Abstract
The deficiency of X-inactive specific transcript (XIST) on the inactive X chromosome affects the behavior of female human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), and further chromosomal erosion can occur with continued passaging of these cells. However, X chromosome instability has not been identified in other species. In the present study, we investigated three female rabbit ESC (rbESC) lines and found that two of them expressed Xist normally and obtained both Xist RNA coating and H3K27me3 foci, thus defined as XiXistXa. Interestingly, the third female rbESC line lacked Xist expression during ESC maintenance and differentiation. This line showed H3K27me3 foci but no Xist RNA coating in the early passages and was thus defined as Xiw/oXistXa. Similar to Xiw/oXistXa hESCs or hiPSCs, Xiw/oXistXa rbESCs lose H3K27me3 and undergo Xi erosion (Xe) with passaging. Moreover, Xist-deficient rbESCs also exhibit impaired differentiation ability and upregulation of cancer-related genes. By overexpressing OCT4, SOX2, KLF4, and c-MYC in Xist-deficient rbESCs under optimized culture conditions, we successfully obtained mouse ESC-like (mESC-like) cells. The mESC-like rbESCs displayed dome-shaped colony morphology, activation of the LIF/STAT3-dependent pathway, and conversion of disordered X chromosome. Importantly, the defective differentiation potential was also greatly improved. Our data demonstrate that variations in X chromosome inactivation occur in early passage of rbESCs; thus, Xi disorders are conserved across species and are reversible using the proper epigenetic reprogramming and culture conditions. These findings may be very useful for future efforts toward deriving fully pluripotent rbESCs or rabbit iPSCs (rbiPSCs).
Introduction
The rabbit is a classic model animal that shares anatomical, physiological, genetic, and biochemical features that are more closely related to primates than either the mouse or rat; thus, this species is more suitable for modeling human pulmonary, cardiovascular, neurological, and metabolic disorders [1–4]. Although the derivation of germline-competent rbESCs or rbiPSCs would be invaluable for generating precise gene-modified rabbits, neither has been successfully established.
The previously established rbESCs and rbiPSCs exhibited flat colony morphology and bFGF dependence and displayed many characteristics that are similar to those of hESCs [5–11]. It is well known that hESCs are more like mouse epiblast stem cells (mEpiSCs) than mESCs [12]. The mEpiSCs are germline incompetent [12,13], both mEpiSCs and hESCs are considered to be in a primed state, whereas mESCs are considered to be in a naive state. The mESCs that are in a naive pluripotent state have two active X chromosomes, that is, XaXa. In contrast, post-X-inactivation-primed mEpiSCs express Xist resembling somatic cells, that is, XiXISTXa [14]. The XaXa state in mice appears to be a specific hallmark of the naive state. However, the X chromosome status in both hESCs and hiPSCs is unstable even in early passages [15–17]. Both female hESCs and hiPSCs have been grouped into three classes based on their X chromosome status: class I lines initially carry two active X chromosomes (XaXa) and undergo XCI upon differentiation, similar to mESCs; class II lines possess one inactive X coated by XIST RNA (XiXISTXa), similar to mEpiSCs; class III lines have a silent Xi but lack of XIST expression (Xiw/oXISTXa). The Xiw/oXISTXa hESCs or hiPSCs will accordingly lose XIST RNA-dependent repressive chromatin modifications, such as H3K27me3 and DNA methylation, yielding an eroded Xi (Xe) [18,19]. The expression pattern of XIST during differentiation can differentiate these cell lines. The expression of XIST and the gaining of XIST RNA coating can be detected in the XaXa hESCs and hiPSCs during differentiation. The class II cells will remain XiXa state with both XIST expression and XIST RNA coating during differentiation. The class III cells with Xiw/oXISTXa or XeXa will retain XIST deficiency, and neither XIST expression nor XIST RNA coating can be detected in the differentiated cells. Loss of XIST in hESCs and hiPSCs is strongly correlated with the upregulation of oncogenes and differentiation deficiencies in vivo [16,17]. Therefore, X chromosome instability should be taken seriously in both basic research and clinical applications. Recent studies have shown that hESCs can be converted into mESC-like state by overexpression of two (OCT4 and KLF4) or more (OCT4, SOX2, KLF4, c-MYC, and NANOG) transcription factors which also result in reactivation of the inactivated X chromosome (Xi) [20,21]. The resulting hESCs exhibit improved pluripotency and are more amenable to gene targeting [20]. Moreover, evidence from previous studies also showed that the conversion of mEpiSCs to an mESC-like state can improve the germline transmission competency [22,23].
Although rbESCs have been previously derived in several groups, little is known about the X chromosome status of female rbESCs or its correlation with pluripotency. In the present study, we investigated the X chromosome status of three female rbESC lines and found variations in X chromosome inactivation in early passages. Two of the rbESC lines (B28-6 and NJ12) can be defined as class II because they displayed a heterochromatic Xi with Xist RNA coating and H3K27me3 foci. In contrast, the third cell line (NJ10) lacked Xist expression and Xist RNA coating. However, unlike the XaXa state, this cell line did not express Xist during differentiation, and displayed H3K27me3 foci in early passages (passage 8), thus indicated as Xist-deficient XiXa. Moreover, the H3K27me3 foci would further lose in passaging and yield an eroded Xi. Therefore, this cell line is considered to be class III. We further found that the class III rbESCs exhibited relatively high expression levels of some oncogenes, whose orthologous genes were reported to be highly expressed in class III hiPSCs [16]. The class III rbESCs also showed poor differentiation in vivo compared with class II and male rbESCs. Furtherly, we found that the class III rbESCs could be converted into an mESC-like pluripotent state by overexpressing the reprogramming factors OCT4, SOX2, KLF4, and c-MYC. We examined the colony morphology, colony formation ability, transcriptional profiles, and the signaling pathways dependence with pluripotency. We found that the induced rbESCs are more like mESCs, which therefore defined as mESC-like rbESCs. We then confirmed that the mESC-like rbESCs reobtain two active X chromosomes, meanwhile, the oncogene expression is repressed and the impaired differentiation ability in vivo is rescued. Our experiments found for the first time that the X chromosome instability and the impact of Xist deficiency in Xi may be conserved between humans and rabbits. Our data also revealed that forced expression of reprogramming factors under optimized culture condition can reprogram the rbESCs to mESC-like state, which is XaXa state, and even the disorder on eroded X chromosome can be rescued. These data may be informative to other groups who attempt to derive fully pluripotent rbESCs or rbiPSCs.
Materials and Methods
Derivation and maintance of rbESCs
The rbESCs were derived as reported previously [7,8,10]. Three early passage (p<12) female rbESC lines (B28-6, NJ10 and NJ12) were used for this study. Female rabbit fibroblasts and a male rbESC line (P27-1) were used as controls. The female rbESC line B28-6 was derived from a Black-modified rabbit blastocyst, whereas NJ10 and NJ12 cell lines were derived from the F1 blastocysts of New Zealand White rabbit×Japanese White rabbit. Female fibroblasts were derived from a dpc13.5 New Zealand White rabbit embryo. The male rbESC line P27-1 was derived from a Pigmented rabbit blastocyst. All of our study procedures were consistent with the National Institute of Biological Sciences Guide for the care and use of laboratory animals.
All rbESCs were cultured on mitomycin C-treated ICR mouse embryonic fibroblasts in K-D+bFGF medium at 37°C with 5% CO2. Half of the media was replaced daily. Cells were passaged using 1 mg/mL collagenase IV (Invitrogen, 17104019) every 3–4 days.
The K-D+bFGF medium contained KnockOut DMEM (Gibco; 10829018) supplemented with 15% (v/v) KnockOut Serum Replacement (Gibco; 10828028), 5% fetal bovine serum (Hyclone; 3007.03E), 2 mM l-glutamine (Millipore; TMS-002-C), 1% nonessential amino acids (Millipore; TMS-001-C), 0.1 mM β-mercaptoethanol (Invitrogen; 21985023), and 8 ng/mL human basic fibroblast growth factor (bFGF; PeproTech; 100-18B).
Small molecule treatment
To investigate the individual effects of leukemia inhibitory factor (LIF) and bFGF on the maintenance of rbESCs, rbESC culture medium was supplemented with the following: 10 μm of JAK inhibitor I (Merck; 420097) to inhibit the LIF/STAT3 pathway, 10 μm of LY294002 (Selleck; S1105) to inhibit the PI3K-AKT pathway, or 2i containing 1 μm of PD0325901 and 3 μm of CHIR99021 (Stemgent; 04-0006 and Selleck; 1263) to inhibit the MEK/ERK pathway.
Reverse transcription–polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR) analyses
Total RNA was purified using TRIzol reagent (Invitrogen) and reverse transcribed using a reverse transcription system (Promega) according to the manufacturer's suggestions. All qRT-PCRs were performed using SYBR Premix Ex Taq (Takara) and run on an ABI 7500 Real-Time PCR System (Applied BioSystems). Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used as an endogenous control. Primer sequence are present in Supplementary Table S3.
Single-cell RT-PCR
We carried out two rounds of amplification to detect Oct4 and Xist expression in single cells. Single-cell RT-PCR was performed according to the manufacturer's suggestions of QIAGEN OneStep RT-PCR Kit (Qiagen; 210210). Single cell was incubated in 2.5 μL of lysis buffer containing 0.1% NP-40, 0.5 U RNase inhibitor, and 0.5 U RNase-free DNase at 37°C 15 min, 75°C 3 min, and 4°C 5 min. Reverse transcription and first PCR amplification was carried out by adding 10 μL 5×OneStep RT-PCR buffer, 2 μL dNTP (400 μM, final concentration), 2 μL OneStep RT-PCR enzyme Mix, and a mixture of Xist, Oct4, and Gapdh primers (0.5 μM/each, final concentration) in 50 μL reactions. The reactions were incubated at 50°C 30 min, 95°C 15 min for active HotStarTaq DNA polymerase and then 35 cycles of 94°C 30 s and 60°C 30 s and 72°C 30 s, and 72°C for 10 min. About 0.5 μL of the product was used for the second-round PCR amplification.
Xist RNA FISH
To detect the Xist expression and Xist RNA coating on X chromosome in rbESCs, we performed Xist RNA fluorescence in situ hybridization (FISH). The first exon of the Xist gene was used as the RNA targeting probe. The target fragment was cloned into the pGEM(R) -T Easy Vector (Promega; A1360). The vector was linearized using PstI or BstxI (Takara; 1073A and 1027A). The digoxigenin-labeled antisense and sense probes were synthesized using T7 or Sp6 RNA polymerase (Takara; cat No. 2540A and 2520A) and further labeled with digoxigenin-UTP (Roche; 11277073910). The dispersed cells were placed onto glass slides. RNA FISH was performed as previously described [17,24].
Western blotting
Cells were harvested by gentle scrapping and total protein was extracted using a lysis buffer with protease inhibitor cocktail (Roche; 04906845001). Total proteins were separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the resolved proteins were transferred onto nitrocellulose membranes (Millipore; HAHY00010). Each membrane was blocked and incubated with primary antibody at 4°C overnight followed by incubation with HRP-conjugated secondary antibodies for 1 h. The protein signal intensity of three duplicates from each sample was determined using Image J software. The antibodies used were anti-Oct4 (Santa Cruz Biotechnology; SC-5279, 1:1,000), anti-Nanog (Bethyl Laboratories; A300-397A, 1:1,000), anti-STAT3 (Cell Signaling Technology; 9139P, 1:1,000), anti-phospho-STAT3 (Cell Signaling Technology; 9138S, 1:1,000), anti-AKT (Cell Signaling Technology; 2938S, 1:2,000), anti-phospho-AKT (Cell Signaling Technology; 4060S, 1:2,000), anti-ERK (Santa Cruz Biotechnology; sc-101761, 1:1,000), and anti-phospho-ERK (Santa Cruz Biotechnology; sc-135900, 1:1,000). Anti-mouse HRP (GE Healthcare; NA931, 1:10,000) and anti-rabbit HRP (GE Healthcare; NA934, 1:10,000) were used as secondary antibodies. β-Tubulin (Santa Cruz Biotechnology; SC-9104, 1:5,000) served as a loading control.
Conversion of primed rbESCs into an mESC-like state
The plasmids purchased from Addgene and the lentiviral infection procedure were previously described [25]. Briefly, 293T cells were individually transfected with Fuw-Teto-OCT4, SOX2, KLF4, c-MYC, and FUW-rtTA plasmid along with the packaging plasmids psPAX2 and pMD2.G (5:3:2). Virus-containing medium was collected after 48 h. Approximately 30 mL of virus-containing medium from two 150-mm dishes was centrifuged at 20,000 rpm for 1.5 h at 4°C and resuspended in 0.5 mL K-D+bFGF medium containing 5 μg/mL polybrene. On day 0, 105 NJ10 rbESCs were digested and plated on a matrigel-coated 35-mm dishes and infected with the concentrated viruses for 8 h. On day 2, the culture medium was changed to N2B27+LIF+2i medium supplemented with 1 μg/mL doxycycline (DOX). On day 3, the infected rbESCs were digested into single-cell suspensions with 0.05% trypsin and plated onto new feeder cells. Between days 7 and 14, the mESC-like colonies were picked and seeded into a 96-well plate. The cells were further passaged using trypsin and the mESC-like rbESC lines were then established.
The N2B27+LIF+2i medium contained 48% (v/v) DMEM/F12 (Gibco; 10828028), 48% Neurobasal (Gibco; 10828028), 1% N2 (Gibco; 10828028), 2% B27 (Gibco; 10828028), 2 mM l-glutamine, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol, 1,000 U/mL human LIF (Pepro Tech; 300-05), and 2i (1 μm PD0325901 and 3 μm CHIR99021). The culture medium was supplemented with 1 μg/mL DOX or 10 μm forskolin (FK) (Sigma; F6886) as indicated.
In vitro and in vivo differentiation of rbESCs
For spontaneous differentiation in vitro, rbESCs were dissociated with collagenase and then transferred to ultra-low-attachment six-well plates (Costar; 3471) in DMEM (Gibco; 11960) supplemented with 10% FBS and 2 mM l-glutamine to form embryoid bodies (EBs). After 7 days, EBs were trypsinized and plated onto gelatin-coated tissue culture dishes for an additional 7 days.
For in vivo differentiation, 1×106 rbESCs were injected into the groin or renal capsule of severe combined immune deficiency (SCID) mice. Between 8 and 10 weeks after injection, teratomas were dissected and processed for hematoxylin-eosin (HE) staining.
Karyotype analysis
After an initial incubation in culture medium containing 0.25 μg/mL colchicine (Sigma; D1925) for 4 h, the rbESCs were harvested and incubated in 0.4% sodium citrate and 0.4% potassium chloride (1:1, v/v) at 37°C for 5 min. Cells were then fixed twice in a methanol:acetic acid mixture (3:1, v/v). After Giemsa staining, at least 20 chromosome karyoschisis images were examined.
Alkaline phosphatase staining and immunocytochemistry
Alkaline phosphatase staining was performed using the Leukocyte Alkaline Phosphatase Kit (Sigma; 85L3R) according to the manufacturer's instructions.
For immunofluorescence staining, cells growing on gelatin-coated slides were fixed with 4% paraformaldehyde for 15 min and then permeabilized for 15 min with 0.5% Triton X-100. The slides were blocked in 3% bovine serum albumen for 30 min at room temperature and incubated with the following primary antibodies, including Oct4 (1:500), Sox2 (Santa Cruz; sc-17320, 1:500), Nanog (1:500), SSEA1 (1:200) and SSEA3 (Chemicon; SCR002, 1:200), H3K27me3 (Abcam; ab6002, 1:400), AFP (Santa Cruz; Sc-8108, 1:200), SMA (Sigma; A2547, 1:500), Vimentin (Boster; BM0135, 1:100), Tubulin (Santa Cruz; SC-9104, 1:200), Nestin (Santa Cruz; sc-21248, 1:200), and VGlut (Santa Cruz; sc-26027, 1:100) overnight at 4°C. The samples were washed three times and incubated with the appropriate secondary antibodies, including Alexa 488-conjugated goat anti-mouse IgM, Alexa 594-conjugated goat anti-mouse IgG, Alexa 633-conjugated rabbit anti-goat IgG, and Alexa 488-conjugated goat anti-rabbit IgG (all from Invitrogen). Nuclei were stained with DAPI (Molecular Probes; D3571). Stained cells mounted on slides were observed on an LSM 510 META confocal microscope (Zeiss).
Growth curves, cell doubling time, and colony formation assays
Cells were seeded into 48-well plates in triplicate at a density of 1×103 cells per well. The number of cells was counted every day and plotted in 6 days. The cell doubling time was calculated as previously described [26].
For colony formation assays, rbESCs were dissociated into single-cell suspensions using trypsin and then plated onto feeder cells. The number of AP-positive colonies was quantified 24 hs and 48 hs after passaging. The data are presented as the ratio of the number of newly formed colonies over the total number of cells initially seeded.
Statistical analyses
Holm-Sidak test (for ANOVA) or Student's t-test is performed using the software SigmaPlot 11.0 for statistics comparison.
Results
Variations of X chromosome inactivation and Xist expression in early passage of female rbESCs
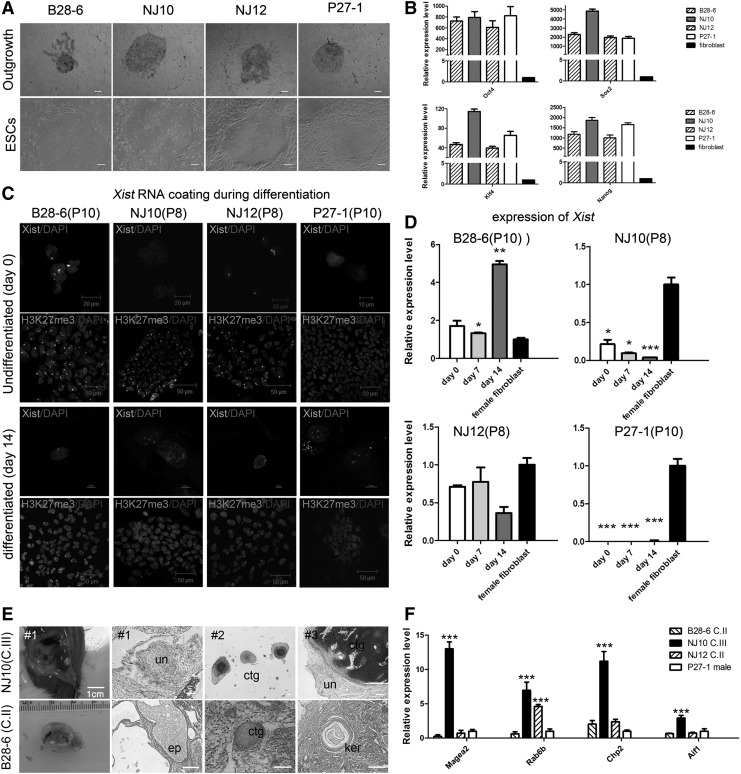
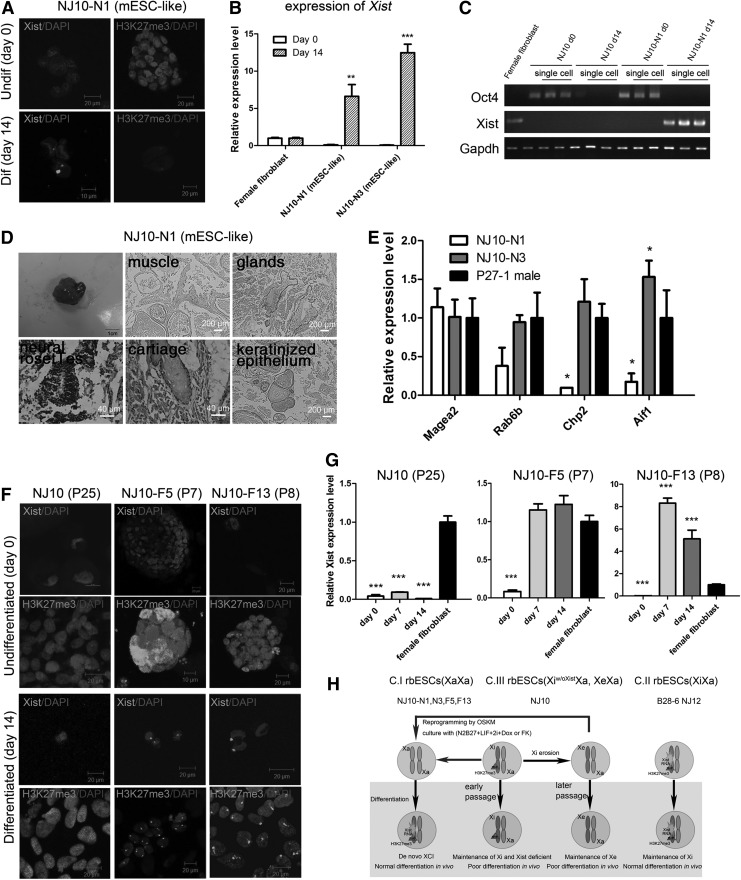
To investigate the X chromosome status in female rbESCs, we derived dozens of rbESC lines from rabbit blastocysts with different genetic backgrounds (Fig. 1A and Supplementary Fig. S1A and S1C; Supplementary Data are available online at www.liebertpub.com/scd). We obtained three female rbESC lines: B28-6, NJ10, and NJ12. A male rbESC line, P27-1, was used as control in X chromosome state and expression analyses. All these rbESC lines were cultured with bFGF and showed flat colony morphology, resembling that of hESCs (Fig. 1A). All these rbESCs expressed high levels of Oct4, Sox2, Klf4, Nanog, and Lin28 (Fig. 1B); maintained normal karyotype; displayed AP-positive staining; and showed Oct4-, Sox2-, and SSEA1-positive in immunofluorescence staining analysis (Supplementary Fig. S1B, S1D and S1E). We further performed RNA fluorescence in situ hybridization (FISH) to examine the Xist RNA coating and used qRT-PCR to examine the expression level of Xist in these rbESCs. Interestingly, the female rbESC lines presented different results. We found that two (B28-6 and NJ12) of the three rbESC cell lines expressed Xist normally comparing with female fibroblasts, and the Xist cloud could be observed in 44%–60% of nuclei (Fig. 1C, 1D and Table 1). The data suggested that B28-6 and NJ12 rbESCs might contain one inactive X coated with Xist RNA. However, in the third line NJ10, no Xist cloud was detected and the expression level of Xist was very low (Fig. 1C, 1D and Table 1). Moreover, the low Xist expression level was not due to the loss of the X chromosome, as a G-banding karyotype analysis indicated that this rbESC line was 44, XX (Supplementary Fig. S1C). The different Xist RNA coating status suggested that the variations of X chromosome inactivation which usually observed in human pluripotent stem cells were also present in female rbESCs. Similar to hESCs and hiPSCs, we assume that female rbESCs may also fall into three defined categories. Class I lines carry two active Xs (XaXa) and resemble mouse ES cells, class II lines possess one inactive X coated with Xist RNA (XiXistXa), and class III lines have deficient Xist expression and still one inactivated X chromosome (Xiw/oXistXa or XeXa). Although both class I (XaXa) and class III (Xiw/oXistXa or XeXa) can be Xist deficient and lack Xist RNA coating. We further used H3K27me3, which is considered a hallmark of an inactive X chromosome, to identify the representative epigenetic modification on inactive X. We found that the H3K27me3 foci were detected in 63% and 93% of nuclei in B28-6 and NJ12 lines, respectively. We also found that 90% of nuclei in NJ10 possess H3K27me3 foci (Fig. 1C and Table 1). Therefore, NJ10 is more like to be in an Xiw/oXistXa state rather than XaXa.
FIG. 1.
(A) Outgrowths and rbESCs derivated from rabbit blastocysts. (B) qRT-PCR analysis for pluripotency gene expression of the primed rbESC lines. (C) Xist RNA FISH (green) and immunofluorescence staining for H3K27me3 (green) in undifferentiated (day 0) and differentiated (day 14) rbESCs. Nuclei are stained with DAPI (blue). (D) Detection of Xist expression in rbESCs on the indicated differentiation day and female fibroblast cells by qRT-PCR. Data were normalized to Gapdh expression levels and comparing with gene expression levels in female fibroblast cells. (E) Teratomas generated by B28-6 (class II rbESCs) or NJ10 (class III rbESCs) as indicated. B28-6 (class II rbESCs) generated teratomas with three germ layers, whereas NJ10 (class III rbESCs) formed much smaller teratomas with mostly undifferentiated tissues. un (undifferentiated tissue), ctg (cartilage), ep (epithelium), kar (keratinocyte). (F) qRT-PCR analysis for oncogene expression in rbESCs. Some oncogenes were highly expressed in NJ10 (class III rbESCs). Data were normalized to Gapdh expression and comparing with gene expression levels of P27-1. All data are shown as the mean±SD (n=3). Statistical analyses were performed using ANOVA. *P<0.05, **P<0.01, and ***P<0.001. rbESCs, rabbit embryonic stem cells; qRT-PCR, quantitative reverse transcription–polymerase chain reaction.
Table 1.
Variations of X Chromosome Inactivation Status of Female rbESCs
| Undifferentiated d0 | Differentiated d14 | ||||||
|---|---|---|---|---|---|---|---|
| Cell lines (passage) | Xist clouds | H3K27me3 | Relative Xist expression | Xist clouds | H3K27me3 | Relative Xist expression | Class designation |
| B28-6 (P10) | 60% (14/23) | 63% (72/114) | 1.700 | 53% (35/66) | 86% (182/211) | 4.592 | II |
| NJ10 (P8) | 0% (0/20) | 90% (78/86) | 0.214 | 0% (0/22) | 41% (70/167) | 0.037 | III |
| NJ12 (P8) | 44% (22/50) | 93% (280/300) | 0.709 | 51% (17/33) | 87% (114/130) | 1.304 | II |
| P27-1 (P10, male) | 0% (0/30) | 0% (1/112) | 0.0003 | 0% (0/20) | 0% (0/72) | 0.059 | — |
| NJ10-N1 (P10) | 0% (0/28) | 0% (0/92) | 0.112 | 28% (6/21) | 17% (15/86) | 6.611 | I |
| NJ10-N3 (P10) | 0% (0/21) | 0% (0/117) | 0.067 | 56% (18/31) | 35% (33/92) | 12.462 | I |
B28-6, NJ10 and NJ12: female rbESCs; P27-1: male rbESCs. NJ10-N1 and N3 were mESC-like rbESCs converted from NJ10 using four transcription factors.
rbESCs, rabbit embryonic stem cells; mESC-like, mouse embryonic stem cell like.
To further confirm this observation, we analyzed the behavior of X in these rbESCs during differentiation. We found that after differentiation both B28-6 and NJ12 could keep the expression of Xist, and the presence of Xist clouds and H3K27me3 foci remained similar to the undifferentiated state (Fig. 1C, D and Table 1). In contrast, the deficiency of Xist in NJ10 cannot be changed in differentiation and the Xist RNA coating cannot be observed in the differentiated NJ10 cells (Fig. 1C, D and Table 1). All these evidence demonstrate that there is no expression of Xist or XCI occurred during the differentiation of NJ10 rbESCs, and the state of X in NJ10 remained as Xiw/oXistXa.
In hESCs and hiPSCs, class III cells are closely related to culture and continuously passaging. We subsequently found that with passaging, the deficiency of Xist exist in passaging and the H3K27me3 foci were lost in NJ10 rbESCs since P12 (Supplementary Fig. S1F and S1G). The loss of both Xist RNA coating and Xist-related histone modification suggested that the rabbit X chromosome was eroded in late passages.
In summary, we demonstrate that the instability of X inactivation as well as X erosion could occur in rbESCs, which resembles hESCs and hiPSCs.
Loss of Xist expression is associated with poor differentiation and the upregulation of cancer-related genes
Since the deficient expression of Xist in hESCs and hiPSCs can greatly affect the differentiation potential [16,17,27], particularly in vivo [16], we further examined the differentiation ability of these three female rbESC lines. In vitro differentiation was firstly performed using EB formation assay. All rbESC lines were able to form EBs in a similar manner and both gene expression and immunofluorescence staining analyses showed that the expression of lineage-specific marker genes was detected in the differentiated cells (Supplementary Fig. S2A, S2B and Table 2).
Table 2.
Differentiation of Female rbESCs In Vitro and In Vivo
| In vitro differentiation | In vivo differentiation | ||||
|---|---|---|---|---|---|
| Class | Cell lines (passage) | EB character | Lineage-specific mark | Teratoma/Injection (SI or RI), size (cm) | Three germ layers |
| II | B28-6 (P10) | Robust | Present | 1/1 (SI), 4.5×2.8 | Present |
| II | NJ12 (P10) | Fair | Present | 1/3 (SI), 4.6×4.0 | Present |
| III | NJ10 (P8-12) | Fair/Robust | Present | 0/3, no teratoma formation by SI. 3/6, 3 teratomas formation by 6 RI. Teratoma #1: 0.4×0.5. Teratoma #2: three of 0.1×0.15. Teratoma #3: 0.2×0.2. |
Endoderm and ectoderm were deficient in all teratomas. Teratoma #1: Primitive and undifferentiated cell mass without any histological structure could be found. Teratoma #2: Contained only cartilage tissues. Teratoma #3: Cartilage tissues covered with some primitive cell mass. |
| — | P27-1 (male, P10) | Fair | Present | 1/2 (SI), 3.0×1.8 | Present |
| I | NJ10-N1 (P8) | Fair | Present | 1/3 (SI), 3.2×3.0 | Present |
| I | NJ10-N3 (P8) | Robust | Present | 2/2 (SI), terotoma #1: 1.8×2.4. terotoma #2: 3.0×4.5 (SI) |
Present |
B28-6, NJ10, and NJ12: female rbESCs; P27-1: male rbESCs. NJ10-N1 and N3 were mESC-like rbESCs converted from NJ10 by four transcription factors.
SI, subcutaneous injection; RI, renal capsule injection.
The rbESCs were then injected into the immune-deficient mouse to analyze the differentiation potential in vivo. Both class II cell lines (B28-6 and NJ12) and the male rbESCs (P27-1) could produce teratomas in normal size and three germ layers were observed in these teratomas (Supplementary Fig. S2C and Table 2). In contrast, traditional subcutaneous injection of NJ10 rbESCs could not form teratoma. We then used renal capsule injection instead, which may enhance the generation of teratoma. From six renal capsule injections, we were able to obtain three small teratomas. All the three teratomas were small in size and cannot exhibit distinctive characteristics of mature differentiated cells, ectodermal and endodermal tissues were absent, and most tissues in these teratomas were cartilage, adipocyte, or undifferentiated cell mass (Fig. 1E, Supplementary Fig. S2C and Table 2). Together, these data indicated that the NJ10 rbESCs possess poor in vivo differentiation potentials, which is in line with previous reports in hESCs or hiPSCs. XIST deficiency in hESCs or hiPSCs might lead to the piecemeal demethylation of X-linked and autosomal genes, thereby inducing the upregulation of oncogenes as well as the downregulation of several tumor suppressors [16,17,28]. Of the 10 genes displaying the greatest abnormal upregulation in class III hiPSCs, 4 were X-linked genes and 7 (including 2 of the X-linked genes) were highly expressed in cancers [16]. We then asked whether the orthologous gene of 7 oncogenes (MAGEA2, MAGEA6, RAB6B, RAS, CHP2, ACP5, and AIF1) in rabbit also display a similar expression pattern in class III rbESCs. Due to the lack of a complete rabbit genome sequence, we designed gene-specific primers based on the human and mouse sequences. Four of these genes were confirmed by qRT-PCR (the remaining three were not detected), and the results revealed that Magea2, Rab6b, Chp2, and Aif1 are more highly expressed in class III rbESCs (NJ10) than in class II rbESCs (B28-6 and NJ12) or male (P27-1) rbESCs (Fig. 1F). In sum, our data suggest that the Xist-expression deficiency in rbESCs might also impair the in vivo developmental potential of these cells and cause the activation of some oncogenes.
Stable mESC-like rbESCs can be established by overexpressing transcription factors
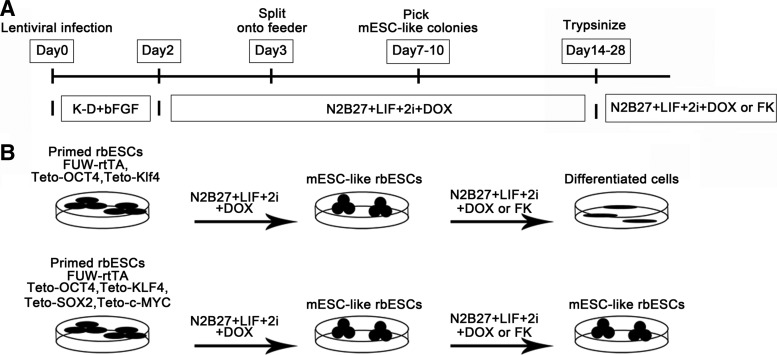
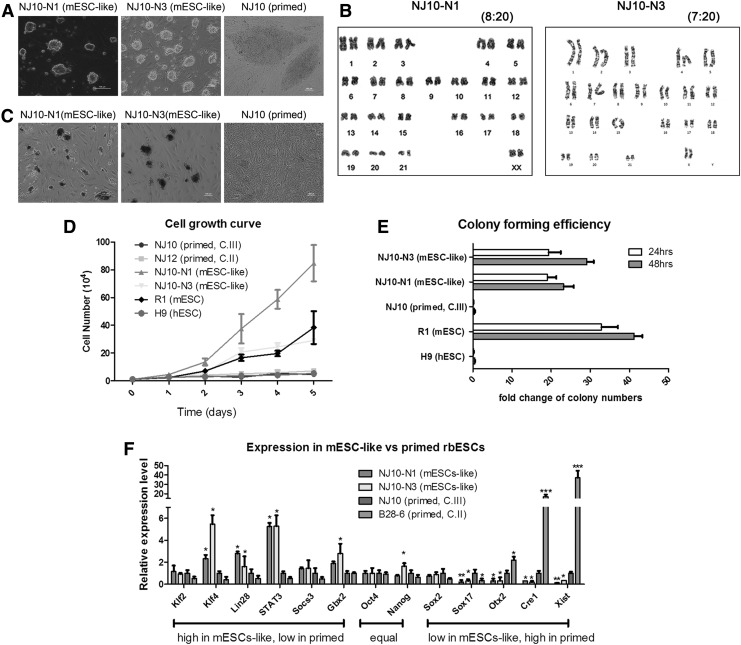
Previous studies have demonstrated that Xist deficiency is caused by epigenetic changes rather than gene deletions [18]. Thus, we attempted to reestablish the correct epigenetic landscapes of class III rbESCs by reprogramming [20], and the reprogramming protocol was described in the schematic diagram (Fig. 2A). Firstly, we attempted to infect the cells with two factors, OCT4 and KLF4, to convert the class III rbESCs (NJ10, P8) into an mESC-like state. Although mESC-like colonies with dome-shaped morphologies appeared 1 week after induction, the colonies were unstable and difficult to maintain, meanwhile no mESC-like cell lines were established using this method (Fig. 2B). Then, we added factors to OCT4, SOX2, KLF4, and c-MYC. Since the culture condition is reported to be important for the generation of XaXa female hESCs and iPSCs, we ameliorate the medium with different combinations of small molecules. After 1 week of induction in N2B27+LIF+2i+DOX medium, mESC-like colonies with dome-shaped morphologies were observed. We successfully established more than 10 mESC-like rbESC lines, all of which were nearly morphologically indistinguishable from mESCs but distant from NJ10 (Fig. 3A); two of the new established rbESCs were further characterized and were referred to as mESC-like rbESCs (NJ10-N1 and NJ10-N3) (Fig. 3A). The mESC-like rbESCs maintained the 44, XX karyotype (Fig. 3B), and displayed AP-positive staining (Supplementary Fig. S3A). We further confirmed that pluripotency markers were expressed in these cells (Supplementary Fig. S3B) and could differentiated normally in EB formation analysis (Supplementary Fig. S3E). As reported, the addition of FK in culture medium is believed to stabilize the naive state by inducing the expression of endogenous Klf2 and Klf4 [20]. The supplementation of DOX helped to keep the expression of exogenous genes. Upon DOX withdrawal, the exogenous genes were silenced (Supplementary Fig. S3C), and the mESC-like rbESC lines could be further passaged with trypsin for more than 25 passages. However, if DOX and FK were withdrawn together, the mESC-like rbESCs lost their mESC morphology quickly and underwent spontaneous differentiation within three passages. After 25 passages, the cells became unstable, and most underwent apoptosis for unknown reasons. To further test which conditions were necessary for the conversion of rbESCs from the primed to the naive state, we adjusted the derivation method. Firstly, we digested NJ10 class III rbESCs into single-cell suspensions and then cultured the cells in N2B27+LIF+2i medium without infection of transcription factors. To improve the single-cell survival efficiency, 10 μm of the Rho-associated coiled-coil kinase inhibitor (ROCKi, Y-27632) was added 24 h before and after digestion. Then, NJ10 cells were infected by 4 factors but cultured in traditional K-D+bFGF medium supplemented with 1 μg/mL DOX only. As expected, mESC-like rbESCs were not established by either of these experiments, which further indicating that successful induction of mESC-like rbESCs requires both the presence of exogenous factors and supplementation with LIF and inhibitors.
FIG. 2.
(A) The strategy for generating mESC-like rbESCs. After 1 week of induction in N2B27+LIF+2i+DOX medium, mESC-like colonies appeared. (B) Stable conversion to mESC-like rbESCs was achieved by induction with OCT4, SOX2, KLF4, and c-MYC, but not with OCT4 and KLF4. mESC-like, mouse embryonic stem cell like; LIF, leukemia inhibitory factor; 2i, MEK and GSK-3β inhibitor; DOX, doxycycline; FK, forskolin.
FIG. 3.
The mESC-like rbESCs display characteristics similar to mESCs. (A) The morphology of primed and mESC-like rbESCs. NJ10 (Primed rbESCs) exhibit classical flat colony morphology while NJ10-N1 and N3 (mESC-like rbESCs) display dome-shaped colony morphology. (B) The karyotype of NJ10-N1 and N3. The number at the top-right corner means the ration of euploid: mitosis count. (C) Twenty-four hours after trypsinized, AP staining of NJ10 (Primed rbESCs) and NJ10-N1 and N3 (mESC-like rbESCs). (D) The cell proliferation rates of primed rbESCs and mESC-like rbESCs. Both hESCs (H9) and mESCs (R1) were used as controls. (E) Single-cell cloning efficiency of different pluripotent stem cell lines; data are shown as the ratio of AP-positive colonies to the number of cells initially seeded at 24 and 48 h. R1 mESCs and H9 hESCs were used as controls. (F) qRT-PCR analysis for pluripotency gene expression. The NJ10-N1 and NJ10-N3 (mESC-like) rbESCs exhibited a unique gene expression profile compared with parental NJ10 or B28-6 (primed rbESCs). Data in B, D and E are represented as the mean±SD (n=3). Statistical analyses were performed using ANOVA. *P<0.05, **P<0.01, and ***P<0.001.
The mESC-like rbESCs share characteristics of mESCs
To define the characteristics of the mESC-like rbESCs, we assessed the cell morphology, growth rates, gene expression profiles, and signaling pathway dependence of these cells. The mESC-like rbESCs display a dome-shaped colony morphology that is similar to mESCs (Fig. 3A). We next compared the growth rates between the primed and the mESC-like rbESCs. The ESC growth curve was plotted and doubling times were calculated using NJ10 (primed), NJ12 (primed), NJ10-N1 (mESC-like), NJ10-N3 (mESC-like), R1 (mESCs), and H9 (hESCs) cells, respectively (Fig. 3D and Supplementary Table S1). As expected, NJ10 and NJ12 (primed rbESCs) exhibit a growth rate similar to hESCs (H9), whereas NJ10-N1 and N3 (mESC-like rbESCs) displayed an accelerated growth rate similar to mESCs (R1) (Fig. 3D). We further performed the colony formation assays with primed and mESC-like rbESCs; hESCs (H9) and mESCs (R1) were used as control. We found that only mESCs and mESC-like rbESCs could survive in single-cell passaging and form AP-positive colonies in this analysis. In contrast, NJ10 and NJ12 (primed rbESCs) generated few colonies within 48 h (Fig. 3C, 3E, and Supplementary Table S2). These results demonstrate that the mESC-like rbESCs adopted an mESC-like proliferation rate. In addition to differences in colony morphology and cell survival rates, primed and mESC-like rbESCs also exhibited numerous differences in their gene expression profiles. The qRT-PCR results demonstrated that the expression levels of Klf4, Gbx2, Lin28, and Stat3 were upregulated in mESC-like rbESCs, while the expression levels of Sox17, Cer1 and Otx2 were downregulated (Fig. 3F). These differences of gene expression between primed and mESC-like rbESCs were consistent with previously described differences in gene expression between human and mouse ESCs [12,13,20].
The mESC-like rbESCs share signaling pathways of mESCs
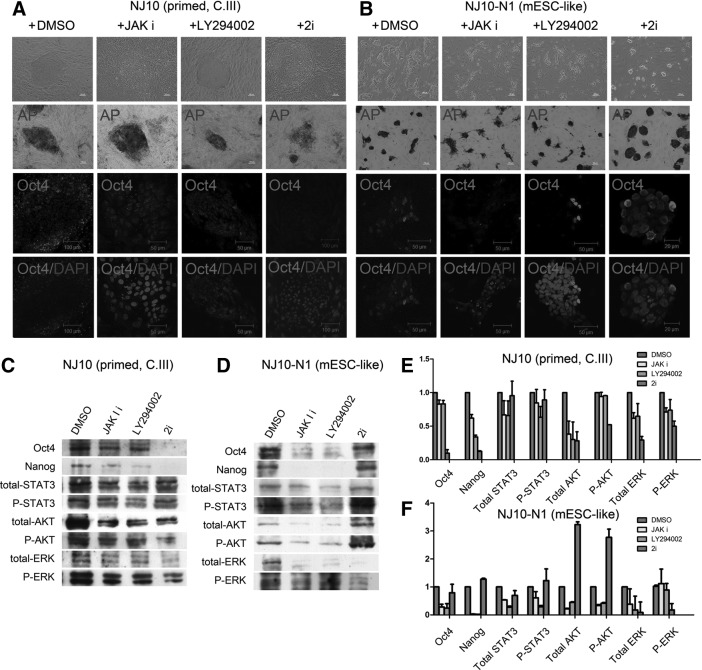
We then tried to characterize the major signaling pathways depended on in the primed and mESC-like rbESCs. Several small molecular inhibitors, including a JAK inhibitor I, LY294002, and 2i (ERK and GSK3β inhibitor), were individually used to treat the primed and the mESC-like rbESCs. The primed rbESCs could maintain the flat colony morphology in JAK inhibitor I and LY294002. But when exposed to 2i, the NJ10 rbESCs lost the clear colony edge quickly, and both cellular morphology and nuclear-cytoplasmic ratio changed greatly (Fig. 4A). In contrast, mESC-like rbESCs (NJ10-N1) displayed dome-shaped colony morphology despite 2i addition, but lost the colony structure after the culture medium was supplemented with JAK inhibitor or LY294002 (Fig. 4B). Both AP staining and Oct4 immunofluorescence staining confirmed these changes were related to pluripotency (Fig. 4A, B).
FIG. 4.
The effects of the bFGF and LIF signaling pathways on primed and mESC-like rbESCs. (A) NJ10 (Primed) rbESCs exhibit the classical flat colony morphology and the strongest AP and Oct4 signals when cultured in control medium supplemented with DMSO, but lose this morphology after 2i treatment. (B) The NJ10-N1 (mESC-like) rbESCs display a dome-shaped colony morphology and the strongest AP- and Oct4-positive signals when cultured in medium containing 2i, but lost this morphology when the LIF pathway was blocked by the presence of the JAK I inhibitor or LY294002. (C) Treatment with 2i induced a dramatic downregulation of Oct4 and Nanog protein levels in NJ10 (primed) rbESCs. (D) The NJ10-N1 (mESCs-like) rbESCs express high levels of Oct4 and Nanog proteins in 2i-containing medium, whereas blocking the LIF signaling pathway caused the downregulation of Oct4 and Nanog expression levels. (E) Image J software was used to quantify the expression levels of the proteins indicated in NJ10 (primed) rbESCs under different culture conditions. There was a strong correlation between Oct4 and Nanog expression levels and ERK and P-ERK expression levels. (F) Image J software was used to quantify the expression levels of the proteins indicated in NJ10-N1 (mESC-like) rbESCs under different culture conditions. There was a strong correlation between Oct4 and Nanog expression levels and STAT3 and P-STAT3 expression levels. Data in E and F are compared with DMSO-treated controls and are represented as the mean±SD (n=3).
Western blot analyses also revealed that primed rbESCs (NJ10) express the highest Oct4 and Nanog protein levels and exhibit constant production of phospho-ERK in control medium. However, 2i supplementation in primed rbESCs resulted in downregulation of phospho-ERK as well as the protein level of Oct4 and Nanog (Fig. 4C, 4E). In contrast, Oct4 and Nanog expression in NJ10-N1 (mESC-like) rbESCs was high when treated with 2i, but dramatically reduced after exposed to the JAK inhibitor or LY294002. Meanwhile, there was also a decrease in phospho-STAT3 and phospho-AKT protein when NJ10-N1 was treated with JAK inhibitor or LY294002 (Fig. 4D, F). These results demonstrate that in mESC-like rbESCs LIF activates the STAT3 and PI3K/AKT signaling pathways and maintains the pluripotency, but in primed rbESCs, bFGF activates the MEK/ERK signaling pathways and maintains the pluripotency.
Reprogramming rescued the disorder of Xist and poor differentiation potential of class III rbESCs
As described previously, reprogramming of rbESCs could convert the primed rbESCs to an mESC-like state. An important question is whether this conversion could rescue the Xist deficiency and disorders of X-inactivation in Class III rbESCs. We further examined the Xist RNA coating and H3K27me3 foci in the mESC-like rbESCs (NJ10-N1 and NJ10-N3). We found that neither of the inactive X markers can be detected in the converted cells at the early passage (Fig. 5A), which is different from the primed NJ10 cells. To differentiate from the XeXa rbESCs (NJ10>P25), which also lack the two markers, we further examined the X state during differentiation. In the differentiated cells, we observed that 28%–56% of the nuclei possess Xist clouds and 17%–35% of the nuclei possess H3K27me3 foci (Fig. 5A and Table 1). The qRT-PCR further revealed that Xist RNA expression in mESC-like rbESCs could be reactivated via differentiation (Fig. 5B and Table 1). To further confirm that the Xist reactivation relied on differentiation, we analyzed the expression of Xist in single cell of undifferentiated and differentiated mESC-like rbESCs. Oct4 is used as an indicator of stem cell differentiation. We found that Xist expression was not detected in either NJ10 or NJ10-N1 cells at pluripotent state when Oct4 expressed. Following differentiation, Oct4 was silenced, but Xist expression was only detected in NJ10-N1 cells (Fig. 5C). All these evidence suggested that the mESC-like rbESCs could express Xist and undergo XCI upon differentiation, and therefore the mESC-like rbESCs (NJ10-N1, N3) obtain two active X, and the Xist expression disorder in class III rbESCs (NJ10) was rescued through the factor-induced reprogramming.
FIG. 5.
(A) Xist RNA FISH and immunofluorescence staining of H3K27me3 during NJ10-N1 (mESC-like rbESCs) differentiation. (B) qRT-PCR analysis for Xist expression in undifferentiated (day 0) or differentiated (day 14) NJ10-N1 and N3 (mESC-like rbESCs). (C) Single-cell RT-PCR, Xist expression was not detected in NJ10, NJ10 differentiated cells, and NJ10-N1, but could be detected in NJ10-N1 differentiated cells. Oct4 was used as pluripotent marker; female fibroblasts were used as controls. (D) Teratomas generate with three germ layers by NJ10-N1 (mESC-like rbESCs). (E) qRT-PCR reveals that X-linked oncogenes are downregulated in the NJ10-N1 and N3 (mESC-like) rbESCs. (F) Xist RNA FISH and immunofluorescence staining for H3K27me3 in undifferentiated (day 0) and differentiated (day 14). NJ10-F5 and F13 (mESC-like) rbESCs: NJ10-F5 and F13 were mESC-like rbESCs converted from Xi erosion NJ10 (P25). (G) An analysis of Xist expression in NJ10-F5 and F13 (mESC-like) rbESCs during differentiation by qRT-PCR. (H) Schematic for X-chromosome inactivation and epigenetic fluidity in rbESCs. The transition from the Xist-/H3K27me3+ stage (NJ10, P8) to the Xist-/H3K27me3- stage occurred within a few passages, which displayed as XCI erosion (P>12). Loss of Xist expression was associated with a poor differentiation potential in vivo. Xi conversion occurred when NJ10 were reprogrammed to mESC-like cells by four transcription factors (under 2i conditions). The resulting mESC-like rbESCs underwent de novo X inactivation upon induction of differentiation and showed normal differentiation potential in vivo. Data were normalized to Gapdh expression and are represented as the mean±SD (n=3). Statistical analyses were performed using ANOVA. *P<0.05, **P<0.01, and ***P<0.001.
Moreover, in contrast to the original primed class III rbESCs (NJ10), the mESC-like rbESCs (NJ10-N1 and NJ10-N3) were able to form mature teratomas after subcutaneous injection into SCID mice (Fig. 5D and Table 2). All three germ layers, including glands (endoderm), cartilage and muscle (mesoderm), as well as keratinized epithelium (ectoderm), were detected in these teratomas (Fig. 5D). We further examined the expression of four X-related oncogenes in rabbit that were previously demonstrated to be related with Xist deficiency in class III rbESCs. We found in the mESC-like rbESCs the expression of these genes was greatly repressed to a similar or lower level compared with male rbESCs (Fig. 5E). In sum, our data demonstrate that the disordered X chromosome status in class III rbESCs, including the Xist deficiency and loss of XCI in differentiation, can be rescued when converted into an mESC-like state by OSKM-mediated induction. Moreover, the in vivo differentiation potential can be significantly improved and the undesirable upregulation of oncogenes was significantly repressed in the mESC-like rbESCs.
Reprogramming rescued the eroded X-chromosome inactivation in class III rbESCs
Previous studies have demonstrated that with passaging the class III NJ10 rbESCs would lose the H3K27me3 foci since P12 and therefore believed to undergo X erosion similar to that observed in hESCs and hiPSCs. Since we found that forced expression of OSKM in the early passage of NJ10 cells with Xiw/oXistXa can generate the mESC-like rbESCs with XaXa. An interesting question is whether the eroded X chromosome can be converted to Xa in a similar manner.
We first identified the characteristics of eroded NJ10 rbESCs that were passaged to P25. Neither Xist expression nor H3K27me3 foci could be detected in these late-passage cells of NJ10 (Supplementary Fig. S1F and S1G). The Xist deficiency was also observed during the differentiation of these cells (Fig. 5F). NJ10 (P25) rbESCs were subjected to reprogramming as described previously. Approximately 1 week after induction, mESC-like cells could also appear and could be isolated for expansion as previously described. Dozens of mESC-like rbESC lines were successfully established, and two of them, NJ10-F5 and NJ10-F13, were used for further analysis. Similar to NJ10-N1 and NJ10-N3, both NJ10-F5 and NJ10-F13 exhibited low Xist expression and absence of H3K27me3 foci staining in the undifferentiated state; these cells underwent XCI during differentiation with significant upregulation of Xist expression and the acquisition of Xist RNA coating and H3K27me3 foci (Fig. 5F, G). These data suggested that the eroded Xi might be restored to Xa during reprogramming.
Discussion
In this study, we reported the variations of X chromosome status in female rbESCs derived from the blastocysts. Xist deficiency in rbESCs may not only result in erosion of Xi but also impact the expression of oncogenes and the in vivo differentiation potential of these cells. By optimizing the culture conditions and expressing exogenous transcription factors, including OCT4, SOX2, c-MYC, and KLF4, we managed to reprogram the primed class III rbESCs into an mESC-like state. The resulting mESC-like rbESCs displayed dome-shaped colony morphology and activated LIF/STAT3 pathway. We also found that Xist deficiency or even eroded Xi chromosome can be restored to Xa. The mESC-like rbESCs were rescued from the Xist deficiency and undergo Xist-mediated X chromosome inactivation following differentiation. More importantly, the defective differentiation potential of these cells was greatly improved and the Xist-related oncogenes were repressed.
Our study also revealed that the XCI in ESCs is conserved between human and rabbit and that the lack of Xist in female rbESCs may contribute to their reduced pluripotency and abnormal epigenetic modifications. Based on the H3K27me3 foci, Xist RNA coating and expression of Xist in both ESC state and differentiation state, we demonstrated that female rbESCs have three different states of X chromosome similar to that in hESCs and hiPSCs (Fig. 1C, 1D, and Table 1). According to the finding of this study, we summarized the X chromosome state in rbESCs (Fig. 5H). We found in class II rbESCs (XiXa) that the expression of Xist would maintain the inactive X during differentiation with both Xist-RNA coating and H3K27me3 enrichment (Fig. 1C, D). But the class III rbESCs (Xiw/oXistXa or XeXa) have deficient Xist expression in both pluripotent and differentiation states (Fig. 1C, D). Interestingly, we found that the class III rbESCs could maintain the H3K27me3 enrichment within limited passages (Supplementary Fig. S1F). As reported, H3K27me3 is recruited by XIST in hESCs and hiPSCs. Our data further suggest that the loss of H3K27me3 foci in female rbESCs could occur much later than the loss of Xist RNA coating, and the presence of H3K27me3 in differentiated cells would be much related to the original ESCs. We also detected the erosion of X during passaging of Xiw/oXistXa rbESCs, which indicated that the in vitro culture condition might impact the instability of X. We further demonstrated that OSKM-mediated reprogramming under optimized culture condition could convert the class III rbESCs to an mESC-like state and the disorder of Xist could be rescued. Further experiment revealed that the mESC-like rbESCs lack of Xist RNA coating and H3K27me3 foci, and are capable of undergoing de novo XCI during differentiation (Fig. 5A–C and Table 1). All these evidence suggest that the mESC-like rbESCs possess two active X chromosomes and indicate that class III rbESCs could be converted to class I ESCs through factor-induced reprogramming. In hESCs or hiPSCs, XIST expression is lost by methylation of the XIST promoter and is accompanied by piecemeal demethylation of X-linked and autosomal genes [18]. Since the lack of rabbit Xist sequence, we obtained the homologous gene of rabbit using the human Xist gene, which is also located in X chromosome. We further analyzed the CG methylation status of newly derived female fibroblasts on the TSS regions (which include the fragment −100 bp to +105 bp around TSS) of this rabbit gene. Unexpectedly, we found that this region was hypermethylated but Xist was expressed normally in newly derivate fibroblasts (Supplementary Fig. S3G). Our data indicated that the sequence analyzed here does not possess the methylation-regulation ability of Xist expression, meanwhile we could not rule out the possibility that Xist expression is regulated by DNA methylation through other regulation regions. Xi erosion in hiPSCs and hESCs is believed to be closely associated with the loss of XIST and may broadly affect the characteristics of these cells [16,27]. These epigenetic modifications not only affect X-linked genes but also impact genome-wide gene expression, resulting in the upregulation of oncogenes and the downregulation of several tumor suppressors, thereby broadly affecting the characteristics of these cells [15–17,27–30]. Previous studies have suggested that female hiPSCs that have undergone XCI erosion should not be used clinically [16]. Therefore, investigating the mechanism of XCI erosion might be helpful to avoid Xi erosion in long-term cultures and to improve the in vitro differentiation potential of the existing hESCs or hiPSCs. Our data revealed that XCI shares a similar behavior in rbESCs and hESCs. The instability of XCI has not been previously described in any species besides humans [18]. Our data suggest two possibilities. First, rabbits and humans may share a similar molecular mechanism of XCI regulation [31]. If so, it will be very interesting to investigate the evolutionary importance of XCI during development. However, it is also possible that the similarities are caused by the current use of improper culture conditions for hESCs and rbESCs, given that culture conditions impact the X chromosome stability in both hESCs and rbESCs. In this case, the rbESCs could be used to identify better derivation and maintenance conditions as it is much easier and more convenient to obtain rabbit blastocysts. Moreover, we demonstrated that the class III rbESCs can be converted into an mESC-like state with XaXa and rescued quality. This fact suggests that epigenetic reprogramming has the potential to correct an eroded X chromosome in human.
To our knowledge this is the first report about the conversion of primed rbESCs into mESC-like state; though another report suggest that the impaired Xist expression cannot be reestablished by in vitro differentiation, physiological oxygen levels, HDAC inhibitors, or even a second reprogramming [16,27]. One possible explanation to account for the the conversion of an eroded X in rbESCs is that the small molecules may help to establish and maintain the “right” histone modifications and DNA methylation status of Xist as well as other important regions related. Further investigations into mESC-like rbESCs might be helpful to explore the mechanism of X reactivation in reprogramming.
The derivation of rbESCs and rbiPSCs was successful; however, to our knowledge no rbESCs or rbiPSCs underwent successful germline transmission, and the derivation remains inefficient (<30%). It is reasonable to speculate that rbESCs require maintenance under specific unknown culture conditions [5,6,8,10]. Small molecules are powerful tools for manipulating cell fate, improving derivation, and reprogramming efficiency [32–34], and for enabling the generation of mouse iPSCs from somatic cell types without gene insertion [35]. Inhibition of the MEK and GSK3β signal pathways using 2i might facilitate derivation of nonpermissive ESCs [32]. Thus, we sought to derive XaXa rbESCs from blastocysts by supplementing the culture medium with LIF/2i; however, the inner cell mass outgrowth lost ESC properties and had a high frequency of spontaneous differentiation into nerve-like cells (data not show). An important work in the future is to identify culture systems that are more suitable for deriving and maintaining mESC-like rbESCs, especially with the addition of small molecules.
The conversion from a class III rbESC into an mESC-like state offers direct evidence that pluripotency factors may regulate the proper expression of Xist and mediate the epigenetic resetting of the X chromosome, as has been proposed in the generation of mouse iPSCs [20,21]. Investigating the mechanism of XCI might be helpful for avoiding Xi erosion in long-term cultures and to improve the in vitro differentiation potential of the existing rbESCs and rbiPSCs, which are promising for generating models to study human diseases.
Supplementary Material
Acknowledgments
The authors are grateful to our colleagues in the laboratory for their assistance with the experiments and in the preparation of this article. This project was supported by the Ministry of Science and Technology (grants 2012CBA01308 and 2010CB944900) and the National Natural Science Foundation of China (31325019 and 91319306).
Author Disclosure Statement
The authors have no potential conflicts of interest.
References
- 1.Weekers F, Van Herck E, Coopmans W, Michalaki M, Bowers CY, Veldhuis JD. and Van den Berghe G. (2002). A novel in vivo rabbit model of hypercatabolic critical illness reveals a biphasic neuroendocrine stress response. Endocrinology 143:764–774 [DOI] [PubMed] [Google Scholar]
- 2.Peh WL, Middleton K, Christensen N, Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu WJ. and Doorbar J. (2002). Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol 76:10401–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan J. and Watanabe T. (2003). Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol Ther 99:261–282 [DOI] [PubMed] [Google Scholar]
- 4.Shiomi M, Ito T, Yamada S, Kawashima S. and Fan J. (2004). Correlation of vulnerable coronary plaques to sudden cardiac events. Lessons from a myocardial infarction-prone animal model (the WHHLMI rabbit). J Atheroscler Thromb 11:184–189 [DOI] [PubMed] [Google Scholar]
- 5.Fang ZF, Gai H, Huang YZ, Li SG, Chen XJ, Shi JJ, Wu L, Liu A, Xu P. and Sheng HZ. (2006). Rabbit embryonic stem cell lines derived from fertilized, parthenogenetic or somatic cell nuclear transfer embryos. Exp Cell Res 312:3669–3682 [DOI] [PubMed] [Google Scholar]
- 6.Honda A, Hirose M, Hatori M, Matoba S, Miyoshi H, Inoue K. and Ogura A. (2010). Generation of induced pluripotent stem cells in rabbits: potential experimental models for human regenerative medicine. J Biol Chem 285:31362–31369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda A, Hirose M. and Ogura A. (2009). Basic FGF and Activin/Nodal but not LIF signaling sustain undifferentiated status of rabbit embryonic stem cells. Exp Cell Res 315:2033–2042 [DOI] [PubMed] [Google Scholar]
- 8.Intawicha P, Ou YW, Lo NW, Zhang SC, Chen YZ, Lin TA, Su HL, Guu HF, Chen MJ, et al. (2009). Characterization of embryonic stem cell lines derived from New Zealand white rabbit embryos. Cloning Stem Cells 11:27–38 [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Shen Y, Yuan X, Chen K, Guo X, Chen Y, Niu Y, Li J, Xu RH, et al. (2008). Dissecting signaling pathways that govern self-renewal of rabbit embryonic stem cells. J Biol Chem 283:35929–35940 [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Tang X, Niu Y, Chen H, Li B, Li T, Zhang X, Hu Z, Zhou Q. and Ji W. (2007). Generation and characterization of rabbit embryonic stem cells. Stem Cells 25:481–489 [DOI] [PubMed] [Google Scholar]
- 11.Zakhartchenko V, Flisikowska T, Li S, Richter T, Wieland H, Durkovic M, Rottmann O, Kessler B, Gungor T, et al. (2010). Cell-mediated transgenesis in rabbits: chimeric and nuclear transfer animals. Biol Reprod 84:229–237 [DOI] [PubMed] [Google Scholar]
- 12.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL. and McKay RD. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448:196–199 [DOI] [PubMed] [Google Scholar]
- 13.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA. and Vallier L. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448:191–195 [DOI] [PubMed] [Google Scholar]
- 14.Nichols J. and Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4:487–492 [DOI] [PubMed] [Google Scholar]
- 15.Dvash T, Lavon N. and Fan G. (2010). Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One 5:e11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, et al. (2012). Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11:75–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva SS, Rowntree RK, Mekhoubad S. and Lee JT. (2008). X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A 105:4820–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papp B. and Plath K. (2013). Epigenetics of reprogramming to induced pluripotency. Cell 152:1324–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, et al. (2012). Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10:620–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW. and Jaenisch R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A 107:9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K. and Geijsen N. (2010). A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K. and Surani MA. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461:1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W. and Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136:1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erwin JA. and Lee JT. (2010). Characterization of X-chromosome inactivation status in human pluripotent stem cells. Curr Protoc Stem Cell Biol Chapter 1:Unit 1B 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, et al. (2009). Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136:964–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DK. (1995). Estimating doubling time of cells in vitro. In Vitro Cell Dev Biol Anim 31:419–420 [DOI] [PubMed] [Google Scholar]
- 27.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A. and Eggan K. (2012). Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 10:595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C. and Lawrence JB. (2008). X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol 216:445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, et al. (2010). Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7:329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD. and Fan G. (2008). X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A 105:4709–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V. and Heard E. (2011). Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472:370–374 [DOI] [PubMed] [Google Scholar]
- 32.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL. and Smith A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135:1287–1298 [DOI] [PubMed] [Google Scholar]
- 33.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, et al. (2008). Germline competent embryonic stem cells derived from rat blastocysts. Cell 135:1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong C, Li P, Wu NL, Yan Y. and Ying QL. (2010). Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467:211–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, et al. (2013). Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341:651–654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.