Abstract
HIV-1 NL4-3 Vpu induces downregulation of cell surface CD155, a ligand for the DNAM-1 activating receptor of NK and CD8+ T cells, to evade NK cell mediated immune response. Here we show that the conserved alanine residues at positions 10, 14 and 18 in the TM domain of Vpu are required for the efficient downregulation of cell surface CD155. In contrast, the CK-2 phosphorylation sites and the second α-helix in the cytoplasmic Vpu domain have no influence on the surface expression of CD155. Thus, compared to Vpu’s effect on CD4, NTB-A and tetherin, the Vpu mediated downregulation of CD155 is an independent Vpu function. We finally show that in contrast to other lentiviral strains, only Vpu and Nef from HIV-1 M NL4-3 potently interfere with CD155 surface expression. Thus, Vpu seems to subvert NK cell responses against HIV-1 infected T cells by modulation of receptors necessary for NK cell activation.
Keywords: Vpu, CD155, HIV-1, SIV, Nef
Introduction
The human immunodeficiency virus type 1 (HIV-1) accessory protein Vpu is a 15–20 kDa oligomeric type 1 integral membrane phosphoprotein (Cohen et al., 1988; Maldarelli et al., 1993; Strebel et al., 1988), which is encoded exclusively by HIV-1 and related simian immunodeficiency viruses (SIVs), but not by the majority of SIVs and HIV-2. Vpu has been shown to induce the degradation of newly synthesized CD4 by retaining it in the endoplasmic reticulum (ER) (Magadan et al., 2010) and thereby targeting CD4 to the ER-associated protein degradation (ERAD)-pathway. This specific function of Vpu depends on the CK-2 phosphorylation sites in its cytoplasmic domain (Binette et al., 2007; Magadan et al., 2010; Schubert et al., 1998; Willey et al., 1992).
Furthermore, it has been shown that Vpu supports HIV-1 virion release by counteracting the cellular restriction factor tetherin (also known as CD317, BST-2 or HM1.24) (Neil et al., 2008; Van Damme et al., 2008). Vpu induces an accumulation of tetherin in the trans-Golgi network (TGN), which results in the downregulation of cell surface tetherin (Dube et al., 2010; Schmidt et al., 2011). Vpu and tetherin interact via their TM domains, and this interaction is required for the efficient downregulation of cell surface tetherin (Banning et al., 2010; Dube et al., 2010; Iwabu et al., 2009; Rong et al., 2009). Recently, it has been demonstrated that several highly conserved residues in the TM domain of Vpu, particularly Ala-10, Ala-14 and Ala-18, are required for efficient downregulation of tetherin from the cell surface (Skasko et al., 2012; Vigan and Neil, 2010).
In addition to its role in tetherin antagonism and CD4 degradation, HIV-1 Vpu induces the downregulation of the co-activating NK-cell receptor NK-cell, T-cell, B-cell antigen (NTB-A) (also termed CD352 or SLAMF6) as well as the activating NK cell receptor CD155 (also termed polio virus receptor (PVR) or NECL-5) from the cell surface to evade NK-cell mediated lysis of HIV-1 infected cells (Matusali et al., 2012; Shah et al., 2010). Moreover, Vpu affects the anterograde transport of newly synthesized NTB-A molecules by retention of NTB-A within the Golgi-compartment (Bolduan et al., 2013). With the exception of HIV-1 group N, the Vpu mediated downregulation of cell surface NTB-A is conserved among diverse HIV-1 and SIV strains (Bolduan et al., 2013). In contrast, it is currently not known how Vpu downregulates CD155 or if this function is conserved among different lentiviral Vpu variants.
CD155 is a nectin-like protein that serves as a ligand for the DNAX accessory molecule-1 (DNAM-1) activating receptor of NK and CD8+ T cells (Takai et al., 2008; Vassena et al., 2013), thereby triggering T and NK cell-mediated cytotoxicity (Matusali et al., 2012; Shibuya et al., 1996). Interestingly, while HIV-1 Nef and Vpu are required for optimal downregulation of cell surface CD155 to avoid DNAM-1 mediated immune response against HIV-1 infected cells (Matusali et al., 2012), HIV-1 Vpr induces an upregulation of cell surface and steady state CD155 levels in HIV-1 infected CD4+ T cells. As a consequence, the effect of Vpr seems to decrease the anti-CD155 activity of HIV-1 Nef (Vassena et al., 2013).
In order to elucidate the importance of the TM domain of Vpu in downregulating cell surface CD155, several well-established Vpu TM mutants, particularly A10N, A14N and A18N, were analyzed for their capability to affect the surface expression of CD155. We show that the mutation of Ala-10, Ala-14 and Ala-18 to asparagine impairs the ability of Vpu to downregulate cell surface CD155. In contrast, the CK-2 phosphorylation sites and the second α-helix in the cytoplasmic domain of Vpu have no influence on the surface expression of CD155. Interestingly, the Vpu Δ23 mutant, which lacks the second cytoplasmic α-helix, exhibits an attenuated ability to downregulate cell surface tetherin and NTB-A (Bolduan et al., 2013; Dube et al., 2009), but was fully active on CD155. Thus, the downregulation of CD155 is an independent function of Vpu and can be separated from its effects on CD4, NTB-A or tetherin. Furthermore, we show that Vpu causes intracellular accumulation of CD155 in perinuclear compartments and a Vpu mutant, which is retained in the ER/cis Golgi, still interferes with CD155 cell surface expression. Altogether, these data indicate that Vpu might inhibit trafficking of CD155 to the cell surface. Finally, our data reveal that Vpu and Nef proteins derived from HIV-1 M NL4-3 are most potent agonists of CD155 cell surface expression, whereas those from other strains, except SIVgor CP2139, were strongly attenuated. Thus, these results are in line with a model suggesting that a variety of Vpu and Nef functions mediate HIV-1 immune evasion and add to the full pathogenic potential of HIV-1.
Results
Ala-10, Ala-14 and Ala-18 in the Vpu TM domain are critical for the downregulation of cell surface CD155
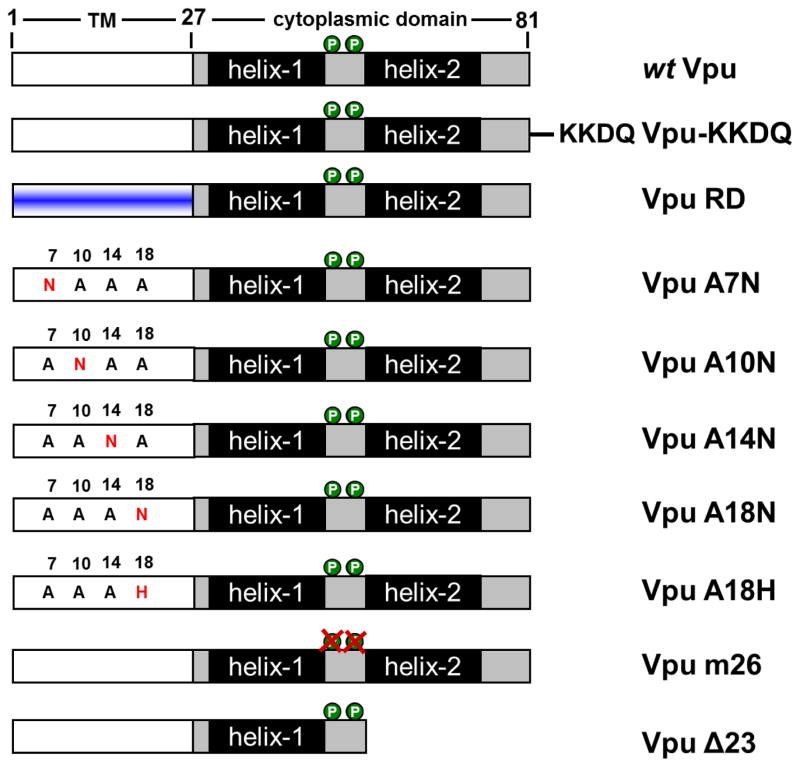
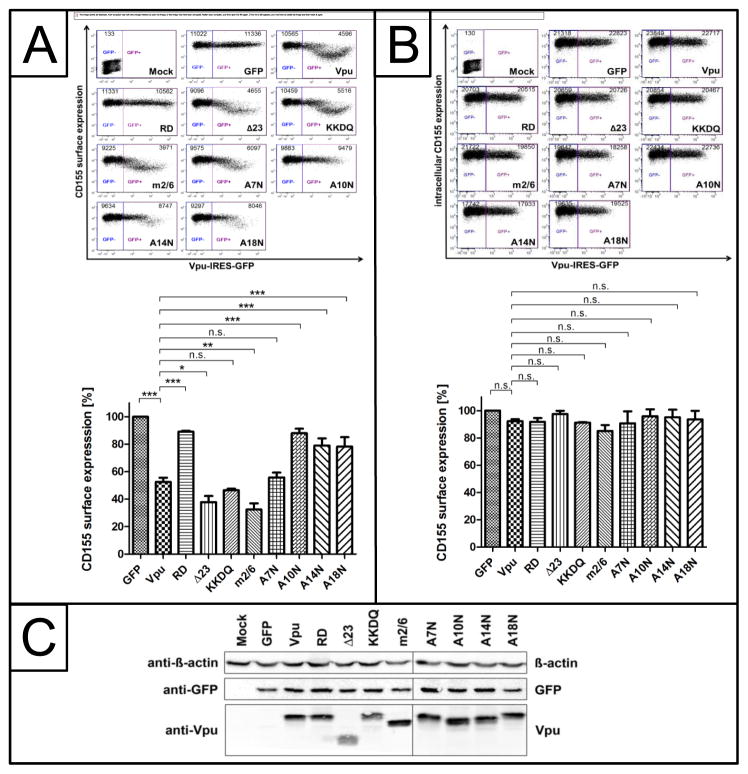
Recently, it was shown that HIV-1 NL4-3 Vpu downregulates cell surface CD155 to evade NK cell mediated immune response against HIV-1 infected cells (Matusali et al., 2012). Moreover, the randomization of Vpu’s TM domain impairs its ability to affect the surface expression of CD155 (Matusali et al., 2012). In order to analyze the effect of Vpu’s TM and cytoplasmic domain on the surface expression of CD155, we analyzed specific Vpu TM and cytoplasmic deletion mutants, which are summarized in Figure 1. HeLa cells, which constitutively express endogenous CD155, were transfected with NL4-3 wt Vpu or mutants translated along with GFP via an internal ribosomal entry site (Vpu-IRES-GFP) to monitor transfected cells. 24 h post transfection cells were harvested, stained for cell surface (Fig. 2A) and intracellular (Fig. 2B) CD155 using a CD155 specific antibody, and analyzed by flow cytometry. The surface expression of CD155 in Vpu expressing cells was calculated as the mean fluorescence intensity (MFI) of CD155 expression in GFP positive cells relative to GFP negative cells. In addition, cells were lysed and the soluble protein fraction was analyzed by Western blot (Fig. 2C). Figure 2A shows that NL4-3 wt Vpu as well as the m2/6 mutant efficiently downregulate cell surface CD155, indicating that the CK-2 phophorylation sites have no influence on the surface expression of CD155. The C-terminal deletion mutant Vpu Δ23, which exhibits a decreased localization in the TGN (Dube et al., 2009; Pacyniak et al., 2005), and the Vpu KKDQ mutant, which is retained in the ER and the cis-Golgi compartment (Shikano and Li, 2003; Skasko et al., 2011; Vigan and Neil, 2011), both downregulate CD155 to the same degree as wt Vpu (Fig. 2A).
Fig. 1.
Schematic representation of wt Vpu (A) or Vpu mutants KKDQ (B), RD (C), A7N (D), A10N (E), A14N (F), A18N (G), A18H (H), m26 (I) or Δ23 (J).
Fig. 2.
The TM domain, particularly A10N, A14N and A18N, of Vpu is required for the efficient downregulation of cell surface CD155. HeLa cells were transfected with either Vpu-IRES-GFP or mutants thereof. 24 h post transfection, cells were harvested and either stained for surface CD155 (A) or intracellular CD155 (B) using a CD155 specific antibody. Subsequently, cells were analyzed by flow cytometry. Summary of relative CD155 surface (A) and intracellular (B) expression from 3 independent experiments is given below. Mock indicates non-infected and unstained control cells. Values from three independent experiments were plotted and assessed for statistical significance by using the GrapPad Prism V5.0 software package (***; p≤0.0005, 1 way- ANOVA with Dunnett’s test). In addition, cells were lysed and analyzed by Western blot using anti-β-actin, anti-GFP and anti-Vpu antibodies (C).
In contrast to the Vpu A7N mutant, the Vpu TM mutants A10N, A14N and A18N were strongly attenuated in their ability to decrease the surface expression of CD155, implying that this stretch of conserved residues is critical for the Vpu induced downmodulation of cell surface CD155. Moreover, the expression levels of all Vpu proteins were indeed comparable (Fig 2C). In Figure 2B, we analyzed the ability of Vpu to affect the intracellular expression of CD155. Here we show that neither wt Vpu nor any of the mutants does alter steady state protein levels of CD155 (Fig. 2B).
Next, we analyzed the impact of Vpu’s TM domain, particularly A18, to affect the surface expression of CD155 in CD4+ SupT1 T cells (Fig. 3). Thus, CD4+ SupT1 cells were infected with either VSV-G pseudotyped Vpu deletion mutant HIV-1 NL4-3 Vpu Del-1, HIV-1 NL4-3 wt, HIV-1 NL4-3 Vpu RD or HIV-1 NL4-3 Vpu A18H, and surface expression of CD155 was analyzed by flow cytometry. As expected, HIV-1 NL4-3 wt was capable of inducing downregulation of CD155 from the cell surface (Fig. 3). However, the Vpu deletion mutant HIV-1 NL4-3 Vpu Del-1 and the Vpu mutants HIV-1 NL4-3 Vpu RD and HIV-1 NL4-3 Vpu A18H lost their ability to induce efficient downregulation of cell surface CD155 (Fig. 3). These results suggest that alanine 18 in the TM domain of Vpu is required to induce downregulation of cell surface CD155 in also in the context of infected CD4+ T cells.
Fig. 3.
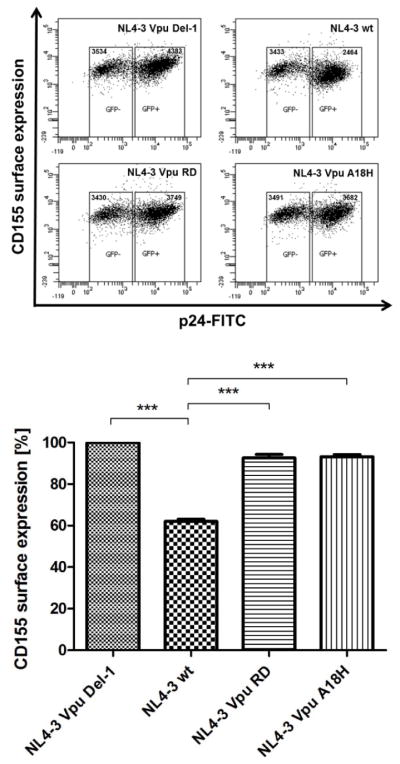
Downregulation of cell surface CD155 by Vpu in CD4+ Sup-T1 T cells. CD4+ Sup-T1 T cells were infected with either VSVG-pseudotyped HIV-1 NL4-3 Vpu Del-1, HIV-1 NL4-3 wt, HIV-1 NL4-3 Vpu RD or HIV-1 NL4-3 Vpu A18H. 2 days after infection the cells were harvested and stained for surface CD155, using a CD155-specific antibody. Afterwards the cells were stained intracellularly for HIV-1 p24 antigen, using anti-p24-FITC specific antibody and analyzed by flow cytometry. Values from three independent experiments were plotted and assessed for statistical significance by using the GrapPad Prism V5.0 software package (***; p≤0.0005, 1 way- ANOVA with Dunnett’s test), which is given below.
In summary, these results suggest that the TM domain of Vpu, particularly Ala-10, Ala-14 and Ala-18, is crucial for cell surface downregulation of CD155, while the CK-2 phophorylation sites and the second α-helix in the cytoplasmic domain of Vpu are dispensable for this phenomenon.
NL4-3 Vpu alters the subcellular localization of CD155 by entrapment in perinuclear compartments
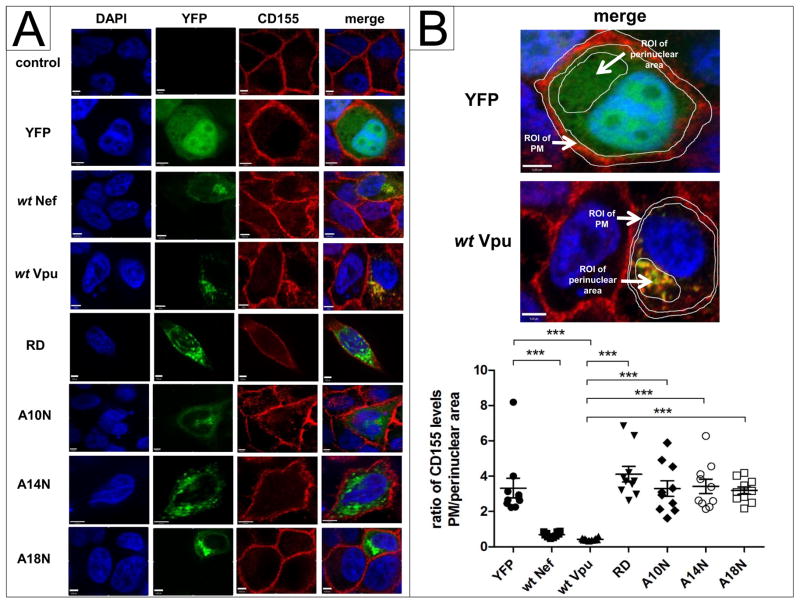
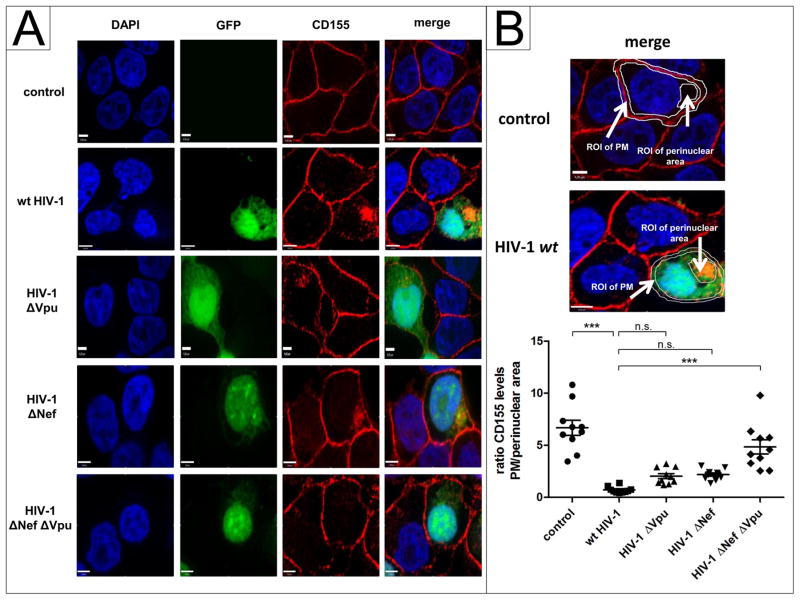
Since NL4-3 wt Vpu induces downregulation of cell surface CD155, we investigated by immunofluorescence whether Vpu has an influence on the subcellular localization of CD155. HeLa cells were transfected with, NL4-3 Nef-YFP or NL4-3 Vpu- fusion proteins (Banning et al., 2010) containing the indicated mutations A10N, A14N, A18N or RD. 24 h post transfection, cells were fixed, permeabilized and stained for CD155. Confirming the results shown in Figure 2A, the A10N, A14N, A18N and RD mutants do not influence the surface expression of CD155 and do not induce intracellular accumulation of CD155 (Fig. 4B). In contrast, NL4-3 Vpu and Nef colocalize with CD155 in the perinuclear compartment (Fig. 4A). Moreover, it was previously shown that HIV-1 requires Vpu and Nef for optimal downregulation of cell surface CD155 (Matusali et al., 2012). To investigate the individual impact of NL4-3 Nef and Vpu on the subcellular localization of CD155 in the context of whole viral genome expression, HeLa cells were transfected with HIV-1 proviral clones harboring single or combined inactivating mutations in vpu and nef (Wildum et al., 2006). 24 h post transfection, cells were fixed, permeabilized and stained for CD155 using anti-CD155 specific antibody. This experiment revealed in NL4-3 HIV-1 wt expressing cells that CD155 is significantly entrapped in perinuclear compartments (Fig. 5A, B). However, in the absence of either Vpu or Nef, CD155 only partially localizes in perinuclear compartments. In contrast, in the absence of Vpu and Nef, CD155 trafficking to the cell surface is not affected (Fig. 5A, B).
Fig. 4.
Sub-cellular distribution of CD155 in presence of NL4-3 wt Nef, wt Vpu or mutants. (A) HeLa cells were transfected with either YFP, wt Nef, wt Vpu-, RD-, A10N, A14N or A18N-YFP fusion proteins (green). 24 h post transfection, cells were fixed, permeabilized and stained for CD155 (red) using anti-CD155 specific antibody. (B) Values for the ratio of CD155 levels at the plasma membrane (PM)/perinuclear area of 10 analyzed cells were plotted and assessed for statistical significance by using the GrapPad Prism V5.0 software package (***; p≤0.0002, 1 way-ANOVA with Dunnett’s test). Scale bars: 4 μm.
Fig. 5.
NL4-3 HIV-1 wt induces an accumulation of CD155 in perinuclear compartments. (A) HeLa cells were transfected with either pBR HIV-1 NL4-3 wt-IRES-GFP, pBR HIV-1 NL4-3 ΔNef-IRES-GFP, pBR HIV-1 NL4-3 ΔVpu-IRES-GFP or pBR HIV-1 NL4-3 ΔNefΔVpu-IRES-GFP (green). 24 h post transfection, cells were fixed, permeabilized and stained for CD155 (red) using anti-CD155 specific antibody. (B) Values for the ratio of CD155 levels at the plasma membrane (PM)/perinuclear area of 10 analyzed cells were plotted and assessed for statistical significance by using the GrapPad Prism V5.0 software package (***; p≤0.0002, 1 way- ANOVA with Dunnett’s test). Scale bars: 4 μm.
Overall, these data show that HIV-1 Vpu and Nef are both required to alter the subcellular localization of CD155 in perinuclear compartments and thus interfere with the efficient transport of CD155 to the cell surface. Furthermore, perinuclear accumulation of CD155 depends on the TM domain, particularly A10N, A14N and A18N, of Vpu.
Lentiviral Vpu and Nef proteins differentially affect the surface expression of CD155
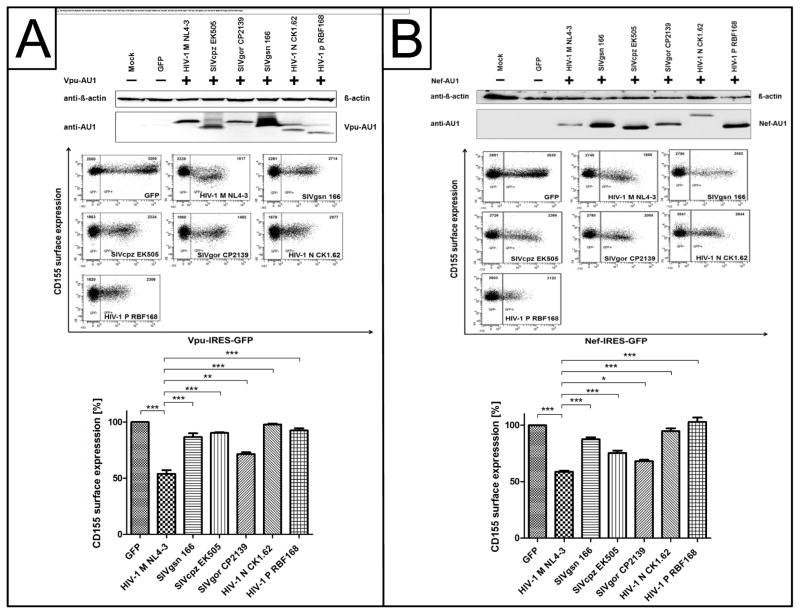
It is currently unknown whether Vpu and Nef mediated downregulation of CD155 is conserved among different lentiviral Vpu and Nef variants. Therefore, HeLa cells were transfected with expression plasmids for AU1-tagged Vpu and Nef derived from HIV-1 M NL4-3, SIVgsn 166, SIVcpz EK505, SIVgor CP2139, HIV-1 N CK1.62 or HIV-1 P RBF168. 24h post transfection, cells were harvested and stained for surface CD155 and analyzed by flow cytometry. In addition, cells were lysed and analyzed by Western blot. In Figure 6A it is shown that Vpu proteins from HIV-1 M NL4-3 and partially SIVgor CP2139 are capable of inducing downregulation of cell surface CD155. In contrast, the Vpu proteins derived from SIVgsn 166, SIVcpz EK505, HIV-1 N CK1.62 and HIV-1 P RBF168 are strongly attenuated in their ability to affect the surface expression of CD155 (Fig. 6A). Although Vpu derived from HIV-1 N CK1.62 and HIV-1 P RBF168 are expressed at slightly lower levels compared to HIV-1 M NL4-3 Vpu (Fig. 6A), it has been previously shown that they are still functional in their anti-CD4 and tetherin activity (Sauter et al., 2009). In addition, Vpu SIVcpz EK505 and SIVgsn 166 are highly expressed but have no detectable effects on cell surface CD155. Moreover, Nef derived from HIV-1 M NL4-3 induces downregulation of cell surface CD155 (Fig. 6B). The HIV-1 Nef precursors SIVcpz EK505 and SIVgor CP2139 exhibit an intermediate activity on the surface expression of CD155. In contrast, the Nef proteins derived from SIVgsn 166; HIV-1 N CK1.62 and HIV-1 P RBF168 are strongly attenuated in CD155 downregulation (Fig. 6B). Thus, we conclude that the Vpu and Nef mediated downregulation of cell surface CD155 is not conserved among lentiviral Vpu proteins in contrast to what is shown for CD4 and NTB-A (Bolduan et al., 2013; Sauter et al., 2009). This further indicates that only HIV-1 M evolved the capacity to evade the NK cell response with high efficiency.
Fig. 6.
Effect of lentiviral Vpu and Nef proteins on the cell surface expression of CD155. (A) HeLa cells were transfected with either AU1-tagged Vpu or Nef of HIV-1 M NL4-3, SIVgsn 166, SIVcpz EK505, SIVgor CP2139 or HIV-1 N CK1.62 HIV-1 P RBF168. 24 h post transfection, cells were harvested and stained for surface CD155 using a CD155 specific antibody and analyzed by flow cytometry. Summary of relative CD155 surface expression in presence of lentiviral Vpu (A) or Nef (B) proteins is given below. Values from three independent experiments were plotted and assessed for statistical significance by using the GrapPad Prism V5.0 software package (***; p≤0.0005, 1 way- ANOVA with Dunnett’s test). In addition, cells were lysed and analyzed by Western blot using anti-β-actin and anti-AU1 antibodies.
Discussion
In this study, we demonstrate that NL4-3 Vpu downregulates the activating NK cell receptor CD155 from the cell surface by the conserved alanine residues Ala-10, Ala-14 and Ala-18 of its TM domain to evade NK cell mediated immune response against HIV-1 infected cells. This is particularly important, because another accessory protein HIV-1 NL4-3 Vpr upregulates the activating NKG2D ligands (ULBP-1, ULBP-2) (Ward et al., 2009) and CD155 (Vassena et al., 2013) to the cell surface, altogether increasing the susceptibility of HIV-1 infected cells for lysis by NK cells.
HIV-1 evolved complex strategies to evade immune response against NK cells by modulation of cell surface molecules that can activate NK cells.. It was previously shown that NL4-3 Vpu downregulates the co-activating NK cell receptor NTB-A from the cell surface, which depends on the TM as well as the second cytoplasmic domain of Vpu, leading to a decreased NK-cell mediated degranulation and cytotoxicity (Bolduan et al., 2013; Shah et al., 2010). Moreover, NL4-3 Nef interferes with the expression of the ligand for the NKp44-activating NK-cell receptor by intracellular retention in HIV-1 infected cells (Fausther-Bovendo et al., 2009). In addition, Nef downregulates HLA-A and –B to evade CTL lysis but not HLA-C and –E, which protect cells from killing by NKs (Cohen et al., 1999). Furthermore, HIV-1 Nef is capable to downregulate the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity (Cerboni et al., 2007; Norman et al., 2011).
Thus, NL4-3 Vpu and Nef are necessary to prevent NK-cell mediated response of HIV-1 infected cells and it has been demonstrated that these two viral proteins are also required for optimal downregulation of cell surface CD155 (Matusali et al., 2012). Interestingly, it was shown that HIV-2, which lacks the vpu gene, is not capable to affect the surface expression of CD155, suggesting an evolutionary pressure on HIV-1 to keep the anti-CD155 activity (Matusali et al., 2012). In contrast to Vpu, the molecular mechanism of the Nef mediated downmodulation of CD155 is better defined. Nef uses the motifs DD175AA or LL165AA to downregulate CD4 molecules from the cell surface, which is distinct from the Nef mediated downregulation of cell surface CD155 (Matusali et al., 2012). However, the domains necessary for CD155 antagonism by Vpu are currently completely unknown as well as the capacity of divergent HIV-1 and SIV Nef and Vpu proteins to downregulate CD155.
We demonstrate that the CK-2 phosphorylation sites and the second cytoplasmic α-helix have no influence on the surface expression of CD155 (Fig. 2A and Table 1), suggesting that the effect of CD155 by Vpu is an independent function compared to CD4, NTB-A and tetherin. Matusali et al., 2012, suggest that the YFP fusion protein NL4-3 Vpu m2/6, containing point mutations of Ser-52 and Ser-56 to Asn in the cytoplasmic domain of Vpu, is attenuated to downregulate cell surface CD155. We could confirm that the Vpu m2/6-YFP fusion protein is slightly attenuated compared to wt Vpu (Fig. S1), but the pCG-Vpu m2/6 mutant exhibits the same anti-CD155 activity as NL4-3 wt Vpu (Fig. 2A). Thus, most likely the large YFP-tag somehow attenuates the anti-CD155 activity of the Vpu m2/6 variant.
Table 1.
Summary of anti-human Tetherin, CD4, NTB-A and CD155 activity of NL4-3 wt Vpu and TM or cytoplasmic mutants (Banning et al., 2010; Bolduan et al., 2013; Bolduan et al., 2011; Dube et al., 2010; Dube et al., 2009; Matusali et al., 2012; Neil et al., 2008; Shah et al., 2010; Skasko et al., 2011; Skasko et al., 2012; Van Damme et al., 2008; Vigan and Neil, 2010, 2011).
| Tetherin | CD4 | NTB-A | CD155 | |
|---|---|---|---|---|
| wt Vpu | + | + | + | + |
| Vpu-KKDQ | − | n.d. | − | + |
| Vpu RD | − | + | − | − |
| Vpu A7N | + | n.d. | n.d. | + |
| Vpu A10N | n.d. | n.d. | n.d. | − |
| Vpu A14N | − | n.d. | n.d. | − |
| Vpu A18N | − | + | n.d. | − |
| Vpu A18H | − | n.d. | − | − |
| Vpu m2/6 | +/− | − | + | + |
| Vpu Δ23 | − | − | − | + |
In addition, our data reveal that the Vpu-KKDQ mutant, which is trapped in the ER/cis-Golgi compartment (Shikano and Li, 2003; Vigan and Neil, 2011), is capable of inducing downregulation of cell surface CD155 (Table 1), indicating that Vpu might inhibit trafficking of CD155 to the cell surface. A previous study demonstrates the TM domain of NL4-3 Vpu is critical to affect the surface expression of CD155 (Matusali et al., 2012). We extend these findings and show that the conserved alanine residues at positions 10, 14 and 18 in the TM domain of NL4-3 Vpu are critical for the downregulation of cell surface CD155 (Fig. 2A and Table 1). This stretch of conserved residues localizes on one face of the α-helix and is required for the interaction and consequently downregulation of cell surface tetherin (Skasko et al., 2011; Vigan and Neil, 2010). In contrast, we could not detect an interaction between NL4-3 Vpu and CD155 via co-immunoprecipitation or FACS-FRET analysis (data not shown). Moreover, we could clearly show by intracellular CD155 staining that Vpu has no influence on the steady state levels of CD155 (Fig. 2B). Thus, the exact role of Ala-10, -14 and -18 to counteract CD155 is still elusive thus far.
NL4-3 wt Vpu and Nef, which affect the surface expression of CD155 (Matusali et al., 2012), induce an accumulation of CD155 in perinuclear compartments (Fig. 4), indicating that Vpu might interfere with the correct transport of CD155 to the cell surface, which agrees with the activity of the Vpu-KKDQ mutant. Consequently, the Vpu mutants RD, A10N, A14N and A18N are inactive to downregulate cell surface CD155 and do not influence its subcellular localization. Since HIV-1 requires Vpu and Nef for optimal downregulation of cell surface CD155 (Matusali et al., 2012), we investigated the individual impact of these proteins on CD155 localization. Our findings reveal that in cells expressing infectious HIV-1 both Vpu and Nef are necessary to interfere with CD155 trafficking and induce perinuclear CD155 accumulation (Fig. 5, B).
Of note, when analyzing a panel of diverse Vpu and Nef variants, isolated from different lentiviral HIV-1 and SIV strains, only NL4-3 HIV-1 M was able to downregulate CD155 (Fig. 6 and Table 2). This finding is in contrast to what is shown for Nef and Vpu mediated modulation of CD4 (Sauter et al., 2009) and Vpu’s activity against NTB-A (Bolduan et al., 2013; Shah et al., 2010) as summarized in Table 2. However, we can formally not exclude the possibility that SIV derived Vpu and Nef proteins are active against their host species derived CD155 proteins as it is the case for viral tetherin antagonism (Sauter et al., 2009). Apart from that, Vpu and Nef from HIV-1 N and P were also inactive against CD155. These are just two isolates and conclusions have to be drawn with high caution. Nevertheless, it is tempting to speculate that only HIV-1 M – the pandemic AIDS virus – has evolved the full capacity to evade NK cell response. Thus, NK cells could critically contribute to HIV-1 control, and viral countermeasures to evade NK cell response might be a determinant of HIV-1 pathogenicity. It will be highly interesting to challenge these hypotheses in future studies.
Table 2.
Summary of anti-human Tetherin, CD4, NTB-A and CD155 activity of Vpu proteins derived from HIV-1 M NL4-3, SIVgsn 166, SIVcpz EK505, SIVgor CP2139 or HIV-1 N CK1.62 HIV-1 P RBF168 isolates (Bolduan et al., 2013; Sauter et al., 2011; Sauter et al., 2009).
| Vpu alleles | Tetherin | CD4 | NTB-A | CD155 |
|---|---|---|---|---|
| HIV-1 M NL4-3 | + | + | + | + |
| SIVgsn 166 | + | + | + | − |
| SIVcpz EK505 | − | + | + | − |
| SIVgor CP2139 | − | + | + | +/− |
| HIV-1 N CK1.62 | + | − | − | − |
| HIV-1 P RBF168 | − | + | n.d. | − |
Conclusion
Taken together, we demonstrate that NL4-3 Vpu downregulates cell surface CD155 by the alanine residues 10, 14 and 18 of its TM domain and induces an accumulation of CD155 in perinuclear compartments. Moreover, only Vpu and Nef derived from HIV-1 M NL4-3 exhibit the ability to downregulate cell surface CD155, suggesting that this phenomenon is not conserved among lentiviral Vpu and Nef proteins of HIV-1 and SIV. HIV-1 interference with CD155 expression in particular and the evasion from the hosts NK cell response in general seem to be important for the full pathogenic potential of HIV-1 M.
Material and methods
Site-directed mutagenesis and plasmid constructions
Mutations of the HIV-1 M NL4-3 Vpu A7N and HIV-1 M NL4-3 Vpu A10N plasmids were introduced by site-directed mutagenesis (Quick change Kit, Stratagene) using oligonucleotides that contain the indicated changes. For cloning of the pCG-HIV-1 M NL4-3 Vpu A7N and pCG-HIV-1 M NL4-3 Vpu A10N mutants, the mutations were introduced by PCR into the previously described pCG-IRES-GFP vector, which expresses GFP from a bicistronic mRNA (Sauter et al., 2009; Schindler et al., 2003). The pCG-HIV-1 M NL4-3 wt Vpu-AU1-IRES-GFP, pCG-HIV-1 M NL4-3 Vpu RD-AU1-IRES-GFP, pCG-HIV-1 M NL4-3 Vpu Δ23-AU1-IRES-GFP, pCG-HIV-1 M NL4-3 Vpu-KKDQ-AU1-IRES-GFP, pCG-HIV-1 M NL4-3 Vpu A18N-AU1-IRES-GFP, pCG-HIV-1 M NL4-3 Vpu m2/6-IRES-GFP, pCG-nefSTOP-IRES-GFP, pCG-SIVgsn 166 Vpu-AU1-IRES-GFP, pCG-SIVcpz EK505 Vpu-AU1-IRES-GFP, pCG-SIVgor CP2139 Vpu-AU1-IRES-GFP, pCG-HIV-1 N CK1.62 Vpu-AU1-IRES-GFP, pCG-HIV-1 P RBF168 Vpu-AU1-IRES-GFP, pCG-HIV-1 M NL4-3 wt Nef-AU1-IRES-GFP, pCG-SIVgsn 166 Nef-AU1-IRES-GFP, pCG-SIVcpz EK505 Nef-AU1-IRES-GFP, pCG-SIVgor CP2139 Nef-AU1-IRES-GFP, pCG-HIV-1 N CK1.62 Nef-AU1-IRES-GFP, pCG-HIV-1 P RBF168 Nef-AU1-IRES-GFP, pBR NL4-3 wt-IRES-GFP, NL4-3 ΔNef-IRES-GFP, NL4-3 ΔVpu-IRES-GFP and NL4-3 ΔNefΔVpu-IRES-GFP, HIV-1 NL4-3 wt, HIV-1 NL4-3 Vpu Del-1, HIV-1 NL4-3 Vpu A18H, HIV-1 NL4-3 Vpu RD plasmids were already described (Banning et al., 2010; Bolduan et al., 2013; Bolduan et al., 2011; Klimkait et al., 1990; Sauter et al., 2009; Schindler et al., 2010; Schindler et al., 2003; Wildum et al., 2006). The pEYFP NL4-3 wt Vpu, Vpu m2/6, Vpu A10N, Vpu A14N, wt NL4-3 Nef, Vpu RD and Vpu A18N plasmids, encoding a Vpu-YFP fusion protein were described elsewhere (Banning et al., 2010; Bolduan et al., 2011).
Cell culture
HeLa cells were cultivated in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat inactivated fetal calf serum (FCS) and 2 mM L-glutamine, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin. SUP-T1 cells were maintained in RPMI 1640-FBS and were fed by replacing 50% of the medium at 2-day intervals.
Western blot analysis
HeLa cells were transiently transfected using Lipofectamine 2000™ (Invitrogen) according to the manufacturer’s protocol. 24h post transfection, cells were lysed in 0.5% NP-40 lysis buffer (150 mM NaCl, 50 mMTris–HCl, 0.5% Nonidet P-40 and protease inhibitor cocktail (Roche)). Cell lysates were cleared by centrifugation at 10.000 rpm at 4 °C for 5 min. NP-40 soluble proteins were separated in 12% SDS/PAA gels according to Laemmli (Laemmli, 1970), transferred onto PVDF membranes (GE Healthcare) and probed with specific antibodies, followed by enhanced chemiluminescence detection. For internal controls, blots were stripped and re-incubated with the appropriate antibody.
HIV-1 infection of Sup-T1 T cells
For each infection, 3 × 106 Sup-T1 T cells were infected with VSV-G pseudotyped HIV-1 NL4-3Vpu Del-1, HIV-1 NL4-3 wt, HIV-1 NL4-3 Vpu RD and HIV-1 NL4-3 VpuA18H. 2 days after infection, cells were analyzed by flow cytometry. Virusstocks were prepared in 293 Tcells transfected with VSV-G pseudotyped HIV-1 NL4-3Vpu Del-1, HIV-1 NL4-3 wt, HIV-1 NL4-3 Vpu RD and HIV-1 NL4-3 Vpu A18H plasmid DNA. Virus-containing supernatants were centrifuged at 1500 g for 5min.
Flow cytometry analysis of Vpu mediated downregulation of CD155
HeLa cells were transfected with the different pCG-Vpu-IRES-GFP and pCG-Nef-IRES-GFP expression plasmids. 24h post transfection, the cells were harvested and stained for surface and intracellular CD155 using an anti-CD155 antibody (Bio Legend) and chicken anti-mouse Alexa 633 conjugated secondary antibody (Molecular Probes, Invitrogen). CD155 surface expression on GFP-positive cells was then analyzed in a FACS LSRII using FACSDiva software (BDBiosciences). Data analysis was performed using FACS Express V3 software (DeNovo). Sup-T1 cells were infected with VSV-G pseudotyped HIV-1 NL4-3Vpu Del-1, HIV-1 NL4-3 wt, HIV-1 NL4-3Vpu RD and HIV-1 NL4-3 VpuA18H. 2 days after infection, cells were harvested and stained for surface and intracellular CD155 using an anti-CD155 antibody (Bio Legend) and chicken anti-mouse Alexa 633 conjugated secondary antibody (Molecular Probes, Invitrogen). Afterwards, the cells were fixed in 2% PFA, permeabilized with 1 % Saponin for 10 min at RT and blocked with 10 % FCS for 20 min at RT. Subsequently, cells were stained with p24-FITC antibody (Fischer Scientific) then analyzed in a FACS LSRII.
Confocal and immunofluorescence microscopy
HeLa cells were transfected with either YFP, NL4-3 Nef, NL4-3 Vpu, A10N, A14N, A18N- or RD-YFP fusion proteins, pBR NL4-3 wt- IRES-GFP, pBR NL4-3 ΔNef-IRES-GFP, pBR NL4-3 ΔVpu-IRES-GFP or pBR NL4-3 ΔNefΔVpu-IRES-GFP. 24 h post transfection, cells were fixed in 3% formaldehyde for 20 min at RT, permeabilized with either 0,1 % Triton X-100 or 1 % Saponin for 10 min. Subsequently, cells were stained for CD155 using a 1:100 dilution of anti-CD155 specific antibody (Pierce) and 1:400 diluted goat anti-mouse Alexa Fluor 633 conjugated secondary antibody (Molecular Probes, Invitrogen). Spinning disc confocal microscopy was performed with either a Nikon TiE equipped with the PerkinElmer UltraView Vox system or Leica TCS SP5 confocal microscope. Images were analyzed with either the Adobe Photoshop software or the Volocity 6.3-software package (Perkin Elmer).
Supplementary Material
Highlights.
Downregulation of CD155 is independent of Vpu’s effects on CD4, NTB-A and tetherin.
Ala residues 10, 14 and 18 in the TM domain of Vpu are required for CD155 modulation.
Vpu induces accumulation of CD155 in perinuclear compartments.
Only HIV-1 M NL4-3 Vpu and Nef efficiently downregulate cell surface CD155.
Acknowledgments
This work was supported by grants to MS from the German Research Council (DFG; SCHI1073/2-1) and the Else Kröner-Fresenius Stiftung (2012_A264), as well as by grants to US from the German Research Council (DFG; SFB643-A1, SFB796-A1, SCHU1125/3 and SCHU1125/5-1), graduate program GRK1071 and NIH grant RO1DK81553. We thank all members of the lab for helpful discussions and critical reading of this manuscript as well as Ute Finkel, Pia Rauch and Kirsten Friedrich for their superior technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sebastian Bolduan, Email: sebastian.bolduan@helmholtz-muenchen.de.
Tatjana Reif, Email: tatjana.reif@viro.med.uni-erlangen.de.
Michael Schindler, Email: michael.schindler@helmholtz-muenchen.de.
Ulrich Schubert, Email: ulrich.Schubert@viro.med.uni-erlangen.de.
References
- Banning C, Votteler J, Hoffmann D, Koppensteiner H, Warmer M, Reimer R, Kirchhoff F, Schubert U, Hauber J, Schindler M. A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One. 2010;5:e9344. doi: 10.1371/journal.pone.0009344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binette J, Dube M, Mercier J, Halawani D, Latterich M, Cohen EA. Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology. 2007;4:75. doi: 10.1186/1742-4690-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduan S, Hubel P, Reif T, Lodermeyer V, Hohne K, Fritz JV, Sauter D, Kirchhoff F, Fackler OT, Schindler M, Schubert U. HIV-1 Vpu affects the anterograde transport and the glycosylation pattern of NTB-A. Virology. 2013;440:190–203. doi: 10.1016/j.virol.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduan S, Votteler J, Lodermeyer V, Greiner T, Koppensteiner H, Schindler M, Thiel G, Schubert U. Ion channel activity of HIV-1 Vpu is dispensable for counteraction of CD317. Virology. 2011;416:75–85. doi: 10.1016/j.virol.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Cerboni C, Neri F, Casartelli N, Zingoni A, Cosman D, Rossi P, Santoni A, Doria M. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol. 2007;88:242–250. doi: 10.1099/vir.0.82125-0. [DOI] [PubMed] [Google Scholar]
- Cohen EA, Terwilliger EF, Sodroski JG, Haseltine WA. Identification of a protein encoded by the vpu gene of HIV-1. Nature. 1988;334:532–534. doi: 10.1038/334532a0. [DOI] [PubMed] [Google Scholar]
- Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- Dube M, Roy BB, Guiot-Guillain P, Binette J, Mercier J, Chiasson A, Cohen EA. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 2010;6:e1000856. doi: 10.1371/journal.ppat.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, Cohen EA. Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. Journal of virology. 2009;83:4574–4590. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausther-Bovendo H, Sol-Foulon N, Candotti D, Agut H, Schwartz O, Debre P, Vieillard V. HIV escape from natural killer cytotoxicity: nef inhibits NKp44L expression on CD4+ T cells. AIDS. 2009;23:1077–1087. doi: 10.1097/QAD.0b013e32832cb26b. [DOI] [PubMed] [Google Scholar]
- Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, Tokunaga K. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem. 2009;284:35060–35072. doi: 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. Journal of virology. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Chen MY, Willey RL, Strebel K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. Journal of virology. 1993;67:5056–5061. doi: 10.1128/jvi.67.8.5056-5061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. Journal of virology. 2012;86:4496–4504. doi: 10.1128/JVI.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, Shen W, Collins KL. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat Immunol. 2011;12:975–983. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak E, Gomez ML, Gomez LM, Mulcahy ER, Jackson M, Hout DR, Wisdom BJ, Stephens EB. Identification of a region within the cytoplasmic domain of the subtype B Vpu protein of human immunodeficiency virus type 1 (HIV-1) that is responsible for retention in the golgi complex and its absence in the Vpu protein from a subtype C HIV-1. AIDS Res Hum Retroviruses. 2005;21:379–394. doi: 10.1089/aid.2005.21.379. [DOI] [PubMed] [Google Scholar]
- Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, Guo F, Liu SL, Wainberg MA, Liang C. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. Journal of virology. 2009;83:7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Hue S, Petit SJ, Plantier JC, Towers GJ, Kirchhoff F, Gupta RK. HIV-1 Group P is unable to antagonize human tetherin by Vpu, Env or Nef. Retrovirology. 2011;8:103. doi: 10.1186/1742-4690-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Rajan D, Banning C, Wimmer P, Koppensteiner H, Iwanski A, Specht A, Sauter D, Dobner T, Kirchhoff F. Vpu serine 52 dependent counteraction of tetherin is required for HIV-1 replication in macrophages, but not in ex vivo human lymphoid tissue. Retrovirology. 2010;7:1. doi: 10.1186/1742-4690-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Wurfl S, Benaroch P, Greenough TC, Daniels R, Easterbrook P, Brenner M, Munch J, Kirchhoff F. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. Journal of virology. 2003;77:10548–10556. doi: 10.1128/JVI.77.19.10548-10556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Fritz JV, Bitzegeio J, Fackler OT, Keppler OT. HIV-1 Vpu blocks recycling and biosynthetic transport of the intrinsic immunity factor CD317/tetherin to overcome the virion release restriction. MBio. 2011;2:e00036–00011. doi: 10.1128/mBio.00036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Bacik I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. Journal of virology. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, Barker E. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe. 2010;8:397–409. doi: 10.1016/j.chom.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, Kitamura T, Nicholl J, Sutherland GR, Lanier LL, Phillips JH. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4:573–581. doi: 10.1016/s1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- Shikano S, Li M. Membrane receptor trafficking: evidence of proximal and distal zones conferred by two independent endoplasmic reticulum localization signals. Proc Natl Acad Sci U S A. 2003;100:5783–5788. doi: 10.1073/pnas.1031748100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skasko M, Tokarev A, Chen CC, Fischer WB, Pillai SK, Guatelli J. BST-2 is rapidly down-regulated from the cell surface by the HIV-1 protein Vpu: evidence for a post-ER mechanism of Vpu-action. Virology. 2011;411:65–77. doi: 10.1016/j.virol.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skasko M, Wang Y, Tian Y, Tokarev A, Munguia J, Ruiz A, Stephens EB, Opella SJ, Guatelli J. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem. 2012;287:58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Klimkait T, Martin MA. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell host & microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena L, Giuliani E, Matusali G, Cohen EA, Doria M. The human immunodeficiency virus type 1 Vpr protein upregulates PVR via activation of the ATR-mediated DNA damage response pathway. J Gen Virol. 2013;94:2664–2669. doi: 10.1099/vir.0.055541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigan R, Neil SJ. Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. Journal of virology. 2010;84:12958–12970. doi: 10.1128/JVI.01699-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigan R, Neil SJ. Separable determinants of subcellular localization and interaction account for the inability of group O HIV-1 Vpu to counteract tetherin. Journal of virology. 2011;85:9737–9748. doi: 10.1128/JVI.00479-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J, Davis Z, DeHart J, Zimmerman E, Bosque A, Brunetta E, Mavilio D, Planelles V, Barker E. HIV-1 Vpr triggers natural killer cell-mediated lysis of infected cells through activation of the ATR-mediated DNA damage response. PLoS Pathog. 2009;5:e1000613. doi: 10.1371/journal.ppat.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildum S, Schindler M, Munch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. Journal of virology. 2006;80:8047–8059. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. Journal of virology. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.