Abstract
PML is a tumor suppressor that promotes apoptosis through both p53-dependent and - independent mechanisms, participates in Rb-mediated cell cycle arrest, inhibits neoangiogenesis, and contributes to maintenance of genomic stability. PML also plays a role in host defense against viruses, conferring antiviral activity. When active, PML localizes to subnuclear structures named PML oncogenic domains (PODs) or PML nuclear bodies (PML-NBs), whereas inactive PML is located diffusely throughout the nucleus of cells, thus providing a morphological indicator. Known activators of PML include arsenicals and interferons, however, these agents induce a plethora of toxic effects, limiting their effectiveness. The objective of the current study was to develop a high content screening (HCS) assay for the identification of chemical activators of PML. We describe methods for automated analysis of POD formation using high throughput microscopy (HTM) to localize PML immunofluorescence in conjunction with image analysis software for POD quantification. Using this HCS assay in 384 well format, we performed pilot screens of a small synthetic chemical library and mixture-based combinatorial libraries, demonstrating the robust performance of the assay. HCS counter-screening assays were also developed for hit characterization, based on immunofluorescence analyses of the subcellular location of phosphorylated H2AX or phosphorylated CHK1, which increase in a punctate nuclear pattern in response to DNA damage. Thus, the HCS assay devised here represents a high throughput screen that can be utilized to discover POD-inducing compounds that may restore the tumor suppressor activity of PML in cancers or possibly promote anti-viral states.
Keywords: PML, POD, nuclear bodies, apoptosis, high content screening
INTRODUCTION
PML oncogenic domains (PODs), also known as PML nuclear bodies (PML-NBs), Kremer bodies, ND10 (nuclear domain 10), or nuclear dots (NDs), are ~0.2–1 μm subnuclear structures present in a wide variety of cell types.1,2 PML is required for the formation of PODs, and more than 30 proteins either transiently or constitutively co-localize with PML in PODs.2 The significance of the PML gene was first noted in acute promyelocytic leukemia (APL), wherein the vast majority of cases are characterized by t(15;17) chromosomal translocations that result in a PML-RARα fusion protein that disrupts PML function by delocalizing PML into microspeckled nuclear structures (the reciprocal RARα-PML fusion protein disrupts RARα function).3 Expression of PML-RARα in the promyelocytic/myeloid compartment of transgenic mice causes leukemia with APL features, underscoring the tumor suppressive activity of PODs.4
PML plays an essential role in both caspase-dependent and caspase-independent cell death.2 pml−/− mice are resistant to apoptosis induced by numerous stimuli, and have an increased tumor incidence.5,6 PML contributes to cell death induced by γ-radiation, the primary treatment modality for a wide variety of tumors.7 Interferons and arsenicals (e.g. As2O3) increase the number and size of PODs per cell, and sensitize and/or induce apoptosis in a variety of tumor cell types.8–10 Interestingly, PML confers direct resistance to many viruses, and numerous viruses have evolved mechanisms for disrupting POD formation.11
Among the proteins that localize to PODs are p53, Daxx, and RecQ DNA helicase. The tumor suppressor p53 requires acetylation by CBP/p300 at PODs for the induction of apoptosis.2,12 Daxx localization to PODs increases PTEN nuclear localization and PTEN tumor suppressor activity by inhibiting HAUSP-mediated PTEN deubiquitinylation.13 PTEN nuclear exclusion is associated with cancer progression, and HAUSP over-expression coincides with PTEN nuclear exclusion in prostate cancer.13–15 Daxx additionally represses the transcription of various anti-apoptotic Rel-B-associated genes, including cIAP2, cFLIP, and Bfl-1 (A1), via histone deacetylase (HDAC) and DNA methyltransferase binding and recruitment.16,17 PODs are required for the maintenance of genomic stability in combination with proteins such as RecQ DNA.18 Notably, PML also suppresses the anchorage-independent growth of transformed cells, is required for hypophosphorylated Rb-mediated cell cycle arrest, and inhibits neoangiogenesis in human and mouse tumors.1,19
The fundamental roles of PODs and POD-related proteins in tumor suppression validate targeting PODs for drug discovery. POD formation is essential in interferon and arsenical cancer therapy, especially for leukemias and multiple myeloma.2,20,21 Interferons and arsenicals, however, induce a plethora of toxic effects, limiting their effectiveness.21,22 The objective of the current study was to develop a high content screening (HCS) assay for the high throughput identification of chemical activators of PODs.
MATERIALS AND METHODS
Cell Culture
HeLa cells were originally obtained from ATCC (Manassas, Virginia) and cultured in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, California), with 10% FBS (Clontech, Mountain View, California) and penicillin-streptomycin (diluted according to manufacturer's specifications, Invitrogen) at 37°C, 5% CO2. PPC-1 cells were cultured similarly, but with RPMI 1640 (Invitrogen) instead of DMEM.
Compounds
The LOPAC1280 collection of 1,280 pharmacologically active single compounds was obtained from Sigma-Aldrich (St. Louis, Missouri). The Torrey Pines Institute for Molecular Studies Combinatorial Libraries are mixture-based libraries in positional scanning format and they were dissolved in DMF.23–25
Immunofluorescence Assay
3150 cells/well (50 μL/well volume) were seeded onto 384-well clear-bottom plates (Greiner Bio-One, Monroe, North Carolina) using the Matrix WellMate liquid dispenser (Thermo Fisher Scientific, Hudson, New Hampshire) and incubated at 37°C (5% CO2). After 24 h, the Biomek FX Laboratory Automation Workstation (Beckman Coulter, Fullerton, California) was used to add IFN-γ (4 U/μL with either 0.1% DMSO or DMF; R&D Systems, Minneapolis, Minnesota), DMSO (0.1% final concentration; Sigma-Aldrich), DMF (0.1% final concentration; Sigma-Aldrich), or compounds. After 12 h, cells were washed with PBS, fixed with 4% formaldehyde (Sigma-Aldrich) for 15 min, washed, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 5 min, washed, incubated with the primary antibody diluted to 0.5 μg/mL in 5% BSA (Thermo Fisher Scientific) for 1 h, washed, incubated with the secondary antibody diluted to 5 μg/mL in 5% BSA for 1 h, washed, and placed in a 100 ng/mL DAPI-PBS solution (Invitrogen) overnight. Each wash step was a multiple fluid change using the MAP-C (aspiration to 10 μL, 50 μL PBS addition, repeated a total of 3 times; Titertek, Huntsville, Alabama).
For PML immunostaining, mouse monoclonal anti-human PML primary antibody (PGM3, Santa Cruz Biotechnology, Santa Cruz, California) was used with an Alexa Fluor 488 chicken anti-mouse IgG secondary antibody (Invitrogen). For phospho-H2AX (Ser139; p-H2AX or γ-H2AX) or phospho-Chk1 (Ser317; pChk1) immunostaining, rabbit polyclonal anti-human phospho-histone H2A.X (Ser139; diluted 1:400; Cell Signaling Technology, Danvers, Massachusetts) or phospho-Chk1 (Ser317; diluted 1:400; Cell Signaling) primary antibodies were used, respectively, with an Alexa Fluor 568 goat anti-rabbit IgG secondary antibody (Invitrogen).
High Content Imaging
Plates were imaged using the Beckman Coulter Cell Lab IC-100 Image Cytometer with a 40x 0.6NA ELWD Plan Fluor dry (air) objective (6 images/well). The images (greater than 200 cells/well) were analyzed using the POD detection algorithm, which was developed based on CytoShop (Beckman Coulter) and MATLAB (MathWorks, Natick, Massachusetts) software.
High content screening (HCS) was performed at the Conrad Prebys Center for Chemical Genomics at Sanford-Burnham Medical Research Institute.
Statistical Analyses
HCS performance was characterized using the equation: Z'-Factor = 1 - (3σpositive + 3σnegative)/|(μpositive−μnegative)|, where σpositive is the standard deviation of the positive control, σnegative is the standard deviation of the negative control, μpositive is the mean of the positive control, and μnegative is the mean of the negative control.26
RESULTS
Development and Optimization of the PML-POD-Localization Assay
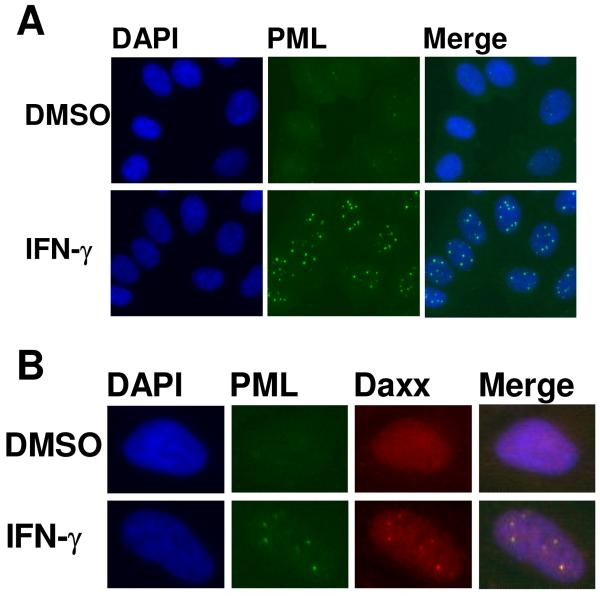
Immunostaining conditions were optimized for detection of PML using a commercially available mouse monoclonal antibody. HeLa cells were seeded in 384-well plates, treated with either DMSO or IFN-γ for 24 h, and immunofluorescently stained for PML to confirm IFN-γ-induced PML localization into PODS (Figure 1A). IFN-γ-induced localization of PML into PODs is accompanied by the localization of various other proteins to PODs, such as Daxx.27 Thus, simultaneous immunofluorescence detection of both PML and Daxx in HeLa cells confirmed co-localization/formation of PODs and validated IFN-γ as a positive control (Figure 1B). IFN-γ-induced PML and Daxx co-localization to PODs was additionally confirmed to occur in PPC-1 cells (observed by immunofluroescence, data not shown).
Figure 1. PML and Daxx localize to PODs after IFN-γγγ treatment.
(A) PML localizes to nuclear bodies. HeLa cells were seeded in 384-well plates (3150 cells/well), treated (12 h) with DMSO (0.1%) or IFN-γ (4 U/μL), immunostained with mouse monoclonal anti-human PML and Alexa Fluor 488 chicken anti-mouse antibodies (green), and incubated in DAPI (nuclear stain; 100 ng/mL; blue). Cells were imaged using the Cell Lab IC-100 Image Cytometer (40× 0.6NA ELWD Plan Fluor objective). (B) PML and Daxx co-localize to PODs. Cells were treated and imaged as in (A), but also immunostained for Daxx (rabbit polyclonal anti-human Daxx primary antibody, Alexa Fluor 568 goat anti-rabbit secondary antibody; red).
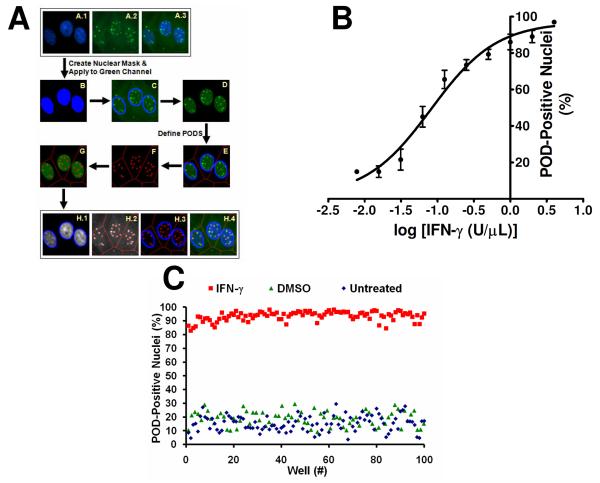
To quantify the extent of POD formation (i.e., the number of PODs per cell, the intensity of PML localization, and the fraction of cells per well with extensive numbers of PODs) in an automated fashion, the “POD detection algorithm” was developed using Beckman Coulter CytoShop and MathWorks MATLAB software (Figure 2A). Firstly, the nuclear image (DAPI stain) was used to produce a “nuclear mask”, which identified all nuclei in an image. This nuclear mask was applied to the PML (“green”) image and all green pixels outside of this nuclear mask were eliminated. Next, PODs were outlined based on the identification of green pixels with higher intensities than their surrounding pixels (using CytoShop's “Aggregate Detection”), with the minimum size of a POD defined based on IFN-γ control wells (CytoShop's “Object Scale”). This number of detected PODs was then reported on a per-nucleus basis, and used to determine the percentage of nuclei per image that were “POD-positive”. The value for the number of detected PODs above which a nucleus is considered POD-positive was determined to be 4.0 by iteratively setting increasing threshold values and determining the Z'-factor for each threshold (on control plates).
Figure 2. High Content Screen (HCS) Development and Optimization.
(A) Algorithm for detecting and quantifying PODs. DAPI (A.1; blue), PML (A.2; green), and merged (A.3) images of IFN-γ-treated (4 U/μL; 12 h) HeLa cells are shown. The nuclear (DAPI) image was used to produce a nuclear mask (B; blue). The nuclear mask (C; blue outline) was applied to the PML image. Green pixels outside of the nuclear mask were eliminated (D). Beckman Coulter CytoShop software was used to estimate cellular area (E; red outline) based on the nuclei. POD outlines (F; red) were identified based on differences in green pixel brightness. The number of detected PODs (G; red) was reported on a per-nucleus basis. The percentage of POD-positive nuclei (>4.0 PODs per cell) was reported. (B) Quantification of IFN-γ-induced POD formation. HeLa cells were seeded in a 384-well plate (3150 cells/well), and treated with increasing concentrations of IFN-γ (12 h) as shown. Cells were immunostained for PODs (mouse monoclonal anti-human PML and Alexa Fluor 488 chicken anti-mouse antibodies), incubated in DAPI (nuclear stain; 100 ng/mL), imaged using the Beckman Coulter Cell Lab IC-100 Image Cytometer (40× 0.6NA ELWD Plan Fluor objective), and quantified using the described computerized algorithm. Mean ± standard deviation are shown (n=4 wells/data point, >200 cells imaged and quantified per well). (C) POD HCS reproducibility assessment. HeLa cells were cultured overnight in a 384-well plate (3150 cells/well seeded using the Matrix WellMate bulk liquid dispenser), treated with IFN-γ (4 U/μL), DMSO (0.1%), or nothing for 12 h, immunostained for PML, imaged, and analyzed (n>85 wells/condition). The Z'-factor is 0.64 or 0.65 using DMSO or untreated cells, respectively, as the negative control.
To both further validate the algorithm, HeLa cells were treated with increasing concentrations of IFN-γ, stained with anti-PML antibody, imaged, and the percentage of POD-positive cells was determined (Figure 2B). Using this automated method, IFN-γ was determined to induce concentration dependent POD formation in HeLa cells. Manual counting of POD-positive cells confirmed the algorithm-defined quantification (data not shown). Examples of whole-field images from single wells (multiple images from single wells) are shown in Supplemental Figure 1.
To characterize the reproducibility of the assay, multiple replicates were prepared (n>80 per condition), where cells were seeded into a 384-well plate using the Matrix Wellmate bulk liquid dispenser (3150 cells/well), treated with IFN-γ (4 U/μL), DMSO (0.1%), or nothing for 12 h, immunostained for PML, imaged, and analyzed. The Z'-factor was determined to be 0.64 or 0.65 using DMSO or untreated cells, respectively, as the negative control (Figure 2C).
High Content Screening (HCS)
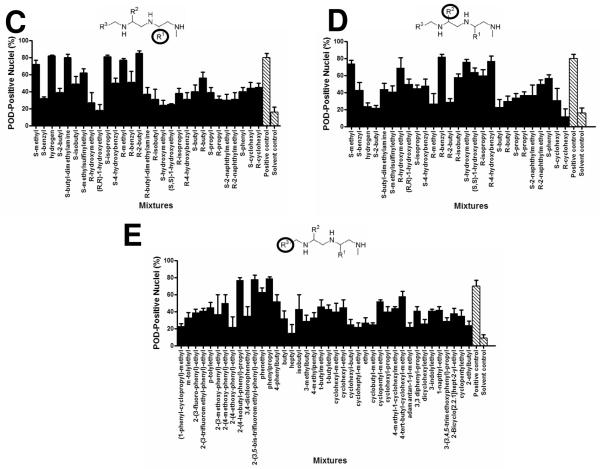
The HCS assay was used to screen the LOPAC1280 library of pharmacologically active compounds for PML activators (Figure 3A). Compounds that increased the number of POD-positive nuclei by at least 50% (relative to the controls), as measured by the POD detection algorithm, were considered “hits”. No hits were identified when the library was screened at 5 μM. Moreover, dose-response curves (serial dilutions performed from 250 μM to 0.25 μM) were generated for the five LOPAC1280 compounds inducing the most POD-positivity, and the highest value observed was less than 40% POD-positive nuclei (data not shown).
Figure 3. High Content Screening (HCS) for Chemical Activators of PODs.
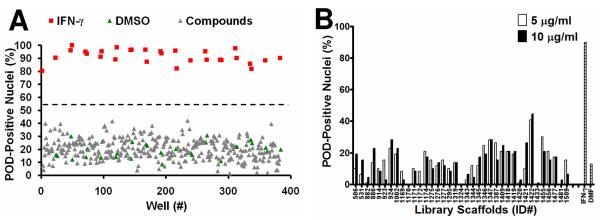
(A) Sample plate from the LOPAC1280 POD localization screen. HeLa cells were seeded in 384-well plates (3150 cells/well) and incubated at 37°C overnight. The cells were then treated for 12 h with IFN-γ (4 U/μL; positive control), DMSO (0.1%; negative control), or LOPAC1280 compounds (5 μM). Plates were immunostained for PML, imaged, and analyzed. The Z'-factor was 0.55. (The dotted line represents the mean value between the positive and negative controls). (B) TPIMS Combinatorial Library POD localization screen. Cells were screened as described previously, but with mixtures from the TPIMS scaffold ranking library (5 μg/mL or 10 μg/mL, representing ~10 μM or ~20 μM, respectively, for MW≈500 g/mol). IFN-γ (4 U/μL) was used as the positive control and DMF (0.1%) was used as the negative control. The Z'-factor was 0.6. (C–E) Combinatorial library deconvolution reveals SAR of N-methyl triamine compounds. Deconvolution of the mixture-based N-methyl triamine library was accomplished by the positional scanning method23–25,28 where one of the substituents used to create the library was fixed at positions R1 (C), R2 (D), or R3 (E), and the other positions were allowed to vary as mixtures of all possible substituents employed in library construction. Compounds were tested at 4 μg/mL (representing ~8 μM for MW≈500 g/mol). IFN-γ (4 U/μL) was used as the positive control and DMF (0.1%) was used as the negative control (striped bar). The Z'-factor was ~0.5.
The TPIMS mixture-based combinatorial libraries were formatted and plated as a “scaffold ranking library”, which was subsequently screened to determine the most active chemical scaffolds before further screening/deconvolution of mixtures.28 The scaffold ranking library was formatted as 38 mixtures (dissolved in DMF), grouped according to scaffold, and represented 5,287,896 compounds (and several million peptides). Each mixture was present in two different wells at two different concentrations. This library was screened at 5 μg/mL (~10 μM for average MW≈500 g/mol) and 10 μg/mL (~20 μM for average MW≈500 g/mol) (Figure 3B). The most active mixture was “1422”, which induced >40% POD-positive cells.
The 1422 library contains compounds with a N-methyl triamine scaffold in which chemical diversity was created at three positions, R1, R2 and R3. Analysis of a collection of N-methyl triamines, at which one of these diversity positions was fixed, allowing the others to vary as a mixture of all possibilities built into the combinational library, revealed substituents at R1, R2 or R3 that induced >50% POD-positive cells when tested at 4 μg/mL (~8 μM) (Figures 3C–E). Successful implementation of the HCS assay for POD inducers for structure activity relation (SAR) analysis of the N-methyl triamine combinatorial library demonstrates the robust performance of the assay.
POD-Inducing Specificity
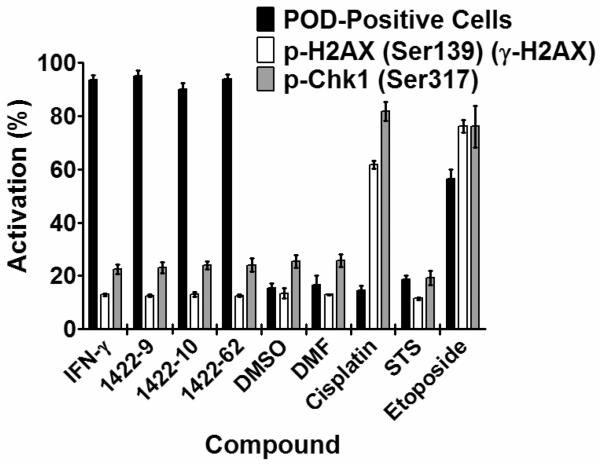
To complement the HCS assay for POD activation, we also devised two HCS counter-screen assays using immunostaining for the DNA-damage/repair-related proteins, H2AX and Chk1. H2AX is phosphorylated primarily by ATM, which “senses” double-stranded DNA breaks, while Chk1 is phosphorylated primarily by ATR, which “senses” single-stranded DNA breaks (a common intermediate found at sites of DNA damage detection and repair pathways).29 Immunofluorescence staining for phospho-H2AX (Ser139) (γ-H2AX) and phospho-Chk1 (Ser317), which are visible as nuclear aggregates, was used as a counter-screen to test the specificity of the most active 1422 compounds. The similarity in antigen redistribution for H2AX and Chk1 compared to PML provides the basis for counter-screens that eliminate false-positive compounds that might affect PML or alter immunostaining patterns in a non-specific manner. In addition, because some types of DNA damaging agents can stimulate POD formation29–33, the phospho-H2AX and phospho-Chk1 immunofluorescence counter-screens serve to eliminate compounds that operate as DNA damaging agents and thus eliminate these from further consideration.
For these counter-screen HCS assays, Cytoshop algorithms were used to quantify the intensity of nuclear phospho-H2AX (Ser139) (γ-H2AX) or phospho-Chk1 (Ser317) immunofluorescence. As shown in Figure 4, IFN-γ (POD-positivity control) induced PML localization to PODs without affecting the markers of DNA damage. Similarly, three active N-methyl triamine (1422) compounds synthesized based on results from the combinatorial library analysis (1422-9, 1422-10, and 1422-62) induced PML localization into nuclear aggregates without altering phospho-H2AX or phospho-Chk1, thus confirming the selectivity of their POD-inducing activity. In contrast, cisplatin (DNA damage control) induced H2AX and Chk1 phosphorylation without affecting POD formation, while etoposide (DNA damage control) simultaneously induced POD formation, H2AX phosphorylation, and Chk1 phosphorylation. Staurosporine (a broad spectrum kinase inhibitor), DMSO (solvent control), and DMF (solvent control) had little effect on POD activation, H2AX phosphorylation, or Chk1 phosphorylation. Altogether, these results validate both the primary HCS assay for detection of POD activators and also the counter-screening assays for elimination of DNA damage-inducing compounds.
Figure 4. POD-inducing N-methyl triamines do not induce DNA damage.
HeLa cells were treated for 12 h with IFN-γ (4 U/μL), DMSO (0.1%), DMF (0.1%), cisplatin (25 μM), staurosporine (25 nM), etoposide (25 μM), or 1422 compounds (10 μM). Plates were then immunostained for PODs (mouse monoclonal anti-human PML, Alexa Fluor 488 chicken anti-mouse antibodies), phospho-H2AX (rabbit polyclonal anti-human phospho-H2AX-Ser139, Alexa Fluor 568 goat anti-rabbit antibodies), or phospho-Chk1 (rabbit polyclonal anti-human phospho-Chk1-Ser317, Alexa Fluor 568 goat anti-rabbit antibodies). POD positive nuclei (%) were quantified as previously described. p-H2AX and p-Chk1 nuclear staining intensity was quantified using CytoShop. Mean and standard deviation are shown (n=4 wells/condition, >200 cells imaged and quantified per well).
DISCUSSION
The current study describes a HCS assay for compounds that induce PML to localize to PODs, subnuclear structures involved in a variety of tumor suppressive pathways and host defense against some types of viruses. After developing algorithms for automated analyses of POD formation, the LOPAC1280 and TPIMS combinatorial libraries were screened. No hits were identified among the 1,280 pharmacologically active LOPAC1280 compounds, possibly providing evidence that the HCS assay is highly specific and does not suffer from promiscuous reactivity. Screening of the TPIMS combinatorial libraries, which consists of mixtures representing 5,287,896 compounds and several million peptides, revealed a bioactive N-methyl triamine mixture. Further SAR evaluations where library deconvolution was performed by the positional scanning method revealed a clear SAR and thus validated the HCS assay for detection of POD-inducing compounds. Further, N-methyl triamines that were active in the POD assay did not impact the distribution of control proteins, H2AX and Chk1, demonstrating specificity.
Some types of DNA damage have been shown to induce the formation of PML-type bodies that are thought to be functionally different from the tumor suppressive and antiviral PODs mentioned in the current study.29–33 Thus, the HCS counter-screen assays described here eliminate DNA-damaging compounds from further consideration.
Active compounds such as the N-methyl triamines could potentially induce PODs via several different mechanisms. First, IFN-α, -β, and -γ induce POD formation by up-regulating PML and various POD-associated proteins.11 Thus, compounds might induce interferon production, a mechanism that can be readily determined by measuring interferon elaboration into culture supernatants (for instance, using immunoassays) or by testing activation of interferoninducible reporter genes (e.g. ISGE-luciferase). Second, arsenicals have been shown to activate PML by directly conjugating cysteines within the zinc fingers of the PML RBCC domain34, and thus compounds that covalently modify these sites define an additional mechanism for POD activation. Third, conjugation of PML by the ubiquitin-like protein SUMO is required for POD formation.35 Hence, compounds that affect SUMOylation represent another potential mechanism, which might include for instance inhibitors of the proteases responsible for de-SUMOylation. Finally, PML-binding proteins such as Daxx are regulated by phosphorylation.36 The protein kinase, ZIPK, for example, has been shown to be required for interferon to induce POD formation.27 Thus, compounds that influence the relevant kinases or phosphatases represent another potential class of POD activators that might be revealed by our HCS assay.
Because of the role of PODs in tumor suppression and host defense against viruses, POD-inducing compounds may have relevance to a wide variety of cancers and infectious diseases. Arsenicals are already being used to treat leukemias (such as APL) and multiple myeloma.2,20 Additionally, adenoviruses, herpes simplex virus-1 (HSV-1), human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), papillomavirus, hepatitis D virus (HDV), human T-lymphotropic virus-1 (HTLV-1), lymphocytic choriomeningitis (LCMV), and rabies viruses disrupt PODs11, suggesting that POD-inducing compounds may also promote antiviral states that could be therapeutically useful. The HCS assay described here enables high throughput screening for compounds with potential medicinal activity based on induction of PODs, thus providing a route to chemical modulators of PML that accommodates the diversity of cellular mechanisms responsible for POD regulation in a manner unachievable with standard biochemical HTS assays.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tessa Siegfried and Melanie Hanaii for assistance with manuscript preparation, as well as Drs. Paul Diaz and Satoshi Ogasawara for technical assistance. Support by DoD grant W81XWH-08-0574 and NIH grant CA-55164.
REFERENCES
- 1.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 2.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 3.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 4.Piazza F, Gurrieri C, Pandolfi PP. The theory of APL. Oncogene. 2001;20:7216–7222. doi: 10.1038/sj.onc.1204855. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 6.Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 7.Salomoni P, Bernardi R, Bergmann S, Changou A, Tuttle S, Pandolfi PP. The promyelocytic leukemia protein PML regulates c-Jun function in response to DNA damage. Blood. 2005;105:3686–3690. doi: 10.1182/blood-2004-09-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- 9.Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 10.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 11.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 12.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 13.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. Nature. 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Puto LA, Reed JC. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 2008;22:998–1010. doi: 10.1101/gad.1632208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croxton R, Puto LA, de Belle I, Thomas M, Torii S, Hanaii F, Cuddy M, Reed JC. Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 2006;66:9026–9035. doi: 10.1158/0008-5472.CAN-06-1047. [DOI] [PubMed] [Google Scholar]
- 18.Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- 19.Khan MM, Nomura T, Kim H, Kaul SC, Wadhwa R, Zhong S, Pandolfi PP, Ishii S. PML RARalpha alleviates the transcriptional repression mediated by tumor suppressor Rb. J Biol Chem. 2001;276:43491–43494. doi: 10.1074/jbc.C100532200. [DOI] [PubMed] [Google Scholar]
- 20.Crowder C, Dahle O, Davis RE, Gabrielsen OS, Rudikoff S. PML mediates IFN-alpha-induced apoptosis in myeloma by regulating TRAIL induction. Blood. 2005;105:1280–1287. doi: 10.1182/blood-2004-04-1614. [DOI] [PubMed] [Google Scholar]
- 21.Berenson JR, Yeh HS. Arsenic compounds in the treatment of multiple myeloma: a new role for a historical remedy. Clin Lymphoma Myeloma. 2006;7:192–198. doi: 10.3816/CLM.2006.n.058. [DOI] [PubMed] [Google Scholar]
- 22.Dunn GP, Ikeda H, Bruce AT, Koebel C, Uppaluri R, Bui J, Chan R, Diamond M, White JM, Sheehan KC, Schreiber RD. Interferon-gamma and cancer immunoediting. Immunol Res. 2005;32:231–245. doi: 10.1385/ir:32:1-3:231. [DOI] [PubMed] [Google Scholar]
- 23.Houghten RA, Pinilla C, Appel JR, Blondelle SE, Dooley CT, Eichler J, Nefzi A, Ostresh JM. Mixture-based synthetic combinatorial libraries. J Med Chem. 1999;42:3743–3778. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]
- 24.Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- 25.Pinilla C, Appel JR, Borras E, Houghten RA. Advances in the use of synthetic combinatorial chemistry: mixture-based libraries. Nat Med. 2003;9:118–122. doi: 10.1038/nm0103-118. [DOI] [PubMed] [Google Scholar]
- 26.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T, Akira S, Reed JC. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol Cell Biol. 2003;23:6174–6186. doi: 10.1128/MCB.23.17.6174-6186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houghten RA, Pinilla C, Giulianotti MA, Appel JR, Dooley CT, Nefzi A, Ostresh JM, Yu Y, Maggiora GM, Medina-Franco JL, Brunner D, Schneider J. Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J Comb Chem. 2008;10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- 29.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 30.Zhong S, Salomoni P, Ronchetti S, Guo A, Ruggero D, Pandolfi PP. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J Exp Med. 2000;191:631–640. doi: 10.1084/jem.191.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seker H, Rubbi C, Linke SP, Bowman ED, Garfield S, Hansen L, Borden KL, Milner J, Harris CC. UV-C-induced DNA damage leads to p53-dependent nuclear trafficking of PML. Oncogene. 2003;22:1620–1628. doi: 10.1038/sj.onc.1206140. [DOI] [PubMed] [Google Scholar]
- 32.Eskiw CH, Dellaire G, Mymryk JS, Bazett-Jones DP. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J Cell Sci. 2003;116:4455–4466. doi: 10.1242/jcs.00758. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T, Halicka HD, Traganos F, Seiter K, Darzynkiewicz Z. Induction of ATM activation, histone H2AX phosphorylation and apoptosis by etoposide: relation to cell cycle phase. Cell Cycle. 2007;6:371–376. doi: 10.4161/cc.6.3.3835. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, Zhang QY, Yang HY, Huang QH, Zhou GB, Tong JH, Zhang Y, Wu JH, Hu HY, de The H, Chen SJ, Chen Z. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 35.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salomoni P, Khelifi AF. Daxx: death or survival protein. Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.