Abstract
Linkage studies of alcoholism have implicated several chromosome regions, leading to the successful identification of susceptibility genes, including ADH4 and GABRA2 on chromosome 4. Quantitative endophenotypes that are potentially closer to gene action than clinical endpoints offer a means of obtaining more refined linkage signals of genes that predispose alcohol use disorders (AUD). In this study we examine a self-reported measure of the maximum number of drinks consumed in a 24-hour period (abbreviated Max Drinks), a significantly heritable phenotype (h2 = 0.32 ± 0.05; P = 4.61 × 10−14) with a strong genetic correlation with AUD (ρg = 0.99 ± 0.13) for the San Antonio Family Study (n = 1,203). Genome-wide SNPs were analyzed using variance components linkage methods in the program SOLAR, revealing a novel, genome-wide significant QTL (LOD = 4.17; P = 5.85 × 10−6) for Max Drinks at chromosome 6p22.3, a region with a number of compelling candidate genes implicated in neuronal function and psychiatric illness. Joint analysis of Max Drinks and AUD status shows that the QTL has a significant non-zero effect on diagnosis (P = 4.04 × 10−3), accounting for 8.6% of the total variation. Significant SNP associations for Max Drinks were also identified at the linkage region, including one, rs7761213 (P = 2.14 × 10−4), obtained for an independent sample of Chinese families. Thus, our study identifies a potential risk locus for AUD at 6p22.3, with significant pleiotropic effects on the heaviness of alcohol consumption that may not be population specific.
Keywords: AUD, alcohol dependence, variance components linkage analysis, pleiotropy, endophenotype ranking value (ERV), SNP association
INTRODUCTION
Alcohol use disorder (AUD) represents patterns of unhealthy drinking (e.g., more than three or four standard drinks per day) combined with occupational or psychosocial disability that can cause substantial morbidity and mortality [Schuckit, 2009]. Evidence from family, twin and adoption studies have consistently shown familial transmission of AUDs, ranging from severe to broad definitions of the phenotype, indicating an important genetic component in its liability [Kendler et al., 1994; McGue, 1999; Nurnberger et al., 2004]. Heritability estimates range from 50–80% [Heath et al., 1997; Knopik et al., 2004; Kendler and Prescott, 2006]. Susceptibility loci have been mapped to various chromosomes using genetic linkage methods [Reich et al., 1998; Foroud et al., 2000; Williams et al., 1999b; Nurnberger et al., 2001; Dick et al., 2002; Prescott et al., 2006; Gelernter et al., 2009], with successful gene localization occurring from the follow-up of some positive linkage regions, most notably GABRA2 and ADH4 [Edenberg et al., 2004, 2006a]. More recently, the first generation of genome-wide, case-control association studies (GWAS) revealed a number of promising SNP associations to AUD [Treutlein et al., 2009; Bierut et al., 2010; Edenberg et al., 2010; Gelernter et al., 2014], although most of the variation in genetic liability remains to be explained.
Genetic analyses of alcoholism have tended to rely on binary measures of dependence, despite the continuum of alcohol-related problems that underlie its symptomatology. Clearly, heaviness of alcohol consumption represents a salient dimension of AUD [American Psychiatric Association, 1994], as it is highly heritable [Heath et al., 1991; Bierut et al., 2002; Edenberg et al., 2006b], with substantial, and in some cases complete, genetic correlation with diagnoses of dependence or abuse [Heath and Martin, 1994; Whitfield et al., 2004; Kendler et al., 2010]. This genetic overlap persists even for AD symptoms of nondependent individuals, suggesting that the genetic determinants of dependence risk are acting in large degree through the heaviness of use [Grant et al., 2009]. Because measures that quantify alcohol consumption are heritable and correlated with dependence, they are ideal candidate endophenotypes [Gottesman and Gould, 2003] that could place genetic analyses closer to the action of genes that predispose alcoholism [Glahn et al., 2012]. Moreover, quantitative traits have improved statistical power in association tests and, in many instances, are more practical to collect from large samples than to recruit psychiatric probands and appropriate controls.
Alcohol consumption may be measured in a variety of ways, including: frequency or quantity in a given reference period (e.g., daily, weekly or annually), frequency of heavy drinking (e.g., 5 or more drinks), and frequency of intoxication. In this study, we focus on the maximum number of drinks consumed in a 24-hour period (denoted as Max Drinks), a highly heritable measure strongly correlated to other patterns of excessive alcohol consumption [Saccone et al., 2000; Grant et al., 2009; Kendler et al., 2010; Agrawal et al., 2012]. Previous linkage analyses of Max Drinks in predominantly European-American families have detected a genome-wide significant quantitative trait locus (QTL) on chromosome 4q in proximity to a cluster of alcohol dehydrogenase (ADH) genes [Saccone et al., 2000], as well as robust linkage signals on chromosomes 2 and 7 [Saccone et al., 2005]. Candidate studies of alcohol metabolism genes, namely the ADH and aldehyde dehydrogenase (ALDH) families, have produced significant associations with various measures of alcohol consumption, including Max Drinks, for various populations, including ones of Asian, European or African ancestry [Edenberg et al., 2006a; Luczak et al., 2006; Macgregor et al., 2009; Bierut et al., 2012].
In this paper, we analyze data obtained from 1,203 Mexican-American individuals from large, randomly ascertained pedigrees in the Genetics of Brain Structure and Function study (GOBS), to test for genome-wide univariate linkage within a variance components framework. Moreover, we explore the genetic relationship of Max Drinks to AUD, computing its genetic correlation and endophenotype ranking value (ERV), as well as conducting bivariate linkage analysis and likelihood ratio tests for localized pleiotropic effects, with the aim of identifying QTLs that influence both alcohol consumption and the risk for alcoholism.
MATERIALS AND METHODS
Participants
From pedigrees originally recruited for the San Antonio Family Heart Study (SAFHS) [Mitchell et al., 1996] and the San Antonio Family Diabetes/Gallbladder Study (SAFDGS) [Hunt et al., 2005], a total of 1,203 Mexican-American individuals, representing 81 families with an average size of 14.2 subjects, were analyzed for the current report [Olvera et al., 2011]. Participants are 64% female and ranged in age from 18 to 87 years (mean 45.6 ± SD 14.9). The cohort was randomly selected from the community with the requirements that they are of Mexican-American ancestry, part of a large family, and live within the San Antonio region. Reported pedigree relationships were empirically verified with autosomal markers. All participants provided written informed consent approved by the Institutional Review Boards at the University of Texas Health Science Center in San Antonio and at Yale University.
Max Drinks and Psychiatric Assessments
The maximum number of alcoholic drinks ever consumed in any 24-hour period (i.e., Max Drinks) was obtained from each participant with the understanding that one alcoholic drink corresponds to either one 5 oz. glass of wine, one 12 oz. beer, or one 1.5 oz. shot glass of “hard liquor” (i.e., 80-proof spirits), alone or as part of a mixed beverage (note: each drink option contains approximately 0.6 oz. of alcohol). In addition, the Mini-International Neuropsychiatric Interview (MINI) [Sheehan et al., 1998], a semi-structured interview augmented with items on lifetime diagnostic history, was administered to all participants by Masters- and doctorate-level research staff with established reliability (κ ≥ 0.85) for addictive disorders. All potential cases of psychopathology or addictive disorders were discussed in conferences that included licensed psychologists or psychiatrists. Lifetime consensus for DSM-IV diagnoses [American Psychiatric Association, 1994], including alcohol dependence (AD) and alcohol abuse (AA), was determined by available medical records, the MINI interviews, and the interviewer’s narrative. Participants diagnosed for either AD or AA were designated AUD in this study, in line with the proposed changes for the upcoming DSM-V that will collapse these two diagnoses into a unified disorder based on a graded scale of clinical severity, a move that is supported by strong evidence for the continuous nature of alcohol-related problems [Helzer et al., 2006]. Although our particular categorization for AUD is binary, our expectation is that risk alleles influence a range of symptoms throughout the Mexican-American sample, and thus we elected to focus our analyses on a state-independent consumption measure, Max Drinks, and its pleiotropic relationship with AUD.
Genotyping
Genome-wide genotyping of 1,203 subjects were performed using Illumina HumanHap550v3, HumanExon510Sv1, Human1Mv1 and Human1M-Duov3 BeadChips, according to the Illumina Infinium protocol (Illumina, San Diego, CA). All BeadChips were scanned using the Illumina® BeadArray™ 500GX Reader and genotypes were determined using the GenomeStudio Software package. In total, 944,565 high quality SNPs were available for analysis following quality control processing that excluded SNPs on the basis of low call rates, minor alleles present in < 10 individuals, and Hardy-Weinberg Equilibrium test statistics with p ≤ 10−4, adjusted for pedigree relationships using the SOLAR software (Texas Biomedical Research Institute, San Antonio, TX) [Almasy and Blangero, 1998]. Missing genotypes were imputed based on available pedigree data using the MERLIN package [Abecasis et al., 2002] and all SNPs were checked for Mendelian consistency with SimWalk2 [Sobel et al., 2002], an application that employs Markov chain Monte Carlo and simulated annealing algorithms to assign probabilities of genotyping error. Allele frequencies were computed using a maximum likelihood method in SOLAR.
For linkage analysis, a subset of 28,219 genotyped SNPs was used. Selection was based on 345 founders, with a target density of approximately 8 SNPs per cM (minimum spacing of 1,000 bp), a minimum minor allele frequency (MAF) of 0.05, and a maximum pairwise r2 of 0.02 within a 100 kb sliding window in order to limit linkage disequilibrium (LD) between markers.
Statistical Methods
All genetic analyses were performed using the SOLAR program [Almasy and Blangero, 1998], which uses a maximum likelihood (ML) variance decomposition approach to parameterize the genetic and environmental contributions to phenotypic traits by modeling the covariance among family members as a function of genetic relation. The overall mean parameters were adjusted for effects of age, age2, sex and their interactions, with the likelihood of the polygenic model calculated by assuming multivariate normal distributions for focal traits within pedigrees.
For linkage analyses, both univariate and bivariate [Williams et al., 1999a], standard variance components models were utilized. The log likelihoods of restricted linkage models in which the locus-specific heritabilities were constrained to zero were compared to models that estimated these components, with the differences distributed as ½:½ mixtures of a chi-square with one degree of freedom and a point mass at zero, which were converted into classical LOD scores [Ott, 1988].
To evaluate Max Drinks as a potential endophenotype to AUD, and thus its utility for identifying shared genetic factors, endophenotype ranking values (ERVs) were computed [Glahn et al., 2012]. The ERV index represents a standardized genetic covariance between the endophenotype and illness, and is defined by the following formula:
where is the heritability of the illness, is the heritability of the endophenotype, and ρg is their additive genetic correlation. Values range between 0 and 1 for candidate endophenotypes, where higher values indicate the stronger influence of a shared genetic component, enabling more focused genetic analyses that delineate the genetic complexity inherent in many clinical diagnoses. The significance of the ERV statistic was evaluated using a likelihood ratio test (LRT) of the restricted variance components model for phenotypic covariance (i.e., ρg constrained to zero) against the alternative model for which ρg is estimated, with the resultant test being asymptotically distributed as a chi-square with a single degree of freedom [Glahn et al., 2012].
To determine if any QTLs identified through linkage analysis of Max Drinks have pleiotropic effects on AUD, formal single degree of freedom LRTs were performed comparing bivariate linkage models at the location of the QTL with the estimated locus-specific heritability for AUD to ones fixed to zero [Almasy et al., 1997].
Association testing was performed on SNP markers under linkage peaks using measured genotype analyses (MGA) in SOLAR [Boerwinkle et al., 1986]. This single degree of freedom test assumes genetic additivity (i.e., minor alleles coded as 0, 1, or 2), and compares a model saturated for both the random effects of kinship, as represented by a residual heritability parameter, and the main effect of a SNP genotype to a null model that constrains the SNP effect to zero.
RESULTS
Descriptive Statistics of Max Drinks
Max Drinks information was collected from 1,203 Mexican-American individuals, with a mean of 12.01 (SD = 16.76). Due to the presence of extreme outliers, high degrees of skewness (g1 = 4.74) and kurtosis (g2 = 35.43) were detected in the data set. To ensure normality, measures above 30 drinks were winsorized, followed by inverse Gaussian transformation (g1 = −0.01; g2 = −0.49). For the winsorized data, the mean consumption score for male participants (15.41 ± 9.33) was found to be significantly higher (P < 0.0001) than the mean of their female counterparts (6.97 ± 6.97). When delineated according to AUD status (Fig. 1), the mean score of affected individuals (17.59 ± 8.42) was substantially higher than that of their unaffected relatives (6.41 ± 6.51; P < 0.0001), as one would expect, with similar differences observed within the sexes. Notably, when the AUD category was broken down by AD and AA diagnoses (Table SI), the mean consumption score was significantly higher among AD subjects (19.59 ± 8.06) than for AA individuals (13.13 ± 7.46), who in turn exhibited significantly higher scores than their unaffected relatives, regardless of sex (P < 0.0001 in each instance), underscoring the gradation in clinical severity across these two DSM-IV conditions as indexed by Max Drinks. Overall, 32.5% of our sample are diagnosed with lifetime AUD, which is comparable to the 30.3% (SE = 0.77) prevalence estimated for the US population based on the 2001–02 National Epidemiologic Survey on Alcohol and Related Conditions (NESARC; n = 43,093), although it is markedly higher than the estimate obtained for US Hispanics (21.0% ± 1.19) [Hasin et al., 2007]. By sex, 19.0% and 56.2% of females and males in our families, respectively, are diagnosed with AUD. In comparison to the NESARC data, for which the prevalence of AUD among females and males in the US are respectively 19.5% ± 0.64 and 42.0% ± 1.00, males in our study show an elevated rate of affection.
FIG. 1.
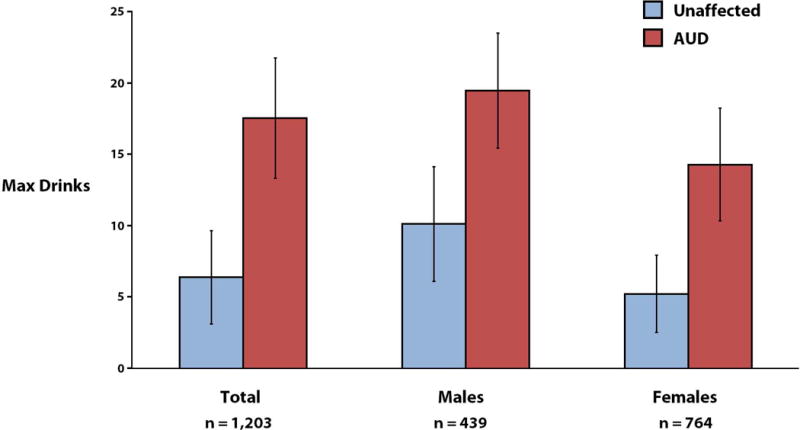
Means and standard deviations of winsorized Max Drinks measures, according to AUD status and sex.
Univariate Linkage Analysis of Max Drinks
We performed a genome-wide, multipoint linkage scan for QTLs influencing the normalized Max Drinks measure (Fig. 2 and Table SII), revealing a highly significant locus (LOD = 4.17; P = 5.85 × 10−6) at chromosome 6p22.3, between 49 to 51 cM units, spanning base pair positions 24,000 kb to 25,000 kb on the GRCh37 assembly build, representing 12 linkage markers. The peak region encompasses twelve genes (Table SIII), some with prior evidence of neuronal and/or cognitive effects, notably NRSN1, DCDC2, ALDH5A1, KIAA0319 and GMNN [De Rango et al., 2008; Jansen et al., 2008; Paracchini et al., 2008; Suzuki et al., 2009; Couto et al., 2009; Scerri et al., 2011; Darki et al., 2012; Pinel et al., 2012]. However, if one considers all LOD scores above 3.0, which are considered genome-wide significant (P-values ≤ 0.0001), the QTL becomes quite extensive, ranging from 49 to 55 cM units, which equates to about 10 Mb. Outside of this QTL, suggestive linkage signals (LOD > 1.5) were detected on chromosomes 3q21, 8p23 and 12q14.
FIG. 2.
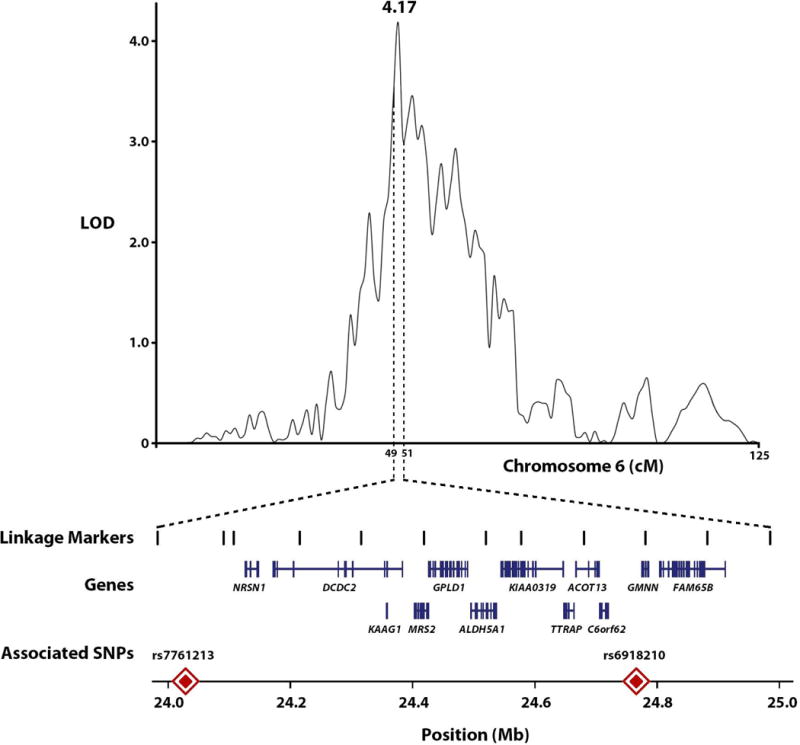
Multipoint linkage results for Max Drinks on chromosome 6, with locations of genes and associated SNPs under the linkage peak.
Endophenotype Relationship of Max Drinks to Alcohol Use Disorder
The estimated heritabilities of Max Drinks and AUD are 0.32 ± 0.05 (P = 4.61 × 10−14) and 0.34 ± 0.10 (P = 4.74 × 10−5), respectively, with a very strong genetic correlation of 0.99 ± 0.13 (P = 1.24 × 10−7) estimated for the two traits. This generated a highly significant ERV score of 0.33 (P = 2.27 × 10−9) for Max Drinks, indicating a substantial shared genetic component with AUD. However, a genome-wide linkage scan of AUD did not yield any significant or suggestive signals (i.e., all LODs < 1.5), including the QTL for Max Drinks on chromosome 6. This negative result likely stems from the dichotomous nature of AUD, as continuous measures are more informative, thus suggesting that the variation of Max Drinks among affecteds and/or among unaffecteds may be driving the signal at 6p22.
Bivariate Linkage Analysis of Chromosome 6
To evaluate whether the chromosome 6p22 Max Drinks QTL has pleiotropic effects on AUD, we performed a bivariate linkage analysis of the two phenotypes for chromosome 6. In these analyses we jointly estimated the effect of the locus on Max Drinks and AUD, with an LRT performed for a null model that constrained the local additive genetic variance of AUD. In these joint analyses of endophenotype and diagnosis, the QTL accounted for a modest proportion of the total variance in AUD (8.6%), however yielding a significant LRT result for a non-zero effect (P = 4.04 × 10−3), indicating that genetic variation in the 6p22 region jointly influence the amount of alcohol consumption in a 24 hour period and the risk of developing AUD.
Association Testing of SNP Markers at Linkage Peak
Association with Max Drinks was tested for each of the 491 SNPs genotyped at the 6p22 QTL (Table I). A significant Bonferroni-corrected association was detected for rs6918210 (P = 7.60 × 10−5; corrected P = 0.037) at nucleotide position (np) 24,753,933, upstream approximately 20 kb of the gene GMNN. The minor, derived allele (G→A) is associated with increased Max Drinks (β = 0.57 ± 0.14), with a ML allele frequency of 0.019 estimated for our families. According to HapMap3 data, the minor allele frequency (MAF) of rs6918210 ranges from 0.18 in African-Americans to its absence among East Asian population samples, with an estimate of 0.020 for Mexican-Americans residing in California. When tested for its association with AUD, rs6918210 shows near significant evidence of an association (P = 0.058), with the minor allele increasing the risk of diagnosis (β = 0.41 ± 0.22). When the two traits are examined jointly, the strong association is maintained (P = 2.94 × 10−4). Of the remaining tested SNPs, none display significant association with Max Drinks when corrected for multiple testing (Table SIV).
TABLE I.
Top Five SNP Association Results (n = 491) for Max Drinks at Chromosome 6p22 QTL
| SNP | Beta (SE) | Chi-Square | P-value | Variance Explained |
|---|---|---|---|---|
| rs6918210 | 0.57 (0.14) | 15.6 | 7.60 × 10−5 | 0.015 |
| rs7749154 | 0.68 (0.24) | 8.1 | 0.0044 | 0.0054 |
| rs2817738 | −0.11 (0.037) | 8.0 | 0.0048 | 0.0096 |
| rs1923185 | −0.10 (0.039) | 6.8 | 0.0091 | 0.0080 |
| rs12393 | −0.23 (0.090) | 6.8 | 0.0093 | 0.0057 |
DISCUSSION
In this study, we report a genome-wide significant QTL at chromosome 6p22.3 for Max Drinks in large Mexican-American pedigrees, with a significant association detected at rs6918210 near the gene GMNN, encoding the nuclear protein geminin, which is a cell cycle regulator and, interestingly, appears to be involved in the timing of neuronal differentiation [Kroll, 2007; Yellajoshyula et al., 2012]. In addition to GMNN, the approximately 1 Mb linkage region contains several other genes with plausible relevance to neurobiology, cognition and/or psychiatric illness. Of these, the gene ALDH5A1 is especially compelling, as it encodes for succinate semialdehyde dehydrogenase (SSADH), an enzyme that plays a key role in the degradation of γ-amino-butyric acid (GABA), the primary inhibitory transmitter in the mammalian brain, and whose inherited deficiency (MIM #271980) can lead to mild to severe neurological defects due to the accumulation of the byproduct γ-hydroxybutyrate (GHB) [Gibson et al., 2003; Pearl et al., 2009]. This metabolic pathway appears to be particularly relevant to our phenotypes of interest, as genetic loci involved in GAGAergic neurotransmission have been convincingly implicated in alcoholism and covarying neuroelectrical measures, most notably the GABAA receptor gene GABRA2 [Porjesz et al., 2002; Covault et al., 2004 ; Edenberg et al., 2004; Fehr et al., 2006]. These receptors are sensitive to ethanol and are believed to mediate many of its effects, including anxiolysis, sedation, and dependence [Buck and Finn, 2001; Ueno et al., 2001]. In a recent study, mutations in a GABA receptor (Gabrb1) were shown to promote alcohol consumption in mutated mouse models, causing spontaneous GABA ion channel opening and increased tonic currents in the nucleus accumbens, a region long associated with alcohol reward, thus identifying a potential mechanistic link to alcohol abuse [Anstee et al., 2013].
Other potential neurological genes located at this QTL include DCDC2 and KIAA0319, two genes involved in neuronal migration, both of which have been associated with dyslexia and reading performance [Meng et al., 2005; Paracchini et al., 2008; Scerri et al., 2011], as well as attention deficit hyperactivity disorder [Couto et al., 2009]. Additionally, NRSN1, which encodes a vesicular membrane protein, appears to be involved in neural organelle transport, nerve signal transduction, and neurite extension and regeneration [Suzuki et al., 2007]; and TTRAP, which codes for a TRAF and TNF receptor associated enzyme, may play a role in the neurodegenerative decline observed in Parkinson’s disease [Vilotti et al., 2012].
To the best of our knowledge, the 6p22 region has not been previously implicated in alcohol consumption and/or AUD. However, given that Mexican-Americans and other Hispanic populations have not been well studied with regards to the genetics of alcoholism, the QTL may be population specific. To investigate this further, we examined association results for 119 SNPs from this linkage region that were genotyped in extended families from northern Hunan Province, China (n = 381), for which a daily Max Drinks measure was collected [Quillen et al., 2013]. Although rs6918210 was not genotyped in this sample, we identified another SNP, rs7761213 at np 24,028,403, upstream of NRSN1 and DCDC2 (Fig. 2), to be significantly associated with Max Drinks after correcting for multiple-testing (P = 2.14 × 10−4; corrected P = 0.025). The minor, derived allele (C→T) is associated with increased drinks (β = 0.24 ± 0.22), with an estimated allele frequency of 0.21, which falls within the MAF range of 0.20 to 0.46 for other global populations based on HapMap3 data. Thus, this finding independently confirms the potential influence of the region on alcohol consumption beyond our Mexican-American sample, although no evidence of an association with alcohol dependence (P = 0.25) was observed at this site in these Chinese pedigrees.
Despite detecting two significant associations at our QTL, the genetic determinants that are responsible for the signal are unclear without fully sequencing the region. Although rs6918210 and rs7761213 are closest to the genes GMNN and NRSN1, respectively, common SNP markers are capable of LD tagging the effects of causal variants as distant as 2.5 Mb away [Dickson et al., 2010]. And given the number of strong candidate genes found in the region, the linkage peak may harbor multiple functional variants from various genes that are distributed along familial lines.
As for its potential influence on AUD risk, our bivariate linkage models reveal significant pleiotropic effects (P = 4.04 × 10−3). This finding underscores the utility of the endophenotype approach as a means of localizing potential disease susceptibility genes, as high noise-to-genetic signal ratios, which typically plague genetic studies of complex, heterogeneous psychiatric disorders, can be reduced by targeting specific components of pathophysiologies through the examination of endophenotypes. One of the most seminal examples of gene discovery using this strategy is GABRA2, whose effects on a neuroelectrical endophenotype (EEG beta frequency) [Rangaswamy et al., 2002] allowed for its chromosomal localization [Porjesz et al., 2002] and eventual association to alcoholism [Edenberg et al., 2004]. However, this approach to disease gene discovery is dependent upon the endophenotype selected for study. According to Gottesman and Gould (2003), for an endophenotype to be informative it should be heritable, quantitative, genetically correlated to the disease of interest, and co-segregate within families. For Max Drinks, these criteria are met, with an ERVscore (i.e., standardized statistic of genetic covariance) of 0.33, a highly significant value (P = 2.27 × 10−9) driven by its strong genetic correlation to AUD.
In conclusion, we report the findings of the first genetic study of Max Drinks in a Hispanic population, identifying a novel, genome-wide significant QTL at chromosome 6p22.3 with potential pleiotropic effects on AUD. Within this linkage region, we observe a significant association for Max Drinks at SNP rs6918210, with a nearly significant effect on AUD risk. For an independent sample of Chinese families, we discover another SNP at this QTL, rs7761213, which is significantly associated with Max Drinks, thus indicating that the genetic variation in the region may have effects on alcohol consumption that is not population specific. Future analyses will examine sequence data under the linkage signal, a region with a number of compelling gene candidates, testing for association to Max Drinks and AUD. This study highlights the distinct advantages of the endophenotype strategy for delineating the complex genetic architectures that underlie most psychiatric illnesses. Although much of the heritability of alcoholism remains unknown, improved success is likely to come from the more intensive interrogation of rare variants, as well as the examination of quantitative endophenotypes, such as Max Drinks, that targets specific components of disease liability.
Supplementary Material
Table SI. Mean Estimates of Winsorized Measures of Max Drinks, Delineated by Clinical Diagnoses and Sex.
Table SII. Genome-Wide, Multipoint Linkage Results for Max Drinks.
Table SIII. Genes Found at the Max Drinks QTL on Chromosome 6p22.
Table SIV. SNP Association Results (n = 491) for Max Drinks at Chromosome 6p22 QTL.
Acknowledgments
Financial support for this study was provided by the National Institute of Mental Health Grants MH0708143 (Principal Investigator [PI]: DCG), MH078111 (PI: JB), and MH083824 (PI: DCG & JB); and NIAAA grants AA015973 and AA11330. Theoretical development of SOLAR is supported by National Institute of Mental Health Grant MH59490 (PI: JB). This investigation was conducted, in part, in facilities constructed with support from Research Facilities Improvement Program Grant Numbers C06 RR13556 and C06 RR017515 from the National Center for Research Resources, National Institutes of Health. We thank participants of the Genetics of Brain Structure and Function Study and the Chinese family study, as well as our research staffs, notably Henry Kranzler. The supercomputing facilities used for this work at the AT&T Genomics Computing Center were supported, in part, by a gift from the AT&T Foundation and by the National Center for Research Resources Grant Number S10 RR029392.
Footnotes
Authors declare no competing financial interests in relation to the described work.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Freedman ND, Cheng Y-C, Lin P, Shaffer JR, Sun Q, Taylor K, Yaspan B, Cole JW, Cornelis MC, et al. Measuring alcohol consumption for genomic meta-analyses of alcohol intake: opportunities and challenges. Am J ClinNutr. 2012;95:539–547. doi: 10.3945/ajcn.111.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. fourth. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Anstee QM, Knapp S, Maguire EP, Hosie AM, Thomas P, Mortensen M, Bhome R, Martinez A, Walker SE, Dixon CI, et al. Mutations in the Gabrb1 gene promote alcohol consumption through increased tonic inhibition. Nat Commun. 2013;4:2816. doi: 10.1038/ncomms3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Saccone NL, Rice JP, Goate A, Foroud T, Edenberg H, Almasy L, Conneally PM, Crowe R, Hesselbrock V, et al. Defining alcohol-related phenotypes in humans. The Collaborative Study on the Genetics of Alcoholism. Alcohol Res Health. 2002;26:208–213. [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. PNAS. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Finn DA. Genetic factors in addiction: QTL mapping and candidate gene studies implicate GABAergic genes in alcohol and barbiturate withdrawal in mice. Addiction. 2001;96:139–149. doi: 10.1046/j.1360-0443.2001.96113910.x. [DOI] [PubMed] [Google Scholar]
- Couto JM, Gomez L, Wigg K, Ickowicz A, Pathare T, Malone M, Kennedy JL, Schachar R, Barr CL. Association of attention-deficit/hyperactivity disorder with a candidate region for reading disabilities on chromosome 6p. Biol Psychiatry. 2009;15:368–375. doi: 10.1016/j.biopsych.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault C, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Darki F, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T. Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo-parietal white matter structure. Biol Psychiatry. 2012;72:671–676. doi: 10.1016/j.biopsych.2012.05.008. [DOI] [PubMed] [Google Scholar]
- De Rango F, Leone O, Dato S, Novelletto A, Bruni AC, Berardelli M, Mari V, Feraco E, Passarino G, De Benedictis G. Cognitive functioning and survival in the elderly: the SSADH C538T polymorphism. Ann Hum Genet. 2008;72:630–635. doi: 10.1111/j.1469-1809.2008.00450.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Nurnberger J, Jr, Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL, et al. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol ClinExp Res. 2002;26:1453–1460. doi: 10.1097/01.ALC.0000034037.10333.FD. [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006a;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006b;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut L, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol ClinExp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, et al. Alcoholism susceptibility loci: Confirmation studies in a replicate sample and further mapping. Alcohol ClinExp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, Farrer L. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, et al. Genomewide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–9. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KM, Gupta M, Pearl PL, Tuchman M, Vezina LG, Snead OC, III, Smit LM, Jakobs C. Significant behavioral disturbances in succinic semialdehyde dehydrogenase (SSADH) deficiency (gamma-hydroxybutyric aciduria) Biol Psychiatry. 2003;54:763–768. doi: 10.1016/s0006-3223(03)00113-6. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW, Jr, Charlesworth JC, Johnson MP, Göring HH, Cole SA, Dyer TD, et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012;71:6–14. doi: 10.1016/j.biopsych.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, et al. Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry. 2009;66:795–800. doi: 10.1016/j.biopsych.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Heath AC, Meyer J, Jardine R, Martin NG. The inheritance of alcohol consumption patterns in a general population twin sample: II. Determinants of consumption frequency and quantity consumed. J Stud Alcohol. 1991;52:425–433. doi: 10.15288/jsa.1991.52.425. [DOI] [PubMed] [Google Scholar]
- Heath AC, Martin NG. Genetic influences on alcohol consumption patterns and problem drinking: results from the Australian NH&MRC twin panel follow-up survey. Ann NY AcadSci. 1994;708:72–85. doi: 10.1111/j.1749-6632.1994.tb24699.x. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: Consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Helzer JE, van den Brink W, Guth SE. Should there be both categorical and dimensional criteria for the substance use disorders in DSM-V? Addiction. 2006;101(Suppl 1):17–22. doi: 10.1111/j.1360-0443.2006.01587.x. [DOI] [PubMed] [Google Scholar]
- Hunt KJ, Lehman DM, Arya R, Fowler S, Leach RJ, Göring HHH, Almasy L, Blangero J, Dyer TD, Duggirala R, et al. Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes. 2005;54:2655–2662. doi: 10.2337/diabetes.54.9.2655. [DOI] [PubMed] [Google Scholar]
- Jansen EE, Struys E, Jakobs C, Hager E, Snead OC, Gibson KM. Neurotransmitter alterations in embryonic succinate semialdehyde dehydrogenase (SSADH) deficiency suggest a heightened excitatory state during development. BMC DevBiol. 2008;8:112. doi: 10.1186/1471-213X-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC. Alcoholism and major depression in women. Arch Gen Psych. 1993;50:690–698. doi: 10.1001/archpsyc.1993.01820210024003. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA. Genes, Environment, and Pscychopathology: Understanding the Causes of Psychiatric and Substance Use Disorders. New York: Guilford Press; 2006. [Google Scholar]
- Kendler KS, Myers J, Dick D, Prescott CA. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol ClinExp Res. 2010;34:1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Madden PA, Bucholz KK, Slutske WS, Nelson EC, Statham D, Whitfield JB, Martin NG. Genetic effects on alcohol dependence risk: Re-evaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34:1519–1530. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- Kroll KL. Geminin in embryonic development: coordinating transcription and the cell cycle during differentiation. Front Biosci. 2007;12:1395–1409. doi: 10.2741/2156. [DOI] [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TL. Meta-analysis of ALDH2 and ADH1B in Asians. Psychol Bull. 2006;132:607–621. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: An integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M. Phenotyping alcoholism. Alcohol ClinExp Res. 1999;23:757–758. doi: 10.1111/j.1530-0277.1999.tb04180.x. [DOI] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O’Reilly-Pol T, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. PNAS. 2005;102:17053–8. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Wiegand R, Bucholz KK, O’Connor S, Meyer ET, Reich T, Rice J, Schuckit M, King L, Petti T, et al. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch Gen Psychiatry. 2004;61:1246–1256. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, Williamson DE, Duggirala R, Blangero J, Glahn DC. Common genetic influences on depression, alcohol, and substance use disorders in Mexican-American families. Am J Med Genet B. 2011;156B:561–568. doi: 10.1002/ajmg.b.31196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J. Analysis of Human Genetic Linkage. Baltimore, MD: The John Hopkins University Press; 1988. [Google Scholar]
- Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, et al. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165:1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- Pearl PL, Gibson KM, Cortez MA, Wu Y, Carter Snead O, III, Knerr I, Forester K, Pettiford JM, Jakobs C, Theodore WH. Succinic semialdehyde dehydrogenase deficiency: Lessons from mice and men. J Inherit Metab Dis. 2009;32:343–352. doi: 10.1007/s10545-009-1034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinel P, Fauchereau F, Moreno A, Barbot A, Lathrop M, Zelenika D, Le Bihan D, Poline JB, Bourgeron T, Dehaene S. Genetic variants of FOXP2 and KIAA0319/TTRAP/THEM2 locus are associated with altered brain activation in distinct language-related regions. J Neurosci. 2012;32:817–825. doi: 10.1523/JNEUROSCI.5996-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, Goate A, Rice JP, O’Connor SJ, Rohrbaugh J, et al. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci USA. 2002;99:3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: Evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Quillen EE, Chen X-D, Almasy L, Yang F, He H, Li X, Wang X-Y, Liu T-Q, Hao W, Deng H-W, et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet Part B. 2013 doi: 10.1002/ajmg.b.32213. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Bauer LO, Rohrbaugh J, O’Connor SJ, Kuperman S, Reich T, et al. Beta power in the EEG of alcoholics. Biol Psychiatry. 2002;52:831–842. doi: 10.1016/s0006-3223(02)01362-8. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Saccone FS, Saccone NL, Neuman RJ, Rice JP. Genetic analysis of the maximum drinks phenotype. BMC Genet. 2005;6(Suppl 1):S124. doi: 10.1186/1471-2156-6-S1-S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, et al. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Scerri TS, Morris AP, Buckingham LL, Newbury DF, Miller LL, Monaco AP, Bishop DV, Paracchini S. DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol Psychiatry. 2011;70:237–245. doi: 10.1016/j.biopsych.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Alcohol-use disorders. Lancet. 2009;373:492–501. doi: 10.1016/S0140-6736(09)60009-X. [DOI] [PubMed] [Google Scholar]
- Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, Hergueta T, Baker R, Dunbar GC. The Mini-International Neuropsychiatric Interview (M.I.N.I): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59:22–33. [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Tohyama K, Nagata K, Taketani S, Araki M. Regulatory expression of Neurensin-1 in the spinal motor neurons after mouse sciatic nerve injury. NeurosciLett. 2007;421:152–157. doi: 10.1016/j.neulet.2007.03.077. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Harris RA, Messing RO, Sanchez-Perez AM, Hodge CW, McMahon T, Wang D, Mehmert KK, Kelley SP, Haywood A, et al. Alcohol actions on GABA(A) receptors: from protein structure to mouse behavior. Alcohol Clin Exp Res. 2001;25:76S–81S. doi: 10.1097/00000374-200105051-00014. [DOI] [PubMed] [Google Scholar]
- Vilotti S, Codrich M, Dal Ferro M, Pinto M, Ferrer I, Collavin L, Gustincich S, Zucchelli S. Parkinson’s disease DJ-1 L166P alters rRNA biogenesis by exclusion of TTRAP from the nucleolus and sequestration into cytoplasmic aggregates via TRAF6. PLoS One. 2012;7:e35051. doi: 10.1371/journal.pone.0035051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Zhu G, Madden PA, Neale MC, Heath AC, Martin NG. The genetics of alcohol intake and of alcohol dependence. Alcohol ClinExp Res. 2004;28:1153–1160. doi: 10.1097/01.alc.0000134221.32773.69. [DOI] [PubMed] [Google Scholar]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999a;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and even-related potentials. Am J Hum Genet. 1999b;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellajoshyula D, Lim JW, Thompson DM, Jr, Witt JS, Patterson ES, Kroll KL. Geminin regulates the transcriptional and epigenetic status of neuronal fate-promoting genes during mammalian neurogenesis. Mol Cell Biol. 2012;32:4549–4560. doi: 10.1128/MCB.00737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table SI. Mean Estimates of Winsorized Measures of Max Drinks, Delineated by Clinical Diagnoses and Sex.
Table SII. Genome-Wide, Multipoint Linkage Results for Max Drinks.
Table SIII. Genes Found at the Max Drinks QTL on Chromosome 6p22.
Table SIV. SNP Association Results (n = 491) for Max Drinks at Chromosome 6p22 QTL.


