Abstract
The maintenance of immune tolerance requires the deletion of self-reactive T cells in the thymus. The expression of tissue-specific antigen genes (TSAs) by thymic epithelial cells is critical for this process and depends on the activity of the Autoimmune Regulator (Aire) protein, however, the molecular mechanism(s) Aire uses to target TSA gene loci are unknown. Here we identified two Aire-interacting proteins – activating transcription factor 7 interacting protein (ATF7ip) and methyl CpG binding protein 1 (MBD1) –that are required for Aire’s targeting of TSA geneloci. Moreover, Mbd1−/− mice developed pathological autoimmunity and had a defect in Aire-dependent thymic TSA gene expression underscoring the critical importance of Aire’s interaction with the ATF7ip-MBD1 protein complex in maintaining central tolerance.
Immunological tolerance to self-antigens is vital to the prevention of autoimmune disease. Central tolerance is established in the thymus and depends on the presentation of tissue-specific antigens (TSAs) to developing T cells for the negative selection of autoreactive T cells1, 2, 3. TSA genes have limited expression and are repressed in the vast majority of tissues except for the thymus which has the unique ability to express thousands of TSAs. Induction of expression of TSAs in the thymus partially relies on the protein Autoimmune Regulator (Aire) which is expressed in specialized medullary thymic epithelial cells (mTECs)4. The importance of Aire function is underscored by the finding that homozygous loss of AIRE in humans results in the Autoimmune Polyglandular Syndrome Type I (APS1) which is characterized by the triad of adrenal insuffuciency, hypoparathyroidism, and mucocutaneous candidias is along with multiple other autoimmune manifestations5, 6. Similarly, Aire deficient mice develop multiorgan autoimmunity4. Despite the clear role of Aire in promoting TSA expression, the molecular mechanisms by which Aire carries out this activity have not been completely worked out.
Studies suggest that Aire functions as a transcriptional regulator, however, it does not seem to act as a typical transcription factor by binding to well-defined consensus sequences within the promoters of target genes to activate transcription7. Multiple functions in transcriptional regulation have been attributed to Aire through the identification of novel Aire-interacting proteins. Initially, Aire was found to interact with the transcription factor, CREB-binding protein (CBP)8, and p-TEFb9 which linked Aire to transcriptional elongation by activating stalled RNA polymerase10. Further evidence that Aire has a role in transcriptional elongation came when it was shown to bind to a complex of proteins centered around DNA-PK11, 12 (DNA-PK, TOP2, PARP-1, FACT, KU) which is thought to enhance transcription by initiating double-strand DNA breaks and removing nucleosomes in front of RNA polymerase13. Aire also appears to interact with a number of splicing factors which enable Aire to enhance pre-mRNA processing11. While these novel Aire-interacting proteins have further confirmed a role for Aire in many aspects of transcription, the exact mechanism Aire uses to target repressed TSA loci has not been fully elucidated.
Initial clues to the mechanism Aire may use to target TSA genes came from the determination that one of the pleckstrin homology domains (PHD) of Aire specifically binds to H3 histone tails that have an unmethylated mark at lysine 414, 15; a histone mark associated with gene repression. Aire’s interaction with this mark has been hypothesized to help target Aire to repressed TSA gene loci in mTECs7, 11. Despite this link, it has become clear that this recognition module is not sufficient for recruitment of Aire to TSA target genes16, and suggests that other epigenetic processes may be involved in driving the specificity of Aire for particular target genes. Additional evidence that epigenetic mechanisms are involved in Aire’s function comes from the observation that the repertoire of genes induced by Aire varies by cell type11, 17, 18. These results suggest that the epigenetic landscape within an individual cell is a major determinant in Aire function11, 17, 18. There are some other characteristic features of the transcriptional targets of Aire in that they often have chromosomal clustering19, are transcribed in a stochastic manner20, 21, and are not transcribed from typical TSA gene associated promoters21. All of these factors together further support the notion that Aire does not act as a typical transcription factor and that it functions in a unique fashion to drive TSA gene expression22.
The functions of several of the structural domains of the Aire protein still remain to be defined, including its SAND domain. This structural domain of approximately 80 amino acids in length is found in a number of nuclear proteins including Aire, Sp10023, 24, Gmeb125, and Deaf126. The SAND domain in several family members has been shown to be a DNA binding domain, however, the Aire-SAND domain is missing a key DNA-binding motif (KDWK) that is observed in the DNA binding family members27. Structural analysis of the SAND domain has also shown that it contains a β pleated sheet, suggesting that it may also participate in protein-protein interactions28. A mutation within the SAND domain, G228W (GW), results in an autosomal dominant autoimmune syndrome which differs from typical APS1 in its inheritance29. Here, we hypothesized that the SAND domain of Aire may mediate an important protein-protein interaction that links Aire to other epigenetic processes.
Using a screening approach, we identified an Aire-SAND domain interacting protein, activating transcription factor 7 interacting protein (ATF7ip). Furthermore, we found that Aire can also form a complex with methyl CpG binding protein 1 (MBD1), a protein previously known to interact with ATF7ip that recognizes methylated CpGs. In genome wide expression analyses, we found that Aire and MBD1 localize to similar target genes. Together, these results help explain how Aire is targeted to a diverse yet select group of TSA genes, allowing it to induce their expression and establish tolerance.
Results
Aire interacts with ATF7ip and MBD1
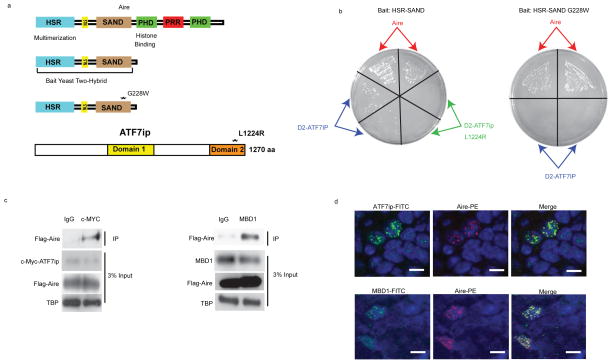
To identify novel Aire-interacting proteins, we devised a yeast two-hybrid screen designed to enrich for physiologically relevant binding partners. Using an Aire-GFP transgenic mouse reporter line, Aire-expressing mTECs were isolated with high purity to generate the screening cDNA library (Supplementary Fig. 1a)30. Focusing on SAND domain interactions, we then probed the library for potential Aire binding partners, using the HSR-SAND domains of Aire (Fig. 1a) as the bait protein. This screening approach yielded several proteins, including full length Aire and a portion of ATF7ip. The identification of Aire provided internal validation of the screen, as Aire is known to multimerize through the HSR domain and to be enriched in the mTEC cDNA library8. ATF7ip, also known as MCAF (MBD1-containing chromatin-associated factor)31, 32 or mAM (mouse ATF associated modulator)33, is an essential cofactor in the generation of the repressive histone mark, H3K9me333. ATF7ip acts in a trimolecular complex with MBD1 and the histone-methyltransferase ESET-SETDB1 to mediate transcriptional repression; this complex is targeted to specific genomic loci through the interaction of MBD1 with methylated CpG’s that are enriched in the regulatory regions of silent genes34, 35.
Figure 1.
Aire interacts with ATF7ip and MBD1. (a) Structural Domains of Aire and ATF7ip. The HSR-SAND domain of Aire was used as bait in the yeast-two hybrid screen. G228W (GW) is a known human mutation of AIRE in which glycine at amino acid 228 is changed to tryptophan. Domain 1 and Domain 2 are known functional domains of ATF7ip. L1224R is a well described point mutation of ATF7ip in which leucine at amino acid 1224 is changed to arginine and results in disruption of ATF7ip binding to MBD1. (b) Representative example of yeast plates from pairwise mating using either HSR-SAND or HSR-SAND GW as bait and Aire or ATF7ip domain 2(D2) as prey. L1224R is a known mutation of ATF7ip in which leucine at amino acid 1224 is changed to arginine. Each mating was performed in duplicate as indicated by arrows. (c) Coimmunoprecipitation (Co-IP) of c-Myc-ATF7ip or endogenous MBD1 with Flag-Aire in HEK 293 cells. Immunoblot shows expression of the various proteins with TATA-binding protein (TBP) used as a loading control. (d) Immunofluorescence of thymic medulla from mice stained for either Aire (red) and ATF7ip (green) or Aire (red) and MBD1(green). Blue indicates nuclear DAPI staining. Scale bar = 10um. Data are representative of two experiments (b) or three experiments (c, d).
The C-terminal region of ATF7ip known as “Domain 2” (D2)31, 32 interacted with HSR-SAND in the two-hybrid screen (Fig. 1a). D2 is required for interacting with the TRD (transcriptional repression domain) of MBD1 and is critical for ATF7ip’s function in transcriptional repression (Supplementary Fig. 1b)31, 32. To map the specific domains that mediate the interaction between Aire and ATF7ip, pairwise yeast two-hybrid matings were performed (Table 1). Pairwise mating using the HSR-SAND domain as bait and D2 as prey confirmed the original interaction seen in our screen (Table 1, Fig. 1b). Introduction of a well-described mutation (L1224R) in D2, which disrupts MBD1 and ATF7ip binding31, 32, abrogated the interaction between HSR-SAND and D2. Similarly, insertion of a known human Aire SAND domain mutation (G228W)29, 36 disrupted the HSR-SAND and D2 interaction, indicating that the SAND domain is required for Aire’s interaction with ATF7ip. The specificity of the SAND-D2interaction was further confirmed with pairwise tests using only the SAND domain or HSR domain as bait with D2 as prey (Supplementary Fig. 1c).
Table 1.
Summary of prey and bait mating results utilized in a pairwise yeast two-hybrid assay. A positive interaction is indicated with the (+) symbol. Data is representative of two experiments.
| BAIT | Prey | Binding |
|---|---|---|
| HSR-SAND | Aire | + |
| HSR-SAND | D2-ATF7ip | + |
| HSR-SAND | D2-ATF7ipL1224R | − |
| HSR-SAND G228W | Aire | + |
| HSR-SAND G228W | D2-ATF7ip | − |
| HSR | Aire | + |
| HSR | D2-ATF7ip | − |
| SAND | Aire | − |
| SAND | D2-ATF7ip | + |
To complement the pairwise mating data, co-immunoprecipitation (Co-IP) was performed in HEK 293 cells transfected with epitope-tagged full-length Aire and ATF7ip (Fig. 1c) and confirmed the interaction of Aire and ATF7ip. To determine if Aire potentially interacted with the ATF7ip-MBD1-ESET protein complex and not just with ATF7ip in isolation, additional Co-IP experiments were performed. Aire interacted with both endogenous and epitope-tagged MBD1 by Co-IP, suggesting that Aire binds to the MBD1-ATF7ip protein complex (Fig. 1c and Supplementary Fig. 1d). Further Co-IP experiments with MBD1 truncation mutants indicated that the CXXC domains of MBD1 are required for the Aire-MBD1 interaction (Supplementary Fig. 1b, d). Finally, immunohistochemistry of thymic tissue showed clear nuclear co-localization of Aire with both ATF7ip and MBD1 in Aire-expressing mTECs (Fig. 1d), providing evidence for the interaction of these protein complex members in vivo.
ATF7ip and MBD1 are required for Aire-dependent gene expression
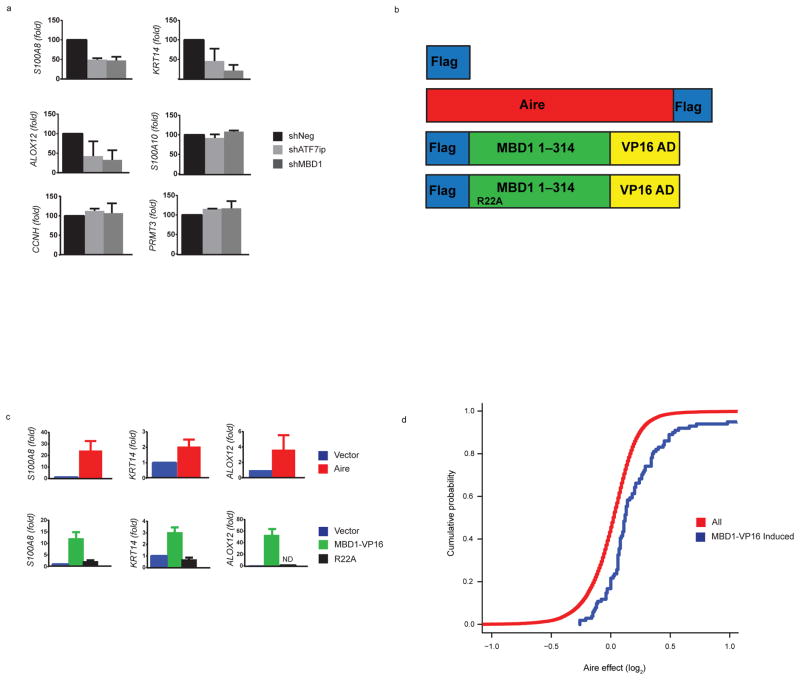
To test the functional relevance of Aire’s interaction with both ATF7ip and MBD1, we next performed shRNA knockdowns of ATF7ip and MBD1 in a cell line model. To facilitate these experiments, a stable cell line expressing epitope-tagged Aire in HEK 293 cells was generated as previously described15. Analysis of the stable cell line confirmed a wild-type speckling pattern of nuclear Aire expression (Supplementary Fig. 2a) as well as the induction of gene expression of the well-characterized Aire-dependent targets S100A8, KRT14, and ALOX12 (Supplementary Fig. 2b)11. shRNA knockdowns of ATF7ip or MBD1 in the Aire-expressing stable cell line resulted in decreased expression of S100A8, KRT14, and ALOX12; however, there was no effect on Aire-dependent gene expression when using a scrambled shRNA (Fig. 2a and Supplementary Fig. 2c). Furthermore, the shRNA knockdowns did not globally alter gene expression as no effect was seen on the previously described11, 15 non Aire-regulated genes S100A10, CCNH, and PRMT3 (Fig. 2a). The observation that ATF7ip and MBD1 were critical for Aire-mediated induction of these selected target genes, led us to hypothesize that Aire’s specificity for TSAs may be broadly explained by the specificity of MBD1 for selected methylated CpG’s in the genome37.
Figure 2.
ATF7ip and MBD1 are required for Aire-induced gene expression and Aire and MBD1-VP16 globally upregulate a similar set of genes. (a) Knockdown of ATF7ip and MBD1 in cells stably expressing Aire. HEK 293 cells that stably express Flag-Aire were transiently transfected with vectors that express scrambled shRNA (shNeg), ATF7ip shRNA (shATF7ip), or MBD1 shRNA (shMBD1) and the effect on Aire-dependent (S100A8, KRT14, ALOX12) and Aire-independent (S100A10, CCNH, PRMT3) gene expression was assessed by qPCR. (b) Schematic of plasmids utilized for gene expression: Flag only (Vector), Aire-Flag (Aire), Flag-MBD1-VP16 (MBD1-VP16), or Flag-MBD1-VP16-R22A (R22A). The Flag-MBD1-VP16 construct has the first 314 amino acids of MBD1 fused to the VP16 transactivation domain at its C terminus. The R22A mutant has arginine at amino acid 22 mutated to alanine and disrupts MBD1 CpG binding.(c) Plasmids outlined in (b) were transiently transfected into HEK 293 cells and Aire-induced gene expression was analyzed normalized to GAPDH. Fold induction was calculated above vector only (Flag) with the vector only control normalized to 1. (d) Effects of Aire on expression of MBD1-VP16-inducible transcripts (>2-fold induction, blue line) versus all transcripts (red line). There is a significant rightward shift in cumulative distribution for MBD1-VP16 transcripts (p < 10−8 by two-tailed Kolmogorov Smirnov tests). (e) Venn Diagram showing the overlap of all genes that are significantly regulated (FDR < 0.05) by Aire and MBD1-VP16. In (a, c) three separate transfections (biologic replicates) were performed. Data from each biologic replicate was combined and the mean +/− s.e.m. are shown.
Aire and MBD1-VP16 induce similar genes
MBD1 is one member of a family of proteins that bind methylated CpGs and includes MeCP2, MBD2, MBD3, and MBD4. MBD1, MeCP2, and MBD2 all contain a TRD domain and recruit different co-repressor complexes to chromatin35. Although some studies support functional redundancy between MBD1 family members38, other reports have highlighted that certain MBD proteins may differentially target specific methylated CpGs based on the surrounding genomic DNA sequence. For example, MeCP2 binds methylated CpG’s enriched in surrounding A/T base pairs39 whereas MBD1 binds more efficiently to a single methylated CpG within the genomic DNA sequences TCpGCA or TGCpGC37. To determine the identity of genes repressed by MBD1 in our cell line model, we utilized a previously described approach whereby the repressive TRD domain of MBD1 is replaced by the strong VP16 transcriptional activation domain (MBD1-VP16) (Fig. 2b)37; this modification results in transcriptional activation rather than repression of MBD1 target genes. Prior work has shown that when MBD1-VP16 was transfected into HeLa cells it induced a specific set of genes that was different from either MECP2-VP16 or MBD2-VP1637. Moreover, the majority of the genes induced by MBD1-VP16 were described as being tissue-specific antigens37.
We found evidence that the MBD1-VP16 fusion could indeed specifically induce the expression of the known Aire-target genes S100A8, KRT14, and ALOX12 (Fig. 2c) and had no effect on the non Aire-regulated gene S100A10 (Supplementary Fig. 2d, e). Furthermore, a MBD1-VP16 fusion carrying an R22A mutation that disrupts methylated CpG binding37 was unable to induce expression of Aire-target genes (Fig. 2c). To extend these findings to the genome-wide level, we next performed microarray analysis on RNA from HEK 293 cells transiently transfected with a control vector, Aire, or MBD1-VP16. On a global level, the overlap of genes regulated by Aire and MBD1-VP16 was striking. Numerous transcripts were induced by both Aire and MBD1-VP16 (Supplementary Fig. 3a, b) and, the cumulative density distribution of the Aire effect on transcription revealed a shift in transcripts induced by MBD1-VP16 versus all transcripts (Fig. 2d), suggesting a large number of shared targets. Finally, a Venn diagram showing the intersection of significant genes (FDR <0.05) for both Aire and MBD1-VP16 demonstrated a substantial overlap (~approximately 80%) (Fig. 2e). The qPCR and microarray results with Aire and MBD1-VP16 indicate that both proteins induce a similar set of genes and thus Aire may target MBD1 for Aire-dependent gene expression.
Aire and MBD1 target Aire-dependent gene promoters
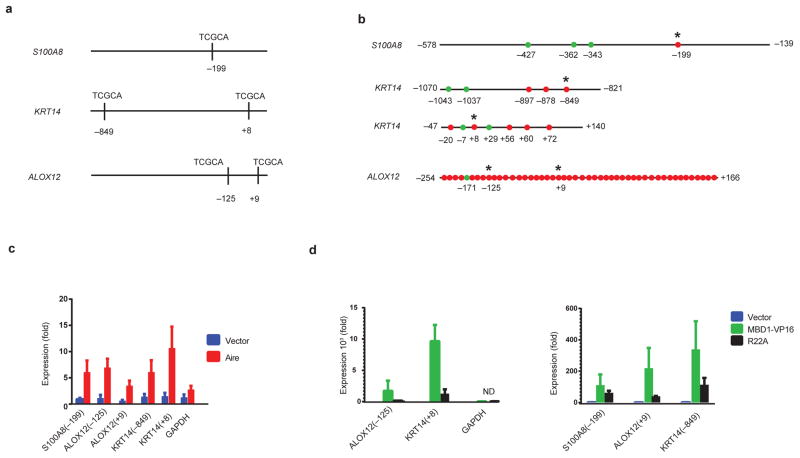
To determine if the substantial overlap in gene induction by Aire and MBD1-VP16 on microarray analysis was secondary to their direct regulation of target genes, chromatin immunoprecipitation (ChIP) with Aire and MBD1-VP16 was performed on selected targets. Previous studies have shown that Aire is located at the promoter of S100A8 in HEK293 cells15. The region encompassed by the ChIP-qPCR primers used in the prior study contained a potential high-affinity MBD1 binding site (TCGCA). Additional analysis of the promoters of the KRT14 and ALOX12 genes showed that both genes contained two potential high-affinity MBD1 binding sites (TCGCA) within approximately 1kb of the transcriptional start site (Fig. 3a). In order for MBD1 to bind to these sites, the cytosine within these sites must be methylated (TCpGCA). To determine the methylation status of the high-affinity sites, bisulfite sequencing of the S100A8, KRT14, and ALOX12 was performed and uncovered that the sites contained methylated cytosines in HEK 293 cells (Fig. 3b).
Figure 3.
Aire and MBD1 are localized at the promoters of Aire-dependent genes. (a) Schematic shows the location of high affinity MBD1 target sites (TCGCA) in the promoters of S100A8, KRT14, and ALOX12 relative to the transcriptional start site. (b) Bisulfite sequencing of the promoters of S100A8, KRT14, and ALOX12 indicate that cytosines within high-affinity MBD1 binding sites are methylated. Red circles indicate methylated CGs while green dots indicate unmethylated CGs. * indicates the location of the CG corresponding to the MBD1 high-affinity binding sequence. Numbers indicate the location of specific CGs with respect to the transcriptional start site. (c) Chromatin immunoprecipitation (ChIP) utilizing epitope tagged Aire. Plasmids expressing Flag only (Vector) or Aire-Flag (Aire), were transiently transfected into HEK 293 cells and ChIP qPCR was performed utilizing primers surrounding the high-affinity MBD1 binding sites. (d) ChIP utilizing epitope tagged MBD1-VP16 or R22A. Plasmids expressing Flag only (Vector), Flag MBD1-VP16 (MBD1-VP16), or Flag MBD1-VP16 R22A (R22A) were transiently transfected into HEK 293 cells and ChIP qPCR was performed utilizing primers surrounding the high-affinity MBD1 binding sites. A Flag immunoprecipitation was utilized to pulldown both Aire and MBD1-VP16 in ChIP assays. In (c, d) three separate transfections (biologic replicates) were performed. Data from each biologic replicate was combined and the mean +/− s.e.m. are shown.
Moreover, ChIP analysis revealed that both Aire (Fig. 3c) and MBD1-VP16(Fig. 3d) were located at the high-affinity MBD1 binding sites within the promoters of S100A8, KRT14, and ALOX12 in HEK 293 cells. ChIP analysis with an MBD1-VP16 fusion carrying an R22A mutation showed reduced binding at these sites. Overall, the magnitude of MBD1-VP16 binding was greater than that of Aire for target sites; this observation could have been due to enhanced expression of MBD1-VP16 (Supplementary Fig. 2e) or secondary to direct DNA binding by MBD1-VP16 compared to indirect binding of Aire to the DNA-protein complex. The substantial overlap in gene induction by Aire and MBD1-VP16 combined with the colocalization of Aire and MBD1 at select methylated CpGs indicates that Aire’s interaction with the MBD1-ATF7ip protein complex targets Aire to repressed TSA loci and leads to their expression.
Mbd1−/− mice develop autoimmunity
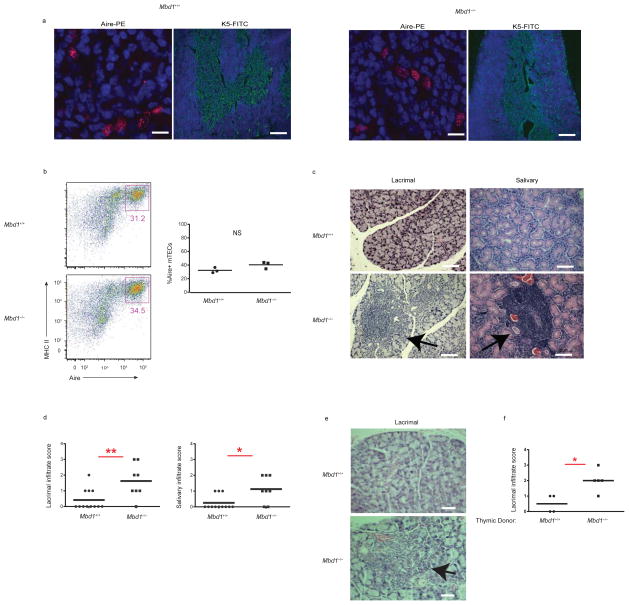
Taken together, the results of the ChIP and microarray experiments indicate that Aire and MBD1 are found at similar locations on genomic DNA in a transfected cell line model system. To extend these findings to an in vivo model, we next analyzed the Mbd1-deficient mouse, which has been previously described to have a neurologic phenotype40. The thymus of Mbd1-deficient mice showed normal architecture with normal numbers of Aire+ mTECs and without alterations in thymocyte subsets or thymic regulatory T cells (Tregs) (Fig. 4a, b and Supplementary Fig. 4). Although the thymus of Mbd1-deficient mice appeared normal, over time these mice spontaneously developed a Sjögren’s-like disease with mononuclear infiltrates of the lacrimal and salivary glands (Fig. 4c, d). Notably, this pattern of autoimmunity is similar to that of a knock in mouse model carrying the G228W mutation of the Aire SAND domain in heterozygosity36, a mutation that also cripples the Aire-ATF7ip interaction (Fig. 1b). To determine if the autoimmunity seen in Mbd1−/− was dependent on the thymus, thymic stroma from Mbd1−/− and Mbd1+/+ mice were transplanted into athymic C57BL/6 mice. Infiltration of the lacrimal gland (Fig. 4e, f) was seen at a higher incidence and greater severity in mice that received thymi from Mbd1−/− donors. These results suggest that that the autoimmunity seen in Mbd1−/− mice tracks with the thymic stroma.
Figure 4.
MBD1 is required for the prevention of autoimmunity. (a) Immunofluorescent staining for Aire (red) (scale bar = 25um) and K5 (green) (scale bar = 200um) in thymic frozen sections from MBD1 wildtype (Mbd1+/+) and MBD1 knockout (Mbd1−/−) mice. (b) Flow cytometry of DAPI-, CD11c-, CD45-, Ly51-, EPCAM+, Class II+, Aire+ mTECs from Mbd1+/+and Mbd1−/− mice. Percentage of Aire+ mTECs within total thymic mTECs from three mice of each genotype (Mbd1+/+and Mbd1−/−). (c) Hematoxylin and eosin (H&E) staining from lacrimal and salivary gland sections from Mbd1+/+and Mbd1−/− mice aged to approximately 30 weeks. Arrows indicate mononuclear infiltrates. Scale bar = 100um. (d) Infiltration scores of lacrimal and salivary glands from a cohort Mbd1+/+and Mbd1−/− mice aged to approximately 30 weeks. (e) Mbd1−/− mice develop spontaneous autoimmune disease that is thymus dependent. Representative H&E staining of the lacrimal gland from C56BL/6 nude mice transplanted with Mbd1+/+ and Mbd1−/− thymi. Arrow indicates infiltrates observed in nude mice receiving Mbd1−/− thymi. Scale bar = 50um. (f) Infiltration scores of the lacrimal gland of thymic transplants aged 8 weeks after transplantation. The genotypes of thymic donors (either Mbd1+/+or Mbd1−/−) are shown. In (b, d, f), each circle or square represents one mouse and the mean infiltrate is shown. * indicates p value < 0.05. ** indicates p-value <0.01. NS = not statistically different.
MBD1 regulates mTEC TSA gene expression in vivo
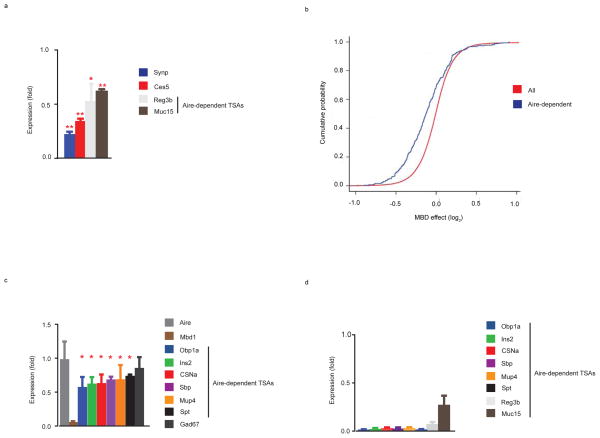
Next, to examine how the loss of MBD1 in vivo affects global TSA gene expression, microarray analysis was performed on flow sorted mTECs from Mbd1-deficientmice and wild-type littermate controls. A table showing the top 20 MBD1 induced genes revealed that similar to Aire4 the majority of the genes fit the criteria of being a TSA and as expected Mbd1 was the most highly differentially expressed gene (Table 2) (Supplementary Table 1). qPCR confirmation of the array was performed in Mbd1-deficient mTECs for several differentially expressed transcripts including the previously described10 Aire-target genes Reg3b and Muc15 (Fig. 5a). On a global level, MBD1 induced genes (Supplementary Table 1) showed a statistically significant enrichment (p=5.06×10−8) of TSA genes by permutation testing with 48.9% of the MBD1 induced genes meeting the criteria of a TSA. Moreover, a cumulative probability plot of the global MBD1 effect on transcription revealed a clear shift to the left of previously published Aire-dependent genes10 (p<2.2 × 10−16) (Fig. 5b), indicating that Aire-dependent gene expression is decreased in Mbd1-deficient mice. To validate that Aire-dependent gene expression was decreased in Mbd1−/− mice, qPCR was performed for selected targets in Mbd1-deficient mTECs and this showed a significant reduction in expression of all of the Aire-dependent TSAs that were tested (Fig. 5c). In parallel, qPCR was performed on Aire-deficient mTECs for several Aire target genes (Fig. 5d). As previously published4, 11, 36, these transcripts were dramatically reduced in Aire-deficient mice. Taken together, these results demonstrate that MBD1 promotes the proper expression of Aire-dependent TSAs in mTECs, and contributes to the maintenance of immune tolerance. Furthermore, our data indicate that Aire may target the MBD1-ATF7ip protein complex for TSA expression in vivo (Supplementary Fig. 5).
Table 2.
Table of the top 20 MBD1 induced genes identified by microarray analysis of flow sorted mTECs from Mbd1+/+and Mbd1−/− mice. Genes are labeled by gene symbol, gene name, gene ID, fold induction, p-value, and tissue specificity. Genes were assigned into one of the following categories for tissue specificity: TSAs (1–5 tissues), housekeeping (> 5 tissues), below the threshold in the database, and not accessible in the database. P values generated using a moderated T statisitic. Data is representative of three experiments.
| Gene Symbol | Gene Name | Gene ID | Mbd1+/+ vs Mbd−/− Fold Induction | P value | Tissue Specificity |
|---|---|---|---|---|---|
| Mbd1 | methyl-CpG binding domain protein 1 | 17190 | 9.145298342 | 2.47E-08 | Housekeeping |
| Mrps35 | mitochondrial ribosomal protein S35 | 232536 | 6.533840784 | 3.99E-06 | Housekeeping |
| Plcz1 | phospholipase C, zeta 1 | 114875 | 3.85141123 | 0.000541754 | TSA |
| Synpr | synaptoporin | 72003 | 3.115568541 | 0.003523747 | TSA |
| NAP043897-1 | 2.725867077 | 0.001398677 | Not accessible | ||
| Hbb-b1 | hemoglobin, beta adult major chain | 15129 | 2.561777232 | 0.003836832 | Housekeeping |
| Arntl2 | aryl hydrocarbon receptor nuclear translocator-like 2 | 272322 | 2.545439487 | 0.003059776 | Below Threshold |
| Hebp1 | heme binding protein 1 | 15199 | 2.347980628 | 0.000348697 | Housekeeping |
| Hddc3 | HD domain containing 3 | 68695 | 2.334891062 | 0.001899169 | TSA |
| NAP046281-1 | 2.324057824 | 0.000201846 | Not accessible | ||
| BC064078* | cDNA sequence BC064078 | 408064* | 2.294147562 | 8.24E-05 | TSA |
| Ces5 | carboxylesterase 5 | 234673 | 2.289398244 | 0.002825006 | TSA |
| Pap (Reg3b) | pancreatitis- associated protein | 18489 | 2.216612695 | 0.003591019 | TSA |
| Klhdc5 | kelch domain containing 5 | 232539 | 2.109659755 | 0.00068541 | Housekeeping |
| Anxa13* | annexin A13 | 69787* | 2.102752679 | 0.001741643 | TSA |
| 2010204N08Rik* | RIKEN cDNA 2010204N08 gene | 69983 | 2.063155969 | 0.000666053 | Not accessible |
| Lrp1b | low density lipoprotein-related protein 1B | 94217 | 2.044290519 | 0.000664668 | TSA |
| Mro* | Maestro | 71263* | 2.038691453 | 0.000202676 | Below Threshold |
| Muc15 | mucin 15 | 269328 | 2.032227953 | 0.000393921 | TSA |
| Apoc3 | apolipoprotein C-III | 11814 | 2.009903405 | 0.000426861 | TSA |
Figure 5.
MBD1 regulates Aire-dependent TSA gene expression in vivo (a) qPCR assays for several transcripts identified by microarray analysis and shown in (Table 2) utilizing RNA prepared from Mbd1+/+ and Mbd−/− mTECs. Expression (fold) is the expression of the indicated targets in Mbd1−/− mTECs relative to Mbd1+/+ mTECs. (b) Cumulative distribution plot of all transcripts with cumulative probability plotted on the Y axis and log2 MBD1 effect on the X axis. Red line indicates all transcripts and blue line shows Aire-dependent gene expression. A two-tailed Kolmogorov Smirnov (K-S) test was used for statistical analysis. (c) qPCR assays utilizing RNA prepared from Mbd1+/+ and Mbd1−/− mTECs for a group of Aire-dependent TSAs and the Aire-independent TSA Gad67. Expression (fold) is the expression of the indicated targets in Mbd1−/− mTECs relative to Mbd1+/+ mTECs. (d) qPCR assays utilizing RNA prepared from Aire+/+ and Aire−/− mTECs for a group of Aire-dependent TSAs. Expression (fold) is the expression of the indicated targets in Aire−/− mTECs relative to Aire+/+ mTECs. Bar graphs represent the mean +/− s.d. * indicates p value < 0.05. ** indicates p value < 0.01. NS = not statistically different. In (a, c), mTECs were sorted from Mbd1+/+and Mbd−/− mice three times (3 biologic replicates). Data from each biologic replicate was combined and the mean +/− s.d. are shown. In (d), data are from one representative experiment of two experiments with similar results.
Discussion
We have identified two novel Aire-interacting proteins, ATF7ip and MBD1 that illuminate a new molecular mechanism for Aire-mediated transcriptional regulation. Although Aire’s selective specificity for inducing the expression of TSAs has been appreciated for some time, the question of how Aire targets such a diverse array of genes has remained difficult to resolve. The previous identification of the Aire-PHD1 domain’s binding to H3K4me0 has implied an epigenetic mechanism for Aire’s recognition of repressed TSA loci14, 15; however, further studies revealed that Aire’s interaction with H3K4me0 alone did not account for Aire’s unique specificity16. Here we provide new insight into the specificity of Aire targeting, whereby Aire coopts the normally repressive MBD1-ATF7ip complex and utilizes the preferential recognition of specific methylated CpGs provided by MBD1 to localize to target TSA loci. Support for this model comes from the large number of shared transcripts induced by Aire and MBD1-VP16 and our ChIP data that localizes Aire and MBD1-VP16 to the promoter regions of Aire-dependent genes that contain the high-affinity MBD1 binding sites. Moreover, the necessity of the MBD1-ATF7ip protein complex is underscored by the requirement for MBD1 for Aire-dependent gene expression in two distinct cell types. In HEK 293 cells, knockdown of either MBD1 or ATF7ip reduces Aire-dependent gene expression. In mTECs, MBD1 expression is required for Aire-dependent TSA gene expression and for the prevention of organ-specific autoimmunity.
Our data supports a model in which the preferential recruitment of Aire to sites of selective repression by the ATF7ip-MBD1 protein complex likely works in concert with the PHD1 domain-mediated recognition of the H3K4me014, 15, 16 motif to identify TSA target loci and thus unifies two distinct epigenetic recognition methods in the Aire targeting mechanism. In fact, such a multivalent strategy appears to be a recurrent theme for many epigenetic complexes 41. Evidence links CpG island recognition with a PHD domain recognition in the histone demethylase KDM2A42 which would parallel our findings here. Furthermore, one can envisage a model for a multivalent recognition pattern for Aire that is similar to the RAG1-RAG2 complex whereby efficient recombination targeting relies on both RAG1’s interaction with the recombination signal sequence and RAG2’s interaction with H3K4me3 via its PHD domain for correct and precise targeting of the complex.
After interacting with repressive epigenetic marks, Aire likely recruits other proteins that promote transcriptional elongation11, such as DNA-PK12, and pre-mRNA processing11 which ultimately leads to TSA gene expression. In vivo support for the role of the DNA-PK in TSA gene expression comes from analysis of DNA-PK deficient mTECs which have a reduction in Aire-dependent TSA gene expression11. These results are similar to Mbd1−/− mice which also have a reduction in TSA gene expression; however, knockout of either Mbd1 or DNA-PK does not completely abolish TSA gene expression in mTECs. As evidenced by the large number of Aire binding partners discovered to date, there is likely functional redundancy in many of its interactions which could explain the milder phenotypes observed. An additional explanation for why MBD1 deficiency did not completely abolish TSA gene expression could be redundancy between MBD family members in vivo or alternatively, other yet to be identified factors that interact with ATF7ip as a co-factor38.
Here, we have also demonstrated a novel function for the Aire-SAND domain in a protein-protein interaction. Previous studies had suggested that the Aire-SAND domain functioned as a DNA binding domain27; however, this function was called into question because the Aire-SAND DNA binding domain lacks the canonical KDWK DNA binding motif found in other Sp100 family members7, 27. Previous structural analysis of the SAND domain revealed that it also contains a β pleated sheet suggesting that the domain may also participate in a protein-protein interaction28. Here, we show that the SAND domain of Aire participates in a protein-protein interaction with ATF7ip. Thus, it will also be of interest to determine if similar interactions can be identified in other SAND domain containing proteins. Furthermore, we also determined that a disease causing mutation in AIRE in the SAND domain (G228W) blocks the ability of AIRE to bind ATF7ip. This finding helps potentially clarify the mechanism by which this patient mutation inhibits AIRE function and further study will be needed to determine if other mutations that disrupt this interaction may also predispose to APS1.
This model alone helps resolve, in part, a perplexing unanswered question in Aire biology specifically related to why Aire targets different genes in different cell types. Aire overexpression inmonocytic U937 cells18, pancreatic β cells17, HEK 293 cells11, or the1C6 TEC line11 induces a different repertoire of genes in each cell type respectively. Furthermore, in pancreatic β cells the repertoire is skewed toward TSA genes, however, they are a different repertoire of TSAs than in mTECs17. Aire’s interaction with the MBD1-ATF7ip protein complex might help explain why Aire induces divergent genes in different cell types because it is well-described that the regulatory regions of repressed genes are enriched in methylated CpGs, and that this methylation pattern varies in a cell-type specific manner43. Therefore, MBD1’s differential binding to specific methylated CpG’s in different cell types may explain the differential target gene induction by Aire.
In summary, we have identified a mechanism Aire exploits to specifically target repressed TSA loci that likely works in a mutualistic fashion with the Aire-H3K4me0 interaction. Whether Aire uses one particular targeting mechanism for a particular TSA gene remains to be determined. The biological relevance of Aire having a dual-targeting mechanism could have been naturally selected for to ensure the continued maintenance of robust central tolerance. Given the unique ability of the thymus to express a wide-array of self-antigens, future studies will aim to identify other novel transcriptional mechanisms that Aire utilizes to drive this important immunologic process.
Full Methods and associated references are available in the online version of this paper.
Materials and Methods
Plasmids and Antibodies
Plasmids expressing epitope tagged Aire, ATF7ip, and MBD1 in HEK 293 were cloned by in frame insertion of a mouse Aire cDNA, mouse ATF7ip cDNA, or human MBD1 cDNA into the following vectors: pCMV-3TAG1 (Flag-Aire, Flag- MBD1 1-314, and Flag-MBD1 1-161), pCMV-3TAG2 (c-Myc-Aire and c-Myc-ATF7ip). MBD1-VP16 was created by cloning the VP16 activation domain in frame to the C terminus of Flag-MBD 1-314. Aire-Flag was a kind gift from Jakub Abramson (Weizmann Institute of Science, Rehovot, Israel). Plasmids used for bait and prey in yeast two-hybrid were cloned using either mouse Aire cDNA or mouse ATF7ip cDNA. Specific bait constructs cloned in frame into pGBKT7 were Aire HSR-SAND, Aire HSR-SAND with glycine at amino acid 228 mutated to alanine (GW), Aire HSR, and Aire SAND. Prey constructs included ATF7ip domain 2 (D2), ATF7ip D2 with leucine at amino acid 1224 mutated to arginine (L1224R), Aire HSR, Aire SAND, and AIRE HSR-SAND. The Aire GW mutation, D2 L1224 mutation, and the MBD1-VP16 R22A mutation were introduced using Agilent’s Quickchange Site-Directed Mutagenesis Kit. pLKO plasmids containing shRNA for the negative control, ATF7ip, or MBD1 were purchased from Sigma.
Antibodies used for coimmunoprecipitation and western blotting were as follows: FLAG M2 (Sigma, F1082), Flag M2-HRP (Sigma, A8592), c-Myc (Sigma, C 3956), c-Myc-HRP (Sigma, A5598), MBD1 (IMGENEX, IMG-306), ATF7ip (Sigma, HPA016578), and TBP(abcam, ab818). Antibodies for immunofluorescence included: Aire (ebioscience, 14-5934-82), ATF7ip (Bethyl, IHC-00187), K5 (abcam, ab53121), DYKDDDDK Tag Antibody (Cell Signaling, #5407), and MBD1 (IMGENEX, IMG-306). All antibodies for flow cytometry and cell sorting were purchased from Biolegend except Aire (ebioscience, 14-5934-82).
Yeast Two-Hybrid
A yeast two-hybrid screen was performed following Clontech’s Matchmaker Two-Hybrid System. Using our Aire-GFP transgenic reporter mice, mTECs were isolated by flow cytometry followed by cell-sorting. A total of twelve mice were used. RNA was isolated using Agilent Technologies Absolutely RNA Kit and a cDNA library was then created using Clontech’s SMART cDNA Library Construction Kit which results in a double-stranded (ds) cDNA library. The ds cDNA library was then cloned into the pGADT7-Rec(2) vector which was used as prey in the yeast two-hybrid screen. The bait and prey plasmids were then transformed into the yeast strain AH109 and the transformed yeast were then mated and plated on double dropout SD/-Leu/-Trp plates. Diploids were selected and grown on quadruple dropout plates (SD/-Leu/-His/-Leu/-Trp/X-a-gal) to identify interacting proteins. Protein interactions were confirmed by pairwise yeast-two hybrid mating following Clontech Matchmaker’s instructions in which the AH109 yeast strain transformed with specific bait and prey constructs were mated and serially selected on double and then quadruple dropout plates.
Cell Culture and Transfection
HEK 293 cells were cultured in DMEM supplemented with 10% FBS, L-glutamate, and Pen/Strep antibiotics in a humidified incubator supplemented with 10%CO2. The day before transfection cells were counted and seeded on either 6 well or 10cm plates. Transfection was performed with the indicated plasmids using Mirus Trans-IT HEK 293 transfection reagent. For the creation of stable cell lines, HEK 293 cells were transfected into 6 well plates. 48 hrs post-transfection media was supplemented with 1.0g/L of G418 and allowed to grow 7–10 days until all cells on the non-transfected plate were killed. Cells were then trypsinized and diluted to 150 cells/ml and seeded in 96 well plates and allowed to grow for 2 weeks in the presence of 1.0g/L G418. Individual colonies were expanded and screened for Aire expression by immunofluorescence.
Mice and Genotyping
Aire-GFP reporter transgenic mice (Adig) have previously been described30. Mbd1+/− mice were rederived from the UC Davis Mutant Mouse Resource Consortium (MMRC). Heterozygous mice were bred to homozygosity and genotyped by PCR using primers supplied by the MMRC. Mbd−/− mice were created in the B6/129 mixed background and were backcrossed to B6 for at least 4 generations. Aire−/− mice have been previously described4. Mice were maintained in SPF facilities following all of the UCSF Institutional Animal Care and Use Committee (IACUC) approved protocols.
Thymic Transplants
Thymi from 1–3 day old neonatal mice were removed and cultured in transwell plates for 7 days in 1.35 mM 2′-deoxyguanosine (2-dG) (Sigma-Aldrich) in complete DMEM-10 to deplete thyme of hematopoietic cells. Thymi were washed in complete DMEM-10 (without 2-dG) 2 hours prior to transplantation into Nude mice in the C57BL/6J background (Jackson Labs). Nude mice were anesthetized and Thymi were transplanted under the kidney capsule. T cell reconstitution was confirmed by the presence of CD4+ and CD8+ T cells in the peripheral blood. Transplanted mice were aged 8 weeks prior to analysis.
Flow Cytometry and Cell Sorting
Thymic stromal cells were isolated from 30 day old mice as previously described36. Briefly, thymi were minced with razor blades and then digested for 1 hr in 100ug/ml DNase I (Roche), 0.125% Collagenase D (Roche), and 100ug/ml Collagenase/Dispase (Roche). Digested cells were then resuspended in PBS containing 0.5% BSA and 2mM EDTA and passed through a 70um cell strainer. Density gradient centrifugation was performed using a three layered Percoll gradient (GE Healthcare) with specific gravities of 1.115, 1.065, and 1.0. Thymic stromal cells were isolated from the light fraction between 1.0 and 1.065 and resuspended in FACS buffer and counted. In preparation for antibody staining, thymic stromal cells were incubated with the Fc blocking antibody 2.4G2 for 10 minutes on ice. Cells were then stained with the indicated antibodies for 30 minutes on ice and washed two times with FACS buffer prior to flow cytometry using the LSR II (BD Biosciences) and analyzed using FloJo data analysis software (Treestar). Medullary thymic epithelial cells (mTECs) were sorted using the same isolation and staining procedure as above, however, cells were isolated with the BD Aria3u cell sorting machine (Beckton Dickinson). mTECs were sorted based on the following markers: DAPI-, CD11c-, CD45-, Ly51-, EPCAM+, Class II+.
Lymphocytes were prepared for FACS analysis by first isolating the indicated organ (thymus, spleen, or lymph node) followed by mashing and filtering the cells through a 70um cell strainer. Spleen samples were further processed by performing red cell lysis in ACK lysis buffer for two minutes. Cells were then resuspended in FACS buffer and incubated with the Fc blocking antibody 2.4G2 for 10 minutes. Lymphocytes were then stained with the indicated antibodies for 30 minutes on ice and washed two times with FACS buffer prior to flow cytometry using the LSR II (BD Biosciences) and analyzed using FloJo data analysis software (Treestar).
Coimmunoprecipitation (Co-IP)
HEK 293 cells were seeded on 10cm plates and transfected with the indicated plasmids. 48 hrs post-transfection cells were lysed as previously described 11. Briefly, cells were first lysed in a hypotonic lysis buffer (0.05% NP-40, 10mM HEPES, 1.5mM MgCL2, 10mM KCL, 5mM EDTA, complete protease inhibitor (Roche). Nuclei were isolated and incubated for 1 hr in nuclear lysis buffer (20 mM HEPES, 300 mM NaCl, 20 mM KCl, EDTA free Protease Inhibitor cocktail containing Nuclease S7 (Roche)). Nuclear lysate was then used in Co-IP utilizing Life Technologies Dynabead’s Protein G Immunoprecipitation Kit. Eluted proteins were loaded onto Novex Life technologies mini-gels, separated, and then transferred to PVDF membranes. Western blotting was performed with the indicated antibodies followed by enhanced chemiluminescence (ECL).
Quantitative Polymerase Chain Reaction (qPCR)
For the analysis of Aire-dependent gene expression in HEK 293 cells, the indicated plasmids were transfected into HEK 293 cells seeded on 6 well plates. Approximately 72 hrs post-transfection, cells were lysed in Trizol Reagent (Life Technologies). RNA was resuspended in DEPC water and 2ug of total RNA was used to create cDNA using Life Technologies Superscript III Reverse Transcriptase and oligo-(dt) primers. Human primers for ALOX12, KRT14, S100A8, S100A10, GAGE1, TRIM49, PRMT3, CCNH, GAPDH, and SPRYD5 were purchased from Applied Biosystems (ABI). qPCR was carried out utilizing ABI 10X master mix on the ABI 7500 Fast Real-Time PCR machine. Transcript levels in each reaction were normalized to GAPDH and results were analyzed by the ABI standard curve method. Fold induction was calculated relative to vector only (Flag) with the vector only control normalized to 1. For the analysis of Aire-dependent gene expression in mTECs, approximately 100,000 mTECs were sorted and RNA was isolated using Qiagen’s RNeasy Micro Plus Kit. RNA was resuspended in DEPC water and 100ng of total RNA was used to create cDNA using Life Technologies Superscript III Reverse Transcriptase and oligo-(dt) primers. Mouse primers for Synpr, Ces5, Reg3b, Muc15, Aire, MBD1, OPB1a, Ins2, Spt1, CSNa, SBP, MUP4, GAD67, and ACTB were purchased from Applied Biosystems (ABI). qPCR was carried out as above with transcript levels normalized to ACTB.
Chromatin Immunoprecipitation (ChIP)
HEK 293 cells were seeded onto 10cm plates and transfected with the indicated plasmids. 72 hrs post-transfection ChIP was carried out using Millipore’s Magna ChIP A/G Immunoprecipitation Kit. Cells were fixed for 15 minutes with 1% formaldehyde and quenched with glycine prior to cell lysis and sonication to 200–800 bp fragments using Diagenode’s BIORUPTER. Approximately one million cells were used for overnight immunoprecipitation, with 1.0 ug of either an irrelevant mouse IgG or Flag M2 (Sigma). ChIP-qPCR was performed using ABI SYBR Green Master Mix on the ABI 7500 Fast Real-Time PCR machine. Fold induction was calculated over the irrelevant IgG control and normalized to the %Input for each sample. S100A8 and GAPDH primers have been previously published15. The following additional primer pairs were utilized:
5′KRT14 (−849) CGAGGCTTCTTGCCATACTC
3′KRT14 (−849) CATCTGCAAAGTGGGAGTCA
5′KRT14 (+8) ATGAAAGCCAAGGGGAATG
3′KRT14 (+8) AGGTGGTCATGGTGCAGAG
5′ALOX12 (−125) TTATCATCCTGCAGCTACGC
3′ALOX12 (−125) AGTTTTAGGCGCTCCCAAGT
5′ALOX12 (+9) CCGCAGACCGGTCCTTTA
3′ALOX12 (+9) GTTGTACGACCCGGAGAAGA
Immunohistochemistry
Frozen thymic sections were cut to 8um sections and fixed 1:1 methanol:acetone for 10 minutes at −20 degrees. After 1 hr of air-drying, thymic sections were blocked with 10% normal goat serum before staining for 1hr with the indicated primary antibodies. Sections were washed three times with PBS, before staining for 1hr with the appropriate FITC or PE conjugated secondary antibody. Staining was visualized with the Leica SP5 confocal microscope acquired with Leica confocal software.
Histology
Organs were harvested and fixed in formaldehyde at room temperature overnight. The following day organs were washed in 30% ETOH for 30minutes and then transferred to 70%ETOH prior to embedding, sectioning, and staining with H&E. Scoring of infiltrates was done as previously performed in a blinded fashion44 with scores of 0, 1, 2, 3, or 4 indicating no, < 25% infiltrate, 25%–50% infiltrate, 50%–75% infiltrate, or >75% infiltrate.
Microarray probe preparation and hybridization
Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility and the Gladstone Genomics Core Facility. RNA was isolated for arrays as described above for qPCR. Total RNA quality was assessed using a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA was amplified using the Sigma whole transcriptome amplification kits, following the manufacturer’s protocol (Sigma-Aldrich, St Louis, MO), and subsequent Cy3-CTP labeling was performed using NimbleGen one-color labeling kits (Roche-NimbleGen Inc, Madison, WI). Labeled Cy3-cDNA was assessed using the Nanodrop ND-8000 (Nanodrop Technologies, Inc., Wilmington DE), and equal amounts of Cy3 labeled target were hybridized to Agilent GE 4×44 mouse for the arrays utilizing mouse mTECs. Hybridizations were performed for 17 hrs, according to the manufacturer’s protocol. Arrays were scanned using the Agilent microarray scanner and raw signal intensities were extracted with Feature Extraction v10.6 software.
A similar protocol was followed for the arrays in HEK 293 cells; however, Affymetrix GeneChip Human gene 1.0 ST arrays were utilized. RNA was isolated and amplified as above for Agilent arrays and then labeled with Biotin using the NuGen Ovation Pico WTA V2 and NuGEN Encore Biotin module. Hybridization was performed following the GeneChip Expression Analysis Technical Manual 701021 Rev. 3 with 2.5ug of cDNA used for hybridization to Affymetrix GeneChip Human gene 1.0 ST. Microarrays were normalized for array-specific effects using Affymetrixs “Robust Multi-Array” (RMA) normalization.
Microarray data analysis
All Microarray data analysis was performed in conjunction with Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility and the Gladstone Genomics Core Facility. All microarray datasets were normalized using a quantile normalization method45. No background subtraction was performed, and the median feature pixel intensity was used as the raw signal before normalization.
A one-way ANOVA linear model was fit to the array data to estimate the mean signal intensity for each group. Moderated t-statistics, B statistics, false discovery rates and p-values were computed for each gene for the comparison of interest. All procedures were carried out using functions in the R package limma in Bioconductor46, 47.
Tissue-specific antigen analysis was performed as previously described30 using previously published expression data across 34 mouse tissues (GSE 1133). Genes with signal intensity greater than 200 in 1–5 tissues were considered a TSA. Each of the following was considered one tissue: 1) small and large intestine, 2) CNS tissues, and 3) eye tissues. Cell lines and embryonic tissues were excluded from analysis. In addition, genes with tissue-specificity for the thymus were excluded.
Bisulfite Sequencing
Genomic DNA from HEK 293 cells was isolated using the Wizard Genomic DNA Purification Kit (Promega). Approximately 500ng of genomic DNA was bisulfite treated with the EZ DNA Methylation Kit (Zymo Research). Primers specific for bisulfite treated DNA were used to generate PCR products that were then subcloned into the pGEM T Easy Vector (Promega) and amplified in bacteria. At least 10 colonies were screened to determine if specific cytosines were methylated. The following primers were used for methylation specific PCR:
5′ALOX12METH – GGGAGGTTTAGGAAGGTTT
3′ALOX12METH – AACCCTATCCCTAATCTTACCCACC
5′KRT14METH – GGGTTAGTTTTATAAGGGGTTTAAA
3′KRT14METH – ACAATCAACTCTAAAAATCCAATAC
5′KRT14METH – TAAGGGGAATGGAAAGTGTTAGAT
3′KRT14METH – CAAAAACCCTTCATAAAACTAAAAAAA
5′S100A8METH – GTTAGTGATTTTGGGGTTTTAG
3′S100A8METH - CTAACCCTCCCCAAAAAAAACC
Statistics
Statistical analysis was performed using Prism 6 (Graphpad) software. Mann-Whitney Rank sum testing was performed for histological analysis with p< 0.05 considered significant. Student’s T-test was performed for mouse mTEC TSA gene analysis with p<0.05 considered significant. Two-tailed Kolmogorov Smirnov (K-S) tests were implemented in R, standard. The permutation test was in-house code performed in conjunction with the SABRE Center Functional Genomics Core Facility. A p-value was generated by determining the probability of choosing X TSA’s randomly, out of a database that is Y% TSA.
Supplementary Material
Acknowledgments
We would like to thank Jeffrey Bluestone, Abul Abbas, and members of the Bluestone and Abbas laboratories for helpful discussions. Mickie Cheng and Taylor LaFlam provided critical feedback on the manuscript. M.W. was supported by the Arthritis Foundation (postdoctoral fellowship) and is currently supported by the Rheumatology Research Foundation (Young Investigator Award). M.S.A is supported by the Burroughs Welcome Fund and the NIH. Microarray and microscopy experiments were supported by a Diabetes and Endocrinology Research Center (DERC) program project grant (DK59958).
Footnotes
Accesion Codes. Microarray Data deposited in Gene Expression Omnibus GSE52548.
Note: Any Supplementary Data is available in the online version of the paper.
Author Contributions: M.W. and M.A.S. wrote the manuscript and designed the experiments; M.W., I.S.K, J.T.C, T.M, U.F., A.G, and M.S. carried out the experiments, K.F. performed histology, M.M.L., J.L.P., and D.J.E. performed microarray analysis.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2(11):1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 2.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23(2):227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4(4):350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298(5597):1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 5.Consortium F-GA. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17(4):399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 6.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17(4):393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 7.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 8.Pitkanen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275(22):16802–16809. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 9.Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27(24):8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. 2012;109(2):535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell. 2010;140(1):123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 12.Liiv I, Rebane A, Saare M, Maslovskaja J, Kisand K, Juronen E, et al. DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim Biophys Acta. 2008;1783(1):74–83. doi: 10.1016/j.bbamcr.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahaney B, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A. 2008;105(41):15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9(4):370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh AS, Kingston RE, Benoist C, Mathis D. Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc Natl Acad Sci U S A. 2010;107(29):13016–13021. doi: 10.1073/pnas.1004436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerau-de-Arellano M, Mathis D, Benoist C. Transcriptional impact of Aire varies with cell type. Proc Natl Acad Sci U S A. 2008;105(37):14011–14016. doi: 10.1073/pnas.0806616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sillanpää N, Magureanu CG, Murumägi A, Reinikainen A, West A, Manninen A, et al. Autoimmune regulator induced changes in the gene expression profile of human monocyte-dendritic cell-lineage. Mol Immunol. 2004;41(12):1185–1198. doi: 10.1016/j.molimm.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202(1):33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci U S A. 2008;105(2):657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci U S A. 2008;105(41):15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venanzi ES, Melamed R, Mathis D, Benoist C. The variable immunological self: genetic variation and nongenetic noise in Aire-regulated transcription. Proc Natl Acad Sci U S A. 2008;105(41):15860–15865. doi: 10.1073/pnas.0808070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloch DB, Nakajima A, Gulick T, Chiche JD, Orth D, Suzanne M, et al. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol Cell Biol. 2000;20(16):6138–6146. doi: 10.1128/mcb.20.16.6138-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci U S A. 1998;95(13):7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen J, Cotmore SF, Tattersall P. Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Mol Cell Biol. 1999;19(11):7741–7750. doi: 10.1128/mcb.19.11.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross CT, McGinnis W. DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO. 1996;15(8):1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 27.Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol. 2001;8(7):626–633. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- 28.Gibson TJ, Ramu C, Gemünd C, Aasland R. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. TIBS. 1998;23(7):242–244. doi: 10.1016/s0968-0004(98)01231-6. [DOI] [PubMed] [Google Scholar]
- 29.Cetani F, Barbesino G, Borsari S, Pardi E, Cianferotti L, Pinchera A, et al. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J Clin Endocrinol Metab. 2001;86(10):4747–4752. doi: 10.1210/jcem.86.10.7884. [DOI] [PubMed] [Google Scholar]
- 30.Gardner JM, Devoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321(5890):843–847. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita N, Watanabe S, Ichimura T, Ohkuma Y, Chiba T, Saya H, et al. MCAF mediates MBD1-dependent transcriptional repression. Mol Cell Biol. 2003;23(8):2834–2843. doi: 10.1128/MCB.23.8.2834-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichimura T, Watanabe S, Sakamoto Y, Aoto T, Fujita N, Nakao M. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J Biol Chem. 2005;280(14):13928–13935. doi: 10.1074/jbc.M413654200. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12(2):475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15(4):595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 35.Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci. 2008;65(10):1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su MA, Giang K, Zumer K, Jiang H, Oven I, Rinn JL, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J Clin Invest. 2008;118(5):1712–1726. doi: 10.1172/JCI34523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clouaire T, de las Heras JI, Merusi C, Stancheva I. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA. Nucleic Acids Res. 2010;38(14):4620–4634. doi: 10.1093/nar/gkq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baubec T, Ivánek R, Lienert F, Schübeler D. Methylation-Dependent and-Independent Genomic Targeting Principles of the MBD Protein Family. Cell. 2013;153(2):480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19(5):667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100(11):6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38(2):179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20(3):274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 44.Jiang W, Anderson MS, Bronson R, Mathis D, Benoist C. Modifier loci condition autoimmunity provoked by Aire deficiency. J Exp Med. 2005;202(6):805–815. doi: 10.1084/jem.20050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 46.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.