Stem cells regulate their fate by binding to, and contracting against, the extracellular matrix. Recently, it has been proposed that in addition to matrix stiffness and ligand type, the degree of coupling of fibrous protein to the surface of the underlying substrate, i.e. tethering and matrix porosity, also regulates stem cell differentiation. By modulating substrate porosity without altering stiffness in polyacrylamide gels, we show that varying substrate porosity did not significantly change protein tethering, substrate deformations, or the osteogenic and adipogenic differentiation of human adipose-derived stromal cells and marrow-derived mesenchymal stromal cells. Varying protein-substrate linker density up to 50-fold changed tethering, but did not affect osteogenesis, adipogenesis, surface-protein unfolding, or underlying substrate deformations. Differentiation was also unaffected by the absence of protein tethering. Our findings imply that the stiffness of planar matrices regulates stem cell differentiation independently of protein tethering and porosity.
The stiffness of ECM has been shown to regulate both shorter- and longer-term cell functions such as cell spreading1 and stem and progenitor cell phenotype changes on planar substrates,2–7 respectively. For example, many types of adult stromal cells grown on substrates of stiffness similar to that of the osteoid or muscle express lineage markers of terminally differentiated cells found in those tissues.3,4,6 Common myosin-based contractile mechanisms are required for matrix-induced differentiation in two-dimensions (2D).3,8–10 However in three-dimensions (3D), a labile11 or degradable matrix,12 which permits cells to first spread and then adhere to the ECM, is required. Similarly, force-mediated protein unfolding of the ECM in vivo regulates cell responses as a function of stiffness.13,14 While creating 3D matrices has become a widespread approach towards understanding how the matrix affects cell fate, the regulatory role of substrate-anchored fibrous protein deformations on stem cell fate in 2D is still unclear.
Recent literature suggests that the mechanical resistance provided by ECM, which opposes myosin-based contractility and results in cell signaling and differentiation, could be due to protein tethers rather than substrate stiffness for planar cultures.15 Since most synthetic planar matrices are not normally cell adhesive, an adhesive layer of matrix protein is attached to the hydrogel surface and covalently “tethered” to the substrate surface at distinct anchoring points. Thus, changing protein-substrate linker density or substrate porosity can vary the length of the fiber segment between two adjacent anchoring points. When a load is applied perpendicularly to the fiber segment, the deflection of the fiber segment is directly related to the load applied, fiber stiffness, and the length of the fiber segment cubed.15,16 If enough resistance were present in tethers, stem cells could differentiate independent of substrate stiffness. However, it is unclear what length these tethers may be and how they compare to substrate deformations17, which have been implicated in mechanotransduction and hence stem cell differentiation.18 Thus it is critical to decouple protein tethering and substrate stiffness and to determine if and how these factors collectively regulate stem cell differentiation.
Tuning Hydrogel Porosity Independent of Stiffness
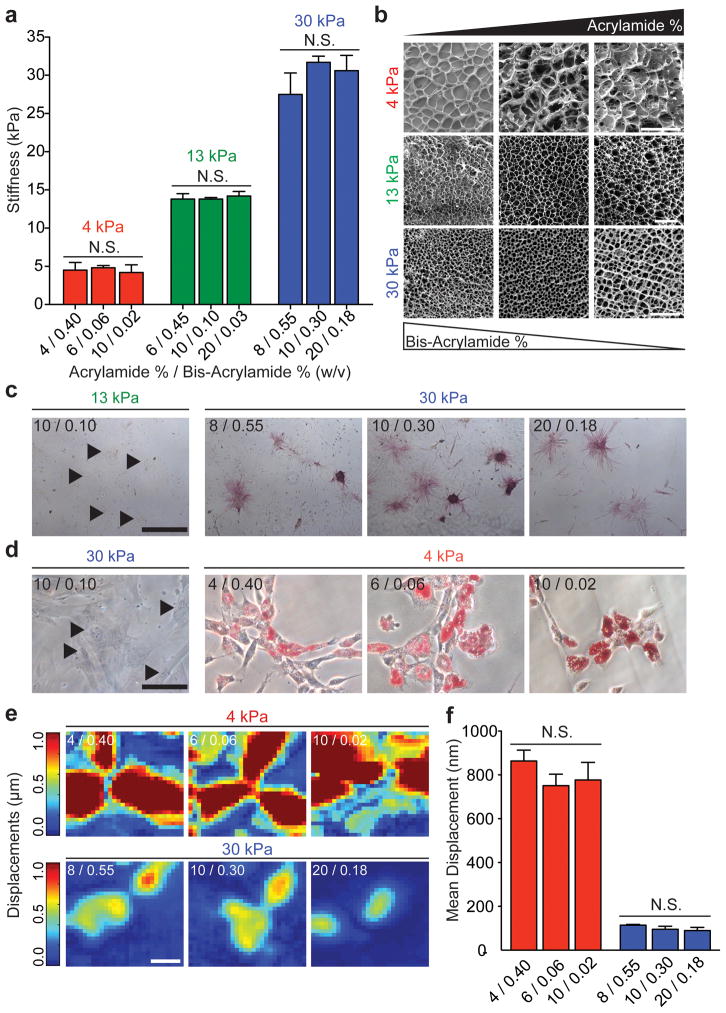
Tuning the acrylamide monomer and bis-acrylamide crosslinker ratio can change polyacrylamide (PA) hydrogel porosity, i.e. the distance between tethering points, independently of stiffness. To accomplish this, three separate acrylamide/bis-acrylamide formulations were polymerized to yield hydrogels of ~4, ~13, and, ~30 kiloPascals (kPa; Fig. 1a), which correspond to the stiffness of adipose tissue, muscle, and osteoid,2,3,6,19–21 respectively. Differences in volume and mass swelling ratios between each of the hydrogels with similar stiffness suggest significant differences in porosity among each substrate subgroup (Supplementary Fig. S1a and S1b). The radius of gyration of extended DNA may be used to estimate the effective maximum pore size of the hydrogel.22 DNA size standards were exposed to an electrophoretic gradient in swollen and unconfined 4 and 30 kPa PA hydrogels to further quantify hydrated pore size. For 30 kPa hydrogels, a 45 nm DNA fragment failed to migrate through the 8/0.55 formulation indicating that the maximum pore size of this formulation is between 23 and 45 nm. Larger DNA fragments migrated through the 10/0.3 and 20/0.15 gel formulations indicating that the approximate pore sizes are between 88 and 166 nm for both formulations; differences in DNA mobility suggest that the two gels have pore sizes that differ within this range. Similarly, differences in DNA mobility suggest that the three 4 kPa formulations yield hydrogels with different pore sizes (Supplementary Fig. S1c). Scanning electron microscopy (SEM) of dried PA hydrogels showed increasing pore sizes with increasing acrylamide and decreasing bis-acrylamide concentrations for 4, 13, and 30 kPa hydrogel formulations (Fig. 1b); these data are consistent with pore size trends in hydrated measurements and together demonstrate that increasing the bis-acrylamide crosslinker concentration decreases relative pore size without substantially changing hydrogel modulus. However, it is important to note here that pore sizes derived from SEM images of freeze-dried hydrogels are likely not representative of actual substrate pore sizes in a hydrated state. Cells interact with hydrated substrates in vitro, and thus SEM images are only provided for relative comparison of pore sizes for the hydrogel formulations reported.
Fig 1. Influence of substrate porosity on ASC differentiation.
a, Elastic modulus measured via AFM (n = 3) for the indicated acrylamide:bis-acrylamide ratios. b, SEM images of PA hydrogels made with varying monomer to crosslinker ratios as indicated (scale bars, 50 μm [top and bottom], 10 μm [middle]). c, Alkaline phosphatase staining of ASCs on 13 and 30 kPa hydrogels of the indicated compositions after 14 days of culture in normal media. Arrowheads indicate stained but yet negative cells (scale bar, 500 μm). d, Oil Red O staining of ASCs on 4 kPa and 30 kPa hydrogels of the indicated compositions after 7 days of culture in adipogenic induction media. Arrowheads indicate stained but yet negative cells (scale bar, 100 μm). e, Displacement maps of embedded fluorescent particles resulting from ASC traction forces on 4 kPa and 30 kPa hydrogels of the indicated compositions (scale bar, 50 μm). f, Quantification of mean displacement was plotted for hydrogels of the indicated composition and stiffness range (n > 20; mean ± S.E.M.; N.S. = not significant).
Differentiation and Deformations are Independent of Porosity
Human ASCs were plated onto 13 and 30 kPa PA hydrogels of the formulations indicated in Fig. 1c. After 14 days of culture in normal growth media, osteogenic differentiation was evident by positive alkaline phosphatase (ALP) staining in sub-confluent cells independent of hydrogel formulation but directly dependent on substrate stiffness, as 13 kPa substrates were negative for ALP (Fig. 1c). Further confirmation of this is demonstrated by positive and nuclear localized RUNX2 immunofluorescence (IF) staining after 7 days in culture on all 30 kPa hydrogels (Supplementary Fig. S2a). The expression of early osteogenic markers ALP and RUNX2 suggest that changes in porosity independent of stiffness have no noticeable effects on differentiation for the range of hydrogel formulations tested. However, allowing cells to reach confluence in normal media on any hydrogel formulation was sufficient to override substrate stiffness-mediated differentiation and induce osteogenesis as previously observed,15 most likely due to other factors including cell-cell signaling and secreted paracrine factors (Supplementary Fig. S3). To avoid complications arising from confluent monolayers and to focus on cell-ECM signaling only, osteogenic differentiation studies were conducted at low cell densities. MSCs, another commonly used cell type in differentiation experiments, also stained positive for ALP after 14 days in culture on the three 30 kPa hydrogel formulations (Supplementary Fig. S3b), implying that substrate porosity has little effect on multiple stem cell types. Additionally, after 14 days in culture in adipogenic induction media, adipogenic differentiation, as assessed by Oil Red O (ORO) presence, was found in over 40% of ASCs on all 4 kPa substrates independent of hydrogel formulation but directly dependent on substrate stiffness as 30 kPa substrates were negative for ORO (Fig. 1d and Supplementary Fig. S2c).
As cell-ECM signaling depends on contractility, and differences in contractility have been shown to regulate differentiation,3,8–10 displacement maps of embedded fluorescent particles resulting from ASC traction forces on all 4 and 30 kPa hydrogel formulations were computed (Fig. 1e, Supplementary Fig. S4) using traction force microscopy (TFM).23 Mean displacements were similar between all formulations of 4 and 30 kPa hydrogels, but different between hydrogels of different stiffness (Fig. 1f). These data indicate that over the range of formulations tested, hydrogel deformations due to cell contractions are similar regardless of porosity but dependent on stiffness (Figs. 1e and 1f). Taken together, these data show that varying porosity alone does not appear to be sufficient to alter the fate of two different adult stem cell sources.
Modulating Protein Tethering by Changing Linker Density
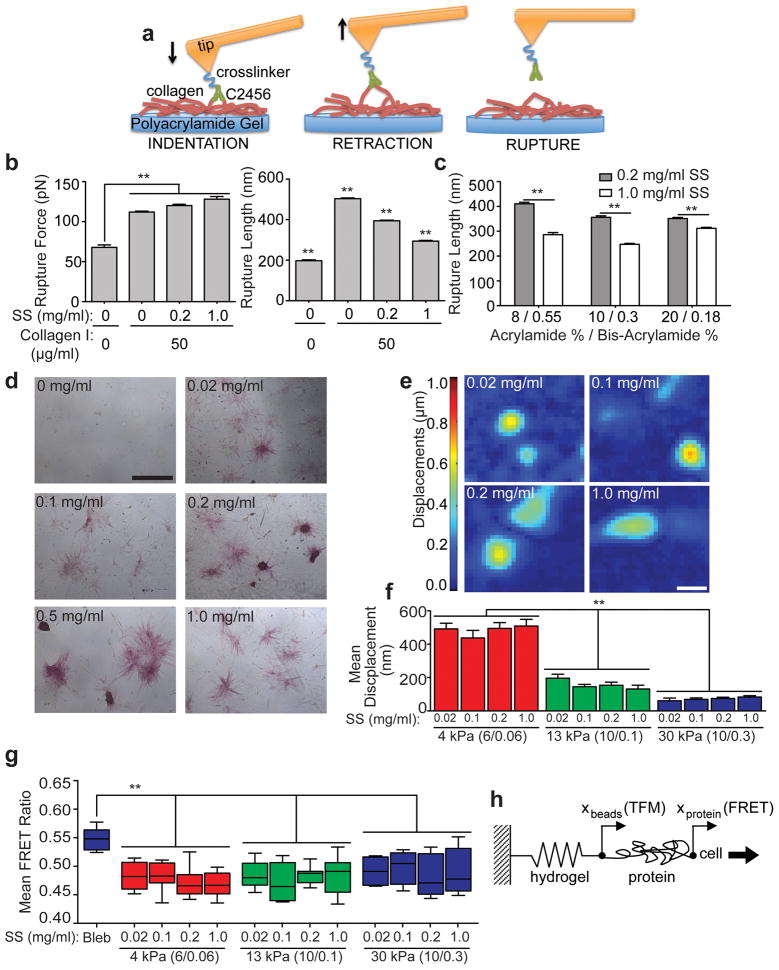
Culturing cells on synthetic hydrogels requires the covalent coupling of a cell-adhesive matrix protein, such as collagen type I, to the hydrogel surface using a protein-substrate linker, such as sulfo-SANPAH.1 Changing the concentration of such linker has been proposed to modulate protein tethering.15 To modulate the tethering of fibrous collagen to PA hydrogels, we tuned the surface density of anchoring points by varying the concentration of sulfo-SANPAH, thus varying the average distance between adjacent anchoring points. To assess possible differences in the physical structure or total amount of bound protein, immunofluorescence staining of collagen covalently coupled to PA substrates activated with varying concentrations of sulfo-SANPAH was performed. Images revealed noticeable surface heterogeneity making quantification of absolute protein amount difficult (Supplementary Fig. S5a); this was further illustrated by collagen pixel intensity histograms for 13 and 30 kPa hydrogels over a range of sulfo-SANPAH concentrations (Supplementary Fig. S5b). Fluorescent detection was unable to quantify surface-bound protein as previously suggested.15 To directly quantify collagen tethering, we obtained individual force spectrograms (Supplementary Fig. S6a) from microindentations of collagen coated PA hydrogels. Substrates were activated with a range of sulfo-SANPAH concentrations using a probe functionalized with an anti-collagen type I antibody (Fig. 2a). As the tip retracts from the surface, the collagen unfolds and/or stretches until the antibody-protein bonds rupture (Supplementary Fig. S6a). Force spectrograms were analyzed to locate rupture events and to determine the force at rupture, i.e., the force required to break a protein-antibody bond, and the rupture length, i.e., the deflection of the collagen fiber segment at rupture. Larger rupture forces and a greater number of rupture events were detected in the presence of collagen I (Fig. 2b, left; Supplementary Fig. S6b) and indicate that the antibody was specifically binding and loading collagen. Decreasing rupture length with increasing sulfo-SANPAH concentration (Fig. 2b, right) confirmed that the number of protein anchoring points scaled with sulfo-SANPAH concentration without substantial changes in rupture force (Fig. 2b). This trend held for all 30 kPa formulations tested despite significant changes in the number of available protein anchoring sites, which is proportional to acrylamide concentration. We observed differences in rupture length between sulfo-SANPAH concentrations across hydrogel formulations (Fig. 2c, gray vs. white bars) indicating that anchoring sites must not be saturated. Furthermore, for a given sulfo-SANPAH concentration, although small differences in average rupture length were detected between the three 30 kPa hydrogel formulations, i.e. < 40 nm, these differences were smaller than the changes in pore size, which were up to 120 nm (Supplementary Fig. S1c). Thus differences in rupture lengths between the hydrogel formulations are not likely due to porosity changes.
Fig 2. Influence of protein tethering on ASC differentiation.
a, Schematic depicting the interaction between an AFM tip (orange) functionalized with a collagen I antibody (C2456; green) and the hydrogel (blue) functionalized with bound collagen I (red). The black arrow indicates the direction of motion. A rupture event occurs following retraction of the tip from the surface. b, Measured rupture force (left) and rupture length (right) for rupture events that occurred on 10/0.3 30 kPa hydrogels activated with the indicated sulfo-SANPAH (SS) and collagen I concentrations (n = 500; mean ± S.E.M.; **p < 0.0001). c, Rupture length was measured for rupture events that occurred on 30 kPa hydrogels with indicated monomer to crosslinker ratios. Hydrogels were activated with either 0.2 mg/ml or 1 mg/ml sulfo-SANPAH. (n = 500; mean ± S.E.M.; **p < 0.0001). d, Images of ASCs stained for ALP expression on 10/0.3 hydrogels as a function of sulfo-SANPAH concentration after 14 days of culture in normal media (scale bar, 500 μm). e, Displacement maps of embedded fluorescent particles resulting from ASC traction forces on 10/0.3 hydrogels for a range of indicated sulfo-SANPAH concentrations (scale bar, 50 μm). f, Quantification of mean bead displacement for the indicated hydrogel stiffness and composition as well as sulfo-SANPAH (SS) concentration (n = 20; mean ± S.E.M; **p < 0.0001). g, Measured fibronectin FRET intensity ratio for ASCs on 4, 14, and 30 kPa hydrogels activated with the indicated concentrations of sulfo-SANPAH (n = 8; *p<0.05). h, Proposed model of a cell on a protein coated substrate attached to a rigid base (glass coverslip) where cell forces are translated through the protein and through the substrate. Deformations of the substrate are measured via TFM and deformations of the protein are measured via FRET.
Differentiation and Deformations do not Depend on Tethering
To investigate whether or not tethering impacts stem cell fate, subconfluent ASCs and MSCs were cultured in normal growth medium on 30 kPa hydrogels over a range of sulfo-SANPAH concentrations and assessed for osteogenic differentiation. Positive ALP and RUNX2 staining was observed on all 30 kPa hydrogels regardless of sulfo-SANPAH concentration, hydrogel formulation, and cell type (Fig. 2d and Supplementary Fig. S7). ASCs were also cultured on 4 kPa hydrogels over a range of sulfo-SANPAH concentrations and ORO expression was observed in over 30% of ASCs regardless of sulfo-SANPAH concentration (Supplementary Fig. S8). Together these data indicate that the degree of collagen tethering to the substrate surface had no observable effect on stem cell fate unlike what has been suggested.15 Myosin contractility deforms the ECM and is required for matrix-induced differentiation;3,8–10 thus to confirm differentiation results, substrate displacements for hydrogels across a range of sulfo-SANPAH concentrations were mapped using TFM (Fig. 2e). Average displacements of beads embedded in hydrogels were independent of sulfo-SANPAH concentration and only dependent on substrate modulus (Fig. 2f), suggesting that for the range of protein-substrate linker concentrations used in this study, the surface density of collagen fiber covalent anchoring points has no impact on how cells deform the underlying substrate.
To determine whether or not differences in rupture lengths, i.e. tethering, detected via force spectrograms could be felt by cells on a molecular scale, a fibronectin FRET sensor14 was covalently attached to hydrogels in place of collagen. Cell-generated forces unfold the protein thus increasing the distance between paired fluorescent probes, which results in a decrease in the FRET ratio (Supplementary Fig. 9a) that can also be shown via chemical denaturation (Supplementary Fig. S9b–c). Changing sulfo-SANPAH concentration has no statistical effect on the FRET ratio of fibronectin underneath spread ASCs regardless of hydrogel formulation, while perturbing myosin contractility using blebbistatin caused a significant increase in the FRET ratio (Fig. 2g and Supplementary Fig. S9d). Thus, molecular conformational changes in protein caused by ASC traction forces are similar regardless of protein-substrate linker concentration implying that ASCs deform the surface protein similarly on all sulfo-SANPAH activated hydrogels. Based on these findings, we propose that cells deform both the adhesive protein on the hydrogel surface well as the underlying polyacrylamide substrate according to the model depicted in Fig. 2h. Cell forces are translated sequentially through the protein layer and the hydrogel. However, our findings suggest that the degree of coupling of the protein to the substrate does not influence substrate deformation and thus differentiation; therefore it was not depicted in Fig. 2h.
Differentiation Occurs in the Absence of Tethering
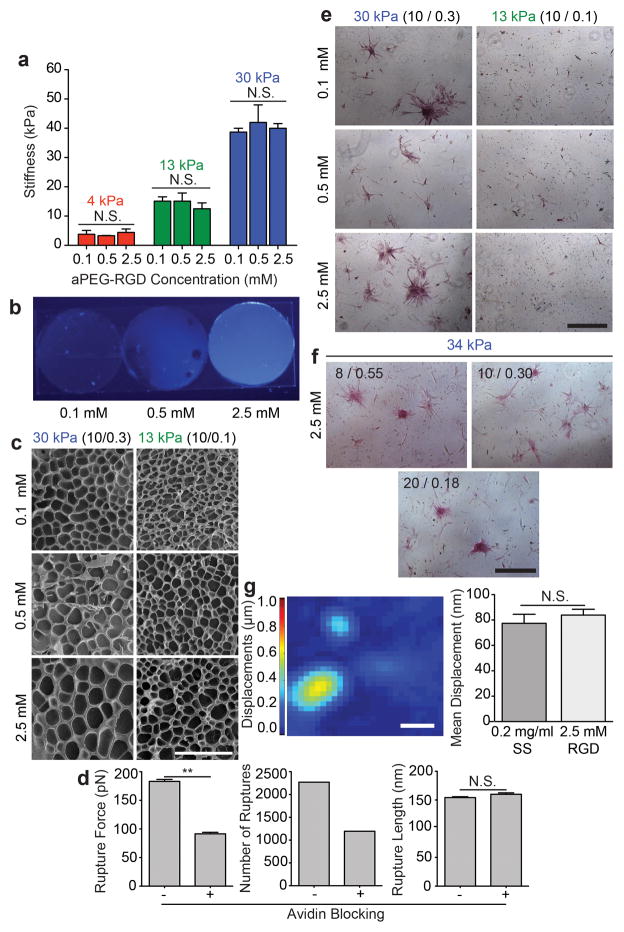
To demonstrate that stiffness-induced differentiation is possible in the absence of fibrous protein tethering, RGD, a short cell-adhesive peptide from fibronectin24, was directly incorporated into the polyacrylamide backbone by including acrylated-PEG-RGD during polymerization rather than tethering an adhesive protein to the substrate. Three separate hydrogel formulations with 0.1, 0.5, and 2.5 mM RGD yielding the same gel stiffness were made for 4, 13, and 30 kPa substrates using the acrylamide/bis-acrylamide ratios listed above (Fig. 3a). A hydroxycoumarin dye-conjugated acrylated-PEG-RGD confirmed that the peptide was incorporated in a dose-dependent manner (Fig. 3b). SEM images of dried hydrogels show similar pore sizes regardless of the concentration of acrylated-PEG-RGD incorporated within each substrate (Fig. 3c). This ensures that differentiation effects can be attributed to changes in adhesive-peptide density and not porosity. Furthermore to ensure that the PEG moiety does not act as a tether, individual force spectrograms were obtained from biotin terminated PEG-coated PA hydrogels and an avidin functionalized probe. Larger rupture forces and a greater number of rupture events were detected on substrates prior to blocking with excess avidin in solution (Figs. 3d, left and middle), which indicate that the avidin functionalized probe was specifically bound to the biotin coated surface. Rupture lengths prior and subsequent to blocking were not statistically different (Fig. 3d, right) and were similar to rupture lengths measured on control PA substrates with no surface coating (Fig. 2b, right). In contrast to collagen coated PA substrates that exhibited significantly greater rupture lengths, the deformations of the PEG moiety are minimal. Thus, PA-PEG-RGD substrates are a valid culture platform absent of protein tethering for the given concentration range and size of PEG tested. ASCs were then cultured for 14 days in normal growth media to determine if differentiation was possible without tethering over the range of peptide concentrations tested. ASCs underwent osteogenic differentiation on 10/0.3 30 kPa hydrogels independent of RGD concentration (Fig. 3e). Furthermore, osteogenic differentiation was seen in ASCs and MSCs cultured on all 30 kPa hydrogel formulations with 2.5 mM RGD (Fig. 3f and Supplementary Fig. S10). Together, these data suggest that differentiation occurs in the absence of fibrous protein tethering over the range of peptide concentrations tested. Cell generated substrate displacements were similar to that of collagen coated hydrogels (Fig. 3g) lending further evidence that matrix-induced differentiation operates through common myosin-based contractile mechanisms given that differentiated cells on these and collagen coated hydrogels were similar.
Fig 3. Direct incorporation of a short adhesive peptide to the PA substrate.
a, Elastic modulus measured via AFM (n = 3; N.S. = not significant). b, aPEG-RGD-dye incorporation is detected under UV light. c, SEM images of PA hydrogels of indicated stiffness made with varying RGD concentration (scale bar, 50 μm). d, Measured rupture force (left), number of events (middle), and rupture length (right) for rupture events that occurred on 10/0.3 30 kPa hydrogels coated with PEG-biotin (n = 1000; mean ± S.E.M.; N. S. = not significant; **p<0.0001). e, ALP staining of ASCs on 13 kPa and 30 kPa hydrogels with low, medium, and high concentrations of RGD (scale bar, 500 μm). f, ALP staining of ASCs on 30 kPa hydrogels of varying monomer to crosslinker ratio and constant high concentration of RGD after 14 days of culture in normal media (scale bar, 500 μm). g, Representative displacement map (left) of embedded fluorescent particles resulting from ASC traction forces on a 30 kPa hydrogel with 2.5 mM RGD. Mean displacement is shown (right) for a collagen coated hydrogel (0.2 mg/ml sulfo-SANPAH and 50 μg/mL collagen I) and a PA-PEG-RGD hydrogel (2.5 mM RGD). (n = 30; mean ± S.E.M; N.S. = not significant; scale bar, 50 μm).
Cell Spread Area on PA and PDMS Substrates
To further support the claim that stiffness mediates cell functions generally, we observed the basic behavior of cell spreading on PA-PEG-RGD hydrogels in the absence of protein tethers. ASC spread area 24 hours after seeding scaled with increasing hydrogel stiffness (Supplementary Fig. S11a). This suggests that stiffness is an important physical factor independent of how adhesive ligands are presented (but not independent of concentration25). In order to determine whether or not this phenomenon is specific to acrylamide-based systems, polydimethylsiloxane (PDMS) substrates were fabricated with base-to-curing ratios of 100:1, 75:1, and 50:1 to modulate stiffness as noted previously.15 These substrates were not functionalized with adhesive protein. Without covalently attaching or tethering ligands to the surface, cell adhesion and spreading was still possible for all substrates due to the well-known fouling properties of PDMS, and furthermore, cell spread area was similar on all substrates (Supplementary Fig. S11b). This observation is in agreement with previous observations that imply stiffness independent cell spreading on PDMS substrates,15 suggesting that cells may sense similar mechanical cues on the three PDMS formulations.
Measuring PDMS Mechanical Properties on a Cell-Sensing Scale
Because of a lack of correlation between cell spread area and PDMS base-to-curing ratio, we independently measured PDMS stiffness by AFM microindentation. The stiffnesses of 50:1 and 100:1 PDMS were found to be 250 and 550 kPa (Supplementary Fig. S12a)—orders of magnitude greater than previously reported.15 Since PDMS has previously been shown to be viscoelastic at higher base-to-curing ratios,26 substrates were instead indented using different indenter geometries and at different indentation speeds. Using two different probes and a wide range of indentation speeds confirmed the viscoelastic behavior of PDMS (Supplementary Fig. S12b) and suggests that different methods of characterization may account for discrepancies in reported values of PDMS stiffness.
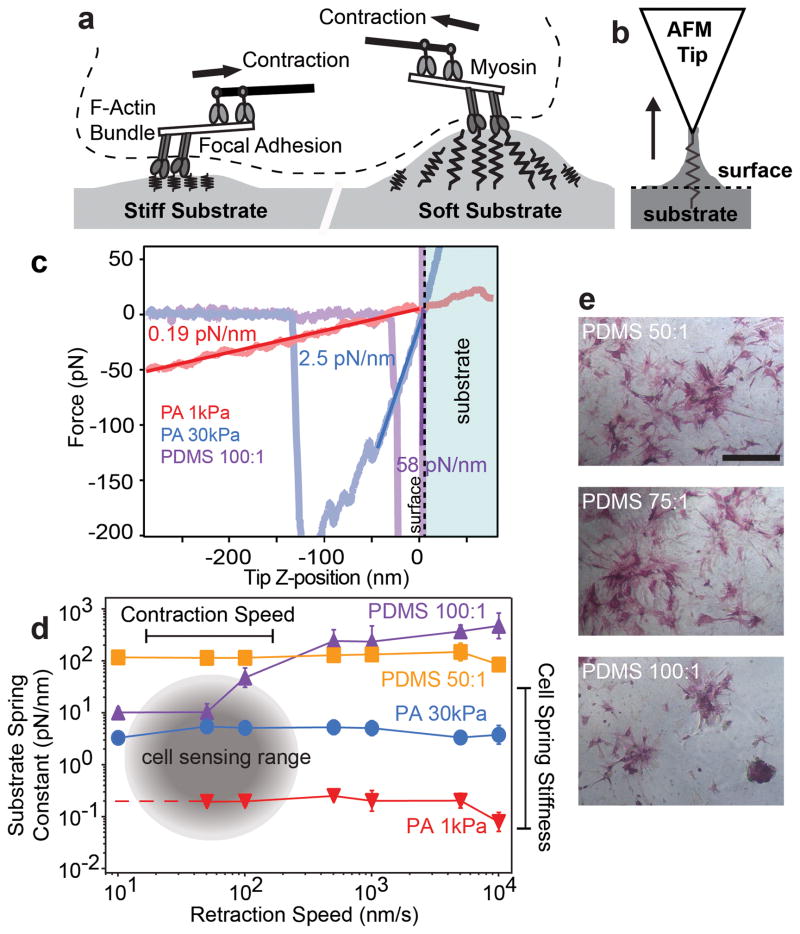
Given the lack of consensus on measuring the mechanical properties of PDMS, it is important to use the most appropriate technique to closely mimic cell-substrate interactions. Cells pull against substrates at 20 to 120 nm/s resulting in deformations that scale inversely with stiffness17,27 (Fig. 4a). We can match AFM tip retraction velocity to the pulling velocity and size of focal adhesions. Consequently, we can simulate these dynamically fluctuating pulling events by analyzing the retraction curves (as opposed to indentation curves) obtained by AFM where the tip has pulled and deformed the material above the surface (Fig. 4b). The substrate stiffness is determined by fitting the linear region beginning at the contact point with the (undeformed) surface (F = 0 pN) to where the force reaches −100 pN (Fig. 4c). 1 and 30 kPa PA hydrogels demonstrated little variation in stiffness over a range of cell relevant strains26 and retraction speeds.17,27 The stiffness of 50:1 and 100:1 PDMS were both significantly higher than the PA hydrogels, and the stiffness of 100:1 PDMS increases 50 fold over the range of retraction velocities tested (Fig. 4d). These data confirm that 100:1 PDMS is highly viscoelastic and 50:1 PDMS is predominantly elastic but both are stiff over cell relevant strains in agreement with prior data.26 Though previous studies have noted lower stiffness values of PDMS for the same cure ratios,15 it is well known that the mechanical properties of PDMS are different at the cellular mechano-sensing scale than at the macroscopic scale.28 At the scale at which a cell mechano-senses,17,27 both 50:1 and 100:1 PDMS substrates are stiffer than 30 kPa PA hydrogels (Fig. 4d). This provides a reasonable explanation as to why cell spreading (Supplementary Fig. S11b) and osteogenic differentiation (Fig. 4e), neither of which changed with cure ratio, were previously reported to be stiffness independent.15 We note here however, that it is possible to decrease the effective stiffness of PDMS by fabricating microposts of identical cure ratios but different heights. Fu et al. found that MSC contractility and differentiation towards adipogenic or osteogenic lineages scaled as a function of effective stiffness pillar height.28 Thus, even in PDMS systems where cure ratio is not directly modulated, effectively modulating stiffness can still yield mechanically-driven differentiation.
Fig 4. Cells sense by contracting against their substrates.
a, Schematic depicting how cells dynamically deform soft and stiff substrates by pulling (and not pushing) via myosin contractions. Softer substrates are deformed to a greater extent than stiffer substrates. b, Schematic depicting the interaction between an AFM tip and the surface of a substrate. The arrow indicates the direction of motion of the tip during retraction. c, Representative retraction curves for 1 and 30 kPa PA hydrogels and a 100:1 PDMS substrate. The dashed line indicates the point at which the tip is no longer indenting into the substrate (shaded light blue). Substrate stiffness is calculated by fitting the linear region of the retraction curve starting at the (undeformed) surface. d, Substrate spring constant determined by the method depicted in c for 1 and 30 kPa PA hydrogels and 50:1 and 100:1 PDMS substrates as a function of AFM tip retraction speed. Cells are sensitive to substrate stiffnesses of 1–100 kPa. Previously measured myosin contraction speeds range from 10–100 nm/s (gray). e, Alkaline phosphatase staining of ASCs on PDMS substrates after 7 days of culture in normal media (scale bar, 500 μm).
PDMS Substrates do not Support Protein Tethering
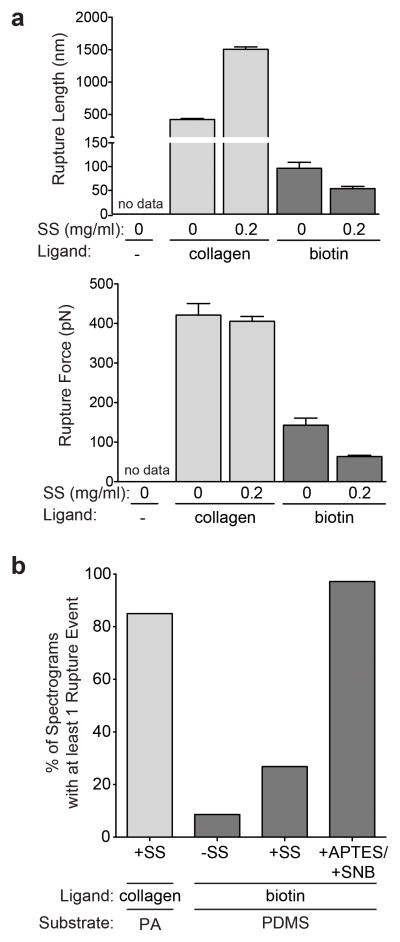
To address the possibility of fibrous protein tethering on PDMS, 50:1 PDMS substrates were examined using force spectroscopy. When 50:1 PDMS substrates were pre-incubated in a collagen solution, rupture events with lengths and forces much greater than that of PA substrates were detected (Fig. 5a) confirming that collagen non-specifically adsorbs to PDMS. Attempting to functionalize PDMS with sulfo-SANPAH prior to collagen incubation15 did not alter rupture force (Fig. 5b). However, rupture length drastically increased from 450 nm to 1.5 μm, which is larger than cell deformations on stiff PA substrates (Fig. 2g and 5a). This observation is opposite of what was seen with PA; treating PA with sulfo-SANPAH increases fibrous collagen tethering to PA, consequently decreasing rupture length (Fig. 2b).
Fig 5. Atomic force spectrography analysis of ligand coated PDMS substrates.
a, Measured rupture length (top) and rupture force (bottom) for rupture events detected on 50:1 PDMS substrates activated with the indicated sulfo-SANPAH (SS) and collagen or NH2-PEG-biotin concentrations. No data indicates that no rupture events were detected. b, Percentage of spectrograms with at least one rupture event on PA and PDMS substrates activated with the linkers and ligands shown.
The increased rupture lengths seen in PDMS may be attributed to the formation of long chains of collagen forming on the PDMS surface as collagen contains many primary amines for sulfo-SANPAH to crosslink, whereas PDMS is void of amines (Supplementary Fig. S13a, left). In this hypothesis, sulfo-SANPAH is not directly coupled to the PDMS surface, but rather only to collagen chains. To test this hypothesis, substrates were functionalized with sulfo-SANPAH prior to incubation in NH2-PEG-biotin, which has only one free primary amine (Supplementary Fig. S13a, right). Rupture lengths and forces obtained from force spectrograms using avidin tips were similar on biotin-coated substrates functionalized with and without sulfo-SANPAH (Fig. 5a).
To further confirm that sulfo-SANPAH does not react with PDMS, amines were covalently bound to PDMS substrate surfaces using the chemistry outlined in Supplementary Fig. S13b. At least one rupture event was detected in more than 90% of force spectrograms obtained from biotin-coated samples. In contrast, rupture events were detected in only 30% of force spectrograms obtained from biotin-coated but not amine-functionalized PDMS samples independent of sulfo-SANPAH (Fig. 5b). Thus it is clear that the sulfo-succinmidyl group requires amines in order to form a covalent bond. Regardless of UV treatment, PDMS surfaces do not display free amines, and thus protein cannot be covalently bound to the surface via sulfo-SANPAH. Prior efforts do not appear to have amine-functionalized PDMS15, and thus it is difficult to attribute fibrous protein tethering on PDMS to cell spreading and differentiation. These results in conjunction with cure ratio-independent stem cell spreading (Supplementary Fig. S11b) and differentiation (Fig. 4e) emphasize the shortcomings of PDMS as a model system to investigate stiffness-dependent behavior over a relevant cell-sensing range.28 Elastic 2D hydrogel systems with controlled stiffness such as polyacrylamide, poly-ethylene glycol,29 hyaluronic acid,30,31 and alginate32 are better suited to investigate these cell behaviors.
Summary
The commonly used PA hydrogel system is easily tuned to modulate substrate porosity, and in combination with different concentrations of sulfo-SANPAH, provides a platform to investigate how substrate stiffness, porosity, and ligand tethering affect stem cell fate. The data presented here provide direct evidence that the mechanical feedback provided by hydrogel deformations on planar matrices regulate osteogenic and adipogenic differentiation of ASCs and MSCs independent of protein tethering and substrate porosity. Furthermore, these data indicate that substrates have fibrous protein tethers as previously hypothesized;15 however, these tethers are not essential for the osteogenic and adipogenic differentiation of ASCs and MSCs. This work further highlights the importance of bulk matrix stiffness as the major mechanical regulator of stem cell differentiation.
METHODS
Polyacrylamide Gels
Glass coverslips were functionalized using 3-(trimethoxysilyl)propyl methacrylate to facilitate covalent attachment of hydrogel substrates to glass. A polymer solution containing acrylamide monomers, crosslinker N,N methylene-bis-acrylamide, Ammunium Persulfate (APS), and N,N,N′,N′-Tetramethylethylenediamine (TEMED) was prepared. The polymerizing solution was sandwiched between a functionalized coverslip and a dichlorodimethylsilane (DCDMS)-treated slide to ensure easy detachment of hydrogels. The ratio of acrylamide%/bis-acrylamide% was varied in order to control hydrogel stiffness and porosity. To allow for cell adhesion and fibrous protein tethering, substrates were incubated in 0.02, 0.1, 0.2, 0.5, or 1 mg/ml N-sulphosuccinimidyl-6-(4′-azido-2′-nitrophenylamino) hexanoate (sulfo-SANPAH), activated with UV light, washed, and then incubated in collagen overnight. For AFM experiments, 0.5 mg/ml amine-PEG3400-biotin was used instead of collagen. Coated hydrogels were UV sterilized prior to use in cell culture.
PA-PEG-RGD Gels
PA-PEG-RGD hydrogels were fabricated by incorporating 0.1 mM, 0.5 mM, or 2.5 mM acrylated-PEG3400-GRGD-amide (aPEG-RGD) into the polymerizing solution described above. In order to visualize RGD concentration differences, a fluorescent hydroxycoumarin dye was conjugated to the peptide.
PDMS Substrates
PDMS was mixed at various elastomer base:curing agent ratios (50:1, 75:1, 90:1, 100:1), thoroughly mixed, and degased under vacuum before pouring directly into multi-well plates or onto coverslips and baked overnight. In certain instances, substrates were functionalized with sulfo-SANPAH and ligand (see Supplementary Information) (Supplementary Fig. S13a). For covalent attachment of moieties to the surface, PDMS substrates were treated with UV/Ozone following an incubation under vacuum in the presence of (3-aminopropyl)triethoxysilane. Surfaces were then incubated in sulfo-NHS-biotin (Supplementary Fig. S13b).
Scanning Electron Microscopy
PA and PA-PEG-RGD solutions were polymerized. Hydrogels were swelled in water overnight, flashed frozen, then lyophilized over night. Lyophilized samples were sputter coated with iridium.
DNA Gel Electrophoresis
DNA ladders were run through polyacrylamide electrophoresis gels in TAE buffer with ethidium bromide at 30V for 14 hours. DNA fragment lengths were converted to radius of gyration described elsewhere.22
Stem Cell Culture
Human ASCs were isolated from freshly aspirated human subcutaneous adipose tissue according to the method described elsewhere33. Commercially available MSCs were purchased. MSCs and ASCs were cultured in Dulbecco’s modified eagle medium with fetal bovine serum and antibiotics. For differentiation experiments, MSCs and ASCs were seeded on PA and PDMS substrates at a density of 1,000 cells/cm2 and on PA-PEG-RGD gels at a density of 2,000 cells/cm2. See Supplementary Information for inductive media formulations.
Immunofluorescence
Cells were fixed, permeabilized, and then stained with rhodamine phalloidin and Hoescht. For osteogenic differentiation studies, cells were stained with RUNX2. To quantify RUNX2 expression, CellProfiler34 (Broad Institute) was used to measure cytoplasmic and nuclear fluorescent intensities using the nuclei and cell outlines as masks to define these regions of interest in the RUNX2 fluorescent channel.
Differentiation Assays
ASCs and MSCs were stained for alkaline phosphatase (ALP) and Oil Red O (ORO) per manufacturer protocols. See Supplementary Information for additional methods.
Atomic Force Microscopy
To determine the mechanical properties of PA hydrogels by indentation and to quantify protein tethering by force spectroscopy, a MFP-3D-Bio atomic force microscope (AFM) was used. Chromium/gold-coated, silicone nitride cantilevers with pyramid-shaped tips with ~50 pN/nm nominal spring constants were used for both methods. Samples were indented at a velocity of 2 μm/s until a trigger of 2 nN was detected using. All AFM data was analyzed using custom written code in Igor Pro to determine the Young’s modulus as previously described35. PDMS substrates were indented with the same cantilevers mentioned above. Additionally a cantilever with a 45μm diameter polystyrene bead tip with 0.03 N/m nominal spring constant was used. For retraction experiments, samples were indented with approach and retraction velocities ranging from 1 nm/s to 10 μm/s. The substrate spring constants were determined by fitting the linear portion of the retraction curve starting at the undeformed surface.
For force spectroscopy, cantilevers were functionalized (Fig. 3a) with an anti-collagen type I antibody, or avidin using a previously established method.36,37 Briefly, cantilevers were cleaned with chloroform and immersed in ethanolamine-HCl in dimethylsulfoxide. Tips were incubated in bis[sulfosuccinimidyl] suberate, rinsed, and then immersed either in an antibody or avidin solution to crosslink the protein to the tip. Force curves were taken in a regular 10x10 array of points spaced ~10 μm apart. To promote binding of the antibody to collagen or avidin to biotin, a dwell time of 1 second was added between approach and retraction cycles. Force curves were converted to force vs. tip Z-position curves (Supplementary Fig. S6a) and then analyzed for rupture events using a previously described algorithm;38 rupture lengths and forces were determined.
Traction Force Microscopy
Traction force microscopy was performed as previously described.23 Briefly, fluorescent 0.2 μm microspheres were added to the pre-polymer solution. Substrates were functionalized and treated as described above. The microspheres underneath selected live cells were imaged with a confocal imaging system. Cells were released with trypsin and the same confocal stacks were acquired. Bead displacements were determined using a particle image velocimetry MATLAB script.
Förster Resonance Energy Transfer
Concentrated fibronectin was denatured in guanidine hydrochloride and dual-labeled with donor and acceptor fluorophores, as previously described.13 Denatured fibronectin was incubated with a molar excess of Alexa Fluor 546 C5 Maleimide and subsequently buffer exchanged into sodium bicarbonate. The single-labeled fibronectin was then incubated with a molar excess of Alexa Fluor 488 succinimidyl ester. Unreacted donor fluorophores were removed using a spin desalting column. The emission spectrum of the dual-labeled fibronectin was characterized in varying concentrations of denaturant by fluorescence spectroscopy. The resulting emission spectrum was measured from 510 to 700 nm (Supplementary Fig. 9b) and the ratio of the maximum acceptor emission (~570 nm) to the maximum donor emission (~520 nm) was determined at each concentration of GdnHCl (Supplementary Fig. 9c). Images of the dual-labeled fibronectin were acquired using a confocal microscope and analyzed using a custom MATLAB script, as previously described.14 The mean FRET ratio within the selected regions was calculated for each cell and then averaged over all the cells in each condition (n = 16 cells per condition) (Fig. 2g).
See Supplementary Information for additional methods.
Supplementary Material
Acknowledgments
The authors thank Matthew Joens, James Kasuboski and James Fitzpatrick at the Waitt Advanced Biophotonics Center at the Salk Institute (supported by NCI P30 Cancer Center Support Grant CA014195-40 and NINDS P30 Neuroscience Center Core Grant NS072031-03A1 and the W.M. Keck Foundation) for technical assistance with microscopy and Drs. William Murphy (University of Wisconsin) and Todd McDevitt (Georgia Tech) for helpful conversations. The National Institutes of Health (DP02OD006460 to A.J.E.), the Human Frontiers Science Program (RGY0064/2010 to A.J.E.), the National Science Foundation Graduate Research Fellowship Program (to J.H.W., L.G.V., and H.T.-W.), the Siebel Scholars Program, and the Achievement Rewards for College Scientists (to L.G.V.) supported this work.
Footnotes
AUTHOR CONTRIBUTIONS
All authors contributed to design of experiments. Y.S.C, K.H., and S.C. characterized the hydrogel substrates. H.T.-W. characterized and performed experiments with the FRET probe. J.H.W., L.G.V., and A.F. conducted all other experiments and performed the data analysis. J.H.W., L.G.V., and A.J.E. wrote the manuscript.
References
- 1.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert PM, et al. Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. science.1191035 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Choi YS, Vincent LG, Lee AR, Dobke MK, Engler AJ. Mechanical derivation of functional myotubes from adipose-derived stem cells. Biomaterials. 2012;33:2482–2491. doi: 10.1016/j.biomaterials.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha K, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. S0006-3495(08)78580-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol Cell Physiol. 2008;295:C1037–1044. 67. doi: 10.1152/ajpcell.67.2008. [pii] [DOI] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 9.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 10.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. 0903269107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci U S A. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith ML, et al. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 16.Rossman JS, Dym CL. Introduction to Engineering Mechanics: A Continuum Approach. CRC Press; 2008. [Google Scholar]
- 17.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holle AW, et al. In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem Cells. 2013 doi: 10.1002/stem.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. nature08602 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guilak F, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. S1934-5909(09)00293-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. 324/5935/1673 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stellwagen NC. Apparent pore size of polyacrylamide gels: comparison of gels cast and run in Tris-acetate-EDTA and Tris-borate-EDTA buffers. Electrophoresis. 1998;19:1542–1547. doi: 10.1002/elps.1150191004. [DOI] [PubMed] [Google Scholar]
- 23.del Alamo JC, et al. Three-dimensional quantification of cellular traction forces and mechanosensing of thin substrata by fourier traction force microscopy. PLoS One. 2013;8:e69850. doi: 10.1371/journal.pone.0069850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 25.Engler A, et al. Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophysical Journal. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. http://dx.doi.org/10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell M, Kamm R, Matsudaira P. Substrate viscosity enhances correlation in epithelial sheet movement. Biophys J. 2011;101:297–306. doi: 10.1016/j.bpj.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bangasser BL, Rosenfeld SS, Odde DJ. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys J. 2013;105:581–592. doi: 10.1016/j.bpj.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khatiwala CB, Peyton SR, Metzke M, Putnam AJ. The regulation of osteogenesis by ECM rigidity in MC3T3-E1 cells requires MAPK activation. J Cell Physiol. 2007;211:661–672. doi: 10.1002/jcp.20974. [DOI] [PubMed] [Google Scholar]
- 30.Young JL, Engler AJ. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32:1002–1009. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chopra A, et al. Reprogramming cardiomyocyte mechanosensing by crosstalk between integrins and hyaluronic acid receptors. J Biomech. 2012;45:824–831. doi: 10.1016/j.jbiomech.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci U S A. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi YS, et al. The alignment and fusion assembly of adipose-derived stem cells on mechanically patterned matrices. Biomaterials. 2012;33:6943–6951. doi: 10.1016/j.biomaterials.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaushik G, et al. Measuring passive myocardial stiffness in Drosophila melanogaster to investigate diastolic dysfunction. J Cell Mol Med. 2012;16:1656–1662. doi: 10.1111/j.1582-4934.2011.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonanni B, et al. Single molecule recognition between cytochrome C 551 and gold-immobilized azurin by force spectroscopy. Biophys J. 2005;89:2783–2791. doi: 10.1529/biophysj.105.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chirasatitsin S, Engler AJ. Detecting cell-adhesive sites in extracellular matrix using force spectroscopy mapping. J Phys Condens Matter. 2010;22:194102. doi: 10.1088/0953-8984/22/19/194102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuhrmann A, Anselmetti D, Ros R, Getfert S, Reimann P. Refined procedure of evaluating experimental single-molecule force spectroscopy data. Physical Review E. 2008;77:031912. doi: 10.1103/PhysRevE.77.031912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.