Abstract
Light and transmission electron microscopy observations are reported on the structure and in vitro cytopathic effect of Acanthamoeba griffini trophozoites isolated from a clinical case. Live trophozoites were moderately active with a remarkable pleomorphism which changed from ovoid to quite elongated shapes. When moving, amoebae formed cytoplasmic projections such as wide lamellae and acanthopodia of diverse size and thickness which contain a significant amount of actin. Ultrastructurally, the cytoplasm showed the main organelles found in other free-living amoebae. Coincubation of trophozoites with MDCK cell monolayers resulted in a local damage to target cells after 24 h of interaction, suggesting that the cytopathic effect is contact-dependent. By transmission electron microscopy, amoebae appeared to engulf small portions of the MDCK cells; however, the cells that were not in contact with trophozoites had an unaltered morphology. When epithelial monolayers were incubated with conditioned medium for 24 h, small areas of cell injury were also observed. The phylogenetical analysis as well as the sequencing of the acquired amplified product for the DF3 region of the amoebae isolate confirmed that it belongs to genotype T3, which includes other pathogenic amoebae; besides the activity of two drugs currently used against Acanthamoeba was tested on A. griffini.
1. Introduction
Free-living amoebae from the genus Acanthamoeba are commonly found in soil and aquatic environments worldwide [1–3]. These amoebae have been isolated from very diverse habitats, including water from the Antarctic [4], bottled water [5], swimming pools [6], dental units [7], eye wash stations [8], and even from dust in the atmosphere [9].
Acanthamoeba spp. are the most common and opportunistic amphizoic protozoa. Acanthamoeba castellanii is one of the etiological agents of chronic granulomatous amebic encephalitis [10] and amoebic keratitis, a progressive and painful sightthreatening eye infection [11–13]. The first description of Acanthamoeba griffini was done by Sawyer [14], but reports on this amoeba on the scientific literature are very limited. Molecular analyses performed in 2003 [15, 16] concluded that this amoeba belongs to the T3 genotype, which is clinically relevant since other pathogenic amoebae are also included in this cluster [17].
By means of light and transmission electron microscopy we present some observations on the morphology of this amoeba isolated from a case of keratitis as well as its cytopathic effect on MDCK epithelial cell monolayers.
2. Material and Methods
2.1. Amoebae
Amoebae were isolated in October 2013 from a severe case of keratitis from both a contact lens and a corneal scrape (Association to prevent Blindness in Mexico, Luis Sánchez Bulnes Hospital, Mexico City, Mexico). After axenization, it was cryopreserved immediately.
2.2. Isolation and Maintenance of Acanthamoeba sp. in Monoxenic Cultures
The technique used for the recovery and maintenance of Acanthamoeba sp. from clinical and environmental sources has been described elsewhere [18, 19]. Briefly, primary isolation was performed by using 1.5% nonnutrient agar plates seeded with heat-killed Enterobacter aerogenes. Clinical samples were then streaked on the agar. Subsequent incubation was performed at room temperature (22–24°C) for 10 days. Upon evidence of growth, cultures were established by transference of a single double-walled cyst to fresh agar medium.
2.3. Axenic Cultures
Monoxenic cultures were selected from areas of profuse amoebic growth. Selected pieces of agar were transferred to axenic culture media such as phosphate-biotriptase-serum glucose medium (PBSGM) and 2% Bacto Casitone (pancreatic digest of casein, Becton-Dickinson, Sparks, MD) medium (DIFCO), which are culture media widely used for growth and amoebic development [9, 20]. Both media were supplemented with 10% fetal bovine serum (Equitech-bio, Kerville, Tex USA) and 1% antibiotics (penicillin, 100 mg/mL; streptomycin, 10 mg/mL). Trophozoites were incubated in both media at 30°C in borosilicate tubes (Pyrex, Mexico), and the medium was changed twice daily for two days and thereafter once daily for three more days. The cultures were considered axenic if no bacterial growth was observed.
Axenized trophozoites were grown and maintained in axenic culture in 2% Bacto Casitone supplemented with 10% fetal bovine serum (Equitech-bio, Kerville, Tex USA) and 1% antibiotics. Cultures were incubated at 30°C and trophozoites were harvested at the end of the logarithmic phase of growth.
2.4. Temperature Tolerance Test
To determine the optimal culture medium and temperature for growth, amoebae were incubated at 25, 30, and 37°C in borosilicate tubes (Pyrex, Mexico). Optimal growth was determined by plotting logarithmic growth phase curves (assays were performed in triplicate). The viability of the trophozoites was determined by trypan blue (0.4%) exclusion.
2.5. Pathogenicity Test
In order to induce granulomatous amoebic encephalitis as a method to evaluate the virulence of the amoebae isolate, intranasal instillation was used in a mouse model of infection. Briefly, axenic cultures of A. griffini trophozoites in the exponential phase of growth (72 h) were chilled at 4°C and concentrated by centrifugation for 5 min at 2500 rpm. 2 × 105 trophozoites were resuspended in 200 μL of fresh culture medium or isotonic solution and instillated into the nostrils of 5 male Balb/C mice [21]. A group of five mice was inoculated with culture medium without amoebae and used as controls. Mice were sacrificed 21 days after amoebae inoculation. The brain, liver, lungs, and kidneys from sacrificed animals were cultured on agar plates with nonnutritive enriched medium (NNE) to retrieve the amoebae. Experimental animals were maintained in optimal conditions according to international standards that regulate the care and management of experimental animals.
2.6. Drugs Activity Assays
The activity of two drugs currently used against Acanthamoeba was tested on Acanthamoeba griffini. Chlorhexidine (Chlorhexidine digluconate, Alfa Aesar, Germany) is a standard antiseptic belonging to the biguanides family which are commonly used in contact lens maintenance solutions; and voriconazole (Sigma, Madrid, Spain) an inhibitor of ergosterol synthesis that has been proven previously to be highly effective against clinical strains of Acanthamoeba [22, 23]. For the sensitivity and activity assays, a type strain from the American Type Culture Collection (ATCC), Acanthamoeba castellanii Neff ATCC 30010, genotype T4 was used as a control.
For the activity assays a previously developed colorimetric 96-well microtiter plate assay, based on the oxide-reduction of Alamar Blue assay [24], was used for the determination of drug efficacy against the trophozoites of the selected Acanthamoeba strains. Subsequently the plates were analyzed, over a period from 72 to 120 h, on a Microplate Reader Model 680 (Biorad, Hercules, CA) using a test wavelength of 570 nm and a reference wavelength of 630 nm. For those strains that were sensitive to the assayed drugs, the percentage of inhibition and 50% inhibitory concentrations (IC50) were calculated by linear regression analysis using a 95% confidence limit. All experiments were performed three times each in duplicate and the mean values were also calculated. A paired two-tailed t-test was used for analysis of the data. Values of P < 0.05 were considered significant. The inhibition curves of the statistical analysis were developed using the Sigma Plot 12.0 software programme (Systat Software Inc.).
2.7. Culture of MDCK Cells
Monolayers of epithelial cells of the established MDCK line of canine kidney origin (Madin Darby Canine Kidney) were grown on 25 cm2 cell culture flasks (Corning Incorporated, NY) in Dulbecco's modified Eagle's medium (Gibco, Grand Islands, NY) supplemented with 10% fetal bovine serum (Equitech-bio, Kerville, Tex USA) and antibiotics in a 5% CO2 atmosphere at 37°C.
2.8. Coincubation of Trophozoites with MDCK Cells
MDCK cell monolayers were trypsinized and grown in round plastic cover slips placed in 24 well styrene plates. Cultures were maintained at 37°C in a 5% CO2 atmosphere, and 24 h later confluent monolayers were obtained. Trophozoites were added in a 1 : 2 target cell : amoeba ratio in a mixture of equal proportions of Bacto Casitone and Dulbecco's modified Eagle's medium (Gibco BRL). Incubations were carried out for different times (6, 12, 16, and 24 h) under the same conditions.
2.9. Incubation of MDCK Monolayers with Conditioned Medium
The conditioned medium was obtained as follows: 6 × 106 trophozoites from a culture in the exponential phase of growth were placed in culture flasks containing 7 mL of fresh Bacto Casitone-DMEM serum-free medium (1 : 1) and incubated at 30°C for 24 h. Trophozoites were chilled on ice for 10 min and centrifuged for 5 min at 2500 rpm. The supernatant was removed, centrifuged, and filtered through a 0.22 μm membrane (Millipore, Bedford, Massachusetts). MDCK cell monolayers were incubated for 24 h with a mixture of conditioned medium and Dulbecco's modified Eagle's medium (Gibco BRL) in equal proportions.
2.10. Confocal Microscopy
Amoebae cultured for 72 h were chilled in an ice-water mixture for 5 min, pelleted by centrifugation, and fixed with 4% paraformaldehyde for 1 h. Samples were then washed with Dulbecco's Phosphate Buffered Saline (DPBS) and blocked for 1 h with 10% fetal bovine serum diluted in DPBS. Afterwards, the cells were washed with DPBS, treated with a 1 : 25 solution of phalloidin-tagged rhodamine complex (Molecular Probes, Eugene, OR, USA) for 20 min at 37°C, and washed exhaustively with DPBS. Samples were mounted with Vectashield (Vector Laboratories Inc., Burlingame, CA, USA) and observed in a LS 700 Laser Scanning Microscope (Carl Zeiss GmbH, Germany).
2.11. Light Microscopy
Observations of live and fixed trophozoites and their interaction with MDCK monolayers were performed using a Zeiss Axiophot photomicroscope equipped with an AxioCam MRc digital camera (Carl Zeiss GmbH, Germany).
2.12. Electron Microscopy
2.12.1. Transmission Electron Microscopy
Samples were fixed at room temperature with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, postfixed with 1% osmium tetroxide in the same buffer, dehydrated in increasing concentrations of ethanol, and embedded in epoxy resins. Thin sections were observed in a JEOL JEM-1011 transmission electron microscope (JEOL Ltd. Tokyo, Japan).
2.12.2. Scanning Electron Microscopy
Samples were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.2, dehydrated with increasing concentrations of ethanol, critical-point dried (31°C and 1100 psi) using a Samdri apparatus (Tousimis Corp., Rockville. MD), and coated with gold particles in an ion sputtering device (JEOL JFC-1100). Samples were then examined with a JEOL-JSM 7100 F scanning electron microscope (JEOL Ltd. Tokyo Japan).
2.13. DNA Extraction and Genotyping of Isolates
Axenic Isolates. Fungi and bacteria-free plates were transferred to axenic culture by placing the amoebae in PYG medium as previously described for further morphological and molecular analyses [17, 25, 26]. Amoebae were grown exponentially (106 cells/mL) for DNA extraction and activity assays.
DNA from amoebic cultures was extracted by placing 1-2 mL of axenic Acanthamoeba cultures directly into the Maxwell 16 Tissue DNA Purification Kit sample cartridge. Acanthamoeba genomic DNA was purified using the Maxwell 16 Instrument as described in the Maxwell 16 DNA Purification Kits Technical Manual number TM284. DNA yield and purity were determined using the NanoDrop spectrophotometer.
rRNA gene amplifications (DF3 region) were also performed as previously described [26, 27]. PCR products were purified using the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced using a MEGABACE 1000 automatic sequencer (Healthcare Biosciences, Barcelona, Spain) in the University of La Laguna Sequencing Services (Servicio de Secuenciación SEGAI, University of La Laguna). The obtained sequences were aligned using Mega 5.0 software program [28]. Phylogenetic analyses were carried out using maximum parsimony, minimum evolution, and maximum likelihood optimality criteria, implemented in Mega 5.0 [28]. Transition : transversion ratios were estimated by maximum likelihood heuristic searches. Estimates of node support were obtained by performing 1000 bootstrap replicates. Genotype identification was based on sequence analysis of DF3 region as previously described by comparison with the available Acanthamoeba DNA sequences in Genbank database [26, 29]. Acanthamoeba castellanii Neff ATCC 30010 DNA was used as a positive control in the PCR reactions. Diagnostic Fragment 3 sequence for the new isolate is deposited in the Genbank database under the accession number: KF914142.
3. Results
3.1. Light Microscopy
Live trophozoites (averaging 25–35 μm) were extremely pleomorphic, moderately mobile and during displacement they exhibited both large lamellae and cytoplasmic acantopodia of varying size and shape. Acantopodia were composed of hyaline cytoplasm of smooth appearance, and many cellular projections were bifurcated (Figures 1(a) and 1(b)). A large round to oval nucleus with a prominent and centrally located nucleolus composed of condensed dark chromatin (Figures 1(c) and 1(d)) as well as abundant vacuoles of different size and content was easily observed (Figure 1(d)).
Figure 1.
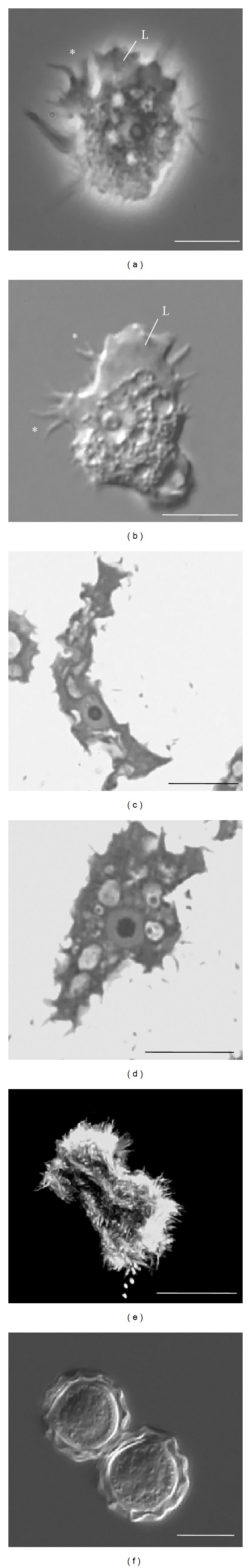
Light and confocal microscopy of Acanthamoeba griffini trophozoites. Phase contrast (a) and differential interference-contrast (b) microscopy of a live A. griffini trophozoite exhibiting an irregular morphology. A large lamellipodium (L) of smooth appearance as well as several thin and slender acanthopodia that are frequently bifurcated ( *) are seen. (c) and (d) Toluidine blue stained thick resin sections of trophozoites. The morphology of the trophozoites is very diverse, ranging from slender to oval. In both amoebae a single large nucleus along with a round dark nucleolus and numerous vacuoles with diverse content are observed. (e) A trophozoite observed by confocal microscopy after treatment with phalloidin-rhodamine complex exhibiting a positive strong reaction to actin. This protein is located in cytoplasmic projections but it is also detected all around the cell body. (f) Differential interference-contrast image of A. griffini cysts with showing clearly defined thick wavy layers located near the rounded endocyst.
The cyst of A. griffini had an average diameter of 14 μm and showed the morphological features that distinguish the amoebae belonging to group II [30]. The ectocyst consisted of a thick wavy layer located near the more or less rounded endocyst. Both layers were found close together around most of the circumference of the cyst and small pores were observed on the cyst surface (Figure 1(f)).
3.2. Confocal Microscopy
Amoebae treated with a phalloidin-rhodamine complex showed a strong reaction to actin, one of the most important structural proteins of cells. Actin was present abundantly in the cytoplasmic projections, although it was also distributed all around the cell body (Figure 1(e)).
3.3. Electron Microscopy
3.3.1. Transmission Electron Microscopy
As observed by light microscopy, trophozoites seen at low magnification by transmission electron microscopy exhibited an amazing pleomorphism. The cell outline was extremely irregular, ranging from thin and slender acanthopodia to broad lamellae. The cytoplasm presented areas of granular appearance as well as abundant vacuoles with different content most of which corresponded to digestive vacuoles. Occasionally a water expulsion vacuole was observed (Figure 2(a)). The cell membrane was composed of the characteristic trilaminar structure and was approximately 8.0 nm thick (Figure 2(a) insert). This amoeba had a large round to oval nucleus with a well-defined centrally located nucleolus of compact appearance (Figure 2(a)). At higher magnification, the nuclear envelope was clearly visible, composed of a double membrane enclosing a narrow perinuclear space of approximately 0.03 μm. Numerous ribosomes were present on the external side of the envelope (Figure 2(b)). Cytoplasmic organelles such as the rough endoplasmic reticulum and mitochondria were a regular finding (Figure 2(c)). The rough endoplasmic reticulum was clearly defined, usually in close relation with mitochondria and was observed as short tubular segments with arrays of ribosomes on its surface; these structures were frequently located near the edge of the cell. Mitochondria were limited by a double membrane and had an oval to round outline with a dense homogeneous matrix of granular aspect, and some of them presented electron-dense granules. The cristae were observed as tubules of irregular shapes, occasionally branched. In the inner side of the tubules a granular content was present. The Golgi system comprised a roughly saucer-shaped stack of smooth flattened membranous sacs. Some membrane-bound vesicles were observed in the vicinity of the concave side of this system (Figure 2(d)).
Figure 2.
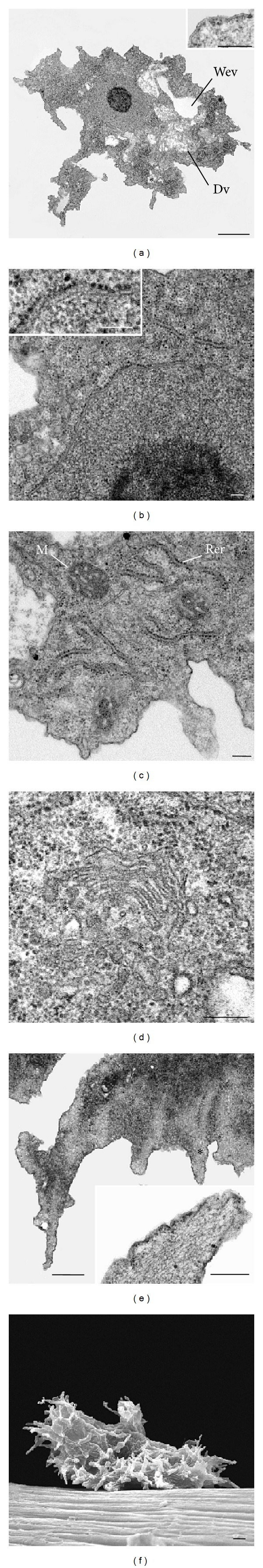
Transmission electron microscopy ((a)–(e)) and scanning electron microscopy (f) of A. griffini trophozoites. (a) Low magnification of an amoeba which shows an extremely irregular profile and a large clearly defined granular round nucleus with a circular electron-dense nucleolus. Numerous digestive vacuoles (Dv) with fibrogranular content and a water expulsion vacuole (Wev) are also observed. The plasma membrane presents its classical three-layered structure (insert). Bar = 2 μm. Insert = 0.1 μm. (b) The nuclear envelope is composed of a double membrane with abundant ribosomes on the external side of the envelope. Bar = 0.2 μm. Insert = 0.1 μm. (c) Mitochondria (M) and numerous profiles of rough endoplasmic reticulum (Rer) located near the edge of the cell were clearly defined. Bar = 0.2 μm. (d) Golgi system with various stacked cisternae. Some membrane-bound vesicles were present in the concave area (asterisks). Bar = 0.2 μm. (e) Fibrogranular matrix located in the periphery of a trophozoite. The asterisk indicates a similar region observed at high magnification where abundant actin filaments are observed. Bar = 1.0 μm. Insert = 0.2 μm. (f) A trophozoite in a lateral view is shown in a displacing position with a portion of the cell body facing upwards. The rough and extremely uneven cell surface exhibits abundant cytoplasmic projections, mostly in the manner of filopodia. Bar = 1.0 μm.
When the trophozoites were tagged with a phalloidin-rhodamine complex, a profuse reaction with actin was found in the areas located around the cells profile, particularly in those domains involving cell movement. Figure 2(e) illustrates a low magnification thin section corresponding to the edge of one trophozoite where the cytoplasm lacks organelles and is constituted by a fibrogranular matrix. At high magnification it was possible to observe abundant actin filaments (Figure 2(e) insert).
3.3.2. Scanning Electron Microscopy
A. griffini trophozoites observed by scanning electron microscopy presented a quite uneven rough surface and plenty of cell surface projections such as filopodia of diverse length; sometimes some of them were bifurcated. A common finding was the presence of flat and smooth lamellae, and occasionally amebostomes were also found (Figure 2(f)).
3.4. Temperature Tolerance Test
Analysis of growth curves revealed that A. griffini grows best from 25°C to 30°C, reaching exponential phase at 72 h, with trophic population and a viability of 100% as determined by trypan blue exclusion. Amoebae incubated at 37°C did not divide. These trophozoites had a round morphology and presented large vacuoles in the cytoplasm and encysted rapidly.
3.5. Incubation of MDCK Monolayers with A. griffini Trophozoites and Conditioned Medium
Monolayers incubated for 24 h in a mixture of equal proportions of Bacto Casitone and Dulbecco's modified Eagle's medium were structurally well preserved (Figure 3(a)). After 24 h of interaction between A. griffini trophozoites and MDCK cell monolayers an evident damage to the epithelial cell monolayer was seen; there were areas of the substrate where MDCK cells were missing and only trophozoites were observed (Figure 3(b)). When epithelial monolayers were incubated with conditioned medium for 24 h, rounded areas of diverse size lacking cells were also seen (Figure 3(c)). In transversal sections observed by transmission electron microscopy, trophozoites were seen in close relation with the apical surface of the cell monolayer; some of them penetrated below the monolayer and detached the epithelial cells, ingesting portions of the cells by means of phagocytic structures of different sizes. The presence of phagocytic structures in the cytoplasm of trophozoites was a regular finding (Figures 3(d) and 3(e)).
Figure 3.
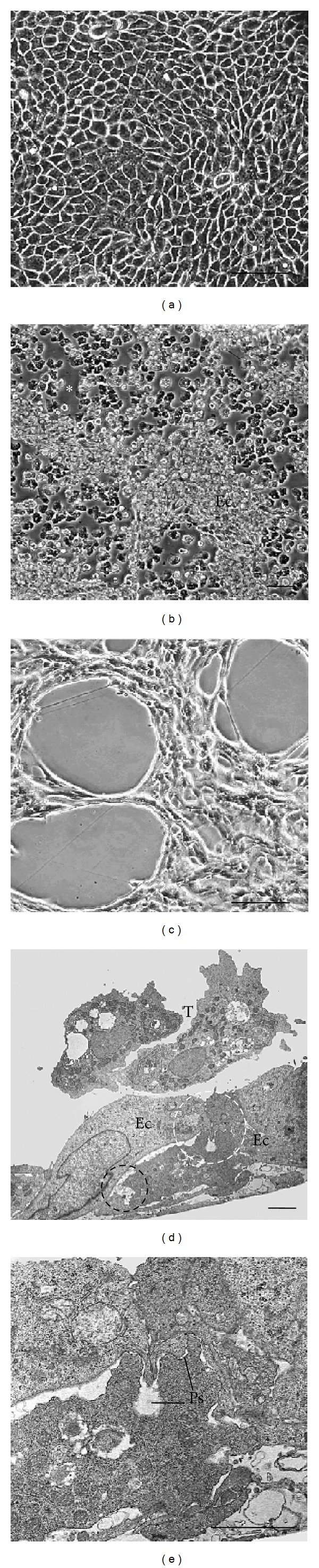
((a)–(c)) Light microscopy analysis of the interaction between A. griffini trophozoites and MDCK monolayers. (a) Control of MDCK epithelial cells monolayer incubated for 24 h in a mixture of equal proportions of Bacto Casitone and Dulbecco's modified Eagle's medium. Note the confluent appearance of the monolayer and the regular morphology of the epithelial cells. Bar = 2.0 μm. (b) After 24 h of coincubation between trophozoites and the epithelial cells, a noticeable damage is evident. Some areas of the substrate clearly lacking epithelial cells (asterisk) as well as remaining portions of the altered cell monolayer (Ec) are seen. Bar = 1.0 μm. (c) When MDCK monolayers were incubated with conditioned medium for 24 h rounded areas of varying size missing cells were seen. Bar = 2 μm. ((d),(e)) Thin sections of coincubations between amoebae and MDCK epithelial cells. (d) Trophozoites (T) were found in close contact with the surface of epithelial cells (Ec) and also under the cell layer. Epithelial cells appear undamaged; nevertheless an amoeba is initiating an injury process (white dotted circle) and is also engulfing cell debris (black dotted circle). Bar = 2 μm. (e) High magnification of the previous image where two phagocytic structures (Ps) start to engulf a portion of the epithelial cell. Bar = 2 μm.
3.6. Pathogenicity Test
Mice infected with the amoeba showed evidence of illness, as manifested by ruffled fur and aimless wandering, but they were able to recover in few days. Mice were sacrificed 21 days after inoculation. Fragments of the brain, lungs, liver, and kidney were freshly macerated and it was possible to observe only a few trophozoites in the brain and lungs. However, numerous cysts were detected in brain which may be the result of a rapid encystment.
Fragments of extracted organs were cultured at 30°C in agar plates with nonnutritive enriched medium (NNE) to recover the amoebae. Again scarce trophozoites were recovered from brain and lungs and numerous cysts were observed.
3.7. Phylogenetic Analysis
After sequencing of the obtained amplified product for the DF3 region of the new isolate and performing the phylogenetical analysis, it was concluded that the new isolate belonged to genotype T3 (Figure 4).
Figure 4.
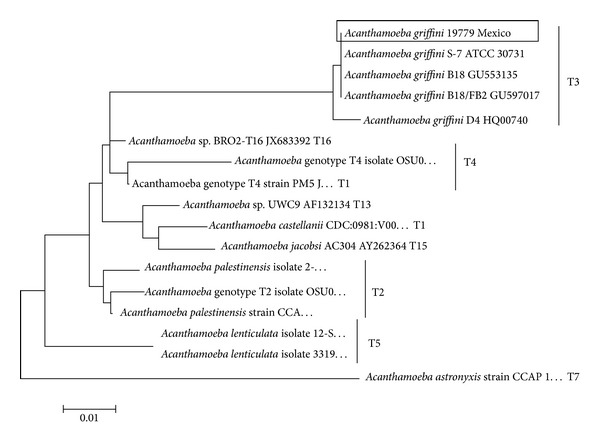
18S rRNA DF3 linearized Neighbour-Joining tree. The phylogenetic analysis was performed using the Kimura two-parameter distance algorithm in MEGA 5.0. The isolate obtained in the present study is identified in the tree (box). The type sequences were taken from GenBank and are presented under the following numbers: A. astronyxis strain CCAP 1534/1 genotype T7 Accession number AF239293, A. castellanii strain genotype T1 CDC:0981:V006 Accession number U07400, A. griffini S-7 ATCC 30731 genotype T3 Accession number U07412, A. griffini isolate B18 genotype T3 Accession number GU553135, A. griffini isolate B18/FB2 genotype T3 Accession number GU597017, A. griffini isolate D4 genotype T3 Accession number HQ00740, A. lenticulata isolate 12-SO #KC694184, A. lenticulata isolate 33195463 Accession number KC438381, Acanthamoeba sp. isolate OSU09-002 Accession number JQ669657, Acanthamoeba sp. genotype T2 Isolate OSU09-006 number JQ669661, Acanthamoeba palestinensis isolate TW-2 Accession number KC694193, A. palestinensis strain CCAP 1547-1 number AF239296, Acanthamoeba sp. isolate BRO2-T16 Accession number JX683392, Acanthamoeba sp. UWC9 genotype T13 Accession number AF132134, Acanthamoeba sp. PM5 genotype T4 Accession number JX494395, Acanthamoeba sp. genotype T4 Accession number JQ669660, and Acanthamoeba jacobsi AC304 Accession number AY262364.
3.8. Drugs Activity Assays
All A. griffini and A. castellanii Neff were sensitive to chlorhexidine and voriconazole assayed microscopically and colorimetrically. Activity assays with chlorhexidine and voriconazole and the IC50 data for each strain indicated that both compounds were able to inhibit the growth of these amoebae and in the case of voriconazole, even at low concentrations with IC90. However, in this study voriconazole activity was ten times higher than the one observed for chlorhexidine (Figure 5).
Figure 5.
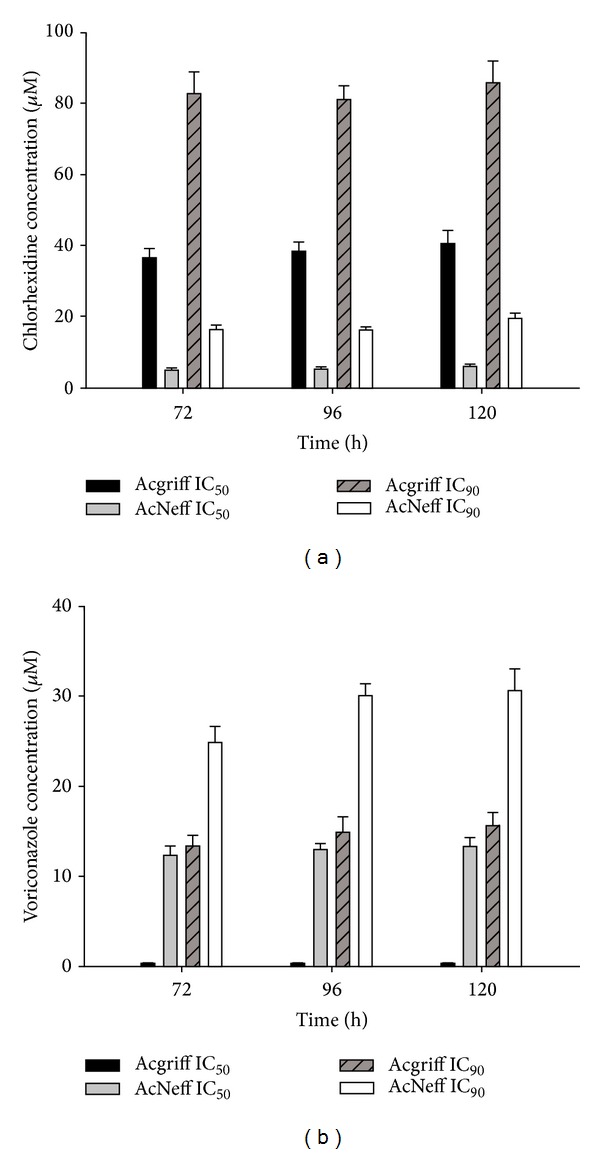
In vitro activity of chlorhexidine (a) and voriconazole (b) against the Acanthamoeba griffini strain. Acanthamoeba castellanii Neff ATCC 30010 was used as a control strain. Activities and concentrations (IC50 and IC90) of the tested compounds are shown at 72, 96, and 120 h.
4. Discussion
In contrast to other free-living amoebas, the morphology of Acanthamoeba griffini is extremely variable, ranging from rounded to elongated shapes. The cytoplasm of A. griffini trophozoites shows the main cell organelles; a large nucleus with a centrally located electron-dense nucleolus clearly defined profiles of rough endoplasmic reticulum, mitochondria, and a Golgi system as well as vacuoles with diverse content and the presence of abundant actin filaments. Actin is described as one of the most profuse and highly conserved proteins in eukaryotic cells, which is implicated in many vital cellular functions such as cell motility. During cell movement and phagocytosis, prearranged meshes of actin filaments were found assembled close to the plasma membrane, providing a framework that allowed fast changes in morphology by means of cell protrusions such as lamellipodia, filopodia, and endocytic structures. Similar observations were done by our group in other free-living amoebae such as Acanthamoeba castellanii [31] and Acanthamoeba royreba [32].
Our results show that A. griffini has a low in vitro cytopathogenicity, which is in agreement with a previous report [33] concerning the cytopathic effect of this parasite on Vero cell cultures. In interactions between MDCK cells and A. griffini trophozoites, the cytopathic effect on the monolayer was evident by light microscopy after 24 h. As observed by transmission electron microscopy, practically all cells exhibited a normal morphology. Nevertheless, trophozoites produced some focal injury on the cells by means of phagocytic structures of diverse size through which they engulfed portions of cells, a behavior that was previously observed in other Acanthamoebae such as A. castellanii and A. royreba [34, 35]. Interestingly, MDCK monolayers incubated with conditioned medium were also slightly injured, suggesting that the trophozoites produce some soluble lytic factor.
Although A. griffini was not able to cause an infection in the CNS in the murine GAE model, their invasive capacity was demonstrated, which correlates with a minor infection easily resolved in the patient from which it was isolated, as well as the assays with MDCK cells, which showed a cytopathic effect in less proportion than reported in other free-living amoebae.
To date, Acanthamoeba spp. isolates belonging to seven different genotypes (T2, T3, T4, T5, T6, T11, and T15) have been found to be associated with Acanthamoeba keratitis (AK) [27, 29, 36–39]. However, the most prevalent genotype in AK cases worldwide is the T4 genotype [39].
Previous studies have reported Acanthamoeba genotype T3 in AK cases. However this is only the report number nine of genotype T3 as a causal agent of AK and to the best of our knowledge, the second report of this genotype in a clinical case in the American continent. Other authors have isolated genotype T3 from clinical cases in a low percentage ranging from 1% to 13% in various countries including China, France, Hong Kong, Japan, Spain, Sweden, Iran, UK, and the United States [27, 29, 36, 40–44]. Furthermore, these observations are not surprising since as mentioned before genotype T4 has been reported to be the most abundant genotype in both clinical and environmental sources [39, 45], being the causative agent of around 80% of the reported infection cases worldwide [42].
Regarding environmental isolation of genotype T3, the number of isolates is also low and therefore, the low number of AK cases due to T3 could be also related to this genotype being less abundant in the environment that genotype T4 and other common genotypes in the environment genotypes such as T5 or T7. Genotype T3 has been isolated from the environment in many countries including Brazil [46], Chile [47], Egypt [17], Hong Kong [27], Iran [48], Philippines [49], Malaysia [50], and Taiwan [51]. Further epidemiological studies and pathogenicity capacity of genotype T3 isolated should be carried out worldwide in order to reach further conclusions about the status of genotype T3 as a potential pathogen in both the environment and clinical cases. Nevertheless, a recent systematic analysis of Acanthamoeba genotype frequency correlated with source and pathogenicity concluded that T4 is in fact the genotype most often associated with human disease, even after its abundance in the general environment is taken into account. Furthermore, T3 and T11 are closest relatives to T4 and they are the second and third most often associated with AK [52]. Further epidemiological studies on the pathogenicity capacity of genotype T3 should be performed to confirm the pathogenic status of genotype T3 in both the environment and clinical cases.
Finally, we also tested the isolate sensitivity to first line treatments currently used in AK cases. Our results showed that voriconazole inhibited the growth of A. griffini at very low concentrations, making it a potential first line treatment against AK cases. Other recent studies have reported similar activities of voriconazole against potentially pathogenic strains of clinical origin [23, 53] and even a successful treatment was achieved using voriconazole for the treatment of a patient suffering from AK in Spain [22].
In summary, our observations showed that the ultrastructure of Acanthamoeba griffini does not differ significantly from that of other Acanthamoeba species, Also, the in vitro cytopathic effect is similar to other low virulence free-living amoebae as Acanthamoeba royreba and is possibly the result of a low production of lytic factors by the trophozoites. Besides, this amoeba was isolated from a clinical case and belongs to genotype T3 which has clinical relevance since other pathogenic species are also included in this group. Also it was important to find that A. griffini is sensitive to drugs used at present in the treatment for AK. Further research with biochemical and molecular biology methods is necessary to better understand the mechanism of pathogenesis of this pathogen.
Acknowledgments
This work was partially supported by the grants RICET (Project no. RD12/0018/0012 of the Programme of Redes Temáticas de Investigación Cooperativa, FIS), Spanish Ministry of Health, Madrid, Spain, and the Project FIS PI10/01298 “Protozoosis emergentes por amebas de vida libre: aislamiento y caracterización molecular, identificación de cepas transportadoras de otros agentes patógenos y búsqueda de quimioterapias efectivas” from the Instituto de Salud Carlos III. Carmen M. Martín-Navarro was supported by a postdoctoral grant from the Fundación Canaria Manuel Morales, La Palma, Canary Islands. Maria Reyes-Batlle was funded by CEI Canarias, Campus Atlántico Internacional. Jacob Lorenzo-Morales was supported by the Ramón y Cajal Subprogramme from the Spanish Ministry of Economy and Competivity RYC-2011-08863. The authors also gratefully thank Dr. Mónica González-Lázaro for critical reading, helpful comments, and correcting the paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.de Jonckheere JF. Ecology of acanthamoeba. Reviews of Infectious Diseases. 1991;13(5):S385–S387. doi: 10.1093/clind/13.supplement_5.s385. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Zaragoza S. Ecology of free-living amoebae. Critical Reviews in Microbiology. 1994;20(3):225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- 3.Szénási Z, Endo T, Yagita K, Nagy E. Isolation, identification and increasing importance of “free-living” amoebae causing human disease. Journal of Medical Microbiology. 1998;47(1):5–16. doi: 10.1099/00222615-47-1-5. [DOI] [PubMed] [Google Scholar]
- 4.Brown TJ, Cursons RTM, Keys EA. Amoebae from antarctic soil and water. Applied and Environmental Microbiology. 1982;44(2):491–493. doi: 10.1128/aem.44.2.491-493.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivera F, Galvan M, Robles E, Leal P, González L, Lacy AM. Bottled mineral waters polluted by protozoa in Mexico. Journal of Protozoology. 1981;28(1):54–56. doi: 10.1111/j.1550-7408.1981.tb02803.x. [DOI] [PubMed] [Google Scholar]
- 6.Muñoz V, Reyes H, Toche P, Cárcamo C, Gottlieb B. Aislamiento de amebas de vida libre en piscinas públicas de Santiago de Chile. Parasitología Latinoamericana. 2003;58(3-4):106–111. [Google Scholar]
- 7.Barbeau J, Buhler T. Biofilms augment the number of free-living amoebae in dental unit waterlines. Research in Microbiology. 2001;152(8):753–760. doi: 10.1016/s0923-2508(01)01256-6. [DOI] [PubMed] [Google Scholar]
- 8.Paszko-Kolva C, Yamamoto H, Shahamat M, Sawyer TK, Morris G, Colwell RR. Isolation of amoebae and Pseudomonas and Legionella spp. from eyewash stations. Applied and Environmental Microbiology. 1991;57(1):163–167. doi: 10.1128/aem.57.1.163-167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera F, Roy-Ocotla G, Rosas I, Ramirez E, Bonilla P, Lares F. Amoebae isolated from the atmosphere of mexico city and environs. Environmental Research. 1987;42(1):149–154. doi: 10.1016/s0013-9351(87)80016-6. [DOI] [PubMed] [Google Scholar]
- 10.Martínez AJ. Is Acanthamoeba encephalitis an opportunistic infection? Neurology. 1980;30(6):567–574. doi: 10.1212/wnl.30.6.567. [DOI] [PubMed] [Google Scholar]
- 11.Butler TKH, Males JJ, Robinson LP, et al. Six-year review of Acanthamoeba keratitis in New South Wales, Australia: 1997–2002. Clinical and Experimental Ophthalmology. 2005;33(1):41–46. doi: 10.1111/j.1442-9071.2004.00911.x. [DOI] [PubMed] [Google Scholar]
- 12.Illingworth CD, Cook SD. Acanthamoeba keratitis. Survey of Ophthalmology. 1998;42(6):493–508. doi: 10.1016/s0039-6257(98)00004-6. [DOI] [PubMed] [Google Scholar]
- 13.Niederkorn JY, Ubelaker JE, McCulley JP, et al. Susceptibility of corneas from various animal species to in vitro binding and invasion by Acanthamoeba castellani . Investigative Ophthalmology and Visual Science. 1992;33(1):104–112. [PubMed] [Google Scholar]
- 14.Sawyer TK. Acanthamoeba griffini, a new species of marine amoeba. The Journal of Protozoology. 1971;18:650–654. [Google Scholar]
- 15.Ledee DR, Booton GC, Awwad MH, et al. Advantages of using mitochondrial 16s rDNA sequences to classify clinical isolates of Acanthamoeba. Investigative Ophthalmology & Visual Science. 2003;44(3):1142–1149. doi: 10.1167/iovs.02-0485. [DOI] [PubMed] [Google Scholar]
- 16.Seal DV. Acanthamoeba keratitis update—Incidence, molecular epidemiology and new drugs for treatment. Eye. 2003;17(8):893–905. doi: 10.1038/sj.eye.6700563. [DOI] [PubMed] [Google Scholar]
- 17.Lorenzo-Morales J, Ortega-Rivas A, Martínez E, et al. Acanthamoeba isolates belonging to T1, T2, T3, T4 and T7 genotypes from environmental freshwater samples in the Nile Delta region, Egypt. Acta Tropica. 2006;100(1-2):63–69. doi: 10.1016/j.actatropica.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Neff RJ. Purification, axenic cultivation, and description of a soil amoeba. Acanthamoeba sp. Journal of Protozoology. 1957;4:176–182. [Google Scholar]
- 19.Visvesvara GS. Pathogenic and opportunistic free-living amebae. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. 7th edition. Washington, DC, USA: ASM Press; 1999. pp. 1383–1390. [Google Scholar]
- 20.Rivera F, Lares F, Gallegos E, et al. Pathogenic amoebae in natural thermal waters of three resorts of Hidalgo, México. Environmental Research. 1989;50:289–295. doi: 10.1016/s0013-9351(89)80010-6. [DOI] [PubMed] [Google Scholar]
- 21.Culbertson CG, Smith JW, Cohen HK, Minner JR. Experimental infection of mice and monkey by Acanthamoeba . American Journal of Pathology. 1959;35:185–197. [PMC free article] [PubMed] [Google Scholar]
- 22.Arnalich-Montiel F, Martín-Navarro MC, Alió JL, et al. Successful monitoring and treatment of intraocular dissemination of Acanthamoeba . Archives of Ophthalmology. 2012;130:1474–1475. doi: 10.1001/archophthalmol.2012.2376. [DOI] [PubMed] [Google Scholar]
- 23.Martín-Navarro CM, López-Arencibia A, Arnalich-Montiel F, Valladares B, Piñero JE, Lorenzo-Morales J. Evaluation of the in vitro activity of commercially available moxifloxacin and voriconazole eye-drops against clinical strains of Acanthamoeba . Graefe's Archive for Clinical and Experimental Ophthalmology. 2013;251:2111–2117. doi: 10.1007/s00417-013-2371-y. [DOI] [PubMed] [Google Scholar]
- 24.Martín-Navarro CM, Lorenzo-Morales J, Cabrera-Serra MG, et al. The potential pathogenicity of chlorhexidine-sensitive Acanthamoeba strains isolated from contact lens cases from asymptomatic individuals in Tenerife, Canary Islands, Spain. Journal of Medical Microbiology. 2008;57(11):1399–1404. doi: 10.1099/jmm.0.2008/003459-0. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzo-Morales J, Ortega-Rivas A, Foronda P, Martínez E, Valladares B. Isolation and identification of pathogenic Acanthamoeba strains in Tenerife, Canary Islands, Spain from water sources. Parasitology Research. 2005;95(4):273–277. doi: 10.1007/s00436-005-1301-2. [DOI] [PubMed] [Google Scholar]
- 26.Niyyati M, Lorenzo-Morales J, Rezaie S, et al. Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Experimental Parasitology. 2009;121(3):242–245. doi: 10.1016/j.exppara.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Booton GC, Kelly DJ, Chu YW, et al. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. Journal of Clinical Microbiology. 2002;40(5):1621–1625. doi: 10.1128/JCM.40.5.1621-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Booton GC, Visvesvara GS, Byers TJ, Kelly DJ, Fuerst PA. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. Journal of Clinical Microbiology. 2005;43(4):1689–1693. doi: 10.1128/JCM.43.4.1689-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page FC. Culture Collections of Algae and Protozoa. Cumbria, UK: Freshwater Biological Association; 1988. A new key to freshwater and soil gymnamoebae with instructionsfor culture; pp. 92–96. [Google Scholar]
- 31.González-Robles A, Castañón G, Hernández-Ramírez VI, et al. Acanthamoeba castellanii: identification of actincytoskeleton. Experimental Parasitology. 2008;119:411–417. doi: 10.1016/j.exppara.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 32.González-Robles A, Salazar-Villatoro L, González-Lázaro M, Omaña-Molina M, Martínez-Palomo A. Vahlkampfia sp: structural observations of cultured trophozoites. Experimental Parasitology. 2012;130:86–90. doi: 10.1016/j.exppara.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 33.de Jonckheere JF. Growth charactertistics, cytopathic effect in cell culture, and virulence in mice of 36 type strains belonging to 19 different Acanthamoeba spp. Applied and Environmental Microbiology. 1980;39(4):681–685. doi: 10.1128/aem.39.4.681-685.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.González Robles A, Castañón G, Cristóbal Ramos AR, et al. Acanthamoeba castellanii: structural basis of the cytophatic mechanisms. Experimental Parasitology. 2006;114:133–140. doi: 10.1016/j.exppara.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 35.González-Robles A, Salazar-Villatoro L, Omaña-Molina M, Lorenzo-Morales J, Martínez-Palomo A. Acanthamoeba royreba: morphological features and in vitro cytopathic effect. Experimental Parasitology. 2013;133(4):369–375. doi: 10.1016/j.exppara.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Maghsood AH, Sissons J, Rezaian M, Holder D, Warhurst D, Khan NA. Acanthamoeba genotype T4 from the UK and Iran and isolation of the T2 genotype from clinical isolates. Journal of Medical Microbiology. 2005;54(8):755–759. doi: 10.1099/jmm.0.45970-0. [DOI] [PubMed] [Google Scholar]
- 37.Spanakos G, Tzanetou K, Miltsakakis D, Patsoula E, Malamou-Lada E, Vakalis NC. Genotyping of pathogenic Acanthamoebae isolated from clinical samples in Greece-report of a clinical isolate presenting T5 genotype. Parasitology International. 2006;55(2):147–149. doi: 10.1016/j.parint.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Lorenzo-Morales J, Morcillo-Laiz R, Martín-Navarro CM, et al. Acanthamoeba keratitis due to genotype T11 in a rigid gas permeable contact lens wearer in Spain. Contact Lens and Anterior Eye. 2011;34(2):83–86. doi: 10.1016/j.clae.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Lorenzo-Morales J, Martín-Navarro CM, López-Arencibia A, Arnalich-Montiel F, Piñero JE, Valladares B. Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends in Parasitology. 2013;29(4):181–187. doi: 10.1016/j.pt.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Niyyati M, Lorenzo-Morales J, Rezaie S, et al. First report of a mixed infection due to Acanthamoeba genotype T3 and Vahlkampfia in a cosmetic soft contact lens wearer in Iran. Experimental Parasitology. 2010;126(1):89–90. doi: 10.1016/j.exppara.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Rahman MM, Yagita K, Kobayashi A, et al. Genetic characterization of clinical Acanthamoeba isolates from Japan using nuclear and mitochondrial small subunit ribosomal RNA. Korean Journal of Parasitology. 2013;51:401–411. doi: 10.3347/kjp.2013.51.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Risler A, Coupat-Goutaland B, Pélandakis M. Genotyping and phylogenetic analysis of Acanthamoeba isolates associated with keratitis. Parasitology Research. 2013;112(11):3807–3816. doi: 10.1007/s00436-013-3572-3. [DOI] [PubMed] [Google Scholar]
- 43.Sharifi N, Botero-Kleiven S, Öhman D, Barragan A, Winiecka-Krusnell J. Genotypic characterization of Acanthamoeba spp. causing ocular infections in Swedish patients: identification of the T15 genotype in a case of protracted keratitis. Scandinavian Journal of Infectious Diseases. 2010;42(10):781–786. doi: 10.3109/00365548.2010.490563. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Sun X, Wang Z, et al. Identification of 18S ribosomal DNA genotype of Acanthamoeba from patients with keratitis in north China. Investigative Ophthalmology and Visual Science. 2004;45(6):1904–1907. doi: 10.1167/iovs.03-1073. [DOI] [PubMed] [Google Scholar]
- 45.Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba . Parasites and Vectors. 2012;5, article 6 doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caumo K, Rott MB. Acanthamoeba T3, T4 and T5 in swimming-pool waters from Southern Brazil. Acta Tropica. 2011;117(3):233–235. doi: 10.1016/j.actatropica.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Astorga B, Lorenzo-Morales J, Martín-Navarro CM, et al. Acanthamoeba belonging to T3, T4, and T11: genotypes isolated from air-conditioning units in Santiago, Chile. Journal of Eukaryotic Microbiology. 2011;58(6):542–544. doi: 10.1111/j.1550-7408.2011.00584.x. [DOI] [PubMed] [Google Scholar]
- 48.Solgi R, Niyyati M, Haghighi A, et al. Thermotolerant Acanthamoeba spp. isolated from therapeutic hot springs in northwestern Iran. Journal of Water and Health. 2012;10(4):650–656. doi: 10.2166/wh.2012.032. [DOI] [PubMed] [Google Scholar]
- 49.Rivera WL, Adao DE. Identification of the 18S-ribosomal-DNA genotypes of Acanthamoeba isolates from the Philippines. Annals of Tropical Medicine and Parasitology. 2008;102(8):671–677. doi: 10.1179/136485908X337544. [DOI] [PubMed] [Google Scholar]
- 50.Chan L-L, Mak J-W, Low Y-T, Koh T-T, Ithoi I, Mohamed SM. Isolation and characterization of Acanthamoeba spp. from air-conditioners in Kuala Lumpur, Malaysia. Acta Tropica. 2011;117(1):23–30. doi: 10.1016/j.actatropica.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang S-W, Hsu B-M. Isolation and identification of Acanthamoeba from Taiwan spring recreation areas using culture enrichment combined with PCR. Acta Tropica. 2010;115(3):282–287. doi: 10.1016/j.actatropica.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Maciver SK, Asif M, Simmen MW, Lorenzo-Morales J. A systematic analysis of Acanthamoeba genotype frequency correlated with source and pathogenicity: T4 is confirmed as a pathogen-rich genotype. European Journal of Protistology. 2013;49(2):217–221. doi: 10.1016/j.ejop.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Cabello-Vílchez AM, Martín-Navarro CM, López-Arencibia A, et al. Voriconazole as a first-line treatment against potentially pathogenic Acanthamoeba strains from Peru. Parasitology Research. 2014;113(2):755–759. doi: 10.1007/s00436-013-3705-8. [DOI] [PubMed] [Google Scholar]


