Abstract
Motivation: Predicting trajectories of signaling networks under complex perturbations is one of the most valuable, but challenging, tasks in systems biology. Signaling networks are involved in most of the biological pathways, and modeling their dynamics has wide applications including drug design and treatment outcome prediction.
Results: In this paper, we report a novel model for predicting the cell type-specific time course response of signaling proteins under unseen perturbations. This algorithm achieved the top performance in the 2013 8th Dialogue for Reverse Engineering Assessments and Methods (DREAM 8) subchallenge: time course prediction in breast cancer cell lines. We formulate the trajectory prediction problem into a standard regularization problem; the solution becomes solving this discrete ill-posed problem. This algorithm includes three steps: denoising, estimating regression coefficients and modeling trajectories under unseen perturbations. We further validated the accuracy of this method against simulation and experimental data. Furthermore, this method reduces computational time by magnitudes compared to state-of-the-art methods, allowing genome-wide modeling of signaling pathways and time course trajectories to be carried out in a practical time.
Availability and implementation: Source code is available at http://guanlab.ccmb.med.umich.edu/DREAM/code.html and as supplementary file online.
Contact: gyuanfan@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Protein signaling networks, especially those involving phosphoproteins, play a critical role in diverse cellular functions and are related to almost all pathways (Barrios-Rodiles et al., 2005; Hanahan and Weinberg, 2011; Olayioye et al., 2000; Sachs et al., 2005). Understanding these networks under defined perturbations, e.g. inhibitions or stimulations on certain gene(s), has wide applications in many medical fields, especially cancers, which are heterogeneous in their genetics (Dillon et al., 2007; Hochgräfe et al., 2010; Meric-Bernstam and Gonzalez-Angulo, 2009; Taylor et al., 2009).
Experimental methods that quantify the dynamics of the signaling phosphoproteins tend to be expensive and time-consuming, making them unaffordable to be extended to the genome scale and tested under a large number of perturbations. Mass spectrometry-based methods are now the major approach used to quantify dynamic changes in phosphoproteins, i.e. trajectories of these proteins over time (Aebersold and Mann, 2003; Ong and Mann, 2005). They are vitally important for inferring signaling networks, drug responses and treatment outcome (di Bernardo et al., 2005; Gardner et al., 2003; Luan et al., 2007; Morris et al., 2011; Steffen et al., 2002). Knockdown coupled with phosphoproteomics can help determine the causal relationships between signaling elements (Bansal et al., 2007; Maetschke et al., 2013; Marbach et al., 2010). However, these experiments are labor-intensive and unlikely to be carried out for a large number of perturbations or a large number of knockdown genes. Thus, computational modeling becomes vitally important for predicting time course signaling network dynamics under unseen situations.
Generally, cell line-specific signaling network dynamics under several known perturbations are observed. The observed data are time course data describing phosphorylation levels of phosphoproteins at different time points and under several different perturbations. The task is to predict time course responses if new drugs/perturbations come in (with the knowledge of their targeted proteins). Thus, given phosphorylation-level data for a group of proteins (k in 1 to N) sampled at a series of time points under perturbations … , we are interested in the phosphorylation levels for the same group of proteins under a new, unobserved perturbation for the same time course.
Computationally predicting the response of signaling networks under unseen perturbations remains a challenging task. First, current models for reconstructing directional signaling networks tend to be time-consuming (Hill et al., 2012; Yu et al., 2004), preventing them to be applied to the genome scale. The algorithms that are capable of reconstructing directional networks are usually designed to search the prohibitively large space of all possible network structures through temporal models. This basic approach renders the problem to be insolvable at the genome scale, even they are explored using an efficient probabilistic algorithm. For example, Molinelli et al. (Molinelli et al., 2013) recently proposed belief propagation (BP)-based searching method, which is three orders of magnitude faster than previous standard Monte Carlo methods used in this field, but still requires minutes to solve a network with 40 proteins. However, the human genome contains about 500 protein kinases and an estimated 6000 proteins modified by kinases (Manning et al., 2002; Whisenant et al., 2010). Because the time required to reconstruct the network is exponentially growing with the number of proteins, these methods are impossible to be applied to the genome of a typical mammalian species.
Second, the performance of most established algorithms is not satisfactory. As quoted from the DREAM 3 report (Prill et al., 2010), ‘Overall, a handful of best-performer teams were identified, while a majority of teams made predictions that were equivalent to random.’ DREAM 3 challenges focused on the same topic of the most recent DREAM 8: signaling cascade identification and signaling response prediction. It is indeed repeatedly observed from community blind assessments that the actual performance of most submitted algorithms are not far from random guesses (Prill et al., 2010). Thus, a more accurate mathematical model different from what has already been established must be sought.
Last but not least, even if these networks can be established at a smaller scale, it is still challenging to use these networks to accurately predict trajectories for a time course under unseen situations. Most of the previous endeavors have focused on inferring the network structures, but not predicting its changes over time. Thus, we developed a novel mathematical model to solve the above challenges in signaling network modeling. We formulate the network inference problem into a standard-form regularization problem, and the solution to this discrete ill-posed problem was found using truncated singular value decomposition (TSVD). This new method developed here has four advantages: (i) Highly accurate: In the 2013 DREAM 8 subchallenge: time course predictions, this algorithm was the top performing one when tested on experimental data; it also achieved the top performance in aggregated experimental and in silico results. Furthermore, it was identified as the most consistent performer, defined as robustly ranked first when evaluated with a subset of data. (ii) Highly computationally efficient: Both the network inference and the trajectory prediction steps of this algorithm take linear time to the number of proteins, which allow us to achieve magnitudes of improvement in speed over previous methods, making its application to the genome scale possible. (iii) Capable of de novo prediction: This algorithm is capable of inferring network structures and predicting trajectories without any prior knowledge of signaling interactions. (iv) Able to incorporate unseen perturbations: By manipulating the solution to the regularization problem (the regression coefficient matrix), the levels of phosphoproteins under unseen perturbations can be predicted.
2 METHODS
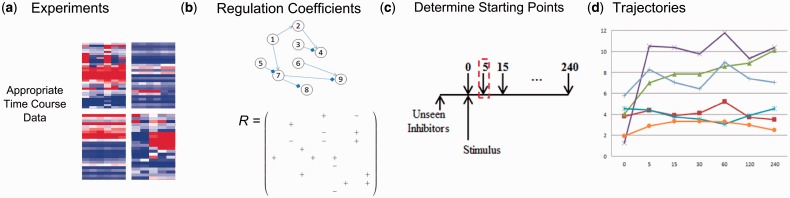
In this study, we developed an algorithm for reconstructing signaling networks through formulating the problem into a standard regularization problem and estimating its solution with TSVD. The algorithm then predicts the trajectory of phosphoproteins under new inhibitors by manipulating the regression coefficients of the networks. This trajectory predication method contains four steps (Fig. 1): (i) Collect time course phosphorylation data under several separate perturbations. (ii) Determine the regression coefficient matrix representing the influence level (positive and negative) of protein i on protein j. (iii) Estimate the starting phosphorylation levels of the proteins under unseen perturbations. (iv) Iteratively predict the phosphorylation level across a time course under new inhibitors.
Fig. 1.
Workflow of time course prediction of the network trajectory under unseen perturbations. (a) Time course dynamic phosphorylation levels under several observed perturbations are collected. (b) Regression coefficient matrix representing the influence level (positive and negative) between proteins are calculated based on the observed data. (c) Starting phosphorylation levels of the proteins under unseen perturbation are estimated using observed data. (d) Regression coefficient matrix is adjusted according to the new perturbation and phosphorylation levels are predicted iteratively across a time course
2.1 Assumption
We make the first-order Markov and stationary assumption (Friedman et al., 1998; Husmeier, 2003): every variable at a given time point ti only depends on the variables at the previous time point ti-1. Furthermore, we assume that the value of a variable remains stable without the influence from any other variables (including self-regulation). Equation 1 is the foundation of our method:
| (1) |
Where represents the phosphorylation value for protein k at the ith time point, is the regression coefficient indicating how much protein j will affect protein k. N is the number of proteins and ε is the error caused by uncontrollable factors such as measurement errors.
2.2 Inferring matrix R
Combining Equation 1 for all proteins, all time points and all inhibitors, we can form the following equation:
| (2) |
For the convenience of description, we assign an abbreviation for each matrix in Equation 2:
| (3) |
Where each element in T is the observed phosphorylation value for a phosphoprotein at a given time point, whereas the corresponding element in T+ is the observed value for the same protein at the next time point. Both T and T+ are obtained from the observed time course data and R is the unknown regression coefficient matrix we are interested in.
The problem can be solved by:
| (4) |
T-1 in Equation 4 is the pseudo-inverse of matrix T. There are numerous pseudo-inverse methods, such as singular value decomposition (SVD) (Golub and Reinsch, 1970), QR method, L1-regulation and L2-regulation. To handle the noise ε in Equation 1, we used truncated SVD (Hansen, 1990; Henry and Hofrichter, 2010), a variant of SVD, which has been shown to have nice properties in denoising the data in other domains of application (Hansen 1998; Hansen et al., 1992). Truncated SVD (TSVD) allows us to reduce the effect of noise and calculate the pseudo-inverse matrix. In TSVD, the first step is to decompose the observed phosphorylation matrix into:
| (5) |
Where U and V are unitary matrices, VT is the transpose matrix of V and ∑ is diagonal matrix. A convention is to order the diagonal matrix ∑ in a decreasing order and the diagonal entries of ∑ are known as the singular values of original matrix T. Elements on the diagonal matrix ∑ are non-negative real numbers.
As there may be zero-value elements on the diagonal matrix Σ, inverse matrix does not exist. In traditional SVD, the is calculated using following equation:
| (6) |
Note that this is a pseudo-inverse operation that ∑∑−1 ≠ I.
Because the small values caused by noise in the diagonal matrix may result in extremely large values in ∑−1, the noise is emphasized and dominates the inverses diagonal matrix using traditional SVD. A truncating step is therefore used to prevent the amplification of noise. A truncation parameter is defined, which specifies the number of the largest elements to be kept in the diagonal matrix ∑, while the rest of the elements will be set to zero in ∑−1. Thus, the amplification to the observation noise is restrained. In TSVD, Equation 6 is written as:
| (7) |
Finally, based on Equations 4, 5 and 7, the regression coefficient matrix can be calculated as:
| (8) |
2.3 Determine the starting phosphorylation levels
Under an unseen inhibitor, there is no access to the phosphorylation levels at the first time point. A protein’s initial phosphorylation level was therefore set to its average value at the time 0 under observed inhibitors. Furthermore, proteins, when targeted by an inhibitor(s), tend to have different initial phosphorylation levels compared with the levels when they are not targeted. Thus, we excluded the data points for inhibited proteins in the training data when estimating the starting points.
2.4 Adjust the influence of inhibited proteins
The activities of the phosphorylation proteins targeted by inhibitors are expected to be much lower than what are calculated in the matrix R in Equation 8. To model the situation of a complete inhibition, the contribution of inhibited proteins toward other proteins was set to 0 when predicting the unseen trajectory of phosphorylation levels.
| (9) |
Equation 9 is applied to both inferring the R matrix (Equation 2) and predicting the dynamics of levels of phosphorylation under unseen situation.
During the model training, time course data under different inhibitors can be combined together in Equations 2, 3, 4 and 8, i.e. multiple observed trajectories under different perturbations could be combined to predict a single regression coefficient matrix R (see Supplementary Material).
2.5 Iteratively predict the phosphorylation levels across a time course
After estimating the phosphorylation levels of each protein at the first time point and adjusting the regression coefficient matrix R based on the new, unseen inhibitors, Equation 1 is used iteratively to predict the time course phosphorylation levels at the rest of the time points under this specific perturbation.
3 RESULTS
This trajectory prediction method is a regression-based method using the first-order Markov and stationarity assumption. It first estimates the regression coefficients between phosphoproteins and determines the starting phosphorylation levels under unseen perturbation, then predicts the trajectory one time point by one time point. In the following sections, we first demonstrate the accuracy of this algorithm using simulation data. Second, we evaluate the prediction accuracy under different assumptions, such as different levels of noise and regression coefficient matrix density. Third, we examine the accuracy of this method on experimental data in breast cancer cell lines.
3.1 Simulation model
A simulation study was performed using 20 phosphoproteins, seven time points with five different inhibitions. The simulation model is constructed following the description in previously studies (de Jong et al., 2003; Hill et al., 2012; Yu et al., 2004). Briefly, the simulation contains six steps: (i) A random regression coefficient matrix R with a non-zero element density of 15% is generated and its non-zero values are replaced by values in a uniform distribution of ([−0.2, −0.02] ∪ [0.02, 0.2]). These values represent the effect of one phosphoprotein on the phosphorylation value of another protein, where positive values represent activation relationships and negative values represent inhibition relationships. A zero-element indicates that the first phosphoprotein has no effect on the second phosphoprotein. (ii) Proteins’ initial phosphorylation levels are randomly generated from a uniform distribution over [1, 10]. (iii) Randomly select one (or two) inhibited protein(s) and adjust the corresponding elements to 0 in the regression coefficient matrix R, for simulating phosphorylation dynamics under defined inhibitors. (iv) Equation 1 is used to iteratively generate the observed time course data. (v) A normally distributed noise or or is added to each phosphoprotein at every time point. (vi) Repeat step 3 and 5 to generate time course trajectories under five different inhibitors.
To evaluate the performance of our method, we simulated 5-fold cross-validation for 100 repeated runs. Thus, this prediction method uses time course data under four inhibitors as training data and data under the remaining inhibitor are used as the test set and withheld from model training.
3.2 Evaluation on reconstructing the signaling networks
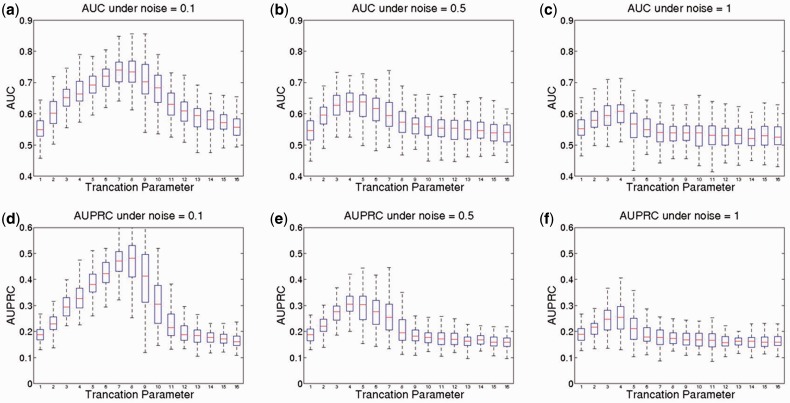
As this prediction method predicts the regression coefficient matrix before predicting the trajectories over a time course, we first evaluated how well it can recover the true regression coefficient matrix. Figure 2 shows the area under receiver operating characteristic curve (AUC) and area under precision recall curve (AUPRC) under low (0.1), medium (0.5) and high (1.0) noise levels with different denoising truncation parameters (as described in Equation 7). The performance under each of the noise level and parameter combination was an average of 100 simulations.
Fig. 2.
Accuracy of recovering the singaling networks. This figure shows AUC and AUPRC under different truncation parameters and noise levels, which indicate how well the regression coefficient matrix estimated by truncated SVD recovers the true influence matrix. The box plots are generated from 100 runs, with the density of non-zero elements in the signaling network to be 15%. A truncation parameter equals to N represents that the first N elements in diagonal matrix will be kept, while other elements are set to 0
We found that the denoising step (truncation in TSVD) is critical for recovering the original signaling networks (Fig. 2). In the case that the noise is small, the performance reaches maximum (AUC = 0.74, AUPRC = 0.48, compared with a baseline of 0.5 and 0.15) with a truncation parameter of 8 (Equation 7). As the noise level increases, a stronger denoising step is required, represented by less retained values in the diagonal matrix. As expected, the performance slightly drops with the increase of noise level (AUC = 0.7 for medium level of noise, and AUC = 0.67 for very high level of noises), i.e. the signal-to-noise ratio (SNR, defined as the average of the original phosphorylation levels divided by the SD of noise) is ∼14 (medium level) to 6.6 (high level). This performance was achieved without any prior knowledge.
3.3 Influence of truncation parameters
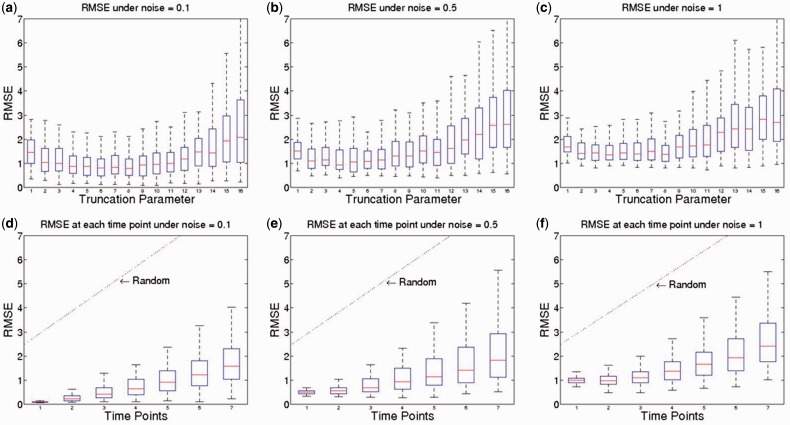
To evaluate the accuracy of this algorithm in predicting the phosphorylation dynamics across a time course under new perturbations, we calculated root-mean-square error, or RMSE, to measure the difference between predicted trajectory values and the withheld values during the cross validation. For each protein and each time point, the predicted value will be compared against the simulated values and differences between these values are considered as the prediction error.
One critical parameter in this prediction method is the truncation parameter (Equation 7), where a strict parameter and thus less retained elements in the diagonal matrix indicate stronger denoising effort. The accuracy in terms of RMSE under different parameters and different noise levels is therefore evaluated (Fig. 3). Under the noise level of 0.1 (low noise level), this method remarkably recovers the shadowed trajectory with a RMSE equals to 0.8, where the RMSE for random prediction is >4 (truncation parameter = 8). Even if the noise level is increased to 1.0, this method is still able to deliver a close prediction with a RMSE <1.5. As expected, loose parameters (>10), could barely estimate the hidden trajectory because of the amplified noise effect using traditional SVD and result in a high level of errors. Furthermore, when the noise level is high, a strict truncation parameter (=3) performs better in terms of RMSE (Fig. 3c) because of its strong denoising effect.
Fig. 3.
RMSE under different truncation parameters and noise levels. This figure indicates how truncation parameter influences the RMSE under different noise levels. Subfigures (a), (b) and (c) demonstrate the overall RMSE under different truncation parameters from 1 to 16 and different noise levels. Subfigures (d), (e) and (f) show the RMSE values at different time points under a truncation parameter of 8. The RMSE baseline of random prediction is also depicted. RMSE was evaluated under 100 repeated runs
Furthermore, the simulation results indicate that the number of protein and sparsity of signaling network does not change the optimal truncation parameter, while, as we expected, noise level could influence the truncation parameter (see Supplementary Figs S1–S3 for more details). Based on the RMSE in the cross-validation, the optimal truncation parameters are 3–10, depending on the noise level, i.e. the optimal truncation parameter is 5–10 under low noise level (SNR ≥ 14) and is 3–5 under high noise level (SNR < 14).
3.4 Influence of noise level
Subfigures (d), (e) and (f) of Figure 3 show the RMSE at each time point under different noise levels. A parameter of eight elements retained in the diagonal matrix is used in these subfigures. The predicted phosphorylation levels at the first point after the starting time point is extremely accurate, with a RMSE value almost the same as the noise level, indicating that we could almost perfectly recover the real phosphorylation levels. The prediction error starts to increase in the following time points, as noise accumulates during the iterative prediction steps. The RMSE predicted by our method is much smaller than random prediction baseline. Taking the low noise level as an example, the predicted trajectory has a RMSE of 0.1 at the first point (which is the same as the noise level) and 0.253 at the second time point, the RMSE increases to 1.60 at the last time point due to the accumulation of the prediction error in each iteration. At the same time, the random prediction has RMSEs ranging from 3.91 to 9.55, which are 5 to 38 folds larger than this prediction model.
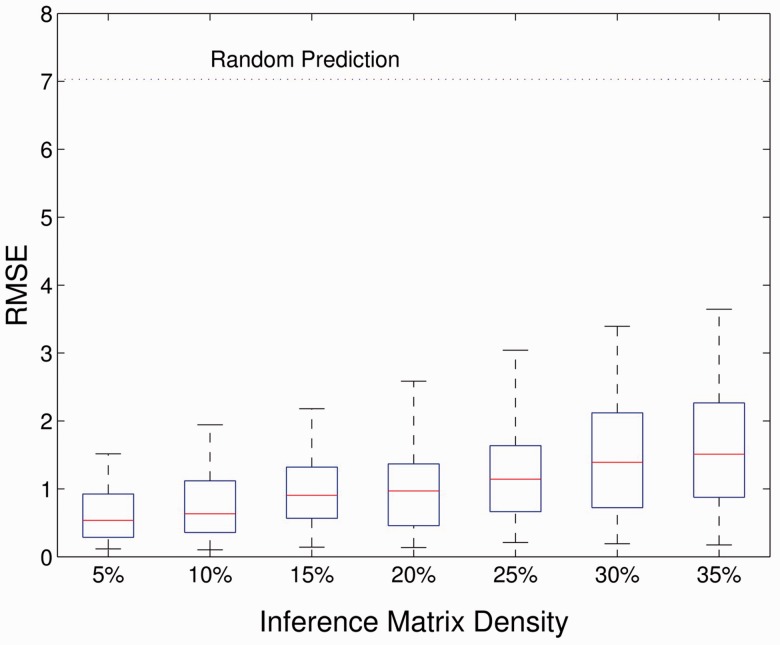
3.5 Influence of sparsity of signaling network
To assess the robustness of this prediction method, we have evaluated the prediction error against different influence matrix densities of the signaling networks (Fig. 4). We found that better performance is achieved when the influence matrix is sparse. With the increased density of non-zero elements and thus more complex network structure, the performance is slightly reduced, but the RMSE values are always <1.5 (compared with a RMSE of 7.02 for random prediction), indicating that this algorithm can robustly perform well even for networks with very complex structures, i.e. many interactions.
Fig. 4.
Influence of matrix densities. This figure illustrates the performance of the prediction method under different densities of the influence matrix. This result is evaluated under the low noise level (0.1) with a truncation parameter, which keeps eight largest elements in the diagonal matrix. The RMSE is evaluated from 100 runs
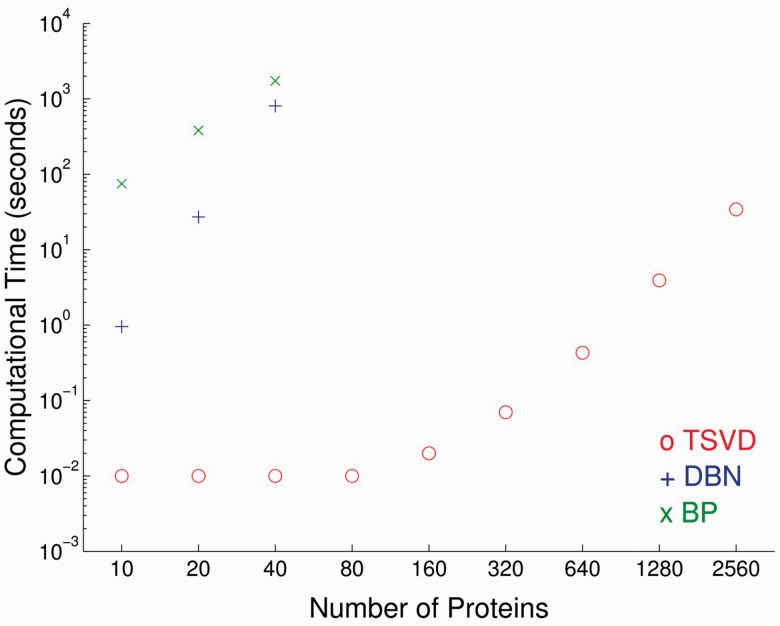
3.6 Computational time
Figure 5 shows the computational time required for this method and other two state-of-the-art network inference methods: dynamic Bayesian network (DBN) (Hill et al., 2012) and BP (Molinelli et al., 2013). The computational time is evaluated on a PowerEdge R410 processor, with four cores and 48 GB memory.
Fig. 5.
Computation time. This figure shows the computational time required to reconstruct the signaling network. The computational cost is evaluated under 100 repeated measurements. DBN and BP are only measured under 10, 20 and 40 proteins because of their exponential computational time required
This new algorithm takes <0.01 s for 20 proteins, while DBN costs ∼1 s and BP takes >1 min. When dealing with 40 proteins, both DBN and BP need >10 min, whereas TSVD only takes 0.01 s. This algorithm has a linear computational time requirement, and the computational time required for 2560 proteins is only 22.7 s. Compared with DBN and BP, TSVD are consistently hundreds of folds faster. Therefore, we conclude that this new algorithm offers tremendous computational performance advantage over the state-of-the-art methods, and make the genome-scale reconstruction of signaling networks possible.
3.7 Validation using experimental data
We further validated this method using time course phosphorylation data in the breast cancer cell line MDA-MB-468 (Hill et al., 2012). These data contain eight time points from three replicates, under different levels of EGF stimulus (0, 5 and 10 ng), for 20 proteins. In the validation, we used data under 0 and 10 ng EGF stimulus as training data and evaluated how well it can recover the phosphorylation levels under 5 ng EGF stimulus. Data from replicates are treated as separate time course observations.
Considering that the signal-to-noise ratio, which is determined by the average of the phosphorylation levels divided by the SD between replicates, is 14.5 (close to the median level of noise and larger than the low level of noise in simulation), a truncation parameter = 6 is used in TSVD. This model is robust that truncation parameters ranging from 5 to 10 all deliver predictions with similar accuracy.
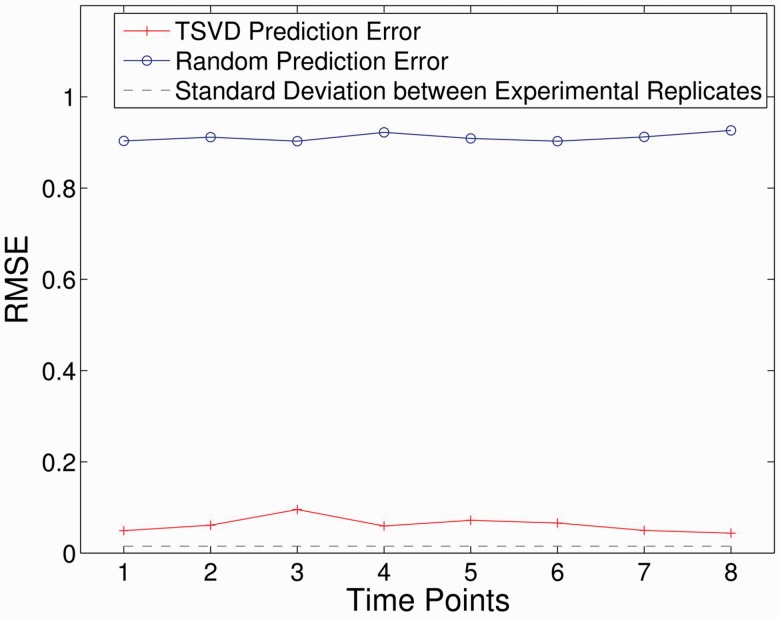
Figure 6 shows the predicting accuracy for experimental data. The average RMSE for the eight time points is 0.06, while random prediction baseline (for 100 runs) is 0.91, indicating 15-fold reductions in error. Notably, the average SD of phosphorylation levels between experimental replicates is 0.015. This indicates that the prediction performance (RMSE = 0.06) almost approximates the performance of real perturbation experiments affected by noises such as measurement error, which strongly supports the effectiveness of our algorithm.
Fig. 6.
Predicting errors in experimental data. This figure illustrates the accuracy of this trajectory predicting method in recovering experimental data. The predicted time course trajectory (red line) has a much lower RMSE across all the time points compared with random prediction (blue line). Additionally, RMSE of the predicted trajectory (0.06) is comparable with the difference between replicates (0.015, black line), indicating robust performance of the algorithms
4 DISCUSSION
Analyzing time course trajectories under unseen perturbations has important application in model biology, such as predicting dynamic phosphorylation networks, drug responses and treatment outcomes. However, previous research mainly focused on recovering the relationship between phosphoproteins. In this article, we describe a novel framework to predict trajectories under unseen perturbations.
We found that the noise level and the signaling network density have noticeable influence on the prediction accuracy. However, this model is able to handle all the noise levels and signaling network structures with a satisfying performance. The effectiveness of this model is also validated using experimental data on a breast cancer cell line MDA-MB-468. The experimental validation indicates that the predicting error of this model is at the similar level as the difference between experimental replicates.
The effectiveness of this model was demonstrated in the 8th Dialogue for Reverse Engineering Assessments and Methods (DREAM 8: https://www.synapse.org/#!Wiki:syn1929437/ENTITY/63075) challenge as the best performing and most consistent method (judged by RMSE, see Supplementary Materials for more details) in the subchallenge time course prediction (Stolovitzky et al., 2007). In this subchallenge, both experimental data and simulation data have been given and the corresponding prediction accuracy has been evaluated.
This algorithm is extremely computationally time efficient, which could predict the trajectory for 20 variables in <0.01 s, and the number of variables versus computational time is close to linear. This model does not require any prior signaling network information, so it is applicable to any cell type without extra effort if given appropriate time course data. These characteristics promise application of this algorithm to genome-wide de novo network reconstruction and trajectory prediction under unseen perturbations.
Supplementary Material
Acknowledgements
The authors thank the 2013 HPN-DREAM breast cancer network inference challenge organizers for hosting the challenge and financial support.
Funding: This work was supported by the National Institutes of Health [NIH 1R21NS082212-01 to Y.G.].
Conflict of Interest: none declared.
REFERENCES
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Bansal M, et al. How to infer gene networks from expression profiles. Mol. Syst. Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- de Jong H, et al. Genetic network analyzer: qualitative simulation of genetic regulatory networks. Bioinformatics. 2003;19:336–344. doi: 10.1093/bioinformatics/btf851. [DOI] [PubMed] [Google Scholar]
- di Bernardo D, et al. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat. Biotechnol. 2005;23:377–383. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- Dillon R, et al. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- Friedman N, et al. Proceedings of the Fourteenth conference on Uncertainty in artificial intelligence. San Francisco, CA: Morgan Kaufmann Publishers Inc; 1998. Learning the structure of dynamic probabilistic networks; pp. 139–147. [Google Scholar]
- Gardner TS, et al. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- Golub GH, Reinsch C. Singular value decomposition and least squares solutions. Numerische Mathematik. 1970;14:403–420. [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hansen PC. Truncated singular value decomposition solutions to discrete ill-posed problems with ill-determined numerical rank. SIAM J. Sci. Stat. Comput. 1990;11:503–518. [Google Scholar]
- Hansen PC. Rank-Deficient and Discrete Ill-Posed Problems: Numerical Aspects of Linear Inversion. Philadelphia: SIAM; 1998. [Google Scholar]
- Hansen PC, et al. The modified truncated SVD method for regularization in general form. SIAM J. Sci. Stat. Comput. 1992;13:1142–1150. [Google Scholar]
- Henry E, Hofrichter J. Singular value decomposition: application to analysis of experimental data. Essential Num. Comput. Methods. 2010;210:81–138. [Google Scholar]
- Hill SM, et al. Bayesian inference of signaling network topology in a cancer cell line. Bioinformatics. 2012;28:2804–2810. doi: 10.1093/bioinformatics/bts514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgräfe F, et al. Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells. Cancer Res. 2010;70:9391–9401. doi: 10.1158/0008-5472.CAN-10-0911. [DOI] [PubMed] [Google Scholar]
- Husmeier D. Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics. 2003;19:2271–2282. doi: 10.1093/bioinformatics/btg313. [DOI] [PubMed] [Google Scholar]
- Luan D, et al. Computationally derived points of fragility of a human cascade are consistent with current therapeutic strategies. PLoS Comput. Biol. 2007;3:e142. doi: 10.1371/journal.pcbi.0030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetschke SR, et al. Supervised, semi-supervised and unsupervised inference of gene regulatory networks. Brief. Bioinform. 2013;15:195–211. doi: 10.1093/bib/bbt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Marbach D, et al. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl Acad. Sci. USA. 2010;107:6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinelli EJ, et al. Perturbation biology: inferring signaling networks in cellular systems. PLoS Comput. Biol. 2013;9:e1003290. doi: 10.1371/journal.pcbi.1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MK, et al. Training signaling pathway maps to biochemical data with constrained fuzzy logic: quantitative analysis of liver cell responses to inflammatory stimuli. PLoS Comput. Biol. 2011;7:e1001099. doi: 10.1371/journal.pcbi.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye MA, et al. The ErbB signaling network: receptor heterodimerization in development and cancer. The EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry–based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Prill RJ, et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PloS One. 2010;5:e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K, et al. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Steffen M, et al. Automated modelling of signal transduction networks. BMC Bioinformatics. 2002;3:34. doi: 10.1186/1471-2105-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolovitzky G, et al. Dialogue on reverse engineering assessment and methods. Ann. N. Y. Acad. Sci. 2007;1115:1–22. doi: 10.1196/annals.1407.021. [DOI] [PubMed] [Google Scholar]
- Taylor IW, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- Whisenant TC, et al. Computational prediction and experimental verification of new MAP kinase docking sites and substrates including Gli transcription factors. PLoS Comput. Biol. 2010;6:e1000908. doi: 10.1371/journal.pcbi.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.