Abstract
The GNE gene encodes the rate-limiting, bifunctional enzyme of sialic acid biosynthesis, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE). Biallelic GNE mutations underlie GNE myopathy, an adult-onset progressive myopathy. GNE myopathy-associated GNE mutations are predominantly missense, resulting in reduced, but not absent, GNE enzyme activities. The exact pathomechanism of GNE myopathy remains unknown, but likely involves aberrant (muscle) sialylation. Here we summarize 154 reported and novel GNE variants associated with GNE myopathy, including 122 missense, 11 nonsense, 14 insertion/deletions and 7 intronic variants. All variants were deposited in the online GNE variation database (http://www.dmd.nl/nmdb2/home.php?select_db=GNE).
We report the predicted effects on protein function of all variants as well as the predicted effects on epimerase and/or kinase enzymatic activities of selected variants. By analyzing exome sequence databases, we identified three frequently occurring, unreported GNE missense variants/polymorphisms, important for future sequence interpretations. Based on allele frequencies, we estimate the world-wide prevalence of GNE myopathy to be ~ 4–21/1,000,000. This previously unrecognized high prevalence confirms suspicions that many patients may escape diagnosis. Awareness among physicians for GNE myopathy is essential for the identification of new patients, which is required for better understanding of the disorder’s pathomechanism and for the success of ongoing treatment trials.
Keywords: distal myopathy with rimmed vacuoles (DMRV), GNE myopathy, hereditary inclusion body myopathy (HIBM), adult onset muscular dystrophy, N-acetylmannosamine (ManNAc), disease prevalence, sialic acid
Background
GNE myopathy (MIM #605820) is a rare, recessive inherited, degenerative skeletal muscle disorder with early adult onset [Askanas and Engel, 1995; Eisenberg et al., 2001; Huizing and Krasnewich, 2009; Huizing et al., 2014a]. The disease was first described in 1981 in Japanese patients, termed distal myopathy with rimmed vacuoles (DMRV) or Nonaka myopathy (MIM #605820) [Nonaka et al., 1981]. A similar disorder, called vacuolar myopathy sparing the quadriceps, was described in 1984 in Persian-Jewish patients [Argov and Yarom, 1984], later referred to as hereditary inclusion body myopathy (HIBM), or inclusion body myopathy 2 (IBM-2). Linkage analysis studies in 1996 of the Persian-Jewish [Mitrani-Rosenbaum et al., 1996] and later the Japanese [Ikeuchi et al., 1997] patient cohorts localized the gene to 9p1-q1, suggesting that the two diseases were allelic. In 2001, the causative gene, GNE (MIM #603824), was identified [Eisenberg et al., 2001], and subsequent mutation analyses reported GNE sequence variants in patients of highly diverse ethnicities, including the original Persian-Jewish HIBM and Japanese DMRV cohorts, and patients from Asia, Europe, Africa and the Americas (Table 1) [Nishino et al., 2002; Eisenberg et al., 2003; Huizing and Krasnewich, 2009]. Since the multiple names for this disorder can be confusing the disorder was recently renamed “GNE myopathy”.
Table 1.
GNE variants associated with GNE myopathy
| Amino Acid Substitution1 | Nucleotide Substitution2 | GNE exon3 | GNE protein domain4 | Severity Prediction5 | Ethnicity6 | References | |
|---|---|---|---|---|---|---|---|
| hGNE1 NP_005467.1 |
hGNE2 NP_001121699.1 |
mRNA Variant 1 NM_001128227.2 |
|||||
| E2G | p.E33G | c.98A>G | 3 | ep | Severe | European | [Saechao et al., 2010] |
| R8* | p.R39* | c.115C>T | 3 | ep | Severe | Caucasian, Chinese, Japanese | [Saechao et al., 2010; Lu et al., 2011; Mori-Yoshimura et al., 2012] |
| R11W | p.R42W | c.124C>T | 3 | ep | Severe | Indian | [Huizing et al., 2004] |
| C13S | p.C44S | c.131G>C | 3 | ep | Medium | Chinese, Japanese, Korean | [Kim et al., 2006; Lu et al., 2011; Park et al., 2012; Tomimitsu et al., 2004] |
| A26P | p.A57P | c.169G>C | 3 | ep | Mild | Caucasian | [Weihl et al., 2011] |
| P27L | p.P58L | c.173C>T | 3 | ep | Medium | Japanese, Indian | [Mori-Yoshimura et al., 2012; Nalini et al., 2013] |
| P27S | p.P58S | c.172C>T | 3 | ep | Medium | Italian | [Broccolini et al., 2004] |
| I28M | p.I59M | c.177C>G | 3 | ep | Medium | Japanese | [Cho et al., 2013] |
| M29T | p.M60T | c.179T>C | 3 | ep | Medium | Korean, Japanese | [Kim et al., 2006; Cho et al., 2013] |
| M29R | p.M60R | c.179T>G | 3 | ep | Severe | Japanese | [Cho et al., 2013] |
| E35K | p.E66K | c.196G>A | 3 | ep | Medium | Chinese | [Li et al., 2011; Lu et al., 2011] |
| P36L | p.P67L | c.200C>T | 3 | ep | Severe | Italian | [Eisenberg et al., 2003] |
| E40K | p.E71K | c.211G>A | 3 | ep | Medium | Japanese | [Cho et al., 2013] |
| frameshift | frameshift | ins10 bp? | 3 | ep | Severe | Japanese | [Nishino et al., 2002] |
| I51M | p.I82M | c.246A>G | 3 | ep | Medium | Chinese | [Li et al., 2011; Lu et al., 2011] |
| exon 3 del | exon 3 del | ? | 3 | ep | Severe | Japanese | [Cho et al., 2013] |
| M60V | p.M91V | c.271A>G | 4 | ep | Mild | Portuguese | novel |
| R71W | p.R102W | c.304A>T | 4 | ep | Severe | Caucasian | [Saechao et al., 2010] |
| G89R | p.G120R | c.358G>C | 4 | ep | Severe | Thai, Japanese | [Liewluck et al., 2006; Cho et al., 2013] |
| G89S | p.G120S | c.358G>A | 4 | ep | Medium | Japanese | [Mori-Yoshimura et al., 2012; Cho et al., 2013] |
| R101C | p.R132C | c.394C>T | 4 | ep | Severe | Korean | [Park et al., 2012] |
| R101H | p.R132H | c.395G>A | 4 | ep | Medium | Japanese | [Cho et al., 2013] |
| I106T | p.I137T | c.410T>C | 4 | ep | Medium | Chinese | [Lu et al., 2011] |
| I128Ifs*6 | p.I159Ifs*6 | c.476insT | 4 | ep-NES | Severe | Japanese | [Mori-Yoshimura et al., 2012; Cho et al., 2013] |
| R129Q | p.R160Q | c.479G>A | 4 | ep-NES | Medium | Japanese | [Mori-Yoshimura et al., 2012] |
| R129* | p.R160* | c.478C>T | 4 | ep-NES | Severe | Indian | novel |
| H132Q | p.H163Q | c.489C>G | 4 | ep-NES | Medium | Japanese | [Nishino et al., 2002; Tomimitsu et al., 2002] |
| G135V | p.G166V | c.497G>T | 4 | ep-NES | Severe | English, Irish, USA | [Sparks et al., 2005] |
| G136R | p.G167R | c.501G>A | 4 | ep-NES | Severe | Japanese | [Cho et al., 2013] |
| I142T | p.I173T | c.518T>C | 4 | ep | Severe | Caucasian | [Saechao et al., 2010] |
| I150V | p.I181V | c.541A>G | 4 | ep | Medium | European | [No et al., 2013] |
| Y156H | p.Y187H | c.559T>C | 4 | ep | Medium | Indian | novel |
| H157fs | p.H188fs | ? | 4 | ep | Severe | Korean | [Sim et al., 2013] |
| R162C | p.R193C | c.515C>T | 4 | ep | Severe | Italian, Indian | [Del Bo et al., 2003; Nalini et al., 2013] |
| M171V | p.M202V | c.604A>G | 4 | ep | Severe | Italian | [Broccolini et al., 2002] |
| D176V | p.D207V | c.620A>T | 4 | ep | Medium | Chinese, Japanese, Korean | [Nishino et al., 2002; Tomimitsu et al., 2002, 2004; Motozaki et al., 2007; Li et al., 2011; Park et al., 2012] |
| R177C | p.R208C | c.622C>T | 4 | ep | Severe | Japanese | [Nishino et al., 2002; Cho et al., 2013] |
| I178N | p.I209N | c.626T>A | 4 | ep | Severe | Japanese | [Cho et al., 2013] |
| I178M | p.I209M | c.627C>G | 4 | ep | Medium | Japanese | [Cho et al., 2013] |
| L179F | p.L210F | c.628C>T | 4 | ep | Medium | Italian | [Grandis et al., 2010] |
| Y186C | p.Y217C | c.650A>G | 4 | ep | Severe | Pakistani | [No et al., 2013] |
| D187G | p.D218G | c.653A>G | 4 | ep | Severe | Japanese | [Mori-Yoshimura et al., 2012; Cho et al., 2013] |
| N194Tfs*4 | p.N225Tfs*4 | c.674delA | 4 | ep | Severe | Japanese | [Cho et al., 2013] |
| I200F | p.I231F | c.691A>T | 4 | ep | Medium | USA | [Eisenberg et al., 2003] |
| R202L | p.R233L | c.698G>T | 4 | ep | Medium | Greek | [Huizing et al., 2004] |
| W204* | p.W235* | c.705G>A | 4 | ep | Severe | Caucasian | [Saechao et al., 2010] |
| G206S | p.G237S | c.709G>A | 4 | ep | Medium | Italian | [Broccolini et al., 2004] |
| splicing | splicing | c.710-4A>G | in 4 | ep | Splicing? | Japanese | [Cho et al., 2013] |
| G206Vfs*3 | p.G237Vfs*3 | c.710delG | 5 | ep | Severe | Italian | [Broccolini et al., 2004] |
| D208N | p.D239N | c.715G>A | 5 | ep | Mild | Korean | [Sim et al., 2013] |
| D213V | p.D244V | c.731A>T | 5 | ep | Medium | Indian | novel |
| V216A | p.V247A | c.740T>C | 5 | ep | Severe | USA, German, Dutch | [ Vasconcelos et al., 2002; Huizing et al., 2004] |
| Q219K | p.Q250K | c.748C>A | 5 | ep | Medium | Japanese | [Cho et al., 2013] |
| D225N | p.D256N | c.766G>A | 5 | ep | Medium | Bahamas | [Eisenberg et al., 2001] |
| F233S | p.F264S | c.791T>C | 5 | ep | Medium | Japanese | [Mori-Yoshimura et al., 2012] |
| I241S | p.I272S | c.815T>G | 5 | ep | Medium | Chinese, Taiwanese | [Ro et al., 2005; Chu et al., 2007; Li et al., 2011; Lu et al., 2011] |
| R246W | p.R277W | c.829C>T | 5 | ep | Severe | Caucasian, Chinese, Japanese, Italian, USA | [Darvish et al., 2002; Ro et al., 2005; Sparks et al., 2005; Saechao et al., 2010; Stober et al., 2010; Li et al., 2011; Cho et al., 2013] |
| R246Q | p.R277Q | c.830G>A | 5 | ep | Mild | Bahamas, Italian, Taiwanese, Japanese | [Eisenberg et al., 2001; Broccolini et al., 2004; Chu et al., 2007; Saechao et al., 2010; Chai et al., 2011] |
| splicing | splicing | c.862+4A>G | in 5 | ep | Splicing | Japanese | [Nishino et al., 2002; Cho et al., 2013] |
| M261V | p.M292V | c.874A>G | 6 | ep-AR | Mild | Korean | [Park et al., 2012] |
| M261I | p.M292I | c.876G>? | 6 | ep-AR | Mild | Korean | [Sim et al., 2013] |
| M265T | p.M296T | c.887T>C | 6 | ep-AR | Medium | European | [No et al., 2013] |
| I270N | p.I301N | c.902T>A | 6 | ep-AR | Medium | Japanese | [Cho et al., 2013] |
| I270T | p.I301T | c.902T>C | 6 | ep-AR | Medium | Japanese | [Cho et al., 2013] |
| R277C | p.R308C | c.922C>T | 6 | ep-AR | Medium | French, Japanese | [Behin et al., 2008; Cho et al., 2013] |
| R277G | p.R308G | c.922C>G | 6 | ep-AR | Medium | Japanese | [Cho et al., 2013] |
| P283S | p.P314S | c.940C>T | 6 | ep-AR | Medium | Japanese | [Tomimitsu et al., 2004] |
| H293R | p.H324R | c.971A>G | 6 | ep-AR | Medium | Indian | [Kannan et al., 2012] |
| G295D | p.G326D | c.977G>A | 6 | ep-AR | Medium | Japanese | [Mori-Yoshimura et al., 2012] |
| G295R | p.G326R | c.976G>C | 6 | ep-AR | Medium | Japanese | [Cho et al., 2013] |
| M297T | p.M328T | c.983T>C | 6 | ep-AR | Medium | Indian | novel |
| I298T | p.I329T | c.986T>C | 6 | ep-AR | Severe | Asian, Chinese, Indian | [Saechao et al., 2010; Lu et al., 2011] |
| N300K | p.N331K | c.993C>A | 6 | ep-AR | Severe | Italian | [Tasca et al., 2012] |
| C303V | p.C334V | c.1000_1001TG>GT | 6 | ep-AR | Medium | Japanese | [Tomimitsu et al., 2002] |
| C303* | p.C334* | c.1002T>A | 6 | ep-AR | Severe | Indian | [Eisenberg et al., 2001] |
| G304R | p.G335R | c.1003G>A | 6 | ep | Severe | Indian | [Nalini et al., 2013] |
| R306Q | p.R337Q | c.1010G>A | 6 | ep | Medium | Japanese | [Nishino et al., 2002] |
| A310P | p.A341P | c.1021G>C | 6 | ep | Severe | Chinese | [Ro et al., 2005; Stober et al., 2010] |
| V315M | p.V346M | c.1036G>A | 6 | ep | Medium | European | [No et al., 2013] |
| N317D | p.N348D | c.1042A>G | 6 | ep | Severe | European | [No et al., 2013] |
| R321C | p.R352C | c.1054C>T | 6 | ep | Severe | Japanese | [Mori-Yoshimura et al., 2012] |
| splicing | splicing | c.1076-1delG | in 6 | ep | Splicing? | Japanese | [Cho et al., 2013] |
| V331A | p.V362A | c.1085T>C | 7 | ep | Severe | Japanese | [Nishino et al., 2002] |
| H333R | p.H364R | c.1091A>G | 7 | ep | Medium | Caucasian | [Weihl et al., 2011] |
| R335W | p.R366W | c.1096C>T | 7 | ep | Severe | Caucasian | [Fisher et al., 2006; Saechao et al., 2010] |
| L347del; H348N | p.L378del; p.H379N | c.1132_1134 del;c.1135C>A | 7 | ep | Severe | Caucasian | [Fisher et al., 2006] |
| L347P | p.L378P | c.1133T>C | 7 | ep | Severe | Japanese | [Cho et al., 2013] |
| splicing | splicing | c.1163+2dupT | in 7 | ep | Splicing | European, Italian | [Broccolini et al., 2004; No et al., 2013] |
| Y361* | p.Y392* | c.1176T>G | 8 | ep | Severe | Caucasian | [Weihl et al., 2011] |
| V367I | p.V398I | c.1192G>A | 8 | ep | Medium | Iranian | [Krause et al., 2003] |
| I377Tfs*15 | p.I408Tfs*15 | c.1223delT | 8 | ep | Severe | Italian | [Broccolini et al., 2004] |
| D378Y | p.D409Y | c.1225G>T | 8 | UF | Severe | European, Irish, Japanese, USA | [Nishino et al., 2002; Eisenberg et al., 2003; No et al., 2013] |
| L379H | p.L410H | c.1229T>A | 8 | UF | Severe | Tunisian | [Amouri et al., 2005] |
| P390S | p.P421S | c.1261C>T | 8 | UF | Medium | Korean | [Sim et al., 2013] |
| R420* | p.R451* | c.1351C>T | 8 | kin | Severe | Japanese, Indian | [Tomimitsu et al., 2004; Nalini et al., 2013] |
| V421A | p.V452A | c.1355T>C | 8 | kin | Medium | Japanese | [Tomimitsu et al., 2004; Cai et al., 2013] |
| K432Rfs*16 | p.K463Rfs*16 | c.1388delA | 9 | kin | Severe | Indian | [Voermans et al., 2010] |
| Y434C | p.Y465C | c.1394A>G | 9 | kin | Medium | Korean | [Sim et al., 2013] |
| Q436* | p.Q467* | c.1399C>T | 9 | kin | Severe | Taiwanese | [Saechao et al., 2010] |
| C453F | p.C484F | c.1451G>T | 9 | kin | Severe | Japanese | [Cho et al., 2013] |
| A460V | p.A491V | c.1472C>T | 9 | kin | Medium | Japanese | [Kayashima et al., 2002] |
| .L463P | p.L494P | c.1481T>C | 9 | ep | Severe | Korean | [Sim et al., 2013] |
| G469R | p.G500R | c.1498G>A | 9 | kin | Severe | Japanese | [Cho et al., 2013] |
| splicing | splicing | c.1504+5G>A | in 9 | kin | Splicing | Japanese | [Cho et al., 2013] |
| splicing | splicing | c.1505-4G>A | in 9 | kin | Splicing? | Japanese | [Cho et al., 2013] |
| I472T | p.I503T | c.1508T>C | 10 | kin | Severe | Japanese | [Nishino et al., 2002; Yabe et al., 2003] |
| W495* | p.W526* | c.1577G>A | 10 | kin | Severe | Caucasian | [No et al., 2013] |
| L508S | p.L539S | c.1616T>C | 10 | kin | Severe | Chinese | [Li et al., 2011; Lu et al., 2011] |
| H509Y | p.H540Y | c.1618C>T | 10 | kin | Medium | Chinese | [Lu et al., 2011] |
| P511H | p.P542H | c.1625C>A | 10 | kin | Severe | Japanese | [Motozaki et al., 2007] |
| P511L | p.P542L | c.1625C>T | 10 | kin | Severe | Thai | [Liewluck et al., 2006] |
| W513* | p.W544* | c.1632G>A | 10 | kin | Severe | Chinese, Taiwanese, Indian | [Ro et al., 2005; Li et al., 2011; Nalini et al., 2013] |
| V514del | p.V545del | c.1634_1637del | 10 | kin | Severe | Japanese | [Cho et al., 2013] |
| N519S | p.N550S | c.1649A>G | 10 | kin | Medium | Italian | [Broccolini et al., 2004] |
| A524V | p.A555V | c.1664C>T | 10 | kin | Severe | French, Mexican, Thai, Japanese | [Darvish et al., 2002; Liewluck et al., 2006; Behin et al., 2008; Cho et al., 2013] |
| F528C | p.F559C | c.1676T>G | 10 | kin | Severe | German | [Eisenberg et al., 2003] |
| G545Efs*11 | p.G576Efs*11 | c.1727delG | 11 | kin | Severe | Korean | [Park et al., 2012] |
| L556S | p.L587S | c.1760T>C | 11 | kin | Severe | Caucasian | [Saechao et al., 2010] |
| I557T | p.I588T | c.1763T>C | 11 | kin | Medium | Italian, Japanese | [Eisenberg et al., 2003; Tomimitsu et al., 2004] |
| G559R | p.G590R | c.1768G>C | 11 | kin | Severe | Japanese, Greek | [Huizing et al., 2004; Cho et al., 2013;] |
| G559A | p.G590A | c.1769G>C | 11 | kin | Severe | Turkish | novel |
| G568S | p.G599S | c.1795G>A | 11 | kin | Severe | Japanese | [Mori-Yoshimura et al., 2012] |
| G568V | p.G599V | c.1796G>T | 11 | kin | Severe | Indian | [Nalini et al., 2013] |
| V572del | p.V603del | c.1806_1808del | 11 | kin | Severe | Japanese | [Cho et al., 2013] |
| V572L | p.V603L | c.1807G>C | 11 | kin | Medium | Asian, Chinese, Japanese, Korean | [Kayashima et al., 2002; Tomimitsu et al., 2002; Kim et al., 2006; Li et al., 2011; Park et al., 2012] |
| G576E | p.G607E | c.1820G>A | 11 | kin | Severe | USA | [Eisenberg et al., 2001] |
| C579Y | p.C610Y | c.1829G>A | 11 | kin | Severe | Japanese | [Cho et al., 2013] |
| C581R | p.C612R | c.1834T>C | 11 | kin | Severe | Pakistani | novel |
| C586* | p.C617* | c.1850delG | 11 | kin | Severe | Japanese | [Mori-Yoshimura et al., 2012] |
| I587T | p.I618T | c.1853T>C | 11 | kin | Medium | Algerian, Chinese, Italian, Cajun, Japanese, Roma Gypsies | [Tomimitsu et al., 2002; Kalaydjieva et al., 2005; Behin et al., 2008; Grandis et al., 2010; Li et al., 2011; Cho et al., 2013] |
| I587N | p.I618N | c.1853T>A | 11 | kin | Medium | Japanese | [Cho et al., 2013] |
| A591T | p.A622T | c.1864G>A | 11 | kin | Severe | Chinese, Korean | [Kim et al., 2006; Lu et al., 2011] |
| A600E | p.A631E | c.1892C>A | 11 | kin | Severe | Japanese | [Mori-Yoshimura et al., 2012; Cho et al., 2013] |
| A600T | p.A631T | c.1891G>A | 11 | kin | Medium | Italian | [Broccolini et al., 2004] |
| L603F | p.L634F | c.1900C>T | 11 | kin | Medium | Japanese | [Mori-Yoshimura et al., 2012] |
| splicing | splicing | c.1909+5G>A | in 11 | kin | Splicing | Indian | [Boyden et al., 2011] |
| S615* | p.S646* | c.1937C>G | 12 | kin | Severe | Caucasian | [Saechao et al., 2010] |
| A630Lfs*12 | p.A661Lfs*12 | c.1980delA | 12 | kin | Severe | Japanese | [Cho et al., 2013] |
| A630T | p.A661T | c.1981G>A | 12 | kin | Medium | Japanese | [Nishino et al., 2002; Cho et al., 2013] |
| A631T | p.A662T | c.1984G>A | 12 | kin | Severe | Caucasian, Senegalese, USA | [Eisenberg et al., 2001; Behin et al., 2008; No et al., 2013] |
| A631V | p.A662V | c.1985C>T | 12 | kin | Severe | Caucasian, Korean, Chinese, German, Irish, S. African, USA, Japanese | [Nishino et al., 2002; Tomimitsu et al., 2002; Vasconcelos et al., 2002; Eisenberg et al., 2003; Saechao et al., 2010; Li et al., 2011; Weihl et al., 2011; Park et al., 2012] |
| N635K | p.N666K | c.1998T>A | 12 | kin | Severe | Japanese | [Cho et al., 2013] |
| N635K | p.N666K | c.1998T>G | 12 | kin | Severe | Japanese | [Cai et al., 2013] |
| A648V | p.A679V | c.2036C>T | 13 | kin | Medium | German | novel |
| I656N | p.I687N | c.2060T>A | 13 | kin | Severe | Thai | [Liewluck et al., 2006] |
| G669R | p.G700R | c.2098G>A | 13 | kin | Severe | Japanese | [Cho et al., 2013] |
| G669R | p.G700R | c.2098G>C | 13 | kin | Severe | Asian, Indian, Portuguese | [No et al., 2013] |
| Y675H | p.Y706H | c.2116T>C | 13 | kin | Medium | Caucasian | [Darvish et al., 2002; Saechao et al., 2010] |
| V679G | p.V710G | c.2129T>G | 13 | kin | Severe | French | [Behin et al., 2008] |
| V696M | p.V727M | c.2179G>A | 13 | kin | Medium | Algerian, Asian, Chinese, Middle-Eastern, Indian, Pakistani, Thai, Portuguese | [Eisenberg et al., 2001; Huizing et al., 2004; Liewluck et al., 2006; Behin et al., 2008; Saechao et al., 2010; Voermans et al., 2010; Boyden et al., 2011; Lu et al., 2011; No et al., 2013] |
| S699L | p.S730L | c.2189C>T | 13 | kin | Severe | Middle-Eastern | [No et al., 2013] |
| G708S | p.G739S | c.2215G>A | 13 | kin | Severe | Japanese | [Tomimitsu et al., 2004; Cho et al., 2013] |
| M712T | p.M743T | c.2228T>C | 13 | kin | Severe | Egyptian-Muslim, Persian Jewish, Japanese | [Eisenberg et al., 2001; Broccolini et al., 2002; Darvish et al., 2002; Noguchi et al., 2004; Tomimitsu et al., 2004; Amouri et al., 2005; Cho et al., 2013] |
| large deletion | large deletion | del ex2-ex10 (>35.7kb) | 2–10 | ep + kin | Severe | Italian | [Del Bo et al., 2003] |
Bold print: cDNA or protein truncating variants; Italic print + dark gray highlight: ‘Mild’ variants; ?: exact nomenclature could not be extracted from the reference. The DNA numbering system is based on cDNA sequence. Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
Amino acid substitutions are provided in the previously used hGNE1 (NP_005467.1) and in the preferred new hGNE2 (NP_001121699.1) nomenclature [Huizing et al. 2014b]. For some variants, updated nomenclature is provided extracted from the reference.
Nucleotide variants are provided in the mRNA variant 1 nomenclature (NM_001128227.2; longest mRNA spliceform; encoding hGNE2 protein).
Exon numbering according to genomic sequence (NC_000009.12) and as indicated in Figure 1. in = intron.
See text for details about GNE protein domains; ep = UDP-GlcNAc 2-epimerase domain; ep-NES = nuclear export signal; ep-AR: allosteric region; UF = unknown function; kin = ManNAc kinase domain; UF epimerase.
Combined pathogenicity scores, see Supp. Table S1. Intronic variants with predicted splicing effects are listed as “splicing’, and without such effects as “splicing?”, see Supp. Table S2.
Extracted from literature reference.
GNE myopathy typically presents in early adulthood with foot drop caused by weakness of the anterior tibialis muscles. The disease has progressive wasting of distal, then proximal skeletal muscles in the lower, then upper extremities and leads to marked disability within 10–20 years of initial symptoms, including wheelchair use and dependent care [Huizing et al., 2014a]. Severity and age of onset vary, even among siblings [Boyden et al., 2011; Mori-Yoshimura et al., 2012; Cho et al., 2013; Huizing et al., 2014a]. Relative “sparing” of the quadriceps occurs, although these muscles become affected at advanced stages of the disease; neck muscles can also be affected late in the disease [Argov and Yarom, 1984; Sivakumar and Dalakas, 1996]. In some cases, weakness of respiratory muscles appears as a manifestation of advanced stages of the disease [Mori-Yoshimura et al., 2012]. Additionally, dilated cardiomyopathy has been observed in a subset of patients [Chai et al., 2011], and a few cases with cardiac conduction abnormalities and sudden death have been reported [Kimpara et al., 1993; Nishino et al., 2005]. Histopathology of GNE myopathy muscle biopsies typically shows rimmed vacuoles and characteristic filamentous inclusions, generally without signs of inflammation [Yunis and Samaha, 1971; Nonaka et al., 1981; Griggs et al., 1995]. Specialized lectin or antibody staining of affected muscle shows decreased sialylation, presumed to be involved in the pathophysiology of the disease [Noguchi et al., 2004; Tajima et al., 2005; Malicdan et al., 2009; Nemunaitis et al., 2011].
The human GNE gene (Gene ID: 10020; NC_000009) encodes a bifunctional enzyme, uridine diphosphate (UDP)-N-acetylglucosamine (GlcNAc) 2-epimerase/N-acetylmannosamine (ManNAc) kinase (GNE) [Hinderlich et al., 1997; Eisenberg et al., 2001]. The GNE enzyme catalyzes the first two committed, rate-limiting steps in the biosynthesis of 5-N-acetylneuraminic acid (Neu5Ac). In mammals, the end product of sialic acid synthesis, CMP-Neu5Ac, feedback-inhibits the UDP-GlcNAc 2-epimerase activity of GNE by binding to its allosteric site [Kornfeld et al., 1964]. Neu5Ac (henceforth referred to as ‘sialic acid’) is the most abundant mammalian sialic acid and the precursor of most other sialic acids [Varki, 1992]. Sialic acids are typically found as the terminal sugars on glycoproteins and glycolipids, where they function in a variety of cellular signaling pathways [Varki, 1997].
Two distinct human disorders, sialuria (MIM #269921) and GNE myopathy are associated with predominantly missense mutations in GNE. Sialuria is an autosomal dominant disorder characterized by coarse facies, variable developmental delay, hepatomegaly and recurrent infections. To date, only seven sialuria patients are described worldwide. All patients have a heterozygous missense mutation affecting the allosteric site of GNE, leading to loss of feedback-inhibition of GNE-epimerase activity by CMP-sialic acid, resulting in excessive sialic acid production. Other publications describe the details of sialuria and its GNE sequence variants [Seppala et al., 1999; Enns et al., 2001; Leroy et al., 2001; Hinderlich et al., 2013].
GNE myopathy patients have non-allosteric, bi-allelic, predominantly missense mutations in GNE. These mutations result in reduced GNE epimerase and kinase enzymatic activities, which may lead to decreased sialic acid production. Therefore, impaired sialylation appears to be a main contributor to the still elusive disease pathology [Noguchi et al., 2004; Sparks et al., 2005; Malicdan et al., 2007; Malicdan et al., 2009]. While overall sialylation of tissues appears to be normal in GNE myopathy, specific circulating proteins and/or specific muscle glycoproteins or glycolipids appear hyposialylated [Huizing et al., 2004; Tajima et al., 2005; Ricci et al., 2006; Broccolini et al., 2008; Patzel et al., 2013].
The GNE enzyme is a complex, soluble, highly conserved protein whose enzymatic activities and gene expression appear to be highly regulated. The GNE protein localizes to the cytoplasm, the Golgi-region and the cell nucleus [Krause et al., 2005]. Eight human GNE (hGNE) isoforms 1–8 are reported; hGNE1 is the major, ubiquitously expressed isoform, while hGNE2-8 are differentially expressed and may act as tissue-specific regulators of sialylation [Reinke et al., 2009a, 2009b; Yardeni et al., 2011]. GNE can homodimerize to oligomeric structures with different enzymatic activities; expression is influenced by GNE ligands [Ghaderi et al., 2007; Reinke et al., 2009b]. GNE gene transcription is regulated by CpG promotor methylation [Oetke et al., 2003]. GNE mRNA is highest in liver and placenta, while skeletal muscle has low expression [Lucka et al., 1999; Yardeni et al., 2011].
Here we provide an overview of all reported GNE sequence variants associated with GNE myopathy to date, as well as novel variants identified in our NIH patient cohort. We also provide an overview of reported GNE enzyme activities for specific mutations, address genotype-phenotype correlations, and update the estimated prevalence of the disease based on available next-generation sequencing data.
GNE Mutation Nomenclature
The eight different splice variants of the GNE mRNA encode (at least theoretically) for 8 different protein isoforms [Reinke et al., 2009a; Yardeni et al., 2011]. The originally described GNE protein is now called hGNE1 (illogically, encoded by GNE mRNA transcript variant 2, NM_005476) which covers 722 amino acids [Eisenberg et al., 2001]. The hGNE2 isoform is encoded by the longest GNE mRNA transcript (variant 1, NM_001128227) and its protein product contains an additional 31 amino acids at the N-terminus. Human isoforms hGNE3-8 are encoded by shorter mRNA splice variants, and it is unclear whether these isoforms are expressed as proteins; if so, they likely lack either epimerase (hGNE3,6,7,8) or kinase (hGNE4) enzymatic activities due to partial (splice) deletions within these respective domains [Yardeni et al., 2011].
The discovery of the additional N-terminal sequence (and novel exon 1) encoding hGNE2, is potentially confusing since all previous (before 2011) molecular and biochemical studies (including all mutation reports) refer to the hGNE1 isoform, while according to universally adapted gene/protein nomenclature rules the longest mRNA splice form ought to be used for annotating mutations and functional domains (http://www.hgvs.org/mutnomen/refseq.html). Hence, amino acid numbering of previously reported GNE studies, including patient mutation reports, should be supplemented with 31 amino acids to adhere to the current (hGNE2) nomenclature guidelines [Huizing et al., 2014b]. While adaptation to the hGNE2 nomenclature can initially be confusing to the clinicians and patients, we strongly support the adaptation to this nomenclature. Laboratories/researchers not familiar with the GNE myopathy field and disease/gene history will report patients’ mutations and biochemical research tools (mutation reports, antibodies, enzyme activities, siRNA, etc.) according to current universally adapted nomenclature rules. In addition, although there are no variants reported yet in the additional 31 amino acids of hGNE2 (perhaps because this region has not been considered for mutation analysis in many patients), future variants in this region could not be accurately named using hGNE1 as a reference. To accommodate the adaption to this new terminology, we list both hGNE1 (GenBank accession numbers: Protein: NP_005467.1; mRNA: NM_005476.5) and hGNE2 (Protein: NP_001121699.1; mRNA: NM_001128227.2) nomenclatures in this manuscript in all Tables listing GNE sequence variants.
Mutation Spectrum
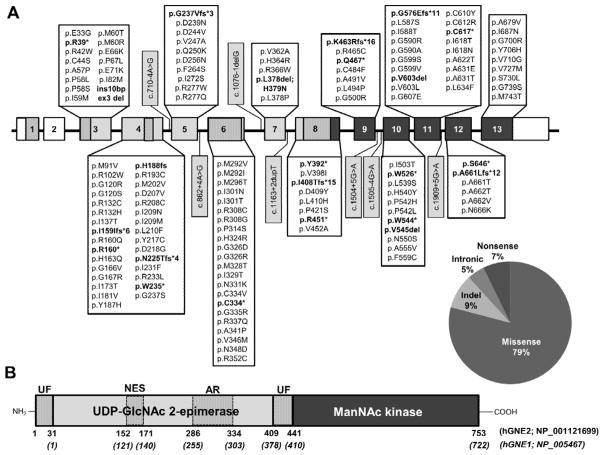
As of January 2014, there were 147 GNE sequence variants reported in the literature associated with GNE myopathy. We identified another 7 variants in our NIH GNE myopathy cohort (Table 1, Fig. 1A). All these variants have been deposited in the online GNE variation database (http://www.dmd.nl/nmdb2/home.php?select_db=GNE). The large majority of GNE sequence variants associated with GNE myopathy results in missense protein variants (122 variants, 79%). The other sequence variants included 11 (7%) nonsense variants, 7 (5%) intronic splice variants, and 14 (9%) insertion/deletions (indel). The variants are scattered throughout the GNE coding region in both the epimerase and the kinase domains of GNE. Note that there are no variants reported in the N-terminal exon 1 (specific for GNE2) and exon 2 (untranslated region specific for the hGNE1 isoform) (Fig. 1B). GNE null mutations have never been identified on both alleles; this would most likely be lethal, due to the critical role of sialic acid in early development, supported by embryonic lethality of Gne ‘knock-out’ mice [Schwarzkopf et al., 2002].
Figure 1. Schematic illustration (not to scale) of the GNE gene and protein and location of sequence variants associated with GNE myopathy.
A: Exon (boxes)-intron (lines) structure of the human GNE gene. White boxes = untranslated region; light gray boxes = encoding UDP-GlcNAc 2-epimerase enzymatic activity; dark gray boxes = encoding ManNAc kinase enzymatic activity; vertical striped boxes = protein domains indicated in (B). Locations and characteristics of all reported human GNE variants associated with GNE myopathy (as of January 2014) are indicated. Variant nomenclature is according to the longest mRNA splice variant (Variant 1; NM_001128227.2) and its translated protein isoform hGNE2 (NP_001121699.1). Exon numbering is according to the GNE genomic DNA sequence (NC_000009.12). Truncating nonsense and indel variants are printed in bold, intronic variants in gray highlight. The large deletion variant del ex 2–10 (>35.7kb) is not displayed. The pie chart visualizes the overall distribution of variants. B: Protein structure of the hGNE2 isoform. Functional domains are indicated as vertical striped regions: UF, unknown function; NES, putative nuclear export signal (NES), AR, experimental allosteric region. Amino acid numbers of hGNE2 and hGNE1 are indicated below the structure. Note that amino acids 1-31 are only present in hGNE2. All other amino acids and protein domains are similar in hGNE1 and hGNE2.
There are a few ethnic GNE founder mutations reported. The first described variant, p.M743T (originally M712T in hGNE1 nomenclature) is predominantly found in patients of Middle Eastern origin [Eisenberg et al., 2001, 2003], p.C44S, p.D207V, and p.V603L (originally reported as C13S, D176V, and V572L in hGNE1) are common in Japanese patients [Nishino et al., 2002; Tomimitsu et al., 2002; Cho et al., 2013], and p.I618T (originally I587T) is found in Roma Gypsies [Kalaydjieva et al., 2005]. Of note, the p.M743T variant has also been reported in some non-Middle Eastern patients [Broccolini et al., 2002; Tomimitsu et al., 2004; Amouri et al., 2005; Grandis et al., 2010; Cho et al., 2013], p.C44S, p.D207V and p.V603L are also described in non-Japanese Asian patients [Kim et al., 2006; Li et al., 2011; Park et al., 2012] and p.I618T is also reported in non-Gypsies [Eisenberg et al., 2003; Grandis et al., 2010; Li et al., 2011; Park et al., 2012]. Some variants are reported more frequently than others (Table 1), including variants frequently found in Asian subjects (p.I272S, p.I329T, p.V727M,) and others frequently reported in subjects of different ethnic origins (p.R277W, p.R277Q, p.D409Y, p.A555V, p.I618T, p.A662V).
Functional Prediction of Variants
There are 123 GNE cDNA sequence variants associated with GNE myopathy leading to 121 missense protein variants (p.N666K and p.G700R can each result from two different cDNA variants; p.H348N results from an indel sequence variant (p.L378del; p.H379N)). The predicted effect of each missense variant on protein function was analyzed using the prediction software programs PolyPhen2 (Polymorphism Phenotyping v2; http://genetics.bwh.harvard.edu/pph2/) [Adzhubei et al., 2010], SIFT (Sort Intolerant From Tolerant human Protein; http://sift.jcvi.org/) [Ng and Henikoff, 2003], Align-GVGD (http://agvgd.iarc.fr/agvgd_input.php) [Tavtigian et al., 2005; Mathe et al., 2006] and PMut (http://mmb2.pcb.ub.es:8080/PMut/) [Ferrer-Costa et al., 2004]. Based on the combined predicted values, we assigned an overall severity prediction score to each variant (Table 1, Supp. Table S1). Please note that these are predicted values of impairment of protein function only, not based on cellular data. Of the 121 missense protein variants, combined severity prediction scores of 55 variants were ‘severe’ (protein damaging) and 60 variants were ‘medium’ (probably protein damaging). We propose that these ‘severe’ and ‘medium’ severity scores can explain the protein damaging capacity of these 115 variants. Two of these missense variants occur at splice junctions: c.709G>A (p.G237S) appears to decrease splice site probability slightly (100% for wild type and 94% for variant sequence) and c.1508T>C (p.I503T) does not influence splice site probability predictions (Supp. Table S2). Tertiary structure predictions have localized many of these variants to essential parts of GNE domains [Penner et al., 2006; Kurochkina et al., 2010]. Our severity prediction analysis predicted only 6 missense variants to be ‘mild’ (tolerated, benign, likely not protein damaging) (p.A57P, p.M91V, p.D239N, p.R277Q, p.M292V, p.M292I). Interestingly, all 6 mild variants localize to the epimerase encoding domain. Note that three of these variants were assigned medium severity prediction scores by one (p.A57P, p.M292V) or two prediction programs (p.R277Q), and may therefore be considered as ‘mild- medium’ severe. Tertiary structure predictions localize p.A57P in an important structural domain, the UDP-binding site [Kurochkina et al., 2010; Yardeni et al. 2011]. Variant p.D239N (c.715G>A) occurs at a splice junction (ex 4/5), but only slightly changes splice site prediction probability (31% for wild type to 30% for variant sequence; Supp. Table S2). None of the other mild variants change prediction scores for nearby splice junctions, and they are not predicted to create new splice junctions (data not shown). Effects on GNE enzyme activities have not been assessed for any of these mild missense variants (Table 2).
Table 2.
Overview of reported GNE enzyme activities related to GNE protein variants
| Allele 11 | Allele 21 | Domain2 | Cell system | GNE-ep3 | GNE-kin3 | References | ||
|---|---|---|---|---|---|---|---|---|
| hGNE1 | hGNE2 | hGNE1 | hGNE2 | |||||
| GNE myopathy patient cells | ||||||||
| G135V | p.G166V | R246W | p.R277W | ep-NES/ep | fibroblasts | 38 % | 72% | [Sparks et al., 2005] |
| V367I | p.V398I | V367I | p.V398I | ep/ep | myoblasts | 40 % | ND | [Salama et al., 2005] |
| D176V | p.D207V | D176V | p.D207V | ep/ep | lymphoblasts | <10% | ND | [Nishino et al., 2002] |
| D176V | p.D207V | V331A | p.V362A | ep/ep | lymphoblasts | <10% | ND | [Nishino et al., 2002] |
| R11W | p.R42W | F537I | p.F568I | ep/kin | myoblasts | 43 % | ND | [Salama et al., 2005] |
| D176V | p.D207V | I472T | p.I503T | ep/kin | lymphoblasts | <10% | ND | [Nishino et al., 2002] |
| D176V | p.D207V | V572L | p.V603L | ep/kin | lymphoblasts | <10% | ND | [Nishino et al., 2002] |
| D176V | p.D207V | A630T | p.A661T | ep/kin | lymphoblasts | <10% | ND | [Nishino et al., 2002] |
| V216A | p.V247A | A631V | p.A662V | ep/kin | fibroblasts | 48 % | 63% | [Sparks et al., 2005] |
| V572L | p.V603L | V572L | p.V603L | kin/kin | lymphoblasts | <10% | ND | [Nishino et al., 2002] |
| M712T | p.M743T | M712T | p.M743T | kin/kin | fibroblasts | 83 % | 55% | [Sparks et al., 2005] |
| M712T | p.M743T | M712T | p.M743T | kin/kin | myoblasts | 75 %4 | ND | [Salama et al., 2005] |
| M712T | p.M743T | M712T | p.M743T | kin/kin | lymphoblasts | 65 %5 | ND | [Hinderlich et al., 2004] |
| In vitro recombinant enzyme - cell expression systems | ||||||||
| R11W | p.R42W | - | - | ep | Sf9 cells | 15 % | 40 % | [Penner et al., 2006] |
| C13S | p.C44S | - | - | ep | COS cells | 20% | 100% | [Noguchi et al., 2004] |
| H132Q | p.H163Q | - | - | ep-NES | COS cells | 5 % | 50 % | [Noguchi et al., 2004] |
| D176V | p.D207V | - | - | ep | COS cells | 18 % | 87 % | [Noguchi et al., 2004] |
| R177C | p.R208C | - | - | ep | COS cells | 10 % | 78 % | [Noguchi et al., 2004] |
| I200F | p.I231F | - | - | ep | Sf9 cells | 90 % | 75 % | [Penner et al., 2006] |
| C303V | p.C334V | - | - | ep-AR | Sf9 cells | 80 % | 60 % | [Penner et al., 2006] |
| C303* | p.C334* | - | - | ep-AR | Sf9 cells | 0 % | 0 % | [Penner et al., 2006] |
| V331A | p.V362A | - | - | ep | COS cells | 16 % | 114 % | [Noguchi et al., 2004] |
| D378Y | p.D409Y | - | - | ep | Sf9 cells | 30 % | 45 % | [Penner et al., 2006] |
| D378Y | p.D409Y | - | - | ep | COS cells | 10% | 100% | [Noguchi et al., 2004] |
| I472T | p.I503T | - | - | kin | COS cells | 48 % | 5 % | [Noguchi et al., 2004] |
| N519S | p.N550S | - | - | kin | Sf9 cells | 40 % | 20 % | [Penner et al., 2006] |
| A524V | p.A555V | - | - | kin | COS cells | 5% | 30 % | [Noguchi et al., 2004] |
| F528C | p.F559C | - | - | kin | Sf9 cells | 70 % | 35 % | [Penner et al., 2006] |
| F537I | p.F568I | - | - | kin | Sf9 cells | 45 % | 60 % | [Penner et al., 2006] |
| V572L | p.V603L | - | - | kin | COS cells | 68 % | 8 % | [Noguchi et al., 2004] |
| G576E | p.G607E | - | - | kin | Sf9 cells | 15 % | 15 % | [Penner et al., 2006] |
| I587T | p.I618T | - | - | kin | Sf9 cells | 55 % | 35 % | [Penner et al., 2006] |
| A630T | p.A661T | - | - | kin | COS cells | 80 % | 40 % | [Noguchi et al., 2004] |
| A631T | p.A662T | - | - | kin | Sf9 cells | 80 % | 75 % | [Penner et al., 2006] |
| A631V | p.A662V | - | - | kin | Sf9 cells | 70 % | 65 % | [Penner et al., 2006] |
| A631V | p.A662V | - | - | kin | COS cells | 75 % | 5 % | [Noguchi et al., 2004] |
| G708S | p.G739S | - | - | kin | COS cells | 48 % | 5 % | [Noguchi et al., 2004] |
| M712T | p.M743T | - | - | kin | Sf9 cells | 100% | 70% | [Hinderlich et al., 2004] |
| Cell free system | ||||||||
| G135V | p.G166V | - | - | ep-NES | cell free | 0.6 % | 7 % | [Sparks et al., 2005] |
| V216A | p.V247A | - | - | ep | cell free | 2 % | 19 % | [Sparks et al., 2005] |
| R246W | p.R277W | - | - | ep | cell free | 0.5 % | 17 % | [Sparks et al., 2005] |
| A631T | p.A662V | - | - | kin | cell free | 3 % | 12 % | [Sparks et al., 2005] |
| M712T | p.M743T | - | - | kin | cell free | 3 % | 8 % | [Sparks et al., 2005] |
Amino acid substitutions are provided in the previously used hGNE1 (NP_005467.1; encoded by mRNA NM_005476.5) and in the preferred new hGNE2 (NP_001121699.1; encoded by mRNA NM_001128227.2) nomenclature [Huizing et al. 2014b]. The numbering system is based on cDNA sequence; nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
Domains correspond to GNE protein domains of Table 1 and Fig. 1. ep = UDP-GlcNAc 2-epimerase domain; ep-NES = nuclear export signal; ep-AR: allosteric region; kin = ManNAc kinase domain.
All enzyme activities are expressed as percentage of wild type activity.
Combined average activity of myoblast cell lines from 9 patients homozygous for M712T (hGNE1 nomenclature).
Combined average of lymphoblast cell lines from 6 patients homozygous for M712T (hGNE1 nomenclature).
Apart from the large group of missense variants, there are 11 nonsense variants reported associated with GNE myopathy (Table 1, Fig. 1). These variants are distributed throughout the entire coding region. It is likely that the alleles containing the nonsense variants undergo nonsense mediated RNA decay and yield no protein product. As mentioned earlier, no nonsense or severely truncating variants are identified on both alleles in GNE myopathy patients.
There are 14 indel sequence variants associated with GNE myopathy. Six of these variants were indels of a single nucleotide, resulting in a frame shift and early termination codon (p.I159Ifs*6, p.H188fs, p.N225Tfs*4, p.G237Vfs*3, p.I408Tfs*15, p.K463Rfs*16, p.G576Efs*11, p.A661Lfs*12). Of interest is that p.G237Vfs*3 (c.710delG) occurs at the exon 4/5 splice junction and splice site prediction software predicts a significant reduction in splice site probability (100% for wild type to 12% for variant; Supp. Table S2). In contrast, p.G576Efs*11 (c.1727delG) also occurs at a splice junction (exon 10/11) but does not appear to decrease splice site probability predictions (Supp. Table S2). Two indels result in in-frame deletion of 1 amino acid (c.1634_1637del; p.V545del and c.1806_1808del; p.V603del), these do not appear to influence splice site prediction probability (data not shown). However, both occur in structurally important sites of the kinase domain: p.V545del is in the vicinity of the site of conformational change upon substrate binding [Kurochkina et al., 2010] and p.V603del is located in the connection region between ROK motifs 1 and 2, which is the predicted vicinity of zinc and substrate binding [Kurochkina et al., 2010] and is also predicted to be the region the GNE dimerization interface [Penner et al., 2006]. One indel results in an in-frame deletion of one amino acid followed by a missense variant (p.L378del; p.H379N). While there are no structural data on p.L378 [Kurochkina et al., 2010], the missense p.H379N is predicted to have ‘medium’ severity (Supp. Table S1). The additional 3 indels are larger insertions or deletions for which the exact sequence data were not provided and/or not retrievable, including insertion of 10-bp in exon 3 [Nishino et al., 2002], exon 3 deletion [Cho et al., 2013], and a large (>35.7 kb) deletion spanning exons 2–10 [Del Bo et al., 2003]. Since these 3 variants delete large regions of GNE and/or are out of frame and likely resulting in an early termination codon and/or nonsense-mediated decay, we assigned them as ‘severe’.
There are 7 intronic variants reported, presumed to affect splicing. In Supp. Table S2 we summarize for each variant whether an experimental splice effect was described, and severity was analyzed in silico with the “Human Splice Site Prediction by Neural Network” program (http://www.fruitfly.org/seq_tools/splice.html) [Reese et al., 1997]. Out of the 7 intronic variants, 3 were experimentally shown to cause missplicing (c.862+4A>G, c.1163+2dupT, c.1909+5G>A) and also in silico predicted to reduce splicing probability compared to the wild type sequence. Of the other variants, c.710-4A>G decreased splice site probability while c.1076-1delG and c.1505-4G>A did not show a decreased probability in silico and need follow-up by experimental testing before assigning pathologic scores to them.
Biological significance: The GNE protein
The functional domains comprising the GNE protein including the location of all identified GNE myopathy-related variants are illustrated in Fig. 1. No sequence variants are located in amino acids 1-31, which comprises an area of unknown function (UF) unique to the hGNE2 isoform. The other GNE region of unknown function (amino acids 409-441, hGNE2 nomenclature) contains 3 missense variants associated with GNE myopathy. Six variants, 4 missense and 2 truncating variants, are reported located in the putative nuclear export signal (ep-NES, amino acids 152-171), of which the missense variants may play a role in nuclear localization of the GNE protein [Krause et al., 2005]. There are no GNE myopathy-associated variants in the same amino acids as those identified to cause sialuria (hGNE2 p.R294 or p.R297, hGNE2 nomenclature) [Seppala et al., 1999; Leroy et al., 2001]. However, 16 variants (15 missense, 1 nonsense) localize to the ‘experimental’ allosteric region (AR; amino acids 286 and 334) which was defined by in-vitro created sialuria-like mutants in cell culture [Yarema et al., 2001] and in-vitro site-directed mutagenesis studies [Penner et al., 2006]. It is still unclear if the allosteric site forms a discrete subdomain or only certain amino acids, integrated in the UDP-GlcNAc 2-epimerase structure, accommodate CMP-sialic acid binding [Hinderlich et al., 2013]. The 15 GNE myopathy associated missense variants in the experimental allosteric region need further research regarding their effect on allosteric feedback inhibition of CMP-sialic acid.
For a large number of GNE variants, tertiary structure predictions are described, based on the crystal structure of ManNAc kinase and sequence comparisons with other enzymes of homologous functions [Penner et al., 2006; Tong et al., 2009; Kurochkina et al., 2010; Martinez et al., 2012]. The effects of selected GNE variants on UDP-GlcNAc 2-epimerase (GNE-ep) and ManNAc kinase (GNE-kin) enzymatic activities in different cellular and cell free systems have been determined, and are summarized in Table 2. First, it was shown that the physical separation of the two GNE enzymatic domains results in enzymes with remaining, but severely decreased, activity [Blume et al., 2004]. Second, it was demonstrated that GNE myopathy-associated missense variants caused reduced, but never absent, enzymatic activities [Hinderlich et al., 2004; Noguchi et al., 2004; Salama et al., 2005; Sparks et al., 2005; Penner et al., 2006]. Third, sequence variants in one enzymatic domain affect not only that domain’s enzyme activity, but also the activity of the other domain [Sparks et al., 2005]. In addition, compared with enzyme activities in a cell-free system, fibroblasts exhibited higher residual activities of both UDP-GlcNAc 2-epimerase and ManNAc kinase, suggesting the presence of additional sugar epimerases and kinases with overlapping substrate specificity [Sparks et al., 2005]. In particular, GlcNAc kinase has a high intrinsic ManNAc kinase activity [Hinderlich et al., 2001].
Clinical Significance: Genotype-Phenotype correlations
Genotype-phenotype correlations for subjects with GNE myopathy are difficult to study because of a lack of systematic natural history data, partly due to the rare nature of the disease, and the heterogeneity of GNE sequence variants. Patients with the same genetic variants can present with variable phenotypes, even within families with multiple affected siblings [Sivakumar and Dalakas, 1996; Ikeuchi et al., 1997; Boyden et al., 2011; Huizing et al., 2014a]. In addition, a few apparently healthy individuals with distinct, biallelic disease-causing (in other individuals) variants in the GNE gene have been identified, indicating incomplete penetrance of the disease [Nishino et al., 2002; Argov et al., 2003]. Variation in age of onset and severity of symptoms among patients with the same mutation, suggest there are epigenetic or environmental factors that contribute significantly to the phenotype. Recent studies in Japanese cohorts suggest that patients homozygous for the p.V603L (originally V572L in hGNE1 nomenclature) GNE kinase domain variant (frequent variant in Japanese population) result in more severe phenotypes with earlier onset and faster progression of the disease. Another common variant in the Japanese population, p.D207V (originally D176V in hGNE1 nomenclature), appears to be a mild variant with relatively late onset of symptoms [Mori-Yoshimura et al., 2012; Cho et al., 2013]. The authors noted that despite the high p.D207V allele frequency the Japanese population, relatively few homozygous patients for this variant are identified, suggesting that homozygotes may not develop an apparent disease [Mori-Yoshimura et al., 2012; Cho et al., 2013].
Prevalence of GNE myopathy and GNE polymorphisms
The worldwide estimated prevalence of GNE myopathy is currently estimated at 1–9/1,000,000 (Orphanet; http://www.orpha.net/). Based on frequency data of the p.M743T (M712T in hGNE1 nomenclature) founder mutation, the prevalence of GNE myopathy in the Persian Jewish community was estimated to be 1:1500 [Argov et al., 1998; Eisenberg et al., 2001].
We examined the occurrence of GNE variants by using data from three exome sequence databases (accessed January 2014), including 1000 Genomes (http://www.1000genomes.org/) [Abecasis et al., 2012], NHLBI GO Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/), and NIH-UDP from the National Institutes of Health (NIH) Undiagnosed Disease Program (UDP) (http://www.genome.gov/27544402) [Gahl and Tifft, 2011]. We assume these databases represent the “general population” diversity. The data are summarized in Table 3 and Supp. Table S3.
Table 3.
Estimated carrier rates and prevalence of GNE myopathy1
| 1000 Genomes | ESP | NIH-UDP | Total | |
|---|---|---|---|---|
| GNE variant/total alleles2 | 10/2184 | 27/13006 | 4/1434 | 41/16624 |
| GNE variant allele frequency | 1/218 | 1/482 | 1/360 | 1/406 |
| Carrier rate (heterozygotes) | 1/109 | 1/241 | 1/180 | 1/203 |
| Predicted Prevalence | 1/47710 | 1/232019 | 1/128522 | 1/164474 |
| Affected per million | 21 | 4 | 8 | 6 |
Calculated according to the Hardy-Weinberg principle. See Supp. Tables S3 for details.
Total alleles containing GNE variants, not including the 3 common GNE SNPs (rs35224402/p.D239E, rs35638832/p.I454V, and rs121908627/p.V727L).
Most GNE variants in the exome sequence databases occurred with very low frequency (mostly only on 1 allele) in the datasets. Both previously identified variants associated with GNE myopathy and novel variants were present (Supp. Table S3). Unexpectedly, there were 3 missense variants/nonsynonymous single nucleotide polymorphisms (SNPs) that occurred with high frequency in all three databases: rs35224402 (c.717T>G; p.D239E), rs35638832 (c.1360A>G; p.I454V) and rs121908627 (c.2179G>T; p.V727L) (all reported in hGNE2 nomenclature). p.D239E even occurred homozygous in one individual (ESP database). Interestingly, these three relatively frequent variants are not reported in any GNE myopathy patients, and importantly, all three are predicted to have a non-damaging/mildly severe effect on GNE protein function (Supp. Table 1). Therefore, we consider them as likely non-disease causing SNPs. These novel SNP data should be considered during future interpretations of GNE myopathy sequence data.
We calculated the prevalence of GNE myopathy according to the Hardy-Weinberg principle of population genetics (p2 + 2pq + q2 = 1; Supp. Table S3, Table 3) [Hardy, 1908; Weinberg, 1908]. To avoid over-estimating GNE variant allele frequencies, we deleted the three frequent variants (likely SNPs) from our calculations. The individual databases predicted the prevalence of GNE myopathy to be 4/1,000,000 (ESP database; based on 13,006 total alleles), 8/1,000,000 (NIH-UDP database; based on 1434 total alleles) and 21/1,000,000 (1000 genomes; based on 2,184 total alleles), considerably higher estimations than originally assumed.
When we combine data of all 3 databases (Table 3), the average variant allele frequency is ~1/406 alleles (ranging between 1/218 and 1/482 alleles), which converts to a carrier (heterozygote) rate of ~ 1/203 individuals (ranging between 1/109 and 1/241). This yields a predicted prevalence of GNE myopathy of ~1/164,474 individuals, converting to approximately 6/1,000,000. When assuming the worldwide (very conservatively estimated) prevalence of GNE myopathy to be 6/1,000,000, this translates to the existence of at least 40,000 GNE myopathy patients worldwide, including ~ 13,000 in Asia (~750 in Japan), ~4000 in Europe and ~ 3000 in North America, even without considering ethnic founder variants. These numbers are dramatically higher than the number of diagnosed/reported GNE myopathy patients so far (only ~800 total reported patients, including founder populations, as per Jan 2014). These numbers confirm suspicions that many patients may go undiagnosed.
Discussion and Future Prospects
There are no approved therapies currently available for GNE myopathy. Past and present clinical trials are based on replacing the defective GNE enzyme (GNE gene therapy) [Nemunaitis et al., 2011], supplementation of precursors or end products of the sialic acid biosynthetic pathway (including supplementation of sialic acid itself), providing intravenous immunoglobulin G (IVIG) as a source of sialic acid, or oral administration of the neutral sialic acid precursor ManNAc (http://clinicaltrials.gov/ identifiers: NCT00195637, NCT01236898, NCT01517880, NCT01830972, NCT01359319, NCT01634750) [Sparks et al., 2007; Huizing et al., 2014a].
To support these ongoing trials, accurate and efficient identification of GNE myopathy patients as well as documenting an accurate course of the disease for each individual patient, are essential. With the initiation of natural history studies [Mori-Yoshimura et al., 2012; Cho et al., 2013; Huizing et al., 2014a] and (http://clinicaltrials.gov/ identifiers: NCT01417533, NCT01784679) it became evident that most GNE myopathy patients experienced a significant delay in diagnosis (around 10 years) after onset of initial symptoms. This is due to the rare nature of the disease and the lack of a conclusive, inexpensive and noninvasive diagnostic test. A significantly delayed diagnosis not only causes emotional hardship for the patient but also delays proper management of the disease and may influence eligibility to enroll in clinical trials and/or response to therapy.
Currently, the diagnosis of GNE myopathy relies upon the presence of muscle weakness, muscle pathology, and, ultimately, the presence of GNE gene mutations. However, non-specific muscle weakness may not directly point to GNE myopathy; histopathology of muscle biopsies may be negative; and GNE mutation analysis is not commonly performed and/or readily available [Huizing et al., 2014a]. The diagnosis of GNE myopathy should be considered in any patient presenting in early adulthood with distal muscle weakness of the lower extremities. The diagnosis should not be delayed until the characteristic clinical finding, sparing of the quadriceps, becomes evident since that occurs late in the disease. In addition, serum creatine phosphokinase (CPK) levels are variable and nerve conduction studies and electromyograms are nonspecific and unhelpful in obtaining a specific diagnosis. With the lack of disease-specific (blood-based) biomarkers, we strongly advocate for early genetic testing since bi-allelic disease-causing mutations in the GNE gene ultimately confirm the diagnosis. Identification of carriers, especially in populations that have higher prevalence of the disease, can assist genetic counseling. Additionally, with the rapid progression of therapeutic trials, genetic newborn screening tests should be considered in the near future.
In this report, we provide an extensive overview of all 154 reported GNE sequence variants associated with GNE myopathy to date (January 2014), which lead to predominantly missense GNE protein variants. Importantly, a combination of prediction programs for severity of missense variants on protein function (PolyPhen, SIFT, Align, and PMut) and tertiary structure reports [Penner et al., 2006; Kurochkina et al., 2010;] predicted that 118 of the 121 reported GNE missense protein variants have deleterious effects on GNE protein function. We also recognized three nonsynonymous SNPs in the GNE gene (rs35224402/p.D239E, rs35638832/p.I454V, and rs121908627/p.V727L) which should be taken into account with GNE myopathy sequence data analysis. Deleterious GNE proteins lead to decreased GNE enzyme activities and underlie GNE myopathy. At this point there are limited or absent correlations between severity of the GNE sequence variants and severities of decreased enzyme activity and severities or age of onset of clinical phenotypes. Identification of more GNE myopathy patients and ongoing natural history studies may reveal such correlations in the future.
We calculated a worldwide prevalence of at least 6/1,000,000 for GNE myopathy, which is encouraging for identification of sufficient numbers of GNE myopathy patients to conduct clinical trials for this rare disease. We expect an increasing awareness of GNE myopathy among physicians, partially triggered by the ongoing clinical trials, natural history studies, and exponentially increased clinical (sequence) reports in the literature. This awareness will increase requests for genetic testing in suspected patients. This GNE mutation overview can serve as a reference for interpretation of the molecular data.
Supplementary Material
Acknowledgments
Grant information: This study was supported by the Intramural Research Program of the National Human Genome Research Institute (NHGRI) and the Therapeutics for Rare and Neglected Diseases (TRND) Program of the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, Bethesda, Maryland, United States.
Footnotes
Disclosure statement
The authors declare no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouri R, Driss A, Murayama K, Kefi M, Nishino I, Hentati F. Allelic heterogeneity of GNE gene mutation in two Tunisian families with autosomal recessive inclusion body myopathy. Neuromuscul Disord. 2005;15:361–363. doi: 10.1016/j.nmd.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Argov Z, Yarom R. Rimmed vacuole myopathy” sparing the quadriceps. A unique disorder in Iranian Jews. J Neurol Sci. 1984;64:33–43. doi: 10.1016/0022-510x(84)90053-4. [DOI] [PubMed] [Google Scholar]
- Argov Z, Eisenberg I, Mitrani-Rosenbaum S. Genetics of inclusion body myopathies. Curr Opin Rheumatol. 1998;10:543–547. doi: 10.1097/00002281-199811000-00006. [DOI] [PubMed] [Google Scholar]
- Argov Z, Eisenberg I, Grabov-Nardini G, Sadeh M, Wirguin I, Soffer D, Mitrani-Rosenbaum S. Hereditary inclusion body myopathy: the Middle Eastern genetic cluster. Neurology. 2003;60:1519–1523. doi: 10.1212/01.wnl.0000061617.71839.42. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK. New advances in the understanding of sporadic inclusion-body myositis and hereditary inclusion-body myopathies. Curr Opin Rheumatol. 1995;7:486–496. doi: 10.1097/00002281-199511000-00005. [DOI] [PubMed] [Google Scholar]
- Behin A, Dubourg O, Laforet P, Pecheux C, Bernard R, Levy N, Eymard B. Distal myopathy due to mutations of GNE gene: clinical spectrum and diagnosis. Rev Neurol (Paris) 2008;164:434–443. doi: 10.1016/j.neurol.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Blume A, Weidemann W, Stelzl U, Wanker EE, Lucka L, Donner P, Reutter W, Horstkorte R, Hinderlich S. Domain-specific characteristics of the bifunctional key enzyme of sialic acid biosynthesis, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biochem J. 2004;384:599–607. doi: 10.1042/BJ20040917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden SE, Duncan AR, Estrella EA, Lidov HG, Mahoney LJ, Katz JS, Kunkel LM, Kang PB. Molecular diagnosis of hereditary inclusion body myopathy by linkage analysis and identification of a novel splice site mutation in GNE. BMC Med Genet. 2011;12:87. doi: 10.1186/1471-2350-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccolini A, Pescatori M, D’Amico A, Sabino A, Silvestri G, Ricci E, Servidei S, Tonali PA, Mirabella M. An Italian family with autosomal recessive inclusion-body myopathy and mutations in the GNE gene. Neurology. 2002;59:1808–1809. doi: 10.1212/01.wnl.0000031808.04545.e0. [DOI] [PubMed] [Google Scholar]
- Broccolini A, Ricci E, Cassandrini D, Gliubizzi C, Bruno C, Tonoli E, Silvestri G, Pescatori M, Rodolico C, Sinicropi S, Servidei S, Zara F, et al. Novel GNE mutations in Italian families with autosomal recessive hereditary inclusion-body myopathy. Hum Mutat. 2004;23:632. doi: 10.1002/humu.9252. [DOI] [PubMed] [Google Scholar]
- Broccolini A, Gidaro T, De Cristofaro R, Morosetti R, Gliubizzi C, Ricci E, Tonali PA, Mirabella M. Hyposialylation of neprilysin possibly affects its expression and enzymatic activity in hereditary inclusion-body myopathy muscle. J Neurochem. 2008;105:971–981. doi: 10.1111/j.1471-4159.2007.05208.x. [DOI] [PubMed] [Google Scholar]
- Chai Y, Bertorini TE, McGrew FA. Hereditary inclusion-body myopathy associated with cardiomyopathy: report of two siblings. Muscle Nerve. 2011;43:133–136. doi: 10.1002/mus.21839. [DOI] [PubMed] [Google Scholar]
- Cai H, Yabe I, Shirai S, Nishimura H, Hirotani M, Kano T, Houzen H, Yoshida K, Sasaki H. Novel GNE compound heterozygous mutations in a GNE myopathy patient. Muscle Nerve. 2013;48:594–598. doi: 10.1002/mus.23862. [DOI] [PubMed] [Google Scholar]
- Cho A, Hayashi YK, Monma K, Oya Y, Noguchi S, Nonaka I, Nishino I. Mutation profile of the GNE gene in Japanese patients with distal myopathy with rimmed vacuoles (GNE myopathy) J Neurol Neurosurg Psychiatry. 2013 doi: 10.1136/jnnp-2013-305587. Epub 2013 Sept 11. [DOI] [PubMed] [Google Scholar]
- Chu CC, Kuo HC, Yeh TH, Ro LS, Chen SR, Huang CC. Heterozygous mutations affecting the epimerase domain of the GNE gene causing distal myopathy with rimmed vacuoles in a Taiwanese family. Clin Neurol Neurosurg. 2007;109:250–256. doi: 10.1016/j.clineuro.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Darvish D, Vahedifar P, Huo Y. Four novel mutations associated with autosomal recessive inclusion body myopathy (MIM: 600737) Mol Genet Metab. 2002;77:252–256. doi: 10.1016/s1096-7192(02)00141-5. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Baron P, Prelle A, Serafini M, Moggio M, Fonzo AD, Castagni M, Bresolin N, Comi GP. Novel missense mutation and large deletion of GNE gene in autosomal-recessive inclusion-body myopathy. Muscle Nerve. 2003;28:113–117. doi: 10.1002/mus.10391. [DOI] [PubMed] [Google Scholar]
- Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, Olender T, Barash M, Shemesh M, Sadeh M, Grabov-Nardini G, Shmilevich I, Friedmann A, et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- Eisenberg I, Grabov-Nardini G, Hochner H, Korner M, Sadeh M, Bertorini T, Bushby K, Castellan C, Felice K, Mendell J, Merlini L, Shilling C, et al. Mutations spectrum of GNE in hereditary inclusion body myopathy sparing the quadriceps. Hum Mutat. 2003;21:99. doi: 10.1002/humu.9100. [DOI] [PubMed] [Google Scholar]
- Enns GM, Seppala R, Musci TJ, Weisiger K, Ferrell LD, Wenger DA, Gahl WA, Packman S. Clinical course and biochemistry of sialuria. J Inherit Metab Dis. 2001;24:328–336. doi: 10.1023/a:1010588115479. [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C, Orozco M, de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004;57:811–819. doi: 10.1002/prot.20252. [DOI] [PubMed] [Google Scholar]
- Fisher J, Towfighi J, Darvish D, Simmons Z. A case of hereditary inclusion body myopathy: 1 patient, 2 novel mutations. J Clin Neuromuscul Dis. 2006;7:179–184. doi: 10.1097/01.cnd.0000211406.94445.f0. [DOI] [PubMed] [Google Scholar]
- Gahl WA, Tifft CJ. The NIH Undiagnosed Diseases Program: lessons learned. Jama. 2011;305:1904–1905. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- Ghaderi D, Strauss HM, Reinke S, Cirak S, Reutter W, Lucka L, Hinderlich S. Evidence for dynamic interplay of different oligomeric states of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase by biophysical methods. J Mol Biol. 2007;369:746–758. doi: 10.1016/j.jmb.2007.03.037. [DOI] [PubMed] [Google Scholar]
- Grandis M, Gulli R, Cassandrini D, Gazzerro E, Benedetti L, Narciso E, Nobbio L, Bruno C, Minetti C, Bellone E, Reni L, Mancardi GL, et al. The spectrum of GNE mutations: allelic heterogeneity for a common phenotype. Neurol Sci. 2010;31:377–380. doi: 10.1007/s10072-010-0248-y. [DOI] [PubMed] [Google Scholar]
- Griggs RC, Askanas V, DiMauro S, Engel A, Karpati G, Mendell JR, Rowland LP. Inclusion body myositis and myopathies. Ann Neurol. 1995;38:705–713. doi: 10.1002/ana.410380504. [DOI] [PubMed] [Google Scholar]
- Hardy GH. Mendelian Proportions in a Mixed Population. Science. 1908;28:49–50. doi: 10.1126/science.28.706.49. [DOI] [PubMed] [Google Scholar]
- Hinderlich S, Stasche R, Zeitler R, Reutter W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- Hinderlich S, Berger M, Keppler OT, Pawlita M, Reutter W. Biosynthesis of N-acetylneuraminic acid in cells lacking UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biol Chem. 2001;382:291–297. doi: 10.1515/BC.2001.036. [DOI] [PubMed] [Google Scholar]
- Hinderlich S, Salama I, Eisenberg I, Potikha T, Mantey LR, Yarema KJ, Horstkorte R, Argov Z, Sadeh M, Reutter W, Mitrani-Rosenbaum S. The homozygous M712T mutation of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase results in reduced enzyme activities but not in altered overall cellular sialylation in hereditary inclusion body myopathy. FEBS Lett. 2004;566:105–109. doi: 10.1016/j.febslet.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Hinderlich S, Weidemann W, Yardeni T, Horstkorte R, Huizing M. UDP-GlcNAc 2-Epimerase/ManNAc Kinase (GNE): A Master Regulator of Sialic Acid Synthesis. Top Curr Chem. 2013 doi: 10.1007/128_2013_464. Epub 2013 Jul 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Rakocevic G, Sparks SE, Mamali I, Shatunov A, Goldfarb L, Krasnewich D, Gahl WA, Dalakas MC. Hypoglycosylation of alpha-dystroglycan in patients with hereditary IBM due to GNE mutations. Mol Genet Metab. 2004;81:196–202. doi: 10.1016/j.ymgme.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Huizing M, Krasnewich DM. Hereditary inclusion body myopathy: a decade of progress. Biochim Biophys Acta. 2009;1792:881–887. doi: 10.1016/j.bbadis.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Malicdan MC, Krasnewich D, Manoli I, Carrillo-Carrasco N. GNE Myopathy. In: Scriver CR, Childs B, Sly WS, Valle D, Beaudet AL, Vogelstein B, Kinsler KW, editors. Scriver’s Online Metabolic and Molecular Bases of Inherited Disease. Chapter 216.1 New York: McGraw-Hill; 2014a. http://www.ommbid.com. [Google Scholar]
- Huizing M, Carrillo-Carrasco N, Malicdan MCV, Noguchi S, Gahl WA, Mitrani-Rosenbaum S, Argov Z, Nishino I. GNE myopathy: New name and new mutation nomenclature. Neuromuscul Disord. 2014b doi: 10.1016/j.nmd.2014.03.004. Epub 2014 Mar 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi T, Asaka T, Saito M, Tanaka H, Higuchi S, Tanaka K, Saida K, Uyama E, Mizusawa H, Fukuhara N, Nonaka I, Takamori M, et al. Gene locus for autosomal recessive distal myopathy with rimmed vacuoles maps to chromosome 9. Ann Neurol. 1997;41:432–437. doi: 10.1002/ana.410410405. [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Lochmuller H, Tournev I, Baas F, Beres J, Colomer J, Guergueltcheva V, Herrmann R, Karcagi V, King R, Miyata T, Mullner-Eidenbock A, et al. 125th ENMC International Workshop: Neuromuscular disorders in the Roma (Gypsy) population, 23–25 April 2004, Naarden, The Netherlands. Neuromuscul Disord. 2005;15:65–71. doi: 10.1016/j.nmd.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Kannan MA, Challa S, Urtizberea AJ, Krahn M, Jabeen AS, Borgohain R. Distal myopathy with rimmed vacuoles and inflammation: a genetically proven case. Neurol India. 2012;60:631–634. doi: 10.4103/0028-3886.105199. [DOI] [PubMed] [Google Scholar]
- Kayashima T, Matsuo H, Satoh A, Ohta T, Yoshiura K, Matsumoto N, Nakane Y, Niikawa N, Kishino T. Nonaka myopathy is caused by mutations in the UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase gene (GNE) J Hum Genet. 2002;47:77–79. doi: 10.1007/s100380200004. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Ki CS, Kim JW, Sung DH, Choi YC, Kim SH. Mutation analysis of the GNE gene in Korean patients with distal myopathy with rimmed vacuoles. J Hum Genet. 2006;51:137–140. doi: 10.1007/s10038-005-0338-5. [DOI] [PubMed] [Google Scholar]
- Kimpara T, Imamura T, Tsuda T, Sato K, Tsuburaya K. Distal myopathy with rimmed vacuoles and sudden death--report of two siblings. Rinsho Shinkeigaku. 1993;33:886–890. [PubMed] [Google Scholar]
- Kornfeld S, Kornfeld R, Neufeld EF, O’Brien PJ. The Feedback Control of Sugar Nucleotide Biosynthesis in Liver. Proc Natl Acad Sci U S A. 1964;52:371–379. doi: 10.1073/pnas.52.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Schlotter-Weigel B, Walter MC, Najmabadi H, Wiendl H, Muller-Hocker J, Muller-Felber W, Pongratz D, Lochmuller H. A novel homozygous missense mutation in the GNE gene of a patient with quadriceps-sparing hereditary inclusion body myopathy associated with muscle inflammation. Neuromuscul Disord. 2003;13:830–834. doi: 10.1016/s0960-8966(03)00140-8. [DOI] [PubMed] [Google Scholar]
- Krause S, Hinderlich S, Amsili S, Horstkorte R, Wiendl H, Argov Z, Mitrani-Rosenbaum S, Lochmuller H. Localization of UDP-GlcNAc 2-epimerase/ManAc kinase (GNE) in the Golgi complex and the nucleus of mammalian cells. Exp Cell Res. 2005;304:365–379. doi: 10.1016/j.yexcr.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Kurochkina N, Yardeni T, Huizing M. Molecular modeling of the bifunctional enzyme UDP-GlcNAc 2-epimerase/ManNAc kinase and predictions of structural effects of mutations associated with HIBM and sialuria. Glycobiology. 2010;20:322–337. doi: 10.1093/glycob/cwp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy JG, Seppala R, Huizing M, Dacremont G, De Simpel H, Van Coster RN, Orvisky E, Krasnewich DM, Gahl WA. Dominant inheritance of sialuria, an inborn error of feedback inhibition. Am J Hum Genet. 2001;68:1419–1427. doi: 10.1086/320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen Q, Liu F, Zhang X, Liu T, Li W, Liu S, Zhao Y, Wen B, Dai T, Lin P, Gong Y, et al. Clinical and molecular genetic analysis in Chinese patients with distal myopathy with rimmed vacuoles. J Hum Genet. 2011;56:335–338. doi: 10.1038/jhg.2011.15. [DOI] [PubMed] [Google Scholar]
- Liewluck T, Pho-Iam T, Limwongse C, Thongnoppakhun W, Boonyapisit K, Raksadawan N, Murayama K, Hayashi YK, Nishino I, Sangruchi T. Mutation analysis of the GNE gene in distal myopathy with rimmed vacuoles (DMRV) patients in Thailand. Muscle Nerve. 2006;34:775–778. doi: 10.1002/mus.20583. [DOI] [PubMed] [Google Scholar]
- Lu X, Pu C, Huang X, Liu J, Mao Y. Distal myopathy with rimmed vacuoles: clinical and muscle morphological characteristics and spectrum of GNE gene mutations in 53 Chinese patients. Neurol Res. 2011;33:1025–1031. doi: 10.1179/1743132811Y.0000000070. [DOI] [PubMed] [Google Scholar]
- Lucka L, Krause M, Danker K, Reutter W, Horstkorte R. Primary structure and expression analysis of human UDP-N-acetyl-glucosamine-2-epimerase/N-acetylmannosamine kinase, the bifunctional enzyme in neuraminic acid biosynthesis. FEBS Lett. 1999;454:341–344. doi: 10.1016/s0014-5793(99)00837-6. [DOI] [PubMed] [Google Scholar]
- Malicdan MC, Noguchi S, Nishino I. Perspectives on distal myopathy with rimmed vacuoles or hereditary inclusion body myopathy: contributions from an animal model. Lack of sialic acid, a central determinant in sugar chains, causes myopathy? Acta Myol. 2007;26:171–175. [PMC free article] [PubMed] [Google Scholar]
- Malicdan MC, Noguchi S, Hayashi YK, Nonaka I, Nishino I. Prophylactic treatment with sialic acid metabolites precludes the development of the myopathic phenotype in the DMRV-hIBM mouse model. Nat Med. 2009;15:690–695. doi: 10.1038/nm.1956. [DOI] [PubMed] [Google Scholar]
- Martinez J, Nguyen LD, Hinderlich S, Zimmer R, Tauberger E, Reutter W, Saenger W, Fan H, Moniot S. Crystal structures of N-acetylmannosamine kinase provide insights into enzyme activity and inhibition. J Biol Chem. 2012;287:13656–13665. doi: 10.1074/jbc.M111.318170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34:1317–1325. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrani-Rosenbaum S, Argov Z, Blumenfeld A, Seidman CE, Seidman JG. Hereditary inclusion body myopathy maps to chromosome 9p1-q1. Hum Mol Genet. 1996;5:159–163. doi: 10.1093/hmg/5.1.159. [DOI] [PubMed] [Google Scholar]
- Mori-Yoshimura M, Monma K, Suzuki N, Aoki M, Kumamoto T, Tanaka K, Tomimitsu H, Nakano S, Sonoo M, Shimizu J, Sugie K, Nakamura H, et al. Heterozygous UDP-GlcNAc 2-epimerase and N-acetylmannosamine kinase domain mutations in the GNE gene result in a less severe GNE myopathy phenotype compared to homozygous N-acetylmannosamine kinase domain mutations. J Neurol Sci. 2012;318:100–105. doi: 10.1016/j.jns.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Motozaki Y, Komai K, Hirohata M, Asaka T, Ono K, Yamada M. Hereditary inclusion body myopathy with a novel mutation in the GNE gene associated with proximal leg weakness and necrotizing myopathy. Eur J Neurol. 2007;14:e14–15. doi: 10.1111/j.1468-1331.2007.01905.x. [DOI] [PubMed] [Google Scholar]
- Nalini A, Gayathri N, Nishino I, Hayashi YK. GNE myopathy in India. Neurol India. 2013;61:371–374. doi: 10.4103/0028-3886.117609. [DOI] [PubMed] [Google Scholar]
- Nemunaitis G, Jay CM, Maples PB, Gahl WA, Huizing M, Yardeni T, Tong AW, Phadke AP, Pappen BO, Bedell C, Allen H, Hernandez C, et al. Hereditary inclusion body myopathy: single patient response to intravenous dosing of GNE gene lipoplex. Hum Gene Ther. 2011;22:1331–1341. doi: 10.1089/hum.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Noguchi S, Murayama K, Driss A, Sugie K, Oya Y, Nagata T, Chida K, Takahashi T, Takusa Y, Ohi T, Nishimiya J, et al. Distal myopathy with rimmed vacuoles is allelic to hereditary inclusion body myopathy. Neurology. 2002;59:1689–1693. doi: 10.1212/01.wnl.0000041631.28557.c6. [DOI] [PubMed] [Google Scholar]
- Nishino I, Malicdan MC, Murayama K, Nonaka I, Hayashi YK, Noguchi S. Molecular pathomechanism of distal myopathy with rimmed vacuoles. Acta Myol. 2005;24:80–83. [PubMed] [Google Scholar]
- No D, Valles-Ayoub Y, Carbajo R, Khokher Z, Sandoval L, Stein B, Tarnopolsky MA, Mozaffar T, Darvish B, Pietruszka M, Darvish D. Novel GNE mutations in autosomal recessive hereditary inclusion body myopathy patients. Genet Test Mol Biomarkers. 2013;17:376–382. doi: 10.1089/gtmb.2012.0408. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Keira Y, Murayama K, Ogawa M, Fujita M, Kawahara G, Oya Y, Imazawa M, Goto Y, Hayashi YK, Nonaka I, Nishino I. Reduction of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase activity and sialylation in distal myopathy with rimmed vacuoles. J Biol Chem. 2004;279:11402–11407. doi: 10.1074/jbc.M313171200. [DOI] [PubMed] [Google Scholar]
- Nonaka I, Sunohara N, Ishiura S, Satoyoshi E. Familial distal myopathy with rimmed vacuole and lamellar (myeloid) body formation. J Neurol Sci. 1981;51:141–155. doi: 10.1016/0022-510x(81)90067-8. [DOI] [PubMed] [Google Scholar]
- Oetke C, Hinderlich S, Reutter W, Pawlita M. Epigenetically mediated loss of UDP-GlcNAc 2-epimerase/ManNAc kinase expression in hyposialylated cell lines. Biochem Biophys Res Comm. 2003;308:892–898. doi: 10.1016/s0006-291x(03)01471-2. [DOI] [PubMed] [Google Scholar]
- Park YE, Kim HS, Choi ES, Shin JH, Kim SY, Son EH, Lee CH, Kim DS. Limb-girdle phenotype is frequent in patients with myopathy associated with GNE mutations. J Neurol Sci. 2012;321:77–81. doi: 10.1016/j.jns.2012.07.061. [DOI] [PubMed] [Google Scholar]
- Patzel KA, Yardeni T, Le Poec-Celic E, Leoyklang P, Dorward H, Alonzi DS, Kukushkin NV, Xu B, Zhang Y, Sollogoub M, Bleriot Y, Gahl WA, et al. Non-specific accumulation of glycosphingolipids in GNE myopathy. J Inherit Metab Dis. 2014;37:297–308. doi: 10.1007/s10545-013-9655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner J, Mantey LR, Elgavish S, Ghaderi D, Cirak S, Berger M, Krause S, Lucka L, Voit T, Mitrani-Rosenbaum S, Hinderlich S. Influence of UDP-GlcNAc 2-epimerase/ManNAc kinase mutant proteins on hereditary inclusion body myopathy. Biochemistry. 2006;45:2968–2977. doi: 10.1021/bi0522504. [DOI] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved Splice Site Detection in Genie. J Comp Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Reinke SO, Eidenschink C, Jay CM, Hinderlich S. Biochemical characterization of human and murine isoforms of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) Glycoconj J. 2009a;26:415–422. doi: 10.1007/s10719-008-9189-6. [DOI] [PubMed] [Google Scholar]
- Reinke SO, Lehmer G, Hinderlich S, Reutter W. Regulation and pathophysiological implications of UDP-GlcNAc 2-epimerase/ManNAc kinase (GNE) as the key enzyme of sialic acid biosynthesis. Biol Chem. 2009b;390:591–599. doi: 10.1515/BC.2009.073. [DOI] [PubMed] [Google Scholar]
- Ricci E, Broccolini A, Gidaro T, Morosetti R, Gliubizzi C, Frusciante R, Di Lella GM, Tonali PA, Mirabella M. NCAM is hyposialylated in hereditary inclusion body myopathy due to GNE mutations. Neurology. 2006;66:755–758. doi: 10.1212/01.wnl.0000200956.76449.3f. [DOI] [PubMed] [Google Scholar]
- Ro LS, Lee-Chen GJ, Wu YR, Lee M, Hsu PY, Chen CM. Phenotypic variability in a Chinese family with rimmed vacuolar distal myopathy. J Neurol Neurosurg Psychiatry. 2005;76:752–755. doi: 10.1136/jnnp.2004.048876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saechao C, Valles-Ayoub Y, Esfandiarifard S, Haghighatgoo A, No D, Shook S, Mendell JR, Rosales-Quintero X, Felice KJ, Morel CF, Pietruska M, Darvish D. Novel GNE mutations in hereditary inclusion body myopathy patients of non-Middle Eastern descent. Genet Test Mol Biomarkers. 2010;14:157–162. doi: 10.1089/gtmb.2009.0157. [DOI] [PubMed] [Google Scholar]
- Salama I, Hinderlich S, Shlomai Z, Eisenberg I, Krause S, Yarema K, Argov Z, Lochmuller H, Reutter W, Dabby R, Sadeh M, Ben-Bassat H, et al. No overall hyposialylation in hereditary inclusion body myopathy myoblasts carrying the homozygous M712T GNE mutation. Biochem Biophys Res Comm. 2005;328:221–226. doi: 10.1016/j.bbrc.2004.12.157. [DOI] [PubMed] [Google Scholar]
- Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R. Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala R, Lehto VP, Gahl WA. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am J Hum Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim JE, Park HJ, Shin HY, Nam TS, Kim SM, Choi YC. Clinical characteristics and molecular genetic analysis of Korean patients with GNE myopathy. Yonsei Med J. 2013;54:578–582. doi: 10.3349/ymj.2013.54.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar K, Dalakas MC. The spectrum of familial inclusion body myopathies in 13 families and a description of a quadriceps-sparing phenotype in non-Iranian Jews. Neurology. 1996;47:977–984. doi: 10.1212/wnl.47.4.977. [DOI] [PubMed] [Google Scholar]
- Sparks SE, Ciccone C, Lalor M, Orvisky E, Klootwijk R, Savelkoul PJ, Dalakas MC, Krasnewich DM, Gahl WA, Huizing M. Use of a cell-free system to determine UDP-N-acetylglucosamine 2-epimerase and N-acetylmannosamine kinase activities in human hereditary inclusion body myopathy. Glycobiology. 2005;15:1102–1110. doi: 10.1093/glycob/cwi100. [DOI] [PubMed] [Google Scholar]
- Sparks S, Rakocevic G, Joe G, Manoli I, Shrader J, Harris-Love M, Sonies B, Ciccone C, Dorward H, Krasnewich D, Huizing M, Dalakas MC, et al. Intravenous immune globulin in hereditary inclusion body myopathy: a pilot study. BMC Neurol. 2007;7:3. doi: 10.1186/1471-2377-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stober A, Aleo A, Kuhl V, Bornemann A, Walter MC, Lochmuller H, Lindner A, Krause S. Novel missense mutation p.A310P in the GNE gene in autosomal-recessive hereditary inclusion-body myopathy/distal myopathy with rimmed vacuoles in an Italian family. Neuromuscul Disord. 2010;20:335–336. doi: 10.1016/j.nmd.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Tajima Y, Uyama E, Go S, Sato C, Tao N, Kotani M, Hino H, Suzuki A, Sanai Y, Kitajima K, Sakuraba H. Distal myopathy with rimmed vacuoles: impaired O-glycan formation in muscular glycoproteins. Am J Pathol. 2005;166:1121–1130. doi: 10.1016/S0002-9440(10)62332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasca G, Ricci E, Monforte M, Laschena F, Ottaviani P, Rodolico C, Barca E, Silvestri G, Iannaccone E, Mirabella M, Broccolini A. Muscle imaging findings in GNE myopathy. J Neurol. 2012;259:1358–1365. doi: 10.1007/s00415-011-6357-6. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimitsu H, Ishikawa K, Shimizu J, Ohkoshi N, Kanazawa I, Mizusawa H. Distal myopathy with rimmed vacuoles: novel mutations in the GNE gene. Neurology. 2002;59:451–454. doi: 10.1212/wnl.59.3.451. [DOI] [PubMed] [Google Scholar]
- Tomimitsu H, Shimizu J, Ishikawa K, Ohkoshi N, Kanazawa I, Mizusawa H. Distal myopathy with rimmed vacuoles (DMRV): new GNE mutations and splice variant. Neurology. 2004;62:1607–1610. doi: 10.1212/01.wnl.0000123115.23652.6c. [DOI] [PubMed] [Google Scholar]
- Tong Y, Tempel W, Nedyalkova L, Mackenzie F, Park HW. Crystal structure of the N-acetylmannosamine kinase domain of GNE. PLoS One. 2009;4:e7165. doi: 10.1371/journal.pone.0007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Sialic acids as ligands in recognition phenomena. Faseb J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- Varki A. Diversity in the sialic acids. Glycobiology. 1992;2:25–40. doi: 10.1093/glycob/2.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos OM, Raju R, Dalakas MC. GNE mutations in an American family with quadriceps-sparing IBM and lack of mutations in s-IBM. Neurology. 2002;59:1776–1779. doi: 10.1212/01.wnl.0000039780.13681.ad. [DOI] [PubMed] [Google Scholar]
- Voermans NC, Guillard M, Doedee R, Lammens M, Huizing M, Padberg GW, Wevers RA, van Engelen BG, Lefeber DJ. Clinical features, lectin staining, and a novel GNE frameshift mutation in hereditary inclusion body myopathy. Clin Neuropathol. 2010;29:71–77. [PMC free article] [PubMed] [Google Scholar]
- Weihl CC, Miller SE, Zaidman CM, Pestronk A, Baloh RH, Al-Lozi M. Novel GNE mutations in two phenotypically distinct HIBM2 patients. Neuromuscul Disord. 2011;21:102–105. doi: 10.1016/j.nmd.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg W. Über den Nachweis der Vererbung beim Menschen. Jahreshefte des Vereins für vaterländische Naturkunde in Württemberg. 1908;64:368–382. [Google Scholar]
- Yabe I, Higashi T, Kikuchi S, Sasaki H, Fukazawa T, Yoshida K, Tashiro K. GNE mutations causing distal myopathy with rimmed vacuoles with inflammation. Neurology. 2003;61:384–386. doi: 10.1212/01.wnl.0000061520.63546.8f. [DOI] [PubMed] [Google Scholar]
- Yardeni T, Choekyi T, Jacobs K, Ciccone C, Patzel K, Anikster Y, Gahl WA, Kurochkina N, Huizing M. Identification, tissue distribution, and molecular modeling of novel human isoforms of the key enzyme in sialic acid synthesis, UDP-GlcNAc 2-epimerase/ManNAc kinase. Biochemistry. 2011;50:8914–8925. doi: 10.1021/bi201050u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarema KJ, Goon S, Bertozzi CR. Metabolic selection of glycosylation defects in human cells. Nat Biotechnol. 2001;19:553–558. doi: 10.1038/89305. [DOI] [PubMed] [Google Scholar]
- Yunis EJ, Samaha FJ. Inclusion body myositis. Lab Invest. 1971;25:240–248. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.