Abstract
Objectives
To determine if alpha-synuclein REP1 genotypes are associated with survival in Parkinson’s disease.
Methods
Investigators from the Genetic Epidemiology of Parkinson’s Disease Consortium provided REP1 genotypes and baseline and follow-up clinical data for cases. The primary outcome was time to death. Cox proportional hazards regression models were used to assess the association of REP1 genotypes with survival.
Results
Twenty-one sites contributed data for 6,154 cases. There was no significant association between alpha-synuclein REP1 genotypes and survival in Parkinson’s disease. However, there was a significant association between REP1 genotypes and age at onset of PD (Hazard Ratio = 1.06, 95% Confidence Interval = 1.01–1.10, p value = 0.01).
Conclusions
In our large consortium study, alpha-synuclein REP1 genotypes were not associated with survival in Parkinson’s disease. Further studies of α–synuclein’s role in disease progression and long-term outcomes are needed.
Keywords: Parkinson’s disease, α-synuclein, gene, survival, association
INTRODUCTION
Genetic studies of familial Parkinson’s disease (PD) discovered rare pathogenic missense and multiplication mutations in the α–synuclein gene (SNCA).1–4 The mechanism by which multiplication mutations cause familial PD is over-expression.5,6 Similarly, we observed that polymorphisms in the promoter region of the SNCA gene confer susceptibility to PD,7 presumably via the same over-expression mechanism.5,6,8–11 Therefore therapies are being developed to reduce α–synuclein in PD, as a method of neuroprotection.12–14
However it is unclear if reduced SNCA expression genotypes or therapies targeting SNCA expression slow progression of PD. Our recent genome-wide study found no evidence of SNCA SNP association with motor and cognitive outcomes of PD at the genome-wide level.15 By contrast, a recent population-based study of 242 PD cases found that SNCA dinucleotide repeat (REP1) allele length variants are associated with rate of motor progression in PD.16 Clinical assessments of motor or cognitive outcomes in PD may be confounded by treatment effects.
Here for the first time the Genetic Epidemiology of Parkinson’s Disease (GEO-PD) consortium conducted a collaborative study to determine whether SNCA genotypes are associated with risk of death in PD (a clear outcome measure).
METHODS
Study subjects
Between June 28, 2010 and November 13, 2011, GEO-PD sites provided the following data for each PD case: SNCA REP1 genotype (bp length/bp length), genotyping laboratory and platform, diagnostic criteria for PD, date of birth, age at disease onset, age at diagnosis, age at the time of study enrollment (baseline), gender, ethnicity, family history of PD, education (years), smoking (ever/never, pack-years), L-DOPA therapy (ever/never, response), date at last follow up, method of last follow up (telephone contact, mail contact, medical records abstraction, death registry, death certificate, other), vital status at last follow up, and date of death. The samples were collected at each site for the purpose of conducting genetic association studies. The samples were not collected specifically for the purpose of a survival analysis.
All studies were approved by the local ethical committees following the procedures of each site.
Genotyping
Each participating GEO-PD site measured SNCA REP1 genotypes using site-specific genotyping platforms (Supplementary Table 1). As in previous studies,17,18 the REP1 score was calculated as the sum of two allele scores, with each 259 bp allele contributing 0 points, each 261 bp allele contributing 1 point, and each copy of a 263 bp allele contributing 2 points, giving a score (sum of the two allele scores) ranging from 0 to 4. In secondary analyses genotypes were coded as: 259 bp allele count (0,1, or 2), 263 bp allele count (0,1, or 2), and 263/263 vs. 259/259 (excluding other genotypes). We evaluated allele frequencies and genotype heterozygosity for each site. We used Pearson χ2 statistics to assess whether genotype distributions for the SNCA REP1 allele-length variants departed from Hardy-Weinberg equilibrium (HWE). Sites with a significant (p< 0.005) deviation from HWE were excluded.
Survival analyses
The primary outcome was time-to-death. A secondary analysis evaluated the association of genotypes with age of onset of PD.. Cox proportional hazard regression was used to assess association between genotypes and outcomes. Because the primary outcome investigated in this study is strongly age-related, age was used as the time scale and left censoring was accounted for by starting analyses at age at enrollment into the study.19 All models were adjusted for the contributing GEO-PD site. Because within each site the samples were ethnically homogeneous (after removing minorities), no further adjustment for race or ethnicity was made. To identify relevant covariates, we performed univariate and stepwise Cox regression analyses to identify demographic or clinical variables (including family history) that were associated with the outcomes (p < 0.05), and the assumption of proportional hazards was evaluated for the covariates using scaled Schoenfeldresiduals.20 For analyses of survival time from age at enrollment into the study until death, the models were adjusted for site, PD duration at baseline, sex, smoking (ever/never), and levodopa therapy (yes/no). When age at onset was the outcome, site, smoking (pack-years) and education were included as covariates in the models. We performed analyses both unadjusted and adjusted for these covariates. A Woolf’s test of homogeneity of hazard ratios (HRs) across sites was performed to assess whether the distribution of HRs across sites is compatible with a common HR.21
All analyses were performed using SAS® version 9.2 (SAS Institute Inc., Cary, NC) and Rversion 2.13 (www.cran.r-project.org).
RESULTS
Sample
Twenty-one GEO-PD global sites contributed a total of 6,154 PD cases. After data cleaning (excluding 18 duplicate subjects, 28 minority race/ethnicity subjects, and 96 carriers of rare alleles), a total of 6,012 PD cases remained. The clinical characteristics of subjects are summarized in Table 1. The median duration of PD at baseline was 6 years (range, 0–54), and the median lag time between baseline and end of follow up was 4.3 years (range 0–20.2). The 6,012 PD cases provided 25,453 person-years of follow-up from enrollment to time to event or censoring. There were 1,228 deaths observed (median age at death, 78.6, range 37.8–98.8). Missing data for variables of all sites is summarized in Supplementary Table 2. This sample size provided ~80% power to detect hazard ratios as small as 1.4 for a dominant effect of the 259 bp allele.22
Table 1.
Demographic and clinical characteristics of subjects by site (based on n=6,012 cases with clean data).
| Site | Study PI | Country | Continent | No. | Male, n (%) | Age at onset, Mean (SD) | Age at study, mean (SD) | No. deaths | Follow-up time, mean (SD) | Family history, n (%) | Diagnostic Criteria |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Aasly | Norway | Europe | 379 | 238 (62.8%) | 58.5 (11.2) | 71 (9.1) | 167 | 3.3 (2.8) | 48 (12.7%) | Gelb |
| 2 | Annesi | Italy | Europe | 185 | 106 (57.3%) | 58.5 (9.9) | 66.6 (8.5) | 60 | 10.1 (2.7) | -- | Gelb |
| 3 | Bentivoglio | Italy | Europe | 99 | 45 (45.5%) | 57.5 (8) | 63.7 (8.9) | 17 | 7.7 (2.3) | 15 (15.2%) | Gelb |
| 4 | Brighina | Italy | Europe | 100 | 58 (58%) | 57.7 (9.3) | 64.9 (9.2) | 11 | 5 (2.5) | 6 (6%) | Gelb |
| 5 | Chartier-Harlin | France | Europe | 221 | 129 (58.4%) | 51 (10.3) | 62.8 (9.4) | 49 | 5.6 (1.8) | 21 (9.5%) | Gelb |
| 6 | Goldwurm | Italy | Europe | 821 | 496 (60.4%) | 56 (10.9) | 63.4 (10.7) | 78 | 3.6 (2) | 141 (17.2%) | Gibb |
| 7 | Hadjigeorgiou | Greece | Europe | 357 | 196 (54.9%) | 63.5 (9.8) | 68.3 (9.9) | 77 | 4 (2.9) | 27 (7.6%) | Bower |
| 8 | Jasinska/Krygowska | Poland | Europe | 134 | 80 (59.7%) | 51.8 (11) | 61 (10.3) | 4 | 4.3 (2.5) | 16 (11.9%) | Gibb |
| 9 | Jeon | Korea | Asia | 248 | 115 (46.4%) | 52.1 (10.2) | -- | 18 | -- | -- | Gibb |
| 10 | Kim | Korea | Asia | 98 | 41 (41.8%) | 60.2 (13.9) | 64.4 (12.8) | 6 | 3.1 (2.3) | 9 (9.2%) | Gelb |
| 11 | Krüger | Germany | Europe | 158 | 96 (60.8%) | 56.2 (11.1) | 65.4 (9.9) | 0 | 1.4 (0.3) | 43 (27.2%) | Gibb |
| 12 | Lesage | France | Europe | 287 | 177 (61.7%) | 47.5 (10.1) | 57.7 (11.4) | 1 | 1.2 (1.1) | -- | Gelb |
| 13 | Maraganore | USA | North America | 1081 | 695 (64.3%) | 61.6 (10.7) | 66.8 (10.3) | 336 | 6.8 (2.8) | 177 (16.8%) | Bower |
| 14 | Markopoulou | Greece | Europe | 59 | 28 (47.5%) | 58.6 (11.3) | 64.8 (11.4) | 7 | 1.4 (2.2) | 19 (32.2%) | Gelb |
| 15 | Mellick | Australia | Australia | 291 | 145 (49.8%) | 59.6 (10.9) | 68.3 (8.8) | 47 | 4.8 (2.8) | 32 (11.3%) | Bower |
| 16 | Morrison | United Kindom | Europe | 574 | 396 (69%) | -- | 70.1 (9) | 111 | 4.8 (2.2) | 94 (16.7%) | Gibb |
| 17 | Puschmann | Sweden | Europe | 102 | 64 (62.7%) | 62 (10.1) | 70.3 (10) | 19 | 2.7 (0.9) | 45 (44.6%) | Gibb |
| 18 | Tan | Singapore | Asia | 182 | 101 (55.5%) | 62.3 (11.5) | 66.4 (11.1) | 0 | -- | 13 (7.1%) | Gibb |
| 19 | Theuns | Belgium | Europe | 429 | 247 (57.6%) | 60.4 (11.5) | 69.7 (10.2) | 136 | 3.3 (2.4) | 74 (19.4%) | Gelb |
| 20 | Wirdefeldt | Sweden | Europe | 94 | 54 (57.4%) | 65.3 (11.2) | 72.4 (8.4) | 60 | 8.9 (3.2) | 9 (9.8%) | Gelb |
| 21 | Wszolek | USA | North America | 113 | 69 (61.1%) | 62 (11.6) | 68.7 (11.4) | 24 | 3.4 (3.5) | 52 (46%) | Gelb |
| Total | 6,012 | 3576(59.5%) | 58.2 (11.6) | 66.5 (10.6) | 1228 | 841 (14.9%) |
Jasinska/Krygowska = short hand for two investigators, Jasinska-Myga and Krygowska-Wajs; Dr. Krygowska-Wajs contributed eight cases.
Bower = Bower JH, et al.. Incidence and distribution of parkinsonism in Olmsted County, Minnesota, 1976–1990. Neurology. 1999; 52(6):1214–1220.
Gelb = Gelb DJ, et al.. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56(1):33–39.
Gibb = Gibb WR, Lees AJ.. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J Neurol Neurosurg Psychiatry. 1988;51(6):745–752
SNCA and survival in PD
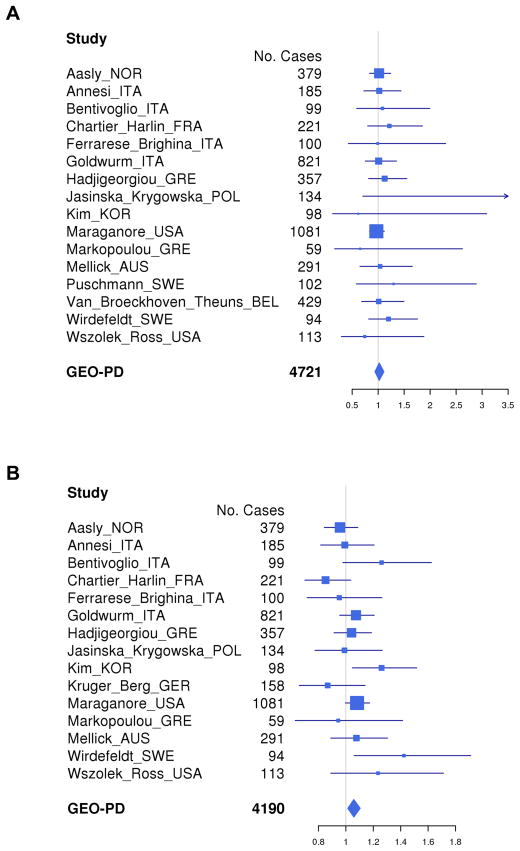
The results of unadjusted analyses for all sites combined and for all four SNCA genotype-coding schemes are illustrated using Kaplan Meier curves (Supplementary Fig. 1). The results of adjusted analyses for each site separately and combined are illustrated using forest plots (Fig. 1A, REP 1 score; and Supplementary Fig. 2, SNCA REP1 259 bp allele count). No significant associations between SNCA genotypes and risk of death were observed for any of the models. In the primary analysis with genotype coded as the REP1 score and with adjustment for site, PD duration at baseline, sex, smoking (ever/never) and levodopa treatment, the HR was 1.02 (95% CI = 0.94–1.11, p = 0.63). Sensitivity analyses with different covariate adjustments and alternative genotype coding schemes also demonstrated no significant association of REP1 score with risk of death (results not shown). Woolf’s test revealed no heterogeneity of HRs between sites.
Figure 1. (A) Forest plot of association hazard ratios (HRs) between SNCA REP1 score and survival in Parkinson’s disease (PD).
Age at study was considered as “Time 0” (T0) and we accounted for left truncation using the start-stop counting process style of input within the Cox regression framework. These analyses assumed a log additive effect and were adjusted for disease duration at baseline, sex, smoking (ever/never) and levodopa treatment. There were no significant associations between SNCA REP1 score and survival in PD. Four sites were excluded from analysis due to missing time to event or covariates information. One site was excluded in per-site analysis due to none or few deaths, but it was included in the overall pooled analysis.
(B) Forest plot of association HRs between SNCA REP1 score and age at onset. These analyses assumed a log additive effect and were adjusted for education and cigarette smoking (pack-years). There was a significant association between SNCA REP1 score and age at onset of PD (HR = 1.06, 95% CI = 1.01–1.10, p value = 0.01).
GEO-PD = Genetic Epidemiology of Parkinson’s Disease
Consortium.Jasinska_Krygowska = two investigators, Jasinska-Myga and Krygowska-Wajs.
SNCA and age at onset of PD
The results of adjusted analyses for each site separately and combined for SNCA REP1 score are illustrated using forest plots (Fig. 1B). There was a significant association between SNCA REP1 genotype and age at onset (adjusted analysis), with higher REP1 scores being associated with earlier age at onset (HR = 1.06, 95% CI = 1.01–1.10, p value = 0.01).
DISCUSSION
In the present study, SNCA REP1 genotypes were not associated with survival in PD but there was some association with age at onset. Multiple studies have demonstrated that SNCA REP1 genotypes are associated with α–synuclein mRNA and protein expression levels; specifically, longer SNCA REP1 alleles are associated with higher expression levels, and shorter REP1 alleles are associated with lower expression levels.5,6,8–11 Moreover, in our previously published collaborative pooled analysis of >5,000 GEO-PD cases and controls, REP1 alleles conferring increased expression (263 bp) were associated with a significantly higher PD risk, while REP1 alleles conferring reduced expression (259 bp) were associated with a significantly lower PD risk.7 Consistent with that study, this present study observed an association between over-expression genotypes and earlier age at onset of PD.23 In aggregate, these studies indicate that the SNCA REP1 genotypes, which are associated with α–synuclein expression levels, are associated with PD susceptibility and onset-age, but that they do not associate with survival in PD. While we did not observe an association between SNCA REP1 and survival in PD, we cannot exclude an association of the genetic variants with disease progression or other outcomes (such as survival free of Hoehn and Yahr stages 4 or 5 or survival free of dementia).
This seemingly paradoxical dissociation between susceptibility, age at onset, and survival suggests that α–synuclein’s role in PD may be complex. In another neurodegenerative disorder, Alzheimer’s disease, a similar dissociation is seen, whereby the inciting pathogenic protein, beta-amyloid42, is associated with susceptibility and age at onset, but not disease progression.24,25 However, our results are contrary to the observations of more rapidly progressing motor and cognitive impairments in families with triplication mutations versus duplication mutations.26,27 It is possible that the level of α–synuclein overexpression in families with multiplication mutations overwhelms cellular systems combating the pathogenic process of α–synuclein aggregation. Our results are also contrary to a recent population-based study reporting that SNCA REP1 over-expression genotypes are associated with faster motor decline in PD.16 However, the sample in that study was small and the duration of follow up was brief, by contrast to our study.
Our study has strengths. Multiple sites from the GEO-PD consortium amassed a large sample size, and with follow up in the tens of thousands of person-years. We assessed a discreet outcome: death. Our study also has limitations. First, while PD cases have a higher risk of death than controls, the median difference in survival is small (about three years).28 Therefore our study may have been underpowered to detect genetic associations underlying limited variability in the death outcome. Second, we considered death from all causes. It was not within the scope of this study to discern causes of death. Third, we did not perform genetic testing to exclude cases with gene mutations known to cause familial parkinsonism (“Mendelian forms”). However, for the three sites with a high frequency of familial PD cases (Markopoulou 32.2%, Puschmann 44.6%, and Wszolek 46%), the number of cases that they contributed to the study was small (n=274 or 4.6% of 6,012 subjects in total). Fourth, the proportion of European sites participating in the study was greater than for other continents. However, the inclusion of additional African, Asian, Australian, North American or South American sites may have introduced population stratification biases in the pooled analyses. Fifth, we only considered REP1 variability in the 5′ core promoter region of the SNCA gene. However, the effects of SNCA REP1 variability on expression levels are well defined,5,6,8–11 by contrast to 3′ SNP variability. We have previously shown that REP1 and 3′ SNPs have separate and equal effects on PD susceptibility (no additive or multiplicative effects).18
Supplementary Material
Acknowledgments
Funding Sources:
The Goldwurm site acknowledges the funding support of the Italian Telethon Foundation (grants n. GTB07001) and the “Fondazione Grigioni per ilMorbo di Parkinson”.
The Maraganore site acknowledges the funding support of the National Institutes of Health (R01 ES10751).
The Morrison site acknowledges the funding support of the Medical Research Council UK, Midlands Neuroscience Teaching and Research Fund, Queen Elizabeth Hospital Birmingham Charity.
The Thuens site acknowledges the funding support of the Interuniversity Attraction Poles program of the Belgian Science Policy Office, the Foundation for Alzheimer Research, the Belgian Parkinson Foundation, the Methusalem Excellence Program of the Flemish Government, the Research Foundation Flanders, the Agency for Innovation by Science and Technology Flanders, and the Special Research Fund of the University of Antwerp, Belgium.
The Wszolek site acknowledges the funding support of the National Institutes of Health (P50 NS072187) and the Mayo Clinic Florida Research Committee CR program.
The Annesi site acknowledges the effort support of M. Gagliardi, P. Tarantino, and A. Quattrone.
The Brighina site acknowledges the effort support of C. Ferrarese.
The Charier-Harlin site acknowledges the effort support of A. Destée and E. Mutez.
The Goldwurm site acknowledges the effort support of S. Duga.
The Hadjigeorgiou site acknowledges the effort support of E. Dardiotis and G. Xiromerisiou.
The Jasinska-Myga site acknowledges the effort support of A. Krygowska-Wajs.
The Jeon site acknowledges the effort support of S.S. Park.
The Krüger site acknowledges the effort support of D. Berg, T. Gasser, He. Huber, A. Hummel, M. Sharma, and O. Riess.
The Lesage site acknowledges the effort support of A. Brice.
The Maraganore site acknowledges the effort support of J.E. Ahlskog and J. Cunningham.
The Morrison site acknowledges the effort support of C.E. Clarke, M. Farrer, J.D. Stockton, and C. Moorby.
The Tan site acknowledges the effort support of Y. Zhao.
The Thuens site acknowledges the effort support of P. Cras, D. Crossiers, P.P. De Deyn, S. Engelborghs P. Pals, B. Pickut, and C. Van Broeckhoven.
The Wszolek site acknowledges the effort support of A. Strongosky and O. Ross.
Footnotes
Financial Disclosures: The authors report no disclosures relevant to the manuscript.
Author Roles
Research Project:
A. Conception: Elbaz, Maraganore
B. Organization: Maraganore
C. Execution: Aasly, Annesi, Bentivoglio, Brighina, Chartier-Harlin, Frigerio, Goldwurm, Hadjigeorgiou, Jasinska-Myga, Jeon, Kim, Krüger, Lesage, Maraganore, Markopoulou, Mellick, Morrison, Puschmann, Tan, Theuns, Wirdefeldt, Wszolek
Statistical Analysis:
A. Design: Anderson, Armasu, Biernacka, Elbaz, Maraganore
B. Execution: Anderson, Armasu, Biernacka, Maraganore
C. Review and Critique: Anderson, Armasu, Biernacka, Elbaz
Manuscript:
A. Writing of the first draft: Chung
B. Review and Critique: Aasly, Brighina, Chartier-Harlin, Elbaz, Frigerio, Goldwurm, Hadjigeorgiou, Jasinska-Myga, Jeon, Kim, Krüger, Maraganore, Markopoulou, Tan, Wirdefeldt
References
- 1.Kruger R, Kuhn W, Muller T, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–8. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 2.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–73. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 4.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.Cronin KD, Ge D, Manninger P, et al. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18:3274–85. doi: 10.1093/hmg/ddp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs J, Tichopad A, Golub Y, et al. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–34. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 7.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–70. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 8.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10:3101–9. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 9.Chiba-Falek O, Touchman JW, Nussbaum RL. Functional analysis of intra-allelic variation at NACP-Rep1 in the alpha-synuclein gene. Hum Genet. 2003;113:426–31. doi: 10.1007/s00439-003-1002-9. [DOI] [PubMed] [Google Scholar]
- 10.Bonsch D, Lederer T, Reulbach U, Hothorn T, Kornhuber J, Bleich S. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum Mol Genet. 2005;14:967–71. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- 11.Linnertz C, Saucier L, Ge D, et al. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One. 2009;16:e7480. doi: 10.1371/journal.pone.0007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis J, Melrose H, Bumcrot D, et al. In vivo silencing of alpha-synuclein using naked siRNA. Mol Neurodegener. 2008;3:19. doi: 10.1186/1750-1326-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormack AL, Mak SK, Henderson JM, Bumcrot D, Farrer MJ, Di Monte DA. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 2010;5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneeberger A, Mandler M, Mattner F, Schmidt W. Vaccination for Parkinson’s disease. Parkinsonism Relat Disord. 2012;18:S11–3. doi: 10.1016/S1353-8020(11)70006-2. [DOI] [PubMed] [Google Scholar]
- 15.Chung SJ, Armasu SM, Biernacka JM, et al. Genomic determinants of motor and cognitive outcomes in Parkinson’s disease. Parkinsonism Relat Disord. 2012;18:881–6. doi: 10.1016/j.parkreldis.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritz B, Rhodes SL, Bordelon Y, Bronstein J. alpha-Synuclein genetic variants predict faster motor symptom progression in idiopathic Parkinson disease. PLoS One. 2012;7:e36199. doi: 10.1371/journal.pone.0036199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brighina L, Frigerio R, Schneider NK, et al. Alpha-synuclein, pesticides, and Parkinson disease: a case-control study. Neurology. 2008;70:1461–9. doi: 10.1212/01.wnl.0000304049.31377.f2. [DOI] [PubMed] [Google Scholar]
- 18.Chung SJ, Armasu SM, Biernacka JM, et al. Common variants in PARK loci and related genes and Parkinson’s disease. Mov Disord. 2011;26:280–8. doi: 10.1002/mds.23376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Therneau TM, Grambsch PM, Fleming TR. Martingale-Based Residuals for Survival Models. Biometrika. 1990;77:147–60. [Google Scholar]
- 20.Grambsch PM, Therneau TM. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika. 1994;81:515–26. [Google Scholar]
- 21.Lumley T. rmeta: Meta-analysis. R package version 2.16. 2009 http://CRAN.R-project.org/package=rmeta.
- 22.David C. Modelling Survival Data in Medical Research. London, UK: Chapman & Hall/CRC; 1999. [Google Scholar]
- 23.Hadjigeorgiou GM, Xiromerisiou G, Gourbali V, et al. Association of alpha-synuclein Rep1 polymorphism and Parkinson’s disease: influence of Rep1 on age at onset. Mov Disord. 2006;21:534–9. doi: 10.1002/mds.20752. [DOI] [PubMed] [Google Scholar]
- 24.Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology. 1992;42:631–9. doi: 10.1212/wnl.42.3.631. [DOI] [PubMed] [Google Scholar]
- 25.Mondragon-Rodriguez S, Basurto-Islas G, Lee HG, et al. Causes versus effects: the increasing complexities of Alzheimer’s disease pathogenesis. Expert Rev Neurother. 2010;10:683–91. doi: 10.1586/ern.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muenter MD, Forno LS, Hornykiewicz O, et al. Hereditary form of parkinsonism--dementia. Ann Neurol. 1998;43:768–81. doi: 10.1002/ana.410430612. [DOI] [PubMed] [Google Scholar]
- 27.Farrer M, Kachergus J, Forno L, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55:174–9. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 28.Elbaz A, Bower JH, Peterson BJ, et al. Survival study of Parkinson disease in Olmsted County, Minnesota. Arch Neurol. 2003;60:91–6. doi: 10.1001/archneur.60.1.91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.