Abstract
Motivation: Recent advances in high-throughput lipid profiling by liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) have made it possible to quantify hundreds of individual molecular lipid species (e.g. fatty acyls, glycerolipids, glycerophospholipids, sphingolipids) in a single experimental run for hundreds of samples. This enables the lipidome of large cohorts of subjects to be profiled to identify lipid biomarkers significantly associated with disease risk, progression and treatment response. Clinically, these lipid biomarkers can be used to construct classification models for the purpose of disease screening or diagnosis. However, the inclusion of a large number of highly correlated biomarkers within a model may reduce classification performance, unnecessarily inflate associated costs of a diagnosis or a screen and reduce the feasibility of clinical translation. An unsupervised feature reduction approach can reduce feature redundancy in lipidomic biomarkers by limiting the number of highly correlated lipids while retaining informative features to achieve good classification performance for various clinical outcomes. Good predictive models based on a reduced number of biomarkers are also more cost effective and feasible from a clinical translation perspective.
Results: The application of LICRE to various lipidomic datasets in diabetes and cardiovascular disease demonstrated superior discrimination in terms of the area under the receiver operator characteristic curve while using fewer lipid markers when predicting various clinical outcomes.
Availability and implementation: The MATLAB implementation of LICRE is available from http://ww2.cs.mu.oz.au/∼gwong/LICRE
Contact: gerard.wong@bakeridi.edu.au or gerard.wong@unimelb.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Supervised feature reduction approaches have commonly been used in genomics to improve computational efficiency and classification performance. For instance, Wong et al. (2012) demonstrated that the omission of uninformative single nucleotide polymorphism (SNP) microarray-derived copy number measurements improved both the classification performance of various cancer subtypes and overall computational efficiency. In the context of SNPs, uninformative features may constitute probesets with probe sequences matching multiple genomic loci (Wong et al., 2010) giving rise to cross-hybridization and spurious copy number measurements. In lipidomics, lipid species experimentally analysed tend to be highly correlated because of similarities in their chemical properties (e.g. lipids that are co-regulated or members of a common metabolic pathway). Thus, feature reduction to minimize the number of correlated lipids in the dataset can be useful to ensure good classification performance by using a limited set of non-redundant but informative features. The advantage of an unsupervised feature reduction approach is that the reduction is not biased towards any sample or outcome classification, and a single reduced dataset can be used for the analysis of multiple outcomes. The use of a limited number of markers also enhances the feasibility for potential translation in a clinical setting for disease risk prediction, diagnosis, prognosis and prediction/monitoring of therapeutic response.
2 METHODS AND IMPLEMENTATION
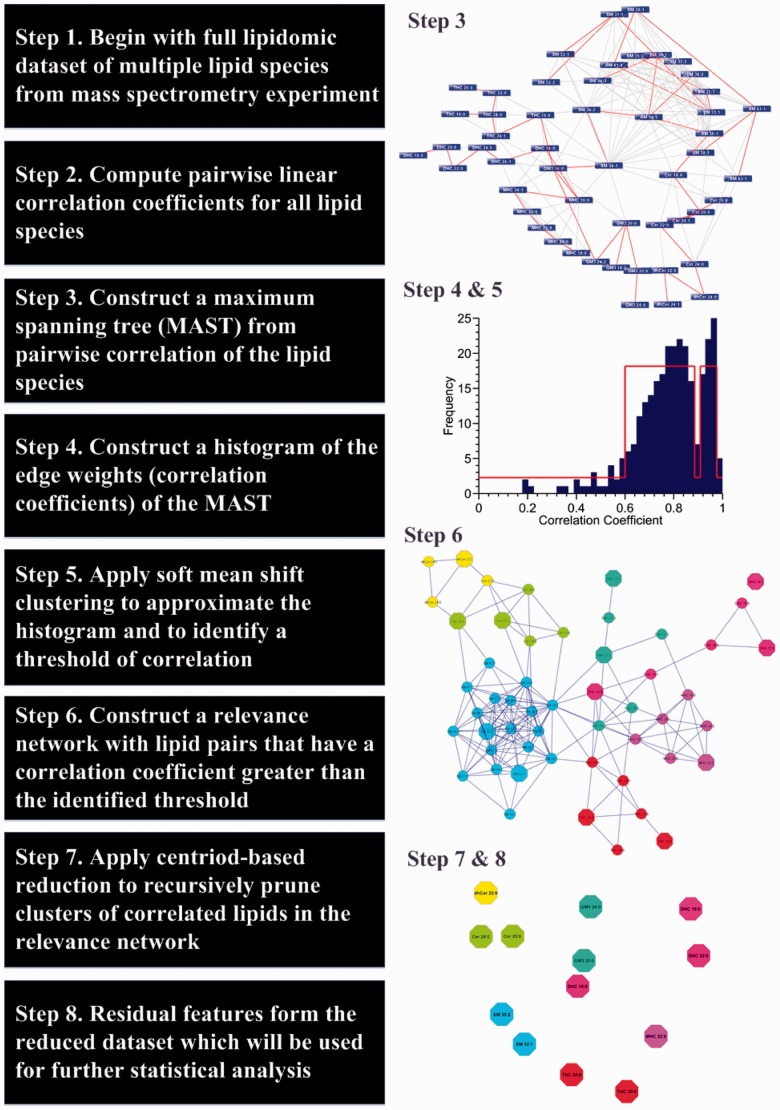
LICRE is an algorithm that reduces the number of highly correlated lipid species in a lipidomic dataset without the need for sample class information. This allows the reduced dataset to be analysed for various outcomes. Our work is motivated by our observation that highly correlated lipid species tend to be members of the same lipid class, i.e. have similar chemical properties and structure or belong to a common metabolic or regulatory pathway. Consequently, the down- or upregulation of these related lipid species can be readily inferred from each other. Unlike other unsupervised methods, e.g. principal components analysis (PCA), the interpretation of the abstracted set of features in LICRE is straightforward, as the original meaning of each feature (lipid) is preserved. In contrast, the principal components in PCA are weighted linear combinations of the original features, which makes their biological interpretation difficult. We have implemented LICRE in MATLAB 2013a as a function that can be applied in the initial pre-processing stage of classification modelling. The challenges presented by this task are: (i) estimating the degree (extent) of correlation of lipids in a dataset, (ii) identifying groups/clusters of lipids (features) that are sufficiently correlated in the dataset, (iii) abstracting each group/cluster of highly correlated lipids with minimal loss of information.
Estimating degree of correlation. To estimate the degree of correlation in the dataset, we create a minimalist network representation of the correlation structure of the data by constructing a maximum spanning tree (MAST) using visual assessment of cluster tendency (VAT) (Bezdek and Hathaway, 2002). VAT is a visual approach used to determine the number of natural clusters in a dataset. Given a correlation matrix of the data, the number of clusters k can identified if an ordering of the rows and columns can be found that results in a sequence of dense diagonal blocks in the rearranged matrix. Each block represents high correlation among its corresponding points and defines a cluster. VAT attempts to discover this ordering by constructing a MAST through all points. In this context, a point is a feature (i.e. a particular lipid species), and the correlation matrix represents all pairwise linear correlation coefficients between features. Starting from the outer edges of a cluster, MAST traversal with VAT will likely visit all nodes in the cluster then cross to the nearest cluster in correlation terms and traverse the nodes within that cluster. Hence, the intra-cluster pairwise correlations are high and the inter-cluster correlation is low. A histogram [bin widths determined by the Freedman–Diaconis rule (Freedman and Diaconis, 1981)] of the correlation coefficients corresponding to the edges of the MAST gives us an estimate of the degree of correlation in the dataset. The distribution obtained is typically multi-modal (Fig. 1, Step 4).
Fig. 1.
A summary of the LICRE algorithm workflow
Identifying threshold of correlation. From the histogram, a cutoff threshold can be identified that delineates higher correlation coefficients from lower correlation coefficients. This enables us to identify clusters of lipids that are sufficiently correlated in the dataset. To determine this threshold, we apply soft means shift clustering (SMSC) (Little and Jones, 2011) to approximate the histogram by discovering levels (Fig. 1, Step 5) in the histogram. The approximation of the histogram allows us to identify breaks between modes in the histogram and potential candidate thresholds of sufficient correlation in the dataset. SMSC is similar to k-means clustering where each level may be considered the equivalent of a centroid in k-means clustering and assigns points in the histogram to the closest level (centroid). Unlike k-means, SMSC automatically finds the number of levels required. Candidate thresholds are determined and ranked by the magnitude of the step increase between the identified discrete levels. The highest ranked threshold is selected. The selected threshold delineates a cluster or clusters of high correlation coefficients from the mass of lower correlation coefficients. Feature pairs that have correlation coefficients beyond this threshold are targets for feature reduction in the next stage of LICRE—feature pruning.
Feature pruning. A relevance network is constructed where an edge between nodes exists if its correlation coefficient exceeds the threshold determined from SMSC. To begin, the mean correlation coefficient of all clusters of degree one in the relevance network is computed. The centroid of the cluster with the highest mean correlation is retained, and all its peers are deleted. This is repeated recursively for the cluster with the next highest mean correlation in the residual network until singletons or pairs of nodes remain in the network. All unlinked nodes (singletons) are retained as features. For exclusive node pairs, the node with the highest median measurement (easy to quantify) is retained while the other is deleted. The set of nodes retained form the reduced dataset that replaces the complete dataset for classification modelling or statistical analysis.
3 CONCLUSION
We have developed an unsupervised feature reduction approach for lipidomic data. The application of LICRE on clinical lipidomic datasets (see Supplementary Material) demonstrates a statistically significant improvement in area under the ROC curve that is achieved with the use of a reduced number of features. This suggests potential utility in identifying concise markers for risk prediction, diagnostic, prognostic models of disease and for monitoring therapeutic response.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jim Bezdek and Andrey Kan for discussions.
Funding: This work was supported by funding from the Dairy Health and Nutrition Consortium, the National Health and Medical Research Council of Australia, the OIS Program of the Victorian Government, Australia and by Award Number 1R01DK088972-01 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, USA and the Australian Research Council’s Discovery Projects funding scheme (project number DP110102621).
Conflicts of Interest: none declared.
REFERENCES
- Bezdek JC, Hathaway RJ. VAT: a tool for visual assessment of (cluster) tendency. Proceedings of the 2002 International Joint Conference on Neural Networks, 2002. IJCNN’02. 2002;Vol. 3 IEEE, Honolulu, HI, pp. 2225–2230. [Google Scholar]
- Freedman D, Diaconis P. On the histogram as a density estimator: L-2 theory. Prob. Theory Rel. Fields. 1981;57:453–476. [Google Scholar]
- Little MA, Jones NS. Generalized methods and solvers for noise removal from piecewise constant signals. I. background theory. Proc. Math. Phys. Eng. Sci. 2011;467:3088–3114. doi: 10.1098/rspa.2010.0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G, et al. Exploiting sequence similarity to validate the sensitivity of SNP arrays in detecting fine-scaled copy number variations. Bioinformatics. 2010;26:1007–1014. doi: 10.1093/bioinformatics/btq088. [DOI] [PubMed] [Google Scholar]
- Wong G, et al. FSR: feature set reduction for scalable and accurate multi-class cancer subtype classification based on copy number. Bioinformatics. 2012;28:151–159. doi: 10.1093/bioinformatics/btr644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.