Abstract
PAX6-null mice exhibit defects in multiple organs leading to neonatal lethality, but the mechanism by which this occurs has not yet fully elucidated. In this study, we generated induced pluripotent stem cells (iPSCs) from Pax6-mutant mice and investigated the effect of PAX6 on cell fate during embryoid body (EB) formation. We found that PAX6 promotes cell migration by directly downregulating miR-124, which is important for the fate transition of migratory cells during gastrulation of embryonic stem (ES) cells. Although several downstream targets of miR-124 have been reported, little is known regarding the upstream regulation of miR-124. When we observed EB formation of iPSCs from Pax6-mutant mice, we found that higher levels of miR-124 in Pax6 homozygous EBs (Homo-EBs) inhibited cell migration, whereas inhibition of miR-124 in Homo-EBs rescued the migratory phenotypes associated with PAX6 deficiency. Further, we found that PAX6 binds to the promoter regions of the miR-124-3 gene and directly represses its expression. Therefore, we propose a novel PAX6-miR-124 pathway that controls ES cell migration. Our findings may provide important information for studies on ES cell differentiation and embryonic development.
Introduction
MicroRNAs (miRNAs) are noncoding small RNAs that posttranscriptionally regulate gene expression [1], thereby regulating diverse biological processes, such as development, fate determination, proliferation, and apoptosis [2–5]. Moreover, it has been shown that specific miRNAs regulate self-renewal, pluripotency, and embryonic stem (ES) cell differentiation [6,7]. Marson et al. reported that key ES cell transcription factors are associated with promoters for most of the miRNAs that are preferentially expressed in ES cells and with promoters for a set of silent miRNA genes, which are expressed in a tissue-specific manner in differentiated cells [8]. One of these miRNAs, miR-124, is thought to be expressed in ES cells and enriched during brain development, as it accounts for 25%–48% of all the brain miRNAs [9]. Several studies have reported that miR-124 is expressed and involved in neurogenesis [10–13]. Lee et al. have reported that miR-124 is important for the fate transition of migratory cells during gastrulation of human ES cells [14]. However, the regulation of miRNA expression by transcription factors is still largely unclear.
The formation of embryoid bodies (EBs) is a principal step in the differentiation of ES cells in vitro [15]. The molecular and cellular morphogenic signals and events that occur within EBs recapitulate numerous aspects of embryonic development and result in cellular differentiation to the three embryonic germ layers (ie, endoderm, mesoderm, and ectoderm), similar to gastrulation of an epiblast-stage embryo in vivo [16]. The specific identity and spatial coordination of the various cell–cell interactions involved in EB formation are thought to influence the course of ES cell differentiation [17,18]. Recently, induced pluripotent stem cells (iPSCs) have been developed as a valuable tool to generate pluripotent stem cells from the somatic cells of patients. The formation of EBs from disease-specific iPSCs can be useful for exploration of disease mechanisms both in vitro and in vivo [19].
PAX6 is a transcription factor essential for the development of tissues, including those of the eyes, central nervous system (CNS), and endocrine glands of vertebrates and invertebrates. PAX6-null mutant mice exhibit defects in multiple organs, including the eye, brain, and pancreas, that lead to neonatal lethality [20–24]. PAX6 regulates the expression of a broad range of molecules, including transcription factors, cell adhesion and short-range cell–cell signaling molecules, hormones, and structural proteins. PAX6 has also been implicated in various key biological processes, including cell proliferation, migration, adhesion, and signaling, both in normal development and in oncogenesis [25]. Our previous study demonstrated that PAX6 deficiency caused downregulation of the Pc1 gene and upregulation of the Pcsk1n gene, which in turn caused diabetes [26,27]. PAX6 appears to be important for neurogenesis in the subventricular zone (SVZ), which is abnormal in PAX6-deficient (Sey/Sey) mice [28], also for genesis and glutamatergic differentiation of late-born neurons [29]. The SVZ defect in the Sey/Sey cortex suggests that CNS development is impaired by Pax6 mutation.
In this study, we generated iPSCs from PAX6-deficient mice and then generated EBs to investigate early embryonic differentiation. Using a spontaneous EB differentiation protocol, we found that cell adhesion/migration of Pax6-homozygous EB (Homo-EB) was dramatically impaired relative to that of wild-type EB (Wt-EB). Many studies have reported that PAX6 regulates cell migration through its downstream molecules, mostly proteins [30,31], but few or no studies have investigated miRNA with respect to this issue, even though it has recently received a great deal of attention because of its important regulatory roles in various cellular processes, including cell migration [32]. The apparent importance of miRNA in cellular regulation, especially the effect of miR-124 on the fate transition of migratory cells [14], prompted us to test whether miRNA mediates the cell migration regulated by PAX6 during early development.
Materials and Methods
Mice
Animals (Pax6 R266Stop mutant mice) were used as previously described [26]. Briefly, a stop-codon mutation at a 266 aa of PAX6 was produced by injecting the chemical mutagen N-ethyl-N-nitrosourea (ENU); the truncated PAX6 could consider being null and the mutation was confirmed to be transmitted to the offspring. Pax6 heterozygous mutant mice show a small-eye phenotype, whereas homozygous mutant embryos lack eyes and die soon after birth. Pax6 heterozygous mutant mice were mated, mouse embryos collected 13.5–14.5 days post-coitus were harvested for mouse embryonic fibroblast (MEF) isolation, and the embryos were genotyped by polymerase chain reaction (PCR) analysis as previously described [26]. All animal studies were performed in accordance with Institutional Animal Care and Use Committee protocols at Peking University.
Cell culture
MEF cells of the three genotypes used for reprogramming in this study were obtained from different embryos from the same uterus. MEF isolation was performed as previously described [33]. Cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Hyclone) containing 10% (v/v) fetal bovine serum (FBS) (Biochrom), and MEFs within three passages were used in this study. Mouse embryonic stem cells and iPSCs were maintained on mitomycin-c-treated MEFs in mouse embryonic stem cell (mESC) medium containing high-glucose DMEM (Hyclone), 20% (v/v) ES-cell-qualified FBS (Biochrom), 2 mM l-glutamine (Hyclone), 0.1 mM nonessential amino acids (Hyclone), 0.1 mM β-mercaptoethanol (Invitrogen), 1× penicillin/streptomycin (Hyclone), and 12.5 ng/mL leukemia inhibitory factor (LIF) (Chemicon). Clones were dissociated using trypsin and passaged at a 1:3–1:5 split ratio every 3–4 days depending on the cell density.
Generation and culture of mouse iPSCs
The generation and structure of doxycycline-controlled Tet-on-inducible lentiviruses expressing mouse-derived Oct4, Sox2, c-Myc, and Klf4 have been described in previous studies [34]. MEF cells of three genotypes in 35-mm tissue culture plates were sequentially infected with lentiviruses two times in the presence of 8 g/mL polybrene (Sigma). After 2 days of viral infection, the culture medium was replaced by mESC medium supplemented with 2 mg/mL doxycycline (Dox) to induce reprogramming. The newly generated mouse iPS colonies were picked up after 6–8 days.
Immunostaining and alkaline phosphatase staining
iPSCs were fixed with 4% (w/v) paraformaldehyde for 30 min and permeabilized with 0.5% (v/v) Triton X-100 in phosphate-buffered saline (PBS) for 15 min at room temperature. After blocking with 5% (w/v) bovine serum albumin for 30 min, the cells were incubated with primary antibodies at 4°C overnight. The cells were washed with PBS and then incubated with secondary antibodies [DyLight™ 594-conjugated AffiniPure goat anti-rabbit immunoglobulin G (IgG) and DyLight™ 488-Conjugated AffiniPure Goat Anti-Mouse IgG; 1:200 dilution] and visualized by fluorescence microscopy (Olympus) or confocal microscopy (Olympus FV1000), after counterstaining with 4,6-diamidino-2-phenylindole. The primary antibodies used for characterization of iPSCs targeted SSEA-1 (1:100; Santa Cruz) and Nanog (1:100; Abcam). Alkaline phosphatase (AP) staining was performed according to the manufacturer's recommendations (Millipore). For EB staining, EBs were plated onto gelatin-coated cover glass (Fisher Scientific); they attached to the glass and expanded spontaneously, and then the slides were stained as described previously. The antibody used in this experiment was the anti-IQGAP1 rabbit polyclonal antibody (1:100; Bioss).
DNA microarray
Total RNA from ES cells, iPSCs, MEFs, or EBs was labeled with biotin. The samples were hybridized to a mouse oligo microarray (Affymetrix) according to the manufacturer's protocol. The arrays were scanned on Affymetrix scanners and the expression value for each gene was calculated using the GENECHIP software (Affymetrix).
Teratoma formation
iPSCs were suspended at 1×107 cells/mL in DMEM containing 10% FBS. We subcutaneously injected 100 μL of the cell suspension (1×106 cells) into the dorsal flanks of severe combined immunodeficient (SCID) mice. At 3–4 weeks after injection, the tumors were explanted, fixed in 4% paraformaldehyde, embedded in paraffin, and examined histologically using hematoxylin and eosin staining.
Spontaneous differentiation and formation of EBs
The three-step spontaneous differentiation protocol for mES cells or iPSCs was adapted from two protocols [35,36], with some modifications. Briefly, our protocol included the depletion of feeder cells, formation of EBs, and spontaneous differentiation. Details for the methods are provided in the Supplementary Data (Supplementary Data are available online at www.liebertpub.com/scd).
In vitro migration assays
All migration experiments with EBs were performed in mES media without LIF on plastic substrates. Culture dishes were handled carefully to keep the EBs at the bottom, and cell migration was evaluated by measuring the distance from the edge of the EB to the nucleus of the most distant cell. Different viewing fields were randomly chosen for counting, and average values were determined. The assays were repeated at least three times.
Transwell migration assay
Transwell migration assays were performed in six-well culture plates with inserts (8-μm pore size; BD Biosciences). iPSCs were transferred to the upper-chamber inserts. In total, 2.5 mL of differentiation medium was added to the lower and upper chambers. The cells were allowed to migrate for 4 days in a humidified CO2 incubator at 37°C. Following incubation, cells on the lower side of the insert filter were quickly fixed and stained. Different viewing fields were randomly chosen for counting, and average values were determined. The assays were repeated three times.
Proliferation and apoptosis assay
Four-day-old EBs were allowed to adhere to gelatin-coated dishes for 18 or 48 h, after which they were dissociated from dishes and digested for cell counting. Cell apoptosis was determined by flow cytometry using propidium iodide and Annexin-V staining.
Western blotting
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (PAGE) and western blotting were performed according to standard procedures [26]. β-Actin was used on the same membrane as the loading control. The primary antibodies used were anti-IQGAP (1:500; Bioss), anti-PAX6 (1:1,000; C-Terminus, Millipore), and anti-β-actin (1:1,000; Santa Cruz). The experiments were repeated three times.
miRNA analysis by quantitative PCR
Total RNA containing miRNAs was extracted from cultured cells or EBs by using the miRNeasy mini kit (Qiagen) according to the manufacturer's protocol. The isolated total RNA (1 μg) containing miRNA was used as starting material for the reverse transcription reaction by using the miScript ii RT kit (Qiagen). The miScript PCR system (Qiagen), which comprises the miScript ii RT Kit, miScript SYBR Green PCR Kit, miScript primer assay, and miScript precursor assay, enables quantification of mRNA, miRNA, and precursor miRNA from the same cDNA. The Mm_miR-124_1 miScript primer assay (MS00029211; Qiagen) was used to detect the expression of mature mmu-mir-124. The Mm_mir-124-1_PR_1 miScript Precursor Assay, Mm_mir-124-1_PR_2 miScript Precursor Assay, and Mm_mir-124-1_PR_3 miScript Precursor Assay (MP00004074, MP00004081, and MP00004088; Qiagen) were used to detect the expression of 3 mmu-mir-124 precursors.
Northern blotting
Total RNA was extracted from iPSCs and EBs by using the miRNeasy mini kit (Qiagen). Northern blotting was performed using the highly sensitive miRNA Northern Blot Assay Kit (NB-1001; Signosis). Briefly, 12 μg of total RNA from each sample was separated on a 15% polyacrylamide gel, transferred to a membrane (Signosis), and UV-crosslinked. The membranes were incubated with a biotin-labeled miR-124 probe (HP-0049; Signosis) and a U6 probe (HP-1001; Signosis). Prehybridization and hybridization were conducted using a hybridization buffer, and the membranes were then exposed using a biotin-based chemiluminescence imaging system.
In situ hybridization
In situ hybridization was performed by following the Exiqon protocol with some modifications. Briefly, EBs were fixed with 4% fresh paraformaldehyde, followed by paraffin embedding. Formalin-fixed paraffin-embedded sections (6-μm thick) were cut and stored at 4°C, and paraffin was melted on the day before the in situ hybridization experiment was conducted. The slides were deparaffinized and pretreated with 10 μg/mL proteinase K for 10 min at room temperature and then dehydrated at room temperature. Subsequently, hybridization was conducted using the hybridization mix at 55°C for 2 h. The probes were labeled using digoxigenin, and in situ hybridization signals were detected using an anti-digoxigenin antibody conjugated with AP (AP-conjugated anti-DIG; Roche). AP was used to convert the soluble substrates 4-nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3′-indolylphosphate (BCIP) into a water- and alcohol-insoluble dark-blue NBT-BCIP precipitate, and the samples were then counterstained with 0.1% Nuclear Fast Red and mounted. A scrambled-sequence probe (LNA™; Exiqon) was used as a control in the hybridization.
Recombinant adenovirus construction
Full-length Pax6 cDNA and a specific Pax6 shRNA (5′-GATCCCCAACTCCATCAGTTCTAATTCAAGAGATTA GAACTGATGGAGTTGG-3′, 5′-AGCTCCAACTCCATC AGTTCTAATCTCTTGAATTAGAACTGATGGAGTT GGG-3′) were subcloned into the adenovirus shuttle vector pDC316-EGFP and pDC316-EGFP-U6, respectively. Recombinant Ad-GFP, Ad-PAX6-GFP, Ad-Pax6-RNAi, and Ad-non-silence were generated as previously described [37,38], using the AdMax system (Microbix Biosystems). The adenovirus was purified using a ViraBind™ Adenovirus Purification Kit (Cell Biolabs). Mouse iPSCs were detached from culture dishes, grown in suspension culture for 1–2 days, and transfected with adenovirus. After 2–3 days, the cells were subjected to western blotting and quantitative PCR (qPCR).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described previously [38] for EBs derived from the mouse ES cell line R1. Briefly, chemical-linked complexes of genomic DNA and PAX6 protein were immunoprecipitated with antibodies specific to PAX6 (C-Terminus; Millipore). The DNA was amplified for 21–25 cycles after ChIP using four pairs of primers: spanning −7,939 to −7,710 bp: forward, 5′-ACTGAGATCTGGCAGGGAGA-3′ and reverse, 5′-AGTGCATGTCATTGGCTCTG-3′; spanning −5,169 to −4,958 bp: forward, 5′-GAGCCTGTTGTGATGCTTGA-3′ and reverse, 5′-CAGTGTGCACGTGGAGTAGG-3′; spanning −4,049 to −3,758 bp: forward, 5′-GAAAGCACACCC TCGGAAT-3′ and reverse, 5′-TCTAACATGGGTGGTCA AGG-3′; spanning −1,754 to −1,475 bp: forward, 5′-GAGG CCTGCC TACTTCTGTG-3′ and reverse, 5′-TCCCCTTTTT CACCCTTTTT-3′.
Luciferase activity
We amplified 2 mmu-mir-124-3 DNA fragments (−1,754 bp/+28 bp, −4,394 bp/−3,432 bp), including putative PAX6-binding sites. According to the manufacturer's instructions, the −1,754 bp/+28 bp DNA fragment was cloned into the pGL3-basic vector (Promega) by using Kpn1 and Nhe1 (New England Biolabs), and the −4,394 bp/−3,432 bp fragment was cloned into the pGL3-promoter vector (Promega) by using Mlu1 and Xho1 (New England Biolabs). Correct insertion was verified by sequencing. The PCR primers were as follows: (−1,754/+28) forward, 5′-AA AGGTACCGAGGCCTGCCTACTTCTGTG-3′ and reverse, 5′-GTGGGCTAGCAATCAAGG TCCGCTGTG AAC-3′; (−4,394/−3,432) forward, 5′-ATCGACGCGTTCCCTGGG TTTTCATCTG AG-3′ and reverse, 5′-ATCGCTCGAGGGC TTGTCATTCCTCCAGAA-3′. The PAX6 and truncated PAX6 (R240Stop mutation) expression vectors (pcDNA3.1-/Pax6 and pcDNA3.1-/Pax6-tru) were constructed as previously described [37]. HEK293 cells (1×105) were seeded in a 24-well plate and transiently transfected with pGL3-basic −1,754/+28 or pGL3-promoter −4,394/−3,432, along with PAX6 (wild or truncated type) at several concentrations (0, 400, and 800 ng/well) (the empty vector was used to supply up to 800 ng/well as control) by using Lipofectamine 2000 (Invitrogen), and pRL-CMV (Promega) expressing Renilla luciferase was transfected as an internal control. Forty hours after transfection, cell lysates were extracted in passive lysis buffer (Promega), and luciferase activity was measured using the dual luciferase reporter assay system (Promega) in a Centro LB960 96-well luminometer (Berthold Technologies). The deleted promoter (−1,533/+28) was amplified using the following primers: forward, 5′-AAAGGTACCCTCCCATCC TCGTAAGCAAG-3′ and reverse, 5′-GTGGG CTAGCAAT CAAGGTCCGCTGTGAAC-3′. This sequence was then cloned into the pGL3-basic vector, and the luciferase assay was performed as described previously for the −1,754/+28 fragment.
Electrophoretic mobility shift assay
Nuclear extracts (NEs) from mES cell R1-derived EBs were prepared using the NE-PER® Nuclear and Cytoplasmic Extraction Reagent (Pierce). Electrophoretic mobility shift assay (EMSA) was conducted using the LightShift® Chemiluminescent EMSA Kit (Pierce) following the manufacturer's protocols. Custom-synthesized 5′ single-strand nucleotides labeled at the 5′-end with biotin (only the sense strands are shown) and covering −1,636 bp/−1,577 bp (F: 5′-CGCTGA TCTCTAGCCCTAAAGACCCAGATCAAGGTAGTCACC CTTCAATAGACTGCAAAG-3′) and −1,570 bp/−1,511 bp (F: 5′-TTCAATTGCAGTCAACTGTGAAGTATGGACTG TGTTTCTCCCATCCTCGTAAGCAAGGAC-3′) were annealed in annealing buffer (Pierce) to form a double-stranded probe. For the competition assay, an unlabeled competitor with the same sequence as the probe was added at 100-fold excess prior to addition of the labeled probe. For supershift EMSA, 4 μg of rabbit anti-PAX6 (Millipore) or normal IgG (Sigma) was incubated with NE prior addition of the labeled probe. Protein–DNA complexes were resolved by nondenaturing 6% PAGE in 0.5× Tris-buffered ethylenediaminetetraaceticacid. Gels were transferred to Biodyne B nylon membranes (Pierce) and protein–DNA complexes were detected using chemiluminescence.
miRNA transfection
An mmu-miR-124 mimic (MSY000134; Qiagen), inhibitor (MIN000134; Qiagen), and negative controls (1027280 and 1027271) were purchased from Qiagen. Transient transfections were performed using Lipofectamine RNAi Max (Invitrogen) according to the manufacturer's instructions. In brief, iPSCs were grown in differentiation medium on low-attachment plates or the upper chambers of transwells for 1–2 days; the EBs were transfected twice on 2 consecutive days with 100 nM miR-124 mimic or inhibitor and then plated on gelatin-coated dishes or transwells for 1–2 days for migration assay.
Statistical analysis
Statistical analysis was performed using an independent sample t-test. The asterisks in each graph indicate statistically significant differences with P-value calculated by t-test and P<0.05 was considered statistically significant.
Results
Generation of iPSCs from PAX6 deficiency mice
To establish a cellular model to address the role of PAX6 in early embryonic development in mice, we generated iPSCs using MEFs from Pax6 R266Stop mutant mice [26]. These ENU-induced mutagenic mice exhibited a C-to-T mutation in the Pax6 gene that generated a polypeptide truncated at residue 266 of the PAX6 protein [26,27]. Because the mutation results in production of a truncated protein lacking the C-terminal domain, the truncated PAX6 is considered functionally inactive. Therefore, mouse homozygous mutants are widely used as Pax6-null mutants [39].
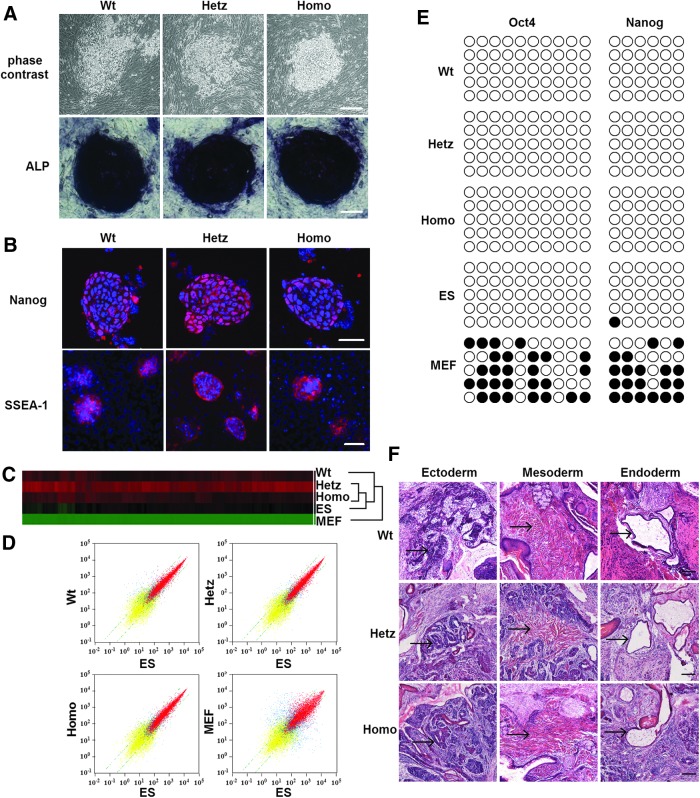
iPSC lines of three different Pax6 genotypes (designated as Wt-iPSCs, Hetz-iPSCs, and Homo-iPSCs) were established from normal siblings as well as heterozygous and homozygous Pax6 mutants; three or four independently validated iPSC clones per genotype were generated and at least two independent clones of each genotype were assayed in each experiment. Representative experimental results are shown here (Fig. 1A; Supplementary Fig. S1A). All iPSC lines exhibited morphological features similar to those of mouse ES cells—strong AP activity (Fig. 1A), expression of pluripotency markers (including Nanog and SSEA-1; Fig. 1B), and normal karyotypes (Supplementary Fig. S1B). Global gene expression was analyzed in Wt-iPSCs, Hetz-iPSCs, Homo-iPSCs, MEFs, and mouse R1 ES cells by using oligonucleotide microarrays. A hierarchical clustering analysis clearly indicated that Wt-iPSCs, Hetz-iPSCs, and Homo-iPSCs clustered closely with R1 ES cells and were distinct from MEFs (Fig. 1C). Analysis of scatter plots further demonstrated that Wt-iPSCs, Hetz-iPSCs, and Homo-iPSCs are highly similar to R1 ES cells and hence completely different from MEFs at the global transcriptional level.
FIG. 1.
Induced pluripotent stem cells (iPSCs) generated from mouse embryonic fibroblast (MEF) cells of wild-type mice and Pax6 mutants exhibit mouse embryonic stem (mES) cell characteristics. (A) Morphological features of iPS (upper line) colonies and their alkaline phosphatase staining (lower line), showing mES-cell-like morphological features distinct from those of MEFs at 5 days after Dox-inducible lentiviral transduction. Scale bar=100 μm. (B) Immunofluorescence staining demonstrates that iPS clones are positive for Nanog and SSEA-1. Nanog scale bar=50 μm, SSEA-1 scale bar=100 μm. (C) Hierarchical cluster analysis of global gene expression microarray data from iPSCs of Wt, Hetz, Homo, mES, and MEF cells. (D) Scatter plot comparing global gene expression profiles among Wt, Hetz, Homo, mES, and MEF cells. (E) Bisulfite genomic sequencing of the promoter regions of Oct4 and Nanog. Open and closed circles indicate non-methylated and methylated CpGs, respectively. (F) Histological sections show differentiation into all three germ layers: neural rosettes (ectoderm), endothelium (endoderm), and muscle (mesoderm). Black arrows heading to typical tissues. Scale bar=100 μm.
To confirm epigenetic remodeling in the reprogrammed cells, we also evaluated the methylation status of CpG dinucleotides in the Oct4 and Nanog promoters. Bisulfite genomic sequencing analysis showed that the Oct4 and Nanog promoter regions were demethylated in Wt-iPSCs, Hetz-iPSCs, and Homo-iPSCs relative to MEFs, whereas their methylation was similar to that of R1 (Fig. 1E). The pluripotent potential of these three types of iPSCs was further indicated by teratoma formation. After iPSCs were transplanted into the hind limbs of SCID mice for 3–4 weeks, hematoxylin and eosin staining (Fig. 1F) and immunohistochemistry staining (Supplementary Fig. S1C) revealed the presence of derivatives of all three germ layers, including neural epithelium (βIII-tubulin), muscle α-smooth muscle actin, and endoderm (GATA4), within the tumors.
Migration of cells from the outer layer of EBs is impaired in Pax6 Homo-EBs
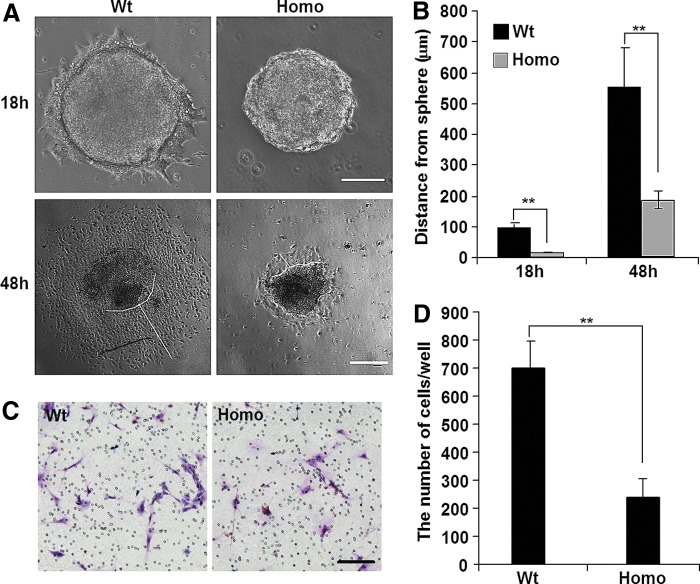
On analyzing EB formation, we detected differences between Wt-iPSCs and Homo-iPSCs. At 18 or 48 h after plating onto the dishes, the number of migrated cells and the distance from the edge of the EB to the most distant migrating cell were significantly reduced in Homo-EBs relative to those in Wt-EBs (Fig. 2A, B). Wt-EBs formed a halo around the clones, whereas Homo-EBs remained largely intact, with far fewer cells migrating. Time-lapse videos also showed that Wt-EBs were flattened and the cells adhered and then migrated quickly, while Homo-EBs remained a status of tight, domed shape and the cells showed significantly decreased migratory capacity (Supplementary Video). Moreover, when we checked the borders of teratomas derived from Wt- and Homo-iPSCs, we found relatively intact borders in homo teratomas, but no clear border was observed in Wt teratomas (Supplementary Fig. S1D).
FIG. 2.
Pax6 mutation limits migration of embryoid body (EB) cells in vitro. (A) Pax6 mutation limited cellular migration away from EBs on a plastic substrate at 18 and 48 h after plating. A representative experiment is shown, and the experiment was repeated five times. 18 h scale bar=100 μm, 48 h scale bar=200 μm. (B) Cellular migration was evaluated by measuring the distance from the edge of the EBs to the nucleus of the most distant cell. Values are the mean±standard deviation (SD). (C) Transwell migration assay indicates that migration of cells from homozygous EBs (Homo-EBs) is blocked. Representative images of cells migrating to the lower chambers (lower cells) are presented. Scale bar=100 μm. (D) Number of migrated cells in transwell migration assays. A representative experiment is shown; the experiment was repeated three times. Each value represents the mean±SD (**P<0.01). Color images available online at www.liebertpub.com/scd
This difference was quantified using a transwell migration assay (Fig. 2C, D). To exclude the possibility that the observed difference in cell number resulted from cell proliferation or apoptosis, we performed cell counting and Annexin V-fluorescence-activated cell sorting analysis and detected no difference between Wt- and Homo-iPSCs and EBs (Supplementary Fig. S2), which is consistent with a previous report; Quinn et al. showed that the proportions of BrdU-labeled proliferating cells were not different between Wt and Pax6−/− EBs [40]. All these findings suggested that cell migration of Homo-EBs is dramatically impaired.
Because PAX6 has been reported to be involved in the progression of differentiation toward neurons (ectoderm) and the pancreas (endoderm) [41], as well as in the migratory behavior of differentiating cells [42], to determine whether the migration defect at day 6 of EB formation resulted from blockade of differentiation progression by Pax6 mutation, we analyzed the spontaneous differentiation of Wt- and Homo-EBs using reverse-transcription PCR, qPCR, and immunofluorescence (Supplementary Fig. S3B–G). Concurrent with the downregulation of pluripotency genes (Oct4, Nanog, and Klf4), the expression of ectoderm (Sox1 and Nestin), mesoderm (Brachyury), and endodermal (Sox17) marker genes was dramatically increased to similar levels in both groups. In addition, cDNA microarray analysis was performed to compare the globe gene expression of the Wt- and Homo-EBs (5–6-day old) (Supplementary Fig. S3H). Although some slight differences were observed, the results confirmed that the expression profiles of ectoderm-, mesoderm-, and endoderm-specific genes were very similar between the two groups. All these observations suggested that PAX6 deficiency did not impair the progression of differentiation during 6 days of EB formation, which is consistent with the normal early embryo development of mice with defective Pax6 [43]. It indicates that at day 6 during the differentiation of EBs, the migration difference induced by Pax6 mutation resulted from the regulation of migration by PAX6 and its downstream molecules.
miR-124 expression is elevated in Homo-EBs
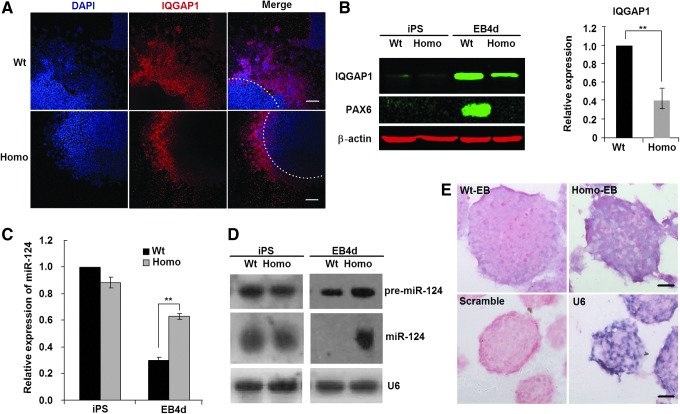
Cell migration from the outer regions of EBs has been reported to be regulated by IQGAP1, which is a Ras GTPase-activating-like protein preferentially located at the outer regions of EBs [14]. To determine IQGAP1 expression patterns in Wt-EBs and Homo-EBs, we plated the EBs on a dish for 1–2 days and then performed immunochemistry and western blotting analyses. IQGAP1 expression decreased in Homo-EBs at the outer region of the EBs (Fig. 3A) or in the whole cell lysate (Fig. 3B). Because IQGAP1 expression is inhibited by miR-124 during EB differentiation [14], we measured the expression of miR-124 in Wt- and Homo-EBs. Consistent with the findings of Lee et al. [14], miR-124 expression decreased significantly during the formation of Wt-EBs, but there was no significant decrease in Homo-EBs. Thus, the expression of miR-124 in Homo-EBs was significantly higher than that in Wt-EBs at day 4 (Fig. 3C). Three genes encode the mature sequence of miR-124—miR-124-1, miR-124-2, and miR-124-3—which are located, respectively, on chromosomes 14, 3, and 2 of the mouse genome [44]. Using Northern blotting analysis (Fig. 3D) and in situ hybridization (Fig. 3E), we further confirmed that the expression of both precursor and mature miR-124 was significantly higher in Homo-EBs than in Wt-EBs. Because the probe for mature miR-124 also recognized all three types of re-miR-124 molecules, which have very similar molecular weights, only one band for pre-miR-124 appears in the Northern blotting results (Fig. 3D, upper line). Based on these data, we hypothesized that the expression of miR-124 was affected by PAX6 during EB formation.
FIG. 3.
miR-124 inhibits migration of cells from Homo-EBs. (A) Immunostaining showing higher expression of IQGAP1 in wild-type EBs (Wt-EBs) (upper line) than in Homo-EBs (lower line). Scale bar=100 μm. (B) Western blotting showing higher expression of IQGAP1 in Wt-EBs than in Homo-EBs; β-actin was used as an internal control. (C) Relative miR-124 expression measured by quantitative polymerase chain reaction (qPCR). Quantitative analyses show that expression of miR-124 decreased in Wt-EBs but did not change in Homo-EBs. (D) Northern blotting showing expression of pre-miR-124 and mature miR-124 in iPS and EB cells. U6 was used as an internal control. (E) In situ hybridization showing miR-124 expression in Wt- and Homo-EBs at day 4 of culture. U6 and Scramble indicate the positive and negative controls, respectively. Scale bar=20 μm. A representative experiment is shown; the experiment was repeated three times. Each value represents the mean±SD (**P<0.01).
Expression of miR-124 is regulated by PAX6
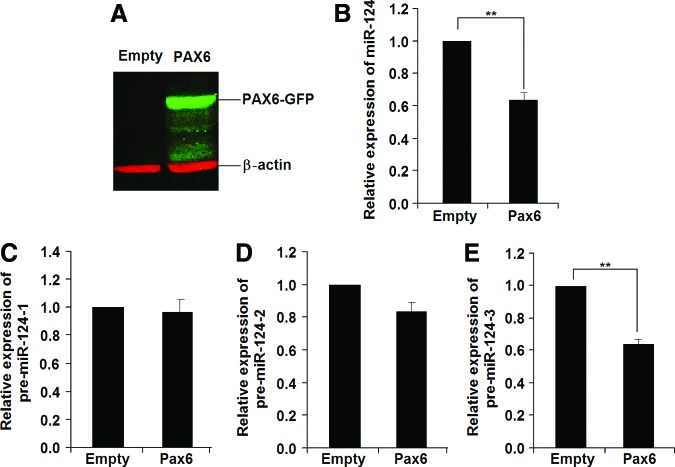
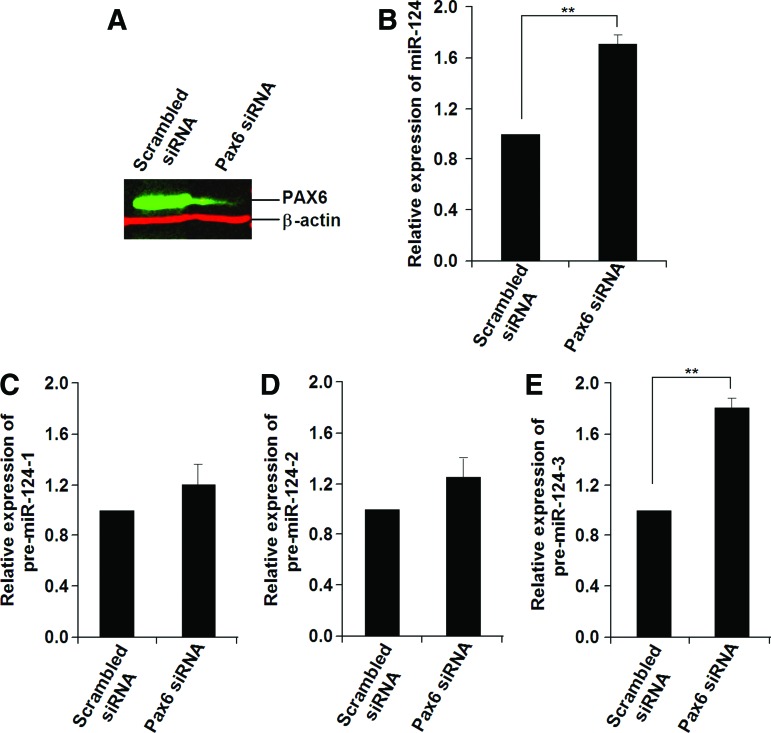
To test this hypothesis, we investigated the changes in expression for mature miR-124 and its three precursor mRNAs in EBs with overexpression or knockdown of Pax6. As shown in Fig. 4, overexpression of Pax6 in Homo-EBs (Fig. 4A) resulted in a decrease in the expression of mature miR-124 (Fig. 4B). To analyze the transcriptional regulation of miR-124, qPCR was used to determine the expression of its precursors, that is, pre-miR-124-1, pre-miR-124-2, and pre-miR-124-3, from three different DNA fragments cleaved and processed to mature miR-124 by a nuclear enzymatic complex consisting of nuclear RNAase III and the adapter protein Drosha [45]. As shown in Fig. 4C–E, pre-miR-124-3, but not pre-miR-124-1 or pre-miR-124-2, was responsible for the decrease in mature miR-124. As expected, knockdown of Pax6 in Wt-EBs resulted in increased expression of pre-miR-124-3 and mature miR-124 (Fig. 5B–E). We confirmed the results using two siRNAs for Pax6 (siRNA ID: s71268 or s71269 Ambion) and two scrambled siRNA9 (Cat. No. AM4611 and AM4613), and used low concentrations (50 nM) to reduce off-target effects. The decrease in PAX6 expression was detected by western blotting analysis (Fig. 5A).
FIG. 4.
Overexpression of Pax6 results in decreased miR-124 expression in Homo-EBs. (A) Western blot result showing increased PAX6 expression after overexpression of Pax6 by Ad-PAX6-GFP infection in Homo-EBs. (B) Overexpression of Pax6 resulted in reduced miR-124 expression. qPCR analysis was performed independently for EBs (n=5). (C–E) Expression of miR-124 precursors was detected by qPCR. Significant changes were not observed for pre-miR-124-1 (C) or pre-miR-124-2 (D), whereas pre-miR-124-3 expression (E) was decreased in Pax6-overexpressing EBs (n=5). A representative experiment is shown; the experiment was repeated three times. Each value represents the mean±SD (**P<0.01). Color images available online at www.liebertpub.com/scd
FIG. 5.
Knockdown of Pax6 results in increased miR-124 expression in Wt-EBs. (A) Western blot result showing decreased PAX6 expression after knockdown of Pax6 by Ad-Pax6-RNAi infection in Wt-EBs. (B) Knockdown of Pax6 led to elevated miR-124 expression. qPCR analyses were performed independently for EBs (n=5). (C–E) Expression of miR-124 precursors was detected by qPCR. Significant changes were not observed for pre-miR-124-1 (C) or pre-miR-124-2 (D), but pre-miR-124-3 expression (E) was increased in Pax6-knockdown EBs (n=5). A representative experiment is shown; the experiment was repeated three times. Each value represents the mean±SD (**P<0.01). Color images available online at www.liebertpub.com/scd
Transcription of miR-124 is directly inhibited by PAX6
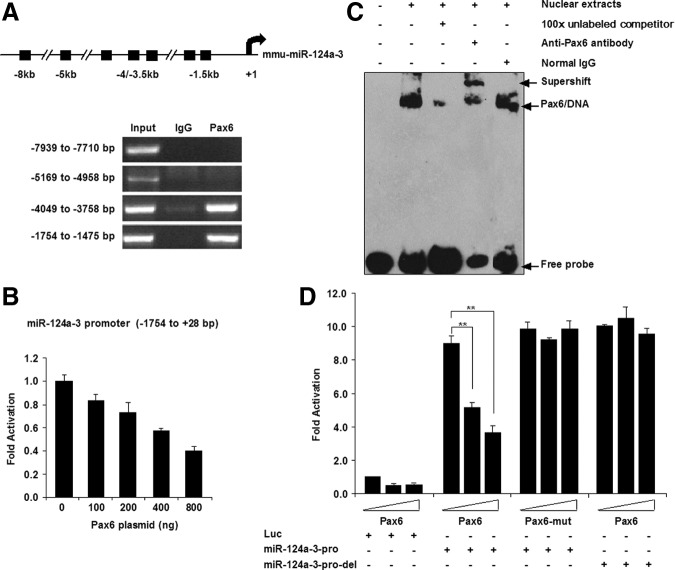
To determine whether miR-124 is the direct inhibitory target of PAX6, we scanned 10-kb sequences upstream of the miR-124-3 gene and found several putative PAX6-binding sites based on the TF SEARCH (Searching Transcription Factor Binding Sites, http://www.cbrc.jp/research/db/TFSEARCH) database (Fig. 6A), and then performed a ChIP assay to evaluate these putative binding sites. Two specific PAX6-binding regions were amplified: one extending from −4,049 to −3,758 bp and the other from −1,754 to −1,475 bp. Using dual-luciferase reporter assay system, we found that PAX6 repressed the mir-124-3 expression via binding to its promoter regions from −1,754 to −1,475 bp (Fig. 6B). Then, the binding ability was further confirmed by EMSA using a probe targeting −1,570 to −1,511 bp of the miR-124-3 promoter (Fig. 6C, lane 2), in which the band disappeared when an excess of unlabeled oligonucleotides with the same sequence was added (Fig. 6C, lane 3). Further, addition of anti-PAX6 antibody to the binding reaction caused the band to super-shift (Fig. 6C, lane 4). These data indicate that PAX6 in EB NEs binds to the miR-124-3 promoter region between −1,570 and −1,511 bp.
FIG. 6.
PAX6 represses miR-124-3 transcription by binding to the promoter region of miR-124-3. (A) Upper panel shows a schematic drawing of 10-kilobase (kb) regions proximal to the miR-124 gene on chromosome 2. Binding was assayed in chromatin immunoprecipitation experiments. Lower panel shows the PCR results. (B) Luciferase activity assays show that PAX6 represses miR-124-3 promoter activity via binding to the region from −1,754 to +28 bp, in a concentration-dependent manner (described in section “Materials and Methods”). (C) Electrophoretic mobility shift assay localized the PAX6 binding to a 60-bp region in the miR-124-3 promoter. (D) Luciferase activity assays indicated that PAX6 acts as a transcriptional repressor of miR-124-3. Overexpression of Pax6, but not truncated Pax6 (Pax6 R240Stop mutation, referred to as Pax6-mut as described in the section “Materials and Methods”), inhibited gene transcription driven by the miR-124-3 promoter. Deletion of the PAX6 binding site in the promoter (miR-124-3-pro-del) abolished the repression effect. The empty plasmid was supplied up to 800 ng/well as control. A representative experiment is shown; the experiment was repeated three times. Each value represents the mean±SD (**P<0.01).
To investigate how mutation of Pax6 leads to higher expression of pre-miR-124-3, we performed a luciferase reporter assay using Pax6 and mutated Pax6. Pax6, but not its mutated form (Pax6-mut), inhibited the expression of pre-miR-124-3 in a concentration-dependent manner (Fig. 6D). In agreement with the ChIP and EMSA data, we found that the inhibitory effect of PAX6 on miR-124-3 promoter activity disappeared when the PAX6 binding site was deleted in the miR-124-3 promoter (miR-124-3-pro-del) (Fig. 6D). We therefore conclude that PAX6 directly represses the expression of pre-miR-124-3, and PAX6 deficiency maintains pre-miR-124-3 expression at a high level.
Regulation of EB cell migration by PAX6 is mediated by miR-124
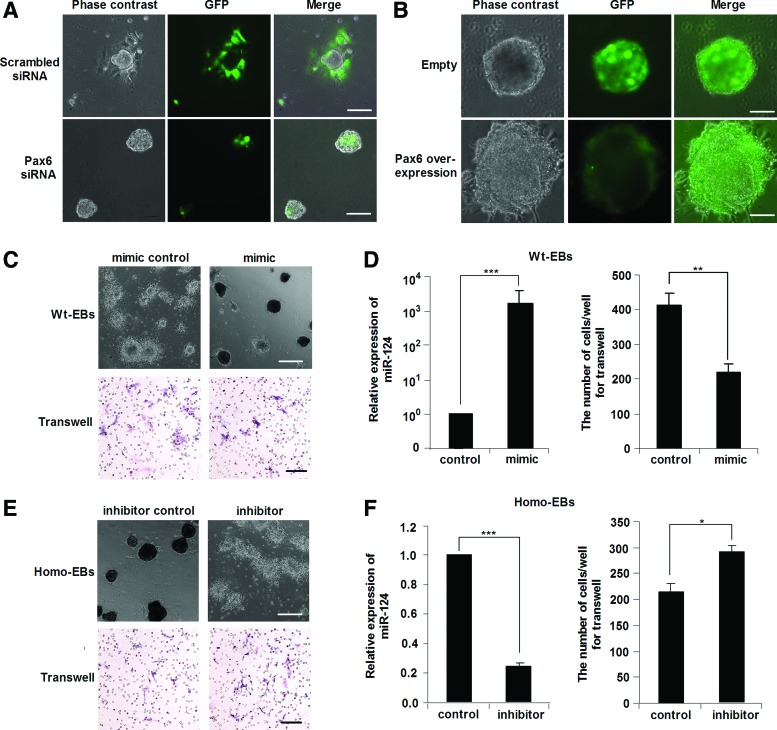
To evaluate the causal role of PAX6-miR-124 in the regulation of EB cell migration, we knocked down Pax6 expression in Wt-EBs or alternatively overexpressed Pax6 in Homo-EBs to determine whether PAX6 expression rescued cell migration in EBs. Consistent with our speculation, the migration of Wt-EBs after Pax6 knockdown was inhibited similarly to that of Homo-EBs (Fig. 7A). In contrast, migration of Homo-EBs after the Pax6 overexpression was enhanced to the Wt-EB level (Fig. 7B). Moreover, we found that the migration of cells from Wt-EBs was blocked when miR-124 mimic was introduced as determined by EB morphology (Fig. 7C, upper lane) and the transwell migration assay (Fig. 7C, lower lane). Figure 7D shows the elevated expression of miR-124 in EBs after transfection of the miR-124 mimic and the statistical results of the transwell migration assay. In contrast, inhibition of miR-124 by miR-124 inhibitor rescued migration in Homo-EBs (Fig. 7E, F). All of these data indicate that the cell migration in EBs regulated by PAX6 is mediated by miR-124 expression.
FIG. 7.
PAX6 regulates cell migration during iPSC differentiation through miR-124. (A) Knockdown of Pax6 inhibited migration of cells in Wt-EBs. Scale bar=200 μm. (B) Overexpression of Pax6 led to increased migration of cells in Homo-EBs. Scale bar=100 μm. (C) Transfection of mmu-mir-124 mimics to Wt-EBs (on days 2 and 3) resulted in decreased migration of cells from Wt-EBs. Upper panel shows the migration of Wt-EB cells on the plate; lower panel shows the migrated cells in the transwell. Scale bar=500 μm; transwell scale bar=100 μm. (D) The left panel shows the relative gene expression of miR-124 after transfection of miR-124 mimics in Wt-EBs; the right panel shows the statistical results of the transwell analysis for wild-type cells. (E) Transfection of mmu-mir-124 inhibitors to Homo-EBs (on days 2 and 3) rescued the migratory activity of cells in Homo-EBs. Upper panel shows the cell migration of Homo-EBs on the plate; lower panel shows the migrated cells in transwell. (F) The left panel shows the relative gene expression of miR-124 after transfection of miR-124 inhibitors in Homo-EBs; the right panel shows the statistical results of the transwell analysis for Homo cells. The experiments were repeated three times. Each value represents the mean±SD (*P<0.05, **P<0.01, ***P<0.001). Color images available online at www.liebertpub.com/scd
Discussion
In this study, using qPCR, in situ hybridization, and Northern blotting analysis, we investigated the expression pattern of miR-124 during EB formation of iPSCs derived from wild-type and Pax6 mutant mice. We show for the first time that miR-124, which is an important regulator of cell migration during gastrulation and neurogenesis, is a direct downstream target of PAX6. Moreover, using gain-of-function and loss-of-function studies, we investigated the role of miR-124 expression in mediating the regulatory effect of PAX6 on cell migration. In the iPS stage, no significant variation of migration-related gene expression was observed from the gene array analysis, but in the EB stage, cell migration ability was impaired markedly in Homo-EBs and migration-related genes, such as IQGAP1, were prominently downregulated; in contrast, PAX6 expression was elevated in Wt-EBs.
Molecular mechanisms that either maintain stemness or precede differentiation of pluripotent stem cells in the early stages of embryonic development remain poorly understood. PAX6 is a key factor in regulating the development of the eyes, CNS, and pancreas [20,25,46,47]. In this study, we generated wild-type iPSCs as well as heterozygous and homozygous Pax6 mutant iPSCs to explore the effects of PAX6 deficiency. Using the formation of cell aggregates in EBs of iPSCs, we investigated cellular behavior during development under PAX6 deficiency. The observation of blocked cell migration in PAX6 deficiency is consistent with other studies, indicating that PAX6 plays important roles in cell migration [25] and neuronal differentiation during development [48,49]. The various cell morphological changes involved in EB formation are considered to recapitulate the course of ES cell differentiation [17,18]. Although previous reports showed that miR-124 is expressed in neural lineages, possibly regulating neurogenesis [10–13], and is a key factor for migratory cell fate during EB formation [14], the mechanism by which miR-124 itself is regulated has not yet been determined clearly. This study provides a tool to characterize the expression of miR-124 regulated by PAX6, which controls cell motility, an essential feature of tissue and organ formation.
We confirmed that miR-124 is downregulated during formation of EBs from ES cells, as previously reported [14], but in Pax6 Homo-EBs, miR-124 was maintained at a higher level similar to that in ES cells (Fig. 3C, D). At the same time, the cell migration capacity of cells in Homo-EBs was obviously lower than that of cells in Wt-EBs. Several other studies have reported that miR-124 is involved in cell migration during neurogenesis [10–13] and gastrulation [14]. We conclude that the regulation of cell migration by PAX6 is probably mediated through effects on the expression of miR-124. Most reported miR-124 targets, including Scp1 [10], Ptbp1 [12], Lamc1 [11], and Iqgap1, which is the target we evaluated here, show reciprocal protein expression with miR-124, which suggests that miR-124 keeps these genes silenced in Homo-EBs. Cells are released from tight adherent junctions through diminished expression of IQGAP1, which subsequently promotes the reorganization of the cytoskeleton for cell polarity and directional cell migration. Thus, PAX6 indirectly regulates downstream genes involved in iqgap1-mediated cell migration, as observed in this study.
Because PAX6 is important for the eye (ectoderm) and pancreas (endoderm) development and brain (ectoderm) patterning in animal studies [41], it remains to be determined whether the progression of differentiation is involved in the migration difference between Wt-EBs and Homo-EBs at day 6 of EB formation. It was reported that compared with transfecting iPSCs prior to EB formation with miRNAs, transfecting EBs can reduce the effects of transfection on the progression of differentiation, because only the cells in the outer layer of EBs were transfected by this way [50]. We transfected 2 (or 3)-day-old EBs with miRNA mimic and inhibitor to minimize the possibility that migration difference due to the progression of differentiation. Consistent with previous reports [51–53], we found that defective PAX6 did not impair the spontaneous differentiation of EBs at day 6 as determined by the expression of markers of the three germ layers (Supplementary Fig. S3). The majority of migrating cells were nestin-positive neural stem cells, and the expression of nestin was not significantly different between Wt- and Homo-EBs (Supplementary Fig. S3D, E), which confirms the observation of a previous study that PAX6 is not necessary for mouse neuroectoderm specification at the early stage [51]. In addition, all-trans retinoic acid treatment induced expression of the mature neuronal marker βIII-tubulin in both Wt- and Homo-EBs, and the further neuronal differentiation did not increase the migration difference between these two groups (Supplementary Fig. S4). These results strongly suggest that progression of neural differentiation did not contribute to the migration difference. A full characterization of three germ layer differentiation of Pax6 mutant versus wild-type cells by microarray (Supplementary Fig. S3H) also indicated that Pax6 deficiency did not impair the progression of differentiation at early stage of EBs. Taking together these results suggest that Pax6 mutation blocked the migration of the cells at the early stage of spontaneous differentiation. We speculated that impaired migration would reflect in the late stage of differentiation, because Pax6 mutant mice have abnormal brain patterns and a disorganized pancreas. Together with PAX6 [23,54,55] and miR-124 [10–12,44] are critical factors in neural (and pancreatic) differentiation, the Pax6-mir-124 pathway may be involved in mouse brain and pancreas development and the function needs to be studied in the future.
The expression of miR-124 is thought to be regulated by RE1-silencing transcription factor (REST) [56], which is a zinc-finger repressor that negatively regulates many neuronal genes in stem cells, progenitor cells, and non-neuronal cell types [57]. Similar to the previously observed upregulation of miR-124 in REST+/− ES cell lines [56,58], in the present study, miR-124 expression significantly increased in Pax6 Homo-EBs. In addition to miR-124, several other downstream targets have been identified to be downregulated by PAX6. VEGFA, which is a growth factor, and MMP2, which is a cellular matrix protein involved in tumor formation and metastasis, were found to be repressed by PAX6 [54,59]. Olig2, which is a transcriptional factor critical for glial cell fate determination during forebrain development, was also found to be directly repressed by PAX6 [55]. Most of these targets encode proteins, but our results indicate that genes encoding miRNAs are novel candidates for target genes downregulated by PAX6. Although the mechanism is not yet clear, some studies have focused on many downstream genes inhibited by PAX6 using cDNA microarray analyses [31,60]. Our finding that miR-124 is downregulated by PAX6 provides a potential target for improving cell migration and differentiation during abnormal development caused by PAX6 deficiency.
Several miRNA polycistrons—those encoding the most abundant miRNAs in ES cells and that are silenced during early cellular differentiation [14,61,62]—are occupied at their promoters by Oct4, Sox2, Nanog, and Tcf3 [8]. The miRNA expression programs directly downstream of Oct4/Sox2/Nanog/Tcf3 could help to prime ES cells for rapid and efficient differentiation. Oct4/Sox2/Nanog/Tcf3 likely contribute to this process through their occupancy of the Let-7g promoter [8]. Similarly, we now report that the promoter of miR-124-3 is occupied by PAX6, which suggests that transcription factors are involved in neural (or pancreatic) differentiation by regulating miRNAs. Indeed, our findings regarding the PAX6-mir-124 pathway are supported by previous studies both in vivo [30,63] and in vitro [64]. Maiorano et al. [63] reported that miR-124 displays three distinct expression levels: low in the VZ [65,66], intermediate in the SVZ, and high in more marginal layers [63]. This pattern of expression is opposite to that of PAX6 in the cortex, suggesting that miR-124 is downregulated by PAX6 during neurogenesis. Sansom et al. [30] analyzed mRNA levels in the E12.5 Pax6 sey/sey cortex relative to those in wild-type littermates by microarray; Mir-124 was one of 938 genes showing reproducible, statistically significant changes in expression. Delaloy et al. [64] observed that the expression pattern of miR-124 is opposite to that of PAX6 during neural differentiation of human ESCs in vitro. All of these reports are consistent with our finding that PAX6 downregulates miR-124 expression during embryonic development.
In conclusion, in addition to supporting early miR-124 expression in EBs and early cortical development, our data also strongly suggest that PAX6 modulates cell migration by regulating miR-24a expression during ES cell differentiation. The current discovery of the relationship between PAX6 and miR-124 is important for functional studies on miRNAs and differential transcriptional factors to uncover the molecular mechanisms that either maintain stemness or precede differentiation of pluripotent stem cells. Further, in light of their key roles in the nervous system and pancreas, our finding may also give new insights into the organogenesis of these tissues.
Supplementary Material
Acknowledgments
This research was supported by the 973 Program (2011CB966203 and 2011CBA01102) and the International S&T Cooperation Program (2011DFA31040 and 2013DFG30680) from the Ministry of Science and Technology of China, the Strategic Research Program of the Chinese Academy of Science (XDA01040408), grants from Shanghai City Committee of Science and Technology (12DZ1910900 and 12DZ1910800), and grants from the National Natural Science Foundation of China (31171417) and Beijing Natural Science Foundation (7142083). The authors also thank Junhua Zou of the Department of Medical Genetics for Peking University for G-banding chromosome analysis, and David Stenger of the Barbara Davis Center for Childhood Diabetes, University of Colorado, Denver, for editorial comments on a late version of the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bartel DP. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 2.Kim KS, Kim JS, Lee MR, Jeong HS. and Kim J. (2009). A study of microRNAs in silico and in vivo: emerging regulators of embryonic stem cells. FEBS J 276:2140–2149 [DOI] [PubMed] [Google Scholar]
- 3.Bang AG. and Carpenter MK. (2008). Development. Deconstructing pluripotency. Science 320:58–59 [DOI] [PubMed] [Google Scholar]
- 4.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, et al. (2007). A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, et al. (2004). Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270:488–498 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Medvid R, Melton C, Jaenisch R. and Blelloch R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melton C, Judson RL. and Blelloch R. (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463:621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, et al. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134:521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W. and Tuschl T. (2002). Identification of tissue-specific microRNAs from mouse. Curr Biol 12:735–739 [DOI] [PubMed] [Google Scholar]
- 10.Visvanathan J, Lee S, Lee B, Lee JW. and Lee SK. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21:744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X, Pfaff SL. and Gage FH. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev 21:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makeyev EV, Zhang J, Carrasco MA. and Maniatis T. (2007). The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27:435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krichevsky AM, Sonntag KC, Isacson O. and Kosik KS. (2006). Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MR, Kim JS. and Kim KS. (2010). miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem Cells 28:1550–1559 [DOI] [PubMed] [Google Scholar]
- 15.Hopfl G, Gassmann M. and Desbaillets I. (2004). Differentiating embryonic stem cells into embryoid bodies. Methods Mol Biol 254:79–98 [DOI] [PubMed] [Google Scholar]
- 16.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H. and Benvenisty N. (2000). Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med 6:88–95 [PMC free article] [PubMed] [Google Scholar]
- 17.Mohr JC, Zhang J, Azarin SM, Soerens AG, de Pablo JJ, Thomson JA, Lyons GE, Palecek SP. and Kamp TJ. (2010). The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials 31:1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messana JM, Hwang NS, Coburn J, Elisseeff JH. and Zhang Z. (2008). Size of the embryoid body influences chondrogenesis of mouse embryonic stem cells. J Tissue Eng Regen Med 2:499–506 [DOI] [PubMed] [Google Scholar]
- 19.Underbayev C, Kasar S, Yuan Y. and Raveche E. (2012). MicroRNAs and induced pluripotent stem cells for human disease mouse modeling. J Biomed Biotechnol 2012:758169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, Stevens JM, Kendall BE, Shorvon SD, Hanson IM, Moore AT. and van Heyningen V. (2001). PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet 28:214–216 [DOI] [PubMed] [Google Scholar]
- 21.Glaser T, Walton DS. and Maas RL. (1992). Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet 2:232–239 [DOI] [PubMed] [Google Scholar]
- 22.Quiring R, Walldorf U, Kloter U. and Gehring WJ. (1994). Homology of the eyeless gene of Drosophila to the small eye gene in mice and aniridia in humans. Science 265:785–789 [DOI] [PubMed] [Google Scholar]
- 23.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR. and German MS. (1997). Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 11:1662–1673 [DOI] [PubMed] [Google Scholar]
- 24.Walther C. and Gruss P. (1991). Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113:1435–1449 [DOI] [PubMed] [Google Scholar]
- 25.Simpson TI. and Price DJ. (2002). Pax6; a pleiotropic player in development. Bioessays 24:1041–1051 [DOI] [PubMed] [Google Scholar]
- 26.Wen JH, Chen YY, Song SJ, Ding J, Gao Y, Hu QK, Feng RP, Liu YZ, Ren GC, et al. (2009). Paired box 6 (PAX6) regulates glucose metabolism via proinsulin processing mediated by prohormone convertase 1/3 (PC1/3). Diabetologia 52:504–513 [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Zhao Y, Tang N, Feng R, Yang X, Lu N, Wen J. and Li L. (2012). Pax6 directly down-regulates Pcsk1n expression thereby regulating PC1/3 dependent proinsulin processing. PLoS One 7:e46934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarabykin V, Stoykova A, Usman N. and Gruss P. (2001). Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development 128:1983–1993 [DOI] [PubMed] [Google Scholar]
- 29.Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, et al. (2004). Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J 23:2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL. and Livesey FJ. (2009). The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet 5:e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauhan BK, Reed NA, Zhang W, Duncan MK, Kilimann MW. and Cvekl A. (2002). Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. J Biol Chem 277:11539–11548 [DOI] [PubMed] [Google Scholar]
- 32.Valastyan S. and Weinberg RA. (2011). Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci 124:999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Cang M, Lee AS, Zhang K. and Liu D. (2011). Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS One 6:e15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu SH. and Lee LT. (2012). Efficient differentiation of mouse embryonic stem cells into insulin-producing cells. Exp Diabetes Res 2012:201295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alipio Z, Liao W, Roemer EJ, Waner M, Fink LM, Ward DC. and Ma Y. (2010). Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. Proc Natl Acad Sci U S A 107:13426–13431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen J, Hu Q, Li M, Wang S, Zhang L, Chen Y. and Li L. (2008). Pax6 directly modulate Sox2 expression in the neural progenitor cells. Neuroreport 19:413–417 [DOI] [PubMed] [Google Scholar]
- 38.Hu Q, Zhang L, Wen J, Wang S, Li M, Feng R, Yang X. and Li L. (2010). The EGF receptor-sox2-EGF receptor feedback loop positively regulates the self-renewal of neural precursor cells. Stem Cells 28:279–286 [DOI] [PubMed] [Google Scholar]
- 39.Osumi N, Shinohara H, Numayama-Tsuruta K. and Maekawa M. (2008). Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26:1663–1672 [DOI] [PubMed] [Google Scholar]
- 40.Quinn JC, Molinek M, Nowakowski TJ, Mason JO. and Price DJ. (2010). Novel lines of Pax6−/− embryonic stem cells exhibit reduced neurogenic capacity without loss of viability. BMC Neurosci 11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi N. and Epstein JA. (2002). Getting your Pax straight: Pax proteins in development and disease. Trends Genet 18:41–47 [DOI] [PubMed] [Google Scholar]
- 42.Suter DM, Tirefort D, Julien S. and Krause KH. (2009). A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells 27:49–58 [DOI] [PubMed] [Google Scholar]
- 43.Ashery-Padan R, Zhou X, Marquardt T, Herrera P, Toube L, Berry A. and Gruss P. (2004). Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol 269:479–488 [DOI] [PubMed] [Google Scholar]
- 44.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA. and Van Obberghen E. (2007). MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem 282:19575–19588 [DOI] [PubMed] [Google Scholar]
- 45.Xu XL, Zong R, Li Z, Biswas MH, Fang Z, Nelson DL. and Gao FB. (2011). FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J Neurosci 31:13705–13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozmik Z, Czerny T. and Busslinger M. (1997). Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J 16:6793–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasuda T, Kajimoto Y, Fujitani Y, Watada H, Yamamoto S, Watarai T, Umayahara Y, Matsuhisa M, Gorogawa S, et al. (2002). PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes 51:224–230 [DOI] [PubMed] [Google Scholar]
- 48.St.-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A. and Gruss P. (1997). Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 387:406–409 [DOI] [PubMed] [Google Scholar]
- 49.Engelkamp D, Rashbass P, Seawright A. and van Heyningen V. (1999). Role of Pax6 in development of the cerebellar system. Development 126:3585–3596 [DOI] [PubMed] [Google Scholar]
- 50.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H. and Verdin E. (2010). miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2:415–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, et al. (2010). Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7:90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liour SS. and Yu RK. (2003). Differentiation of radial glia-like cells from embryonic stem cells. Glia 42:109–117 [DOI] [PubMed] [Google Scholar]
- 53.Liour SS, Kraemer SA, Dinkins MB, Su CY, Yanagisawa M. and Yu RK. (2006). Further characterization of embryonic stem cell-derived radial glial cells. Glia 53:43–56 [DOI] [PubMed] [Google Scholar]
- 54.Zhou YH, Hu Y, Mayes D, Siegel E, Kim JG, Mathews MS, Hsu N, Eskander D, Yu O, Tromberg BJ. and Linskey ME. (2010). PAX6 suppression of glioma angiogenesis and the expression of vascular endothelial growth factor A. J Neurooncol 96:191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang ES. and Goldman JE. (2011). Pax6 expression is sufficient to induce a neurogenic fate in glial progenitors of the neonatal subventricular zone. PLoS One 6:e20894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conaco C, Otto S, Han JJ. and Mandel G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A 103:2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ballas N, Grunseich C, Lu DD, Speh JC. and Mandel G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121:645–657 [DOI] [PubMed] [Google Scholar]
- 58.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H. and Majumder S. (2008). REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayes DA, Hu Y, Teng Y, Siegel E, Wu X, Panda K, Tan F, Yung WK. and Zhou YH. (2006). PAX6 suppresses the invasiveness of glioblastoma cells and the expression of the matrix metalloproteinase-2 gene. Cancer Res 66:9809–9817 [DOI] [PubMed] [Google Scholar]
- 60.Numayama-Tsuruta K, Arai Y, Takahashi M, Sasaki-Hoshino M, Funatsu N, Nakamura S. and Osumi N. (2010). Downstream genes of Pax6 revealed by comprehensive transcriptome profiling in the developing rat hindbrain. BMC Dev Biol 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Houbaviy HB, Murray MF. and Sharp PA. (2003). Embryonic stem cell-specific microRNAs. Dev Cell 5:351–358 [DOI] [PubMed] [Google Scholar]
- 62.Houbaviy HB, Dennis L, Jaenisch R. and Sharp PA. (2005). Characterization of a highly variable eutherian microRNA gene. Rna 11:1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiorano NA. and Mallamaci A. (2009). Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Dev 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN. and Gao FB. (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng LC, Pastrana E, Tavazoie M. and Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12:399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Pietri TD, Calegari F, Fei JF, Nomura T, Osumi N, Heisenberg CP. and Huttner WB. (2006). Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. Biotechniques 41:727–732 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.