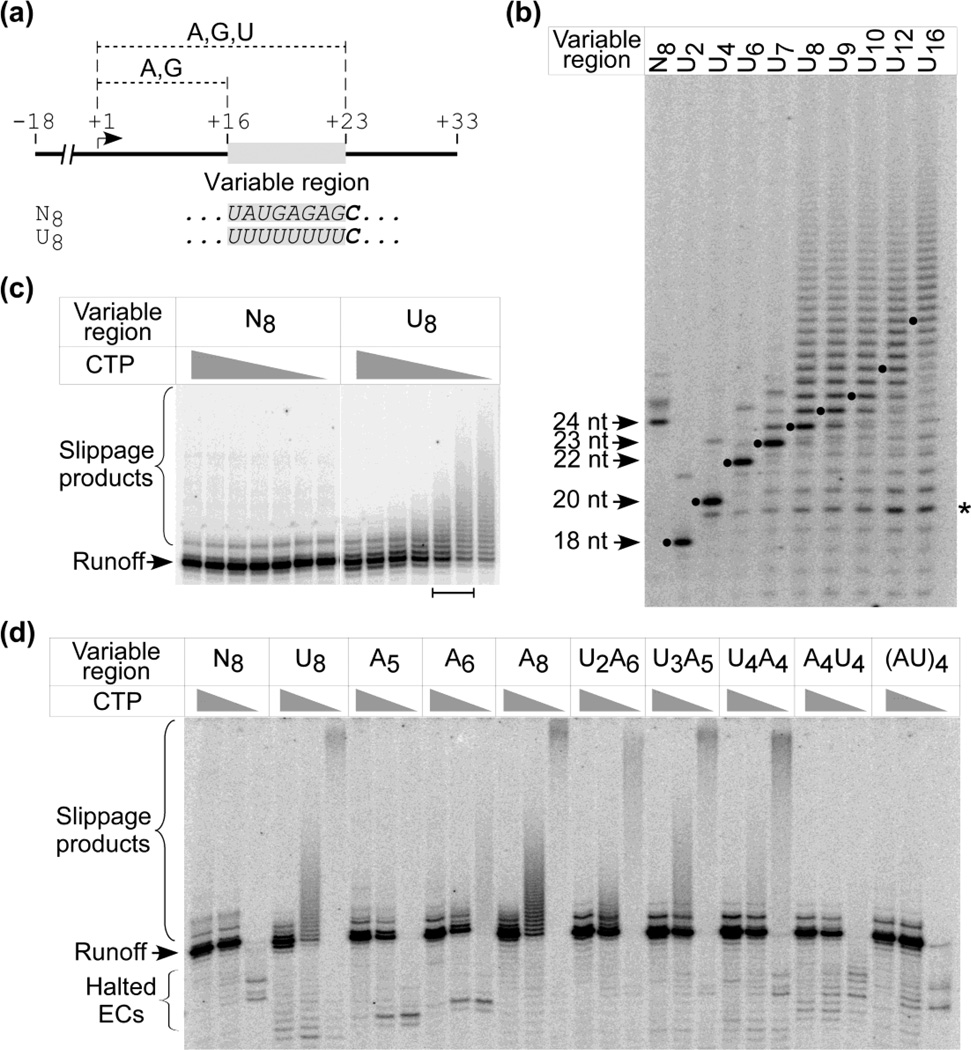
To examine intrinsic transcript slippage in the absence of a stem-loop structure in the RNA (passive slippage), we designed a series of templates in which a test sequence (variable tract) was placed downstream from a T7 promoter in a fixed sequence context (Fig. 2a). In these templates the first incorporation of CMP into the transcript occurs just downstream of the variable region. This feature allowed us to halt RNAP at the end of the variable region (by omission of CTP in the reaction), to alter the time that the elongation complex (EC) spends at the end of the variable tract before proceeding (by varying the concentration of CTP), or to incorporate a single NTP just after the variable tract (using a chain terminating analog of CTP, 3’-dCTP).
Figure 2. Transcript slippage in homopolymeric tracts.
Panel (a). General design of templates. DNA templates were constructed as described in Materials and Methods and Supplemental data. The templates contain a variable region (shaded) commencing 16 bp downstream from the start site (+1, arrow); sequences for the N8 and U8 transcripts are given. The first C in the transcripts occurs just downstream of the variable region (bold). In the reactions shown in panels (b)–(d) the templates were transcribed in the presence of 400 µM GTP, ATP and UTP and either CTP or 3’-dCTP at the concentrations indicated, and the products (labeled with γ-32P-GTP) were resolved by electrophoresis.
Panel (b). Transcript slippage vs length of U-tract. Templates having the variable regions indicated were transcribed in the presence of 50 µM 3’-dCTP. Products that are expected to arise from termination following incorporation of 3’-dCTP are indicated by dots. A band that corresponds to termination or release of a product after incorporation of four consecutive U residues is indicated with an asterisk. Panel (c). Transcript slippage vs the concentration of CTP. Templates having the variable regions indicated were transcribed in the presence of serial two-fold dilutions of CTP (for each template, left to right: 800, 400, 200, 100, 50, 25, 12.5µM); the range of previously reported values for KMapp [CTP]22 are indicated by the bar below the image.
Panel (d). Transcript slippage in other sequence motifs. Templates having the variable regions indicated were transcribed in the presence of CTP at concentrations of 400, 40, or 0 µM (left to right for each template). The expected positions of transcripts that arise from complexes halted at the end of the variable tract due to lack of CTP are indicated; the abundance of these products is low due to the slow turnover rate of these complexes in comparison to that of complexes that have reached the end of the template (see Fig 5c). .
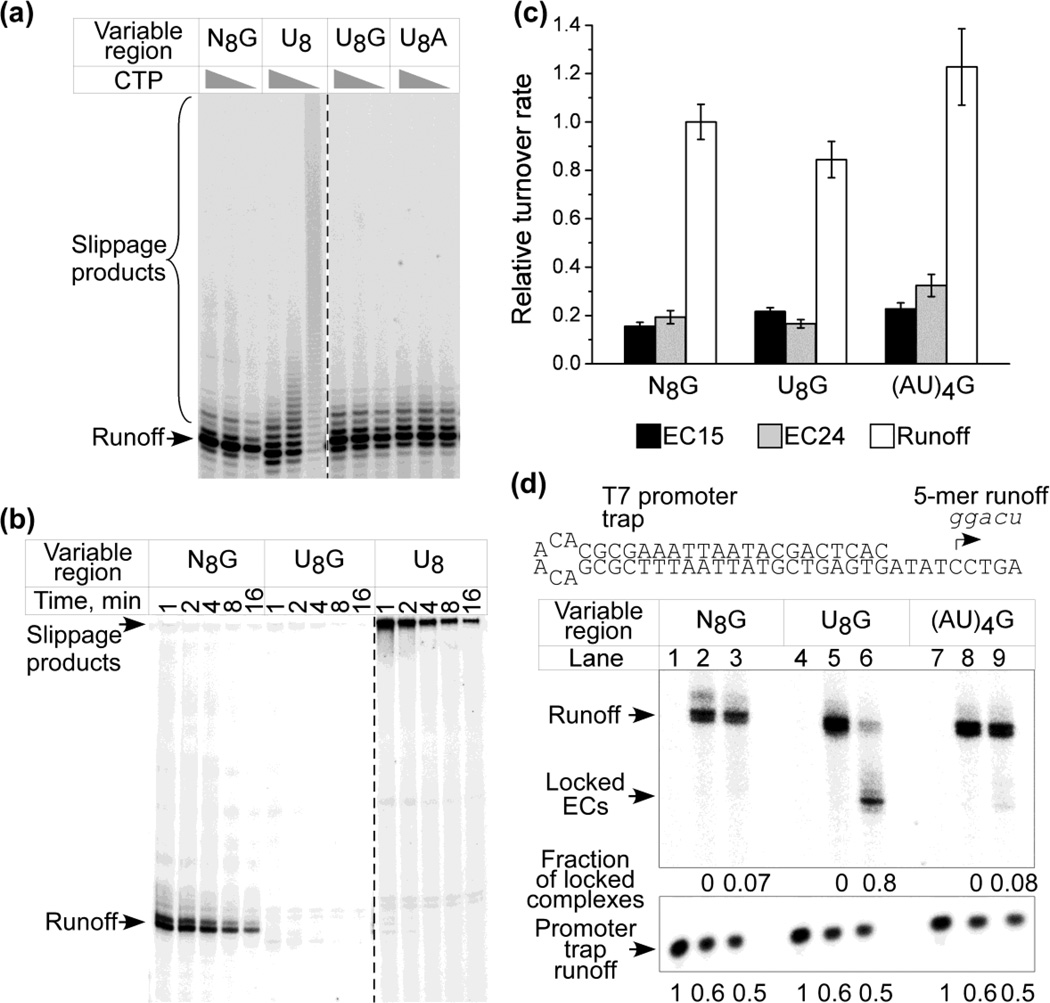
We first explored the effect of changes in the length of a poly(U) tract on the efficiency of slippage (Fig. 2b). In the absence of CTP the efficiency of slippage on some templates is very high, and results in high molecular weight products that migrate near the origin of the gel. To facilitate the detection of discrete slippage products we included low levels of the chain terminating 3’-dCTP nucleotide analog in these reactions. As shown in Fig. 2b some slippage is observed when the U tract has a length of 7 nucleotides (nt) but efficient slippage does not commence until the tract is 8 nt, which corresponds to the length of the RNA:DNA hybrid in the T7 EC17–20. Under these conditions the predominate product corresponds to a transcript that has been terminated just downstream of the poly(U) tract following incorporation of 3’-dCMP (24 nt) but transcripts larger than this size are also observed. As the length of the homopolymeric tract increases, the abundance and heterogeneity of these products increases. A band that would correspond to release of transcripts when only four consecutive UMP residues have been incorporated (asterisk) suggests that the EC may exhibit a decreased stability at this point21. Interestingly, little termination is observed at the end of the U8 tract, suggesting that complexes that are engaged in slippage are rather stable (see below).
Slippage in a U8 tract is remarkably efficient. As the concentration of CTP is decreased the abundance of slipped products increases dramatically. At concentrations of CTP that correspond to reported values of KMapp for CTP (∼30 µM22) most of the transcripts from the U8 template are larger than expected (Fig. 2c). The observation that transcript slippage is competitive with elongation at such levels is consistent with observations of slippage by Escherichia coli RNAP in poly(A) and poly(U) tracts in vivo23.
We also explored slippage in poly(A) tracts and in interrupted poly(A) and poly(U) tracts (Fig. 2d). Efficient slippage was evident in A8 tracts but not in A5 or A6 tracts. However, introduction of a short poly(U) tract just upstream of shorter A tracts (e.g., U2A6, U3A5, U4A4) resulted in efficient slippage. This observation is consistent with the notion that weak rU:dA base pairs in the upstream region of the RNA:DNA hybrid may facilitate “hybrid breathing”, allowing slippage in the remaining short A tract downstream7–9. In support of this, efficient slippage was observed in U4:A4 tracts (where the upstream region of the hybrid is U-rich) but not in A4:U4 tracts (where the upstream region consists of more stable rA:dT bps). We note that these observations were made in the absence of a hairpin, and it has been proposed that steric clash due to the presence of an upstream stem-loop may also disrupt the upstream end of the hybrid7–9, mimicking the effects of an unstable U-rich hybrid in this region and allowing slippage in shorter poly(U) tracts. Little slippage was observed in an alternating (AU)4 tract even though the predicted thermodynamic stability of this hybrid is expected to be less than that of an A8 hybrid28.
Although slippage in G-tracts by T7 RNAP has been observed during initiation24,25, when the RNA:DNA hybrid is 3–4 bps and the organization of the complex is different than that of an EC17,18, we observed no slippage in G8 tracts during elongation26 consistent with previous observations by others that little slippage occurs in poly(C) or poly(G) tracts23,27
Termination at stem-loop signals that include a poly(A) run
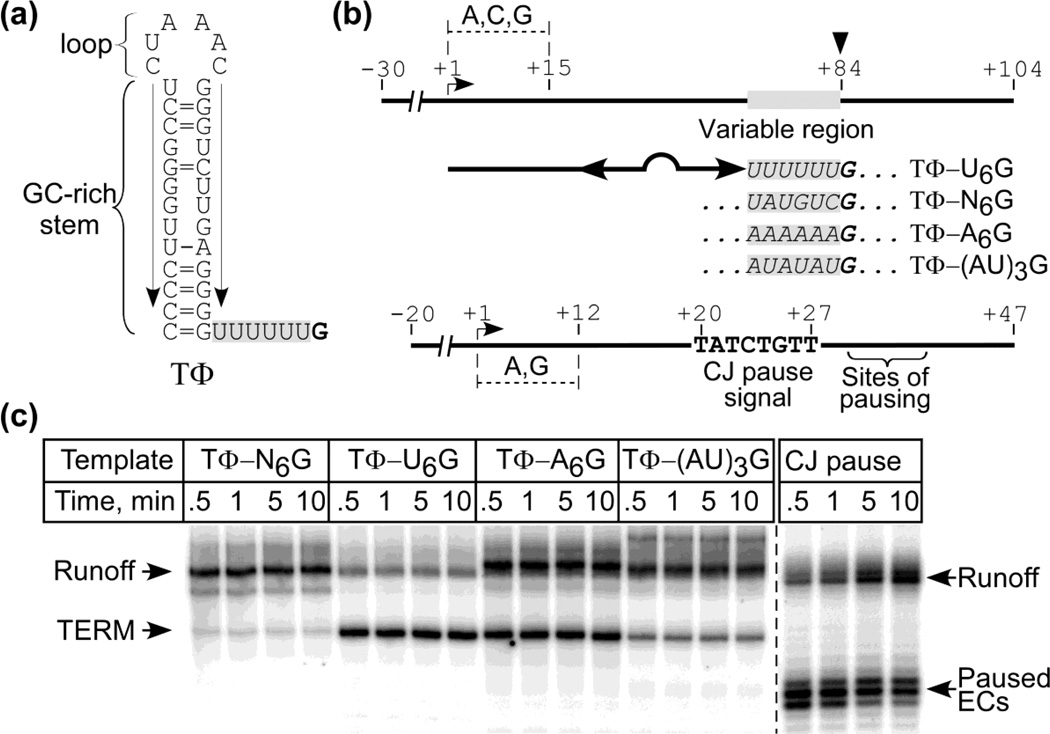
The T7 late termination signal (TΦ) consists of a stable stem loop structure followed by a run of 6 uridine residues, and termination occurs following incorporation of GMP immediately downstream of the U tract (see Fig. 3a). While this U tract is shorter than required for passive slippage (see above), the RNA pullout model suggests that steric clash of the stem-loop structure with the exit pore of the RNAP may facilitate transcript shearing either by inducing shear force and/or by disrupting the upstream end of the hybrid.
Figure 3. Termination at stem-loop poly(A) signals.
Panel (a). Predicted organization of TΦ. Arrows indicate the ascending and descending arms of the stem; the U6 tract downstream is shaded, and the site of termination (G) is in bold.
Panel (b). Template design. Templates encode transcripts having the sequence found in the stem-loop structure in TΦ (double headed arrow) followed by a variable region (boxed sequence); the site of termination is at +84. The signals are identified at the right by noting the nature of the variable region. The lower scheme explains the design of a template containing a CJ pause signal.
Panel (c). Termination at TΦ and its variants. Start up complexes halted at +15 in the presence of GTP, ATP and CTP for 5 min were mixed with a 20-fold excess of promoter trap (to bind free RNAP and prevent reinitiation) and α-[32P]-UTP (to allow extension of the transcripts and subsequent detection). The reactions were stopped after the time intervals indicated and the products were resolved by electrophoresis; positions of terminated and runoff products are indicated to the left. A similar experiment was carried out with a template that contains a sequence-specific pause signal for T7 RNAP found in the concatamer junction (CJ) of replicating T7 DNA29 (right side of panel).
As we had observed that poly(A) tracts allow efficient slippage by T7 RNAP under passive conditions we examined whether replacement of the U6 tract downstream of the stem loop in TΦ with an A6 tract might also result in termination. To accomplish this we constructed a set of templates that carried altered TΦ signals (Fig. 3b). As shown in Fig. 3c and Table I, replacement of the U6 tract in the wild type (WT) signal (here defined as TΦ-U6G) with an A6 tract (TΦ-A6G) resulted in efficient termination. In contrast, the presence of a non-homopolymeric tract (TΦ-N6G) resulted in a much lower level of termination, as did an alternating AU tract (TΦ-(AU)3G) or an interrupted U tract (TΦ-U3GU2G). The appearance of a band that would correspond to termination at the TΦ-A6G signal is not the result of a brief pause, as the abundance of the shorter (terminated) product relative to run off does not change over time (0.5 to 10 min); this is in contrast to the behavior at a known sequence-specific pause signal for T7 RNAP, CJ29 (Fig. 3c).
Table I.
Efficiency of termination at TΦ and its variants
| Signala | % terminationc |
|---|---|
| TΦ-N6G(cu) | 8±1 |
| TΦ-N6G(au) | 5.6±0.1 |
| TΦ-N6G(cg) | 7±1 |
| TΦ- U6G(cu) | 73±2 |
| TΦ-U6G(au) | 58.5±0.1 |
| TΦ-U6G(cg) | 84±1 |
| TΦ-U6A(cu) | 62±1 |
| TΦ-U6A(au) | 55±1 |
| TΦ-U6A(cg) | 50±1 |
| TΦ-A6G(cu) | 35±1 |
| TΦ-(AU)3G(cu) | 16±3 |
| TΦ-U3GU3G(cu) | 16±0.1 |
Signals are identified by the sequence found in the variable region followed by the nt incorporated at the site of termination (underlined) and the sequence of the RNA encoded by the template 2 nt downstream of this site (in parentheses; see Fig. 3b). The wild type signal is in bold.
Termination was analyzed as in Fig. 3c. The efficiency of termination (%T) is expressed as [(terminated RNA)/(terminated RNA + runoff)×100].
Yeast mitochondrial RNAP slips poorly
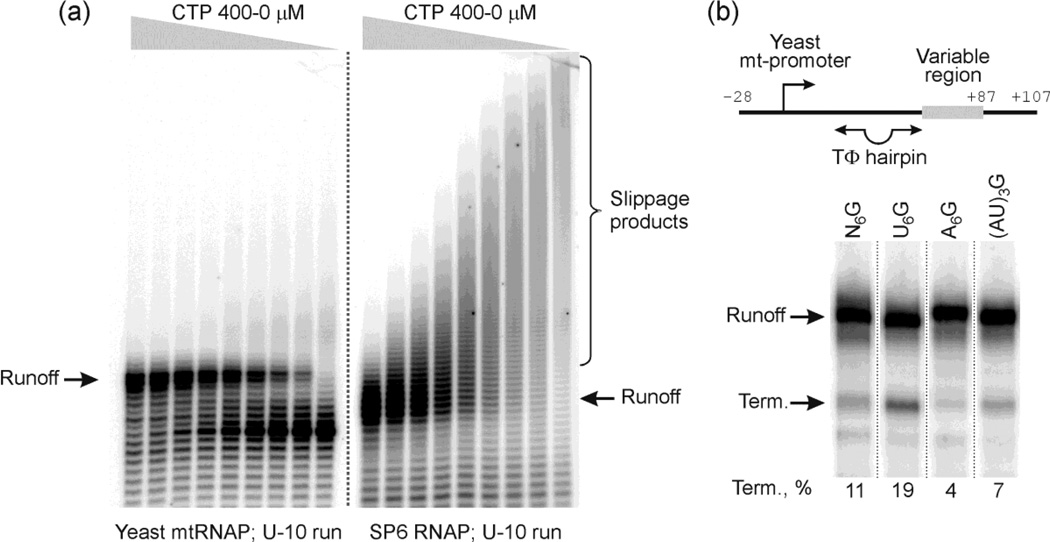
Both T7 and the multisubunit E. coli RNAP have been shown to slip in long poly A and U tracts, and it has been proposed that to prevent frameshifting in vivo the genomes of these organisms have evolved to minimize the average length of such tracts to < 6–7 nt in coding regions1,30. The yeast mitochondrial (mt) genome is highly A:T rich and encodes numerous copies of A- and U-tracts of at least 8 nt in length, but is transcribed by an RNAP that consists of a core subunit (Rpo41p) that resembles T7 RNAP31–33. We were therefore curious to determine if this enzyme is as prone to slippage as T7 RNAP. We also included in our analysis a representative of the SP6 subfamily of phage RNA polymerases, SP6 RNAP, which is similar to, but distinct from, members of the T7 subfamily to determine whether other phage-encoded RNA polymerases can slip on polyU tracts in a similar manner as T7 RNAP. As shown in Fig. 4a, Rpo41p does not exhibit efficient slippage in a U10 tract while SP6 RNAP slips quite efficiently. In the absence of CTP (which is required to escape the U-tract) the predominate transcripts made by Rpo41p correspond to products that are halted at the end of the U-tract,whereas under similar conditionsT7 and SP6 RNAPs yield few such products and slippage products predominate. (c.f., lanes on the right of each set and also see the right lane in Fig. 2c). In view of the discussion above, we examined termination by Rpo41p at TΦ and its variants. As shown in Fig. 4b Rpo41 terminates poorly at TΦ-U6G (∼20% vs ∼75% for T7 RNAP). While this observation supports the notion that termination is linked to slippage, there may be other reasons why the mtRNAP terminates poorly at this signal. The observation that T7 RNAP exhibits greatly reduced, but significant, termination at signals that lack a U6 tract indicates that there must be alternate, less efficient, mechanisms of termination at these signals. This is reminiscent of results with bacterial RNAPs in which alternate pathways for termination exist depending upon the presence of slippage-prone sequences downstream16
Figure 4. The single-subunit yeast mitochondrial RNAP resists slippage on homopolymeric tracts.
Panel (a). Slippage by mtRNAP and SP6 RNAP. Transcription by mtRNAP (left side) and SP6 RNAP (right side) was carried out for 20 min at 37 °C in the presence of ATP, GTP, UTP, α-[32P]-GTP, and CTP added to concentrations of 400, 200, 100, 50, 25, 13, 6, 3, and 0 µM (left to right). The templates encoding U10 runs in the variable region were prepared as in Fig. 2a except a consensus yeast mitochondrial promoter or SP6 promoter were provided for corresponding enzymes. The transcripts formed were resolved by 16 % denaturing PAGE; expected run-off and slippage products are indicated.
Panel (b). Termination by mtRNAP. Templates carrying a WT and altered TΦ termination signal were prepared as in Fig. 3b except a consensus mitochondrial promoter was used upstream of the signals instead of a T7 promoter (see scheme on top). Multiple round transcription reactions with the templates indicated were performed in the presence of Rpo41p (mtRNAP), Mtf1p, ATP, CTP, GTP, UTP, and α-[32P]-GTP in transcription buffer for 15 min at 30 °C and resolved by 20 % PAGE. Run-off transcripts and termination products are marked on the left and the termination efficiency is indicated under each lane.
Incorporation of an additional nucleotide at the 3’ end of a slippage-prone transcript results in the formation of a “locked” complex
In the absence of a stem-loop structure, efficient slippage by T7 RNAP was observed to commence when the homopolymeric A- or U-tract was 8 nt, which corresponds to the length of the RNA:DNA hybrid within the EC17–18. However, termination at TΦ occurs after the synthesis of a shorter, U6 tract, following incorporation of a GMP residue34–35 This suggests that steric clash of the GC-rich RNA hairpin with the exit pore of the RNAP may facilitate transcript shearing in a shorter tract than occurs under passive slippage conditions, perhaps by disrupting the upstream region of the hybrid and shortening its effective length, much as is observed with U3A5 tracts under passive conditions (see Fig. 2d). Under these circumstances, incorporation of GMP at the 3’ end of the transcript may inhibit slippage due to the potential formation of a mismatch in base pairing. However, shearing forces transmitted along the slippage-prone U-tract may alter the position of the 3’ end of the transcript in the active site, inhibiting further extension (see Fig. 6). We note that misalignment of the 3’ end of the transcript has been proposed to account for transcription arrest by multisubunit RNAPs36
Figure 6. A model for termination by T7 RNAP.
Panel (a). Model for the early stages of the termination process. As the RNAP (gray shape) transcribes through the termination signal, annealing of the stem-loop region of the transcript results in steric clash with the exit pore, generating a shearing force that results in a change in the organization of the active site (dashed oval) and the formation of a “locked” complex that represents an early step in the termination pathway. The transmission of the shearing force along the hybrid is facilitated by the presence of homopolymeric tracts that promote transcript slippage under passive conditions. While active slippage (i.e., additional incorporation of UMP residues) is not involved in this model (and in fact, would be expected to decrease the efficiency of termination due to a relief in steric clash, see Discussion) the presence of slippage-prone sequences lowers the thermodynamic barrier to changes in the hybrid. We do not know the organization of the hybrid in such a complex nor the mechanism by which the shearing force is transmitted. A number of pathways have been proposed (for review see1). In scheme 1, extraction of the transcript removes the 3’ end from the active site; the formation of a mismatched base pair may limit extraction to one or a few base pairs. In scheme 2, base misalignment along the hybrid results in extraction of the 3’ end from the active site. In scheme 3, (base sharing, see below) changes in the geometry of the hybrid, without slippage, alter the orientation of the 3’ end of the transcript in the active site.
Panel (b). Base sharing. Cross strand hydrogen bonds may facilitate transcript slippage by base sharing (see1 for review). Structural data indicate that A:T base pairs in homopolymeric (dA:dT)n tracts exhibit a high propeller twist that may be stabilized by cross strand hydrogen bonds between the adenine N-6 amine of a base pair on one strand and the thymine O–4 of the succeeding base pair on the opposite strand (right side)49 This is not observed in non-homopolymeric tracts (left side).
Panel (c). Alternate RNA:DNA hybrid structures; unzipping and slippage of the RNA:DNA hybrid in the polypurine tract of an HIV-RT complex. Structural analysis of HIV-RT complexes in association with the “polypurine tract” reveals a non-canonical form of the RNA:DNA hybrid that involves both slipped and mismatchedbases44. Misaligned (*) and mismatched (**) bases are indicated.
To examine this hypothesis we tested the effects of incorporating a single GMP residue downstream of a U8 tract under passive slippage conditions. As shown in Fig. 5a, incorporation of a G (or A) just after the U8 tract suppressed reiterative addition of U residues. We then asked whether transcription complexes that had been halted in such a context were able to resume transcription (Fig. 5b). Complexes halted at the ends of N8G, U8G, and U8 tracts for two min were challenged with a promoter trap (which binds free RNAP and prevents reinitiation on the test template)37–38. After various intervals, α-[32P]-CTP was added to allow extension and labeling of the products, and the transcripts were resolved by electrophoresis. Complexes halted at the end of the N8G tract (in the absence of slippage) exhibited typical stability profiles for a T7 EC, with a half life of ∼8 min21,39
Figure 5. Transcription complexes halted in a U8G context rapidly loose transcription activity.
Panel (a). Transcript slippage assays. Reactions were carried out as described in Fig. 2d using templates with the variable regions indicated.
Panel (b). Decay in ability of halted complexes to extend transcripts over time. Complexes halted at the end of the variable region were formed by incubation with GTP, ATP, and UTP for 2 min. Promoter trap was then added to prevent reinitiation. After the intervals indicated, α-[32P]-CTP was added and the reactions were incubated for a further 2 min to allow extension of the halted complexes and the products were analyzed by gel electrophoresis.
Panel (c). Comparative turnover rates of halted complexes. The templates indicated were transcribed for 10 min in the presence of GTP and ATP, resulting in complexes halted before entry into the variable region (EC15); with GTP, ATP, UTP, resulting in complexes halted at the end of the variable region (EC24); or all four NTPs, resulting in the synthesis of runoff products (RO). γ-[32P]-GTP was employed as the labeled substrate. The accumulation of products of the sizes indicated are expressed relative to the accumulation of RO products from the N8G template; standard errors (N=3) are indicated. The rates of accumulation under these conditions were linear over a 30 min period.
Panel (d). Rapid inactivation of complexes halted at U8G. Complexes halted before the variable tract were formed in the presence of GTP and ATP and were chased directly to the end of the template by the addition of CTP, α-[32P]-UTP and trap (lanes 2, 5, 8) or were advanced to the end of the variable tract by the addition of UTP, held briefly for 30 sec, and then chased (lanes 3, 6, 9). In control reactions (lanes 1, 4, 7) the promoter trap was added prior to substrate. The design of the trap is given at the top of the panel; the trap gives rise to a 5 nt runoff transcript during the challenge; the level of production of this transcript is expressed as fraction of the control level within each set (bottom of panel).
Surprisingly, complexes halted at the end of the U8 tract (where slippage is occurring) exhibited a similar decay in their ability to extend transcripts as those halted in an N8 context, indicating that transcription complexes that are engaged in slippage are not inherently unstable. The extensive protein:nucleic acid interactions observed in the RNA:DNA hybrid cavity, and/or the topological intertwining of the nucleic acid components, may therefore stabilize an otherwise unstable hybrid configuration17,18,40.
In contrast to the results above, little or no extension was observed from complexes that were halted at the end of the U8G tract, suggesting that these complexes might be particularly unstable and dissociate rapidly, or that they are stable but blocked in their ability to extend. To examine this, we determined the relative turnover rates of halted complexes using a steady state assay (Fig. 5c). In such an assay, product accumulation depends upon the rate at which the polymerase disassociates from the template, reinitiates, and synthesizes transcripts anew (the turnover rate). Since the templates used in these experiments are identical except for the variable region in which the complexes are halted, differences in the turnover rates reflect differences in the stability of these complexes. The rate of accumulation of products from complexes halted at the end of the U8G tract (EC24) was similar to those of complexes halted at the end of N8G and (AU)4G tracts (and about the same as that of complexes halted at +15, prior to entry into the variable region (EC15)), and much lower than turnover rates for complexes that have reached the end of the template (RO), indicating that the U8G complexes remain stably associated with the template.
To examine the fate of complexes halted at the end of a U8G tract in more detail we performed promoter challenge experiments over a shorter period of time and examined the transfer of the RNAP from the test template to the promoter trap during the chase (Fig. 5d). Startup complexes formed in the presence of GTP and ATP were either chased directly to the end of the template by the simultaneous addition of α-[32P]-UTP, CTP and trap (lanes 2, 5, 8) or were advanced to the end of the variable tract by the addition of UTP, held briefly for 30 sec, and then extended by addition of CTP and trap (lanes 3, 6, 9). In the case of an instant challenge (no delay; lanes 2, 5, 8) the N8G, U8G and (AU)4G complexes all showed similar abilities to extend to the end of the template. In contrast, but consistent with the results shown in panel B, complexes halted at the end of the U8G tract for 30 sec exhibited a much lower ability to extend (lane 6).
An indication of release of the RNAP from the test template and its transfer to the trap before and during the chase may be obtained from this experiment. The promoter trap directs initiation of a 5 nt transcript with the sequence 5’-ggacu (see top of panel d). The appearance of this product during the chase reflects the association of the RNAP with the trap (the test template does not yield a labeled product of this size). In control experiments in which template RNAP and trap were mixed together prior to addition of substrates, we observed little or no transcription from the test template and abundant synthesis of the 5 nt trap product during the subsequent 2 min reaction (lanes 1, 4, 7), demonstrating the efficiency of the trap in binding free RNAP and preventing initiation at the promoter in the test templates. In experiments in which the trap was added to halted complexes simultaneously with the missing substrate (lanes 2, 5, 8), or after a 30 sec delay (lanes 3, 6, 9) synthesis of the 5 nt products during the 2 min reaction was reduced to about 50% of the control, reflecting occupancy of the RNAP on the test template during the reaction (and consequent diminished production of the 5 nt transcript from the promoter trap). If complexes halted in the U8G context were less stable than those halted at N8G and (AU)4G, synthesis of the 5 nt product should be greater in these reactions than in reactions in which the complexes were halted at N8G and (AU)4G. However, the level of these products was the same or slightly lower than from the N8G and (AU)4G templates (lanes 7, 8, 9), supporting the conclusion that there is little release of enzyme from the “locked” complexes.
DISCUSSION
Single molecule force studies with bacterial RNAPs16 have indicated that, for these enzymes, the pathway to termination depends upon the nature of the poly(U) tract downstream of the stem-loop. At terminators in which the U tract is uninterrupted, or is interrupted by only one A residue, the predominant pathway to termination involves RNA shearing, whereas at terminators in which the U run is interrupted by two G or C residues, forward translocation appears to be the favored pathway16.
The results reported here support a model for termination by T7 RNAP at TΦ that occurs predominately through transcript shearing The presence of an RNA:DNA hybrid that is prone to slippage (under passive conditions) facilitates this by lowering the thermodynamic barrier to transmission of the shearing forces downstream. In support of this we note that replacement of the U6 tract downstream of the stem loop in the natural TΦ signal with another sequence that is prone to slippage (i.e., an A6 tract) resulted in efficient termination, whereas replacement with less slippery tracts ((e.g.,(AU)3 or U3GU2) did not (Fig. 3). Neverthless, we note that in the absence of a slippery tract (e.g. TΦ-N6G) there remains a low, but significant level of termination, indicating that other, less efficient, pathways must exist.
In a variation of the allosteric model, it has been proposed that formation of the stem-loop structure and its invasion into the RNA exit pore of E.coli RNAP lead to progressive changes in the structure of the enzyme and disruption of the hybrid, culminating in a close approach of the stem-loop to the active site, thereby altering RNAP activity and destabilizing the complex41. In T7 RNAP the RNA exit pore is formed during the transition from an unstable initiation complex to a stable elongation complex, and involves major refolding of elements in the N-terminal domain and the promoter specificity loop17,18 At some point during or after termination the enzyme must return to the initiation-competent state, and so it might be reasonable to assume that clash of the stem-loop with the exit pore in T7 RNAP would result in refolding of the enzyme in the IC and initiate the termination process. However, we have shown that formation of an engineered disulfide crosslink between elements in the exit pore of T7 RNAP that would inhibit this return pathway does not prevent arrest at this site42. Nevertheless, we cannot rule out the possibility that conformational changes resulting from steric clash of the stem loop with elements in the RNAP and/or changes in interactions of the transcript with the exit pore may play a role in the later stages of termination.
While the sequence downstream from the termination site is not expected to contribute to the efficiency of termination in the RNA pullout model, it is expected to contribute to the efficiency of termination in the forward translocation model because translocation requires melting of DNA duplex ahead of the complex. Thus, a lower stability of the downstream duplex (higher A:T content vs G:C content) is expected to facilitate forward translocation and to enhance termination. To examine this in TΦ, we altered the sequence at the predicted termination site and/or the region immediately downstream so as to increase or decrease the G:C content (Table I). In contrast to expectations for the forward translocation model, lowering the G:C content reduced termination efficiency, while increasing the G:C content resulted in a slight enhancement (for example, compare TΦ-U6G(cu) with TΦ-U6G(cg) and TΦ-U6A(au)). This, and the observation that termination at TΦ normally occurs in a G:C rich context (…Gctg… in the non-template strand, where the site of termination is underlined) suggests that forward translocation is not likely to make an important contribution to termination by T7 RNAP (and see6).
Under passive conditions the incorporation of a single G residue at the end of a U8 tract prevents further slippage but results in the formation of a “locked” complex that is poorly extended (Fig. 5). While this might suggest a preference for incorporation of GMP as the 3’ residue during termination, we have found that replacement of this residue with AMP or CMP (i.e., TΦ-U6A or TΦ-U6C) resulted in termination with similar efficiencies35. On the other hand, insertion of an additional U residue (TΦ-U7G, which would not cause a mismatch) also resulted in termination35 We note that in the latter situation unimpeded extraction of the transcript from the active site via slippage should also result in efficient termination. Why then would TΦ encode a G residue that is incorporated at the end of the U6 run during termination? We suggest that under some conditions the rate of intrinsic slippage in the U6 tract might be so high during elongation that the polymerase would incorporate additional U residues before reaching the site of termination, resulting in movement of the hairpin away from the exit pore, a decrease in the likelihood of steric clash, and reduced termination. The incorporation of a non-U residue at the 3’ end of the transcript would inhibit slippage (due to a potential mismatch in the hybrid) but still allow transmission of shearing forces downstream and alterations at the active site, resulting in the formation of a locked complex that allows time for the termination process to proceed.
As a working model (see Fig. 6a) we propose that the incorporation of a non-U residue downstream from the U-run in TΦ results in a “lock” of a misaligned pre-terminated complex. This situation may be analogous to that observed in studies of bacterial RNAPs, in which hybrid shearing in a stem loop terminator having a downstream U-tract resulted in a change in register of the RNA:DNA hybrid of ∼1 nt, and/or to that observed in arrested complexes in which the 3’ end of the complex is misaligned36
Evolutionary and regulatory implications
Ordinarily, one would expect transcript slippage to be a deleterious event, as it would result in an altered reading frame. There would appear to be two ways to circumvent this problem. The first is to select against the occurrence of slippery tracts within coding regions. The other would be to evolve RNAPs that are less prone to slip.
In agreement with the first strategy, there is a significant bias against the occurrence of poly(A) or poly(U) tracts greater than 6–7 nt in coding vs non-coding regions in most bacterial species with A/T rich genomes30. Similarly, there are no poly(A) or poly(U) tracts > 6 nt in the T7 genome, and the only occurrence of a U6 run is at TΦ34.
On the other hand, the observation that the yeast mitochondrial RNAP does not slip as well as the related T7 RNAP indicates that RNAP variants with altered slippage tendencies can exist (see also43).
Why then do slippery tracts and slippage-prone RNA polymerases persist? One answer is that this provides positive advantages for the cell, most likely with regard to gene regulation. While transcript slippage (or stuttering) may be useful for regulatory events that are known to occur during initiation, other means to regulate transcript initiation are available2. Similarly, while slippage appears to play a role in termination at certain signals, other means to terminate are possible. This raises the interesting possibility that there are other phenomena that occur during gene expression that rely upon slippage and its modulation, for example the use of alternate reading frames1.
Importantly, we do not know the structure of the RNA:DNA hybrid or the conformation of the RNAP in a slipped complex or in the putative pretermination complex. Proposed mechanisms of active slippage have included: denaturation of the hybrid along its entire length and reannealing in a new register, propagation of a non-paired base along the length of the hybrid, and base sharing1 (see Fig 6b). The mechanism of transcript slippage may depend upon the ability of the RNAP to accommodate and/or stabilize non-canonical conformations of the hybrid. Structural analysis of complexes of HIV RT (a member of the polI class of nucleotide polymerases that includes T7 RNAP) halted within a polypurine tract reveals an unusual form of the hybrid that involves both misaligned and mismatched bases44 (ee Fig 6c). This structure is not observed in the unbound nucleic acid complex, but is stabilized or induced in the RT complex. This situation is of functional significance, as the hybrid is insensitive to the intrinsic RNaseH activity of RT, allowing the protected RNA to later serve as a primer for DNA synthesis. Further biochemical and structural studies will be needed to explore these questions, Previous attempts to assemble T7 termination complexes on nucleic acid scaffolds indicated that such complexes were unstable6. However, the locked complexes formed under passive conditions appear to be stable (at least as stable as a halted EC; Fig. 5) suggesting that such complexes may be amenable to structural studies.
We suggest that the ability of RNAPs to accommodate or respond to alternate hybrid structures (or slippery tracts) may explain the continued presence of slippery RNAPs during evolution, and further suggest that novel transcription factors that enhance the ability of the RNAP to tolerate or respond to such alternate structures may exist (see also ref1).
MATERIALS AND METHODS
T7 RNA polymerase was purified as previously described45,46 SP6 RNAP was a gift from Dr. Steven Emanuel. Yeast mitochondrial RNAP (Rpo41p) and initiation factor Mtf1p were prepared as described earlier47.
Templates for slippage assays were constructed by annealing two DNA primers followed by extension with DNA polymerase I (Klenow fragment)48 Templates used to study termination were PCR-amplified from plasmid pVM16 (see Supplemental data), which carries a TΦ termination signal. The upstream primers used for amplification contained a T7 RNAP or mtRNAP promoter sequence and the downstream primers encoded variable regions placed in the WT sequence context as noted in the Figures. Sequences of all DNA primers and details on preparation of templates are provided in Supplemental data.
T7 RNAP transcription reactions were carried out in a total volume of 10 µL at 37 °C for 10 min unless indicated otherwise. Transcription buffer contained 20 mM Tris•HCl (pH 7.9), 15 mM MgCl2, and 5 mM β-mercaptoethanol. The concentrations of template and RNAP were 100 nM and 50 nM, respectively. The reactions were initiated by adding NTPs (GE Healthcare) to a final concentration of 400 µM each (unless indicated otherwise) and labeled with 4 µCi of either γ-[32P] or α-[32P] NTP (6000 Ci/mmol−1; PerkinElmer Life Sciences), as noted in the figure legends. Where indicated, reactions contained varying concentrations of CTP, or 50 µM chain terminating 3’dCTP. In single-round transcription reactions start-up complexes were formed by adding GTP, ATP and CTP and chased with UTP mixed with 2 µM promoter trap for 2 min at 25 °C.
Transcription was stopped by adding equal volume of stop buffer (95% formamide, 40 mM EDTA, 0.01% bromophenol blue and 0.01% xylene cyanol) and the products were resolved in denaturing (7M urea) polyacrylamide gels (16% for transcription slippage assays and 20 % for termination assays; 19:1, acrylamide:bisacrylamide) and visualized using a Typhoon 9200 PhosphorImager scanner (GE Healthcare). Data were collected and analyzed using ImageQuant 5.2 software.
SP6 RNAP transcription conditions were the same as used for T7 RNAP; the template used contained a SP6 promoter (see Supplemental data).
Transcription with mtRNAP was carried out in a similar way except the transcription buffer contained 20 mM Tris•HCl (pH 7.9), 50 mM KCl, 10 mM MgCl2, 0.1 % Tween-20, and 5 mM TCEP. Rpo41p and Mtf1p were included at 50 nM each, and the templates (see Supplemental data) were at a concentration of 100 nM.
Supplementary Material
Research Highlights.
T7 RNA polymerase slips efficiently in polyU and polyA tracts
Addition of G residue downstream of slipped RNA results in arrested complex
Termination is enhanced by presence of slippage-prone tract downstream of stable stem-loop structure in RNA
Propose that stem-loop promotes transcript shearing, misaligned active site, and arrest
Related yeast mitochondrial RNA polymerase slips poorly, terminates poorly
ACKNOWLEDGEMENTS
We thank Sergei Borukhov, Dmitry Markov, Steve Emanuel, Dmitry Temiakov, and other members of our group for helpful discussions, and Alexander Kukarin and Maria Savkina for preliminary experiments. This work was supported by grant NIH GM38147 to WTM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Anikin M, Moldotsov V, Temiakov D, McAllister WT. Transcript slippage and recoding. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of decoding rules enriches gene expression. New York: Springer; 2010. pp. 409–432. [Google Scholar]
- 2.Turnbough CL., Jr. Regulation of gene expression by reiterative transcription. Curr Opin. Microbiol. 2011;2:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin FH, Tinoco I., Jr. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Chen C, Russu IM. Dynamics and stability of individual base pairs in two homologous RNA-DNA hybrids. Biochemistry. 2009;48:3988–3997. doi: 10.1021/bi900070f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yager TD, von Hippel PH. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 6.Datta K, von Hippel PH. Direct spectroscopic study of reconstituted transcription complexes reveals that intrinsic termination is driven primarily by thermodynamic destabilization of the nucleic acid framework. J. Biol. Chem. 2008;283:3537–3549. doi: 10.1074/jbc.M707998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 8.Nudler E, Gottesman ME. Transcription termination and anti-termination in. E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 9.Komissarova N, Becker J, Solter S, Kireeva ML, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 10.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 11.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 13.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald LE, Zhou Y, McAllister WT. Termination and slippage by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 15.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 16.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 18.Tahirov TH, et al. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Sousa R. T7 RNA polymerase elongation complex structure and movement. J. Mol. Biol. 2000;303:347–358. doi: 10.1006/jmbi.2000.4150. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Martin CT. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J. Mol. Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 21.Mentesana PE, Chin-Bow ST, Sousa R, McAllister WT. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J. Mol. Biol. 2000;302:1049–1062. doi: 10.1006/jmbi.2000.4114. [DOI] [PubMed] [Google Scholar]
- 22.Arnold S, Siemann M, Scharnweber K, Werner M, Baumann S, Reuss M. Kinetic modeling and simulation of in vitro transcription by phage T7 RNA polymerase. Biotechnol. Bioeng. 2001;72:548–61. [PubMed] [Google Scholar]
- 23.Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin CT, Muller DK, Coleman JE. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 25.Imburgio D, Anikin M, McAllister WT. Effects of substitutions in a conserved DX2GR sequence motif, found in many DNA-dependent nucleotide polymerases, on transcription by T7 RNA polymerase. J. Mol. Biol. 2002;319:37–51. doi: 10.1016/S0022-2836(02)00261-9. [DOI] [PubMed] [Google Scholar]
- 26.Molodtsov V. Transcript slippage as a pathway to termination. Ph.D. Thesis, University of Medicine and Dentistry of New Jersey; 2010. [Google Scholar]
- 27.Parks AR, Court C, Lubkowska L, Jin DJ, Kashlev M, Court DL. Bacteriophage λ N protein inhibits transcription slippage by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;42:5823–5829. doi: 10.1093/nar/gku203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugimoto N, et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 29.Lyakhov DL, et al. Pausing and termination by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1998;280:201–213. doi: 10.1006/jmbi.1998.1854. [DOI] [PubMed] [Google Scholar]
- 30.Baranov PV, Hammer AW, Zhou J, Gesteland RF, Atkins JF. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005;6:R25. doi: 10.1186/gb-2005-6-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 32.Cermakian N, Ikeda TM, Cedergren R, Gray MW. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–54. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW, Cedergren R. On the evolution of the single-subunit RNA polymerases. J Mol Evol. 1997;45:671–81. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 34.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 35.Macdonald LE, Durbin RK, Dunn JJ, McAllister WT. Characterization of two types of termination signal for bacteriophage T7 RNA polymerase. J. Mol. Biol. 1994;238:145–158. doi: 10.1006/jmbi.1994.1277. [DOI] [PubMed] [Google Scholar]
- 36.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–53. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 37.Diaz GA, Rong M, McAllister WT, Durbin RK. The stability of abortively cycling T7 RNA polymerase complexes depends upon template conformation. Biochemistry. 1996;35:10837–43. doi: 10.1021/bi960488+. [DOI] [PubMed] [Google Scholar]
- 38.Gong P, Esposito EA, Martin CT. Initial bubble collapse plays a key role in the transition to elongation in T7 RNA polymerase. J Biol Chem. 2004;279:44277–85. doi: 10.1074/jbc.M409118200. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Navaroli DM, Enuameh MS, Martin CT. Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10352–10357. doi: 10.1073/pnas.0606306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Martin CT. Transcription elongation complex stability: the topological lock. J. Biol. Chem. 2009;284:36262–36270. doi: 10.1074/jbc.M109.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol. Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Ma K, Temiakov D, Anikin M, McAllister WT. Probing conformational changes in T7 RNA polymerase during initiation and termination by using engineered disulfide linkages. Proc Natl Acad Sci USA. 2005;102:17612–7. doi: 10.1073/pnas.0508865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou YN, Lubkowska L, Hui M, Court C, Chen S, Court DL, Strathern J, Jin DJ, Kashlev M. Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J. Biol. Chem. 2013;288:2700–2710. doi: 10.1074/jbc.M112.429464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–61. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He B, et al. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr. Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 46.Temiakov D, et al. Crystallization and preliminary crystallographic analysis of T7 RNA polymerase elongation complex. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:185–187. doi: 10.1107/s0907444902019777. [DOI] [PubMed] [Google Scholar]
- 47.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–40. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma K, Temiakov D, Jiang M, Anikin M, McAllister WT. Major conformational changes occur during the transition from an initiation complex to an elongation complex by T7 RNA polymerase. J. Biol. Chem. 2002;277:43206–43215. doi: 10.1074/jbc.M206658200. [DOI] [PubMed] [Google Scholar]
- 49.Yoon C, Prive GG, Goodsell DS, Dickerson RE. Structure of an alternating B-DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988;85:6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.