Abstract
Summary: Popular methods for 3D protein structure similarity searching, especially those that generate high-quality alignments such as Combinatorial Extension (CE) and Flexible structure Alignment by Chaining Aligned fragment pairs allowing Twists (FATCAT) are still time consuming. As a consequence, performing similarity searching against large repositories of structural data requires increased computational resources that are not always available. Cloud computing provides huge amounts of computational power that can be provisioned on a pay-as-you-go basis. We have developed the cloud-based system that allows scaling of the similarity searching process vertically and horizontally. Cloud4Psi (Cloud for Protein Similarity) was tested in the Microsoft Azure cloud environment and provided good, almost linearly proportional acceleration when scaled out onto many computational units.
Availability and implementation: Cloud4Psi is available as Software as a Service for testing purposes at: http://cloud4psi.cloudapp.net/. For source code and software availability, please visit the Cloud4Psi project home page at http://zti.polsl.pl/dmrozek/science/cloud4psi.htm.
Contact: dariusz.mrozek@polsl.pl
1 INTRODUCTION
Cloud computing has emerged as a result of increased requirements for the public availability of computing power and new technologies for data processing. It has become a mechanism that allows for the control of the development of hardware and software resources by introducing the idea of virtualization. In practice, cloud computing allows the public to run applications and services on a distributed network using a virtualized system and its resources, and at the same time, allows to abstract away from the implementation details of the system itself. Apart from the natural applications of cloud computing architecture in business, the concept is also becoming increasingly popular in scientific research, including life sciences, because the theoretically infinite resources of the cloud allow computationally intensive problems to be solved. Cloud-based solutions were proposed for various areas of bioinformatics, including automated sequence analysis (Angiuoli et al., 2011), identification of peptide sequences from spectra in mass spectrometry (Lewis et al., 2012), mapping next-generation sequence data to the human genome and other reference genomes, for single nucleotide polymorphism discovery, genotyping and personal genomics (Schatz, 2009), 3D ligand binding site comparison and similarity searching of a structural proteome (Hung and Hua, 2013), sequence alignment, clustering, assembly, display, editing and phylogeny (Krampis et al., 2012).
The 3D protein structure similarity searching is a computationally complex and time-consuming process that requires enhanced computational resources. This is the motivation behind efforts to develop new methods in the area and build scalable platforms, such as the presented Cloud for Protein Similarity (Cloud4Psi) platform, that allow for the completion of the task much faster by distributing computation on many machines.
2 IMPLEMENTATION
2.1 Algorithms
Cloud4Psi enables researchers to search for protein structure similarities by using a combination of three algorithms: jCE, jFATCAT-rigid and jFATCAT-flexible (Prlic et al., 2012). These are new versions of the combinatorial extension (CE) (Shindyalov and Bourne, 1998) and flexible structure alignment by chaining aligned fragment pairs allowing twists (FATCAT) (Ye and Godzik, 2003) algorithms, which are well-established among the scientific community. jFATCAT and jCE work on the basis of matching protein structures using aligned fragment pairs. The limitation of the original CE and FATCAT algorithms is that they compute sequence order-dependent alignments. The jCE algorithm, an enhanced version of CE, solves the problem by handling circular permutations (Bliven and Prlic, 2012). The jFATCAT-flexible algorithm, in turn, eliminates the drawbacks of many existing methods that treat proteins as rigid bodies, not flexible structures. The research conducted by the authors of FATCAT has shown that rigid representation causes many similarities, even when strong, to be omitted. The flexible version of jFATCAT allows for the entry of twists in protein structures while matching their fragments, which provides better alignments in a number of cases.
All three algorithms used by Cloud4Psi are publicly available through the Protein Data Bank (PDB) (Berman et al., 2000) Web site for those who want to search for structural neighbors. Moreover, these algorithms are used for precalculated all-to-all 3D-structure comparisons for the whole PDB, which are updated on a weekly basis (Prlic et al., 2010).
2.2 Cloud4Psi in the Microsoft Azure Model
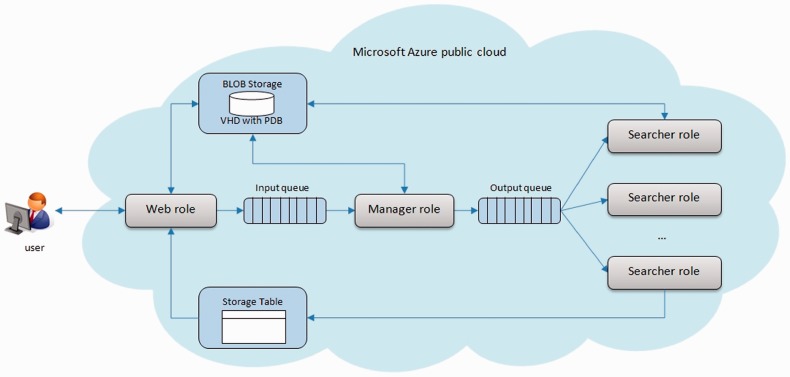
Cloud4Psi was developed for use in the Microsoft Azure public cloud. Any application that runs in the Microsoft Azure model is composed of a set of roles performing some tasks. Basic types of roles include Web roles [for graphical user interface (GUI)] and Worker roles (for computations). Roles use computational units provided by the Microsoft Azure cloud. These computational units have various sizes from A0 to A9, determining their computational capabilities and associated costs of their usage. Description of standard sizes of computational units is provided in the Windows Azure Cloud Specification manual (Microsoft, 2014, http://msdn.microsoft.com/en-us/library/windowsazure/dn197896.aspx). Cloud4Psi consists of several types of roles and storage modules responsible for gathering and exchanging data between roles (Fig. 1).
Fig. 1.
Architecture of the Cloud4Psi: Web role provides a front-end for users of the system; Manager role mediates the distribution of the searching process, which is executed by Searcher roles. Search requests and packages are transferred through Input and Output queues. Roles have access to various storage resources inside the cloud, including Binary large object (BLOB) Storage (VHD with PDB files) and Storage Tables (containing results of similarity searches)
Users can initiate the similarity search and receive the search results through the Web role. The Web role provides a GUI through a user-friendly Web site and also has an additional logical layer. The logical layer is responsible for converting the parameters received from the user into a form of a request message that is placed in the Input queue. The role also has access to the Storage Tables that provide results from the ongoing or finished similarity searches and also access to the virtual hard drive (VHD) for PDB files; this is for when the user decides to send his/her own PDB file for comparison by the Cloud4Psi. Users’ search requests are consumed by the Manager role (Worker role), which schedules distributed computations on many Searcher roles. The Manager role also divides the whole repository of PDB files into packages that are sent to Searcher roles through the Output queue, and manages associated computational loads between Searcher roles. Searcher roles (which are also Worker roles) bear the computational load associated with the process of protein comparison. They perform parallel batch comparisons of a given protein structure with proteins contained in the package retrieved from the Output queue. This role type is scaled out (scaled horizontally) by adding more computational units and scaled up (scaled vertically) by using computational units of different sizes during the similarity searching process. After finishing computations for a package, Searcher roles enter results for their integration into a table in the Storage Tables service of the Microsoft Azure cloud and retrieve another package from the Output queue.
3 RESULTS
Scalability of the Cloud4Psi was tested in the Microsoft Azure cloud by increasing the number of Searcher roles from 1 to 18 (horizontal scaling) and by increasing the size, i.e. computational capabilities, of the Searcher roles from A0 to A4 (vertical scaling). Horizontal scaling was performed with the use of A1-sized Searcher roles. A1 size (formerly small size) represents the cheapest computational unit providing one unshared core and is the smallest sized unit recommended for production workloads (Microsoft, 2014). During the vertical scaling, we gradually enlarged the sizes of the Searcher roles from A0 (formerly extra small, with one shared central processing unit (CPU) core) to A4 (formerly extra large, eight CPU cores). Tests were performed on a repository containing 1000 protein structures from the PDB. Package size was set to 10 protein structures.
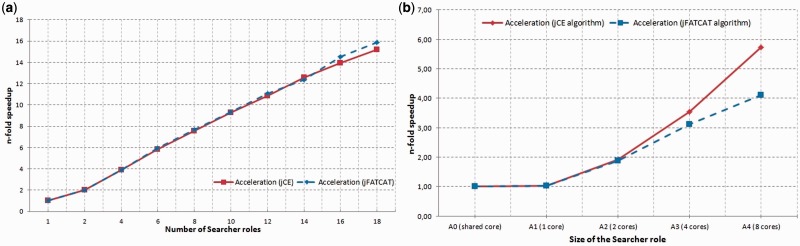
A single comparison of a pair of protein structures takes on average 11 s for jCE and 14 s for jFATCAT (both variants). The time required for computation depends on the size of the given protein structure. In Figure 2a, we have shown the n-fold speedup for jCE and jFATCAT algorithms when scaling the Cloud4Psi system horizontally. We noticed that using two instances of the Searcher role increased the speed of the similarity searching by almost 2-fold. Adding more Searcher roles proportionally accelerated the process. However, when using >8 Searcher roles, the dynamics of the acceleration slowed down. Finally, the acceleration ratio (n-fold speedup) reached the level of 15.17 for jCE, and 15.87 for jFATCAT, when the number of instances of the Searcher roles was increased from 1 up to 18.
Fig. 2.
Acceleration (n-fold speedup) of the similarity searching as a function of (a) the number of Searcher roles, (b) the size of the Searcher roles for jCE (solid line) and jFATCAT (dashed line) algorithms
In Figure 2b, we have shown the n-fold speedup when scaling the system vertically. Good results were obtained for Searcher roles of sizes A1 and A2 (A0 is not recommended for production environments). Above size A2, i.e. two computing units, we noticed that the dynamics of the acceleration significantly slowed down. Acceleration for eight CPU cores (A4 size) reached only 4.12 for jFATCAT and 5.73 for jCE.
When analyzing the results of the scalability tests, we observed that both types of scaling used experienced a slowdown in the acceleration dynamics. For the horizontal scaling, this was caused by several factors, including concurrent accesses to the VHD while retrieving protein structures processed by particular Searchers, and concurrent accesses to the Storage Table while storing partial results of similarity searches. For the vertical scaling, additional slowdown was brought about by sharing the same CPU and additional coordination of computations carried out in parallel on the same computing unit. Results of the performed tests showed that horizontal scaling with the use of A1 roles provided better n-fold speedup than vertical scaling. Moreover, horizontal scaling was easier for deployment and more elastic; the working system could be scaled automatically according to the current needs. For this reason, the available version of the Cloud4Psi runs on A1 roles and is scaled horizontally.
4 DISCUSSION
Although Clou4Psi can be configured to work on various numbers of Searcher roles, results returned by the system are independent of the configuration of the cloud platform. This guarantees reproducibility of results. Regardless of the number of instances of the Searcher role used at a particular moment, the system returns the same set of results for the given protein structure. This applies to the number of results returned, values of similarity measures for returned proteins and the content of the result set. The list of the database proteins returned for the given structure may change only when the repository of macromolecular structures that Searchers operate on is updated. In case of software components updates, the results of searches performed with the previous software version are still available after providing a proper token number. Cloud4Psi provides storage of the results for at least 1 month.
5 CONCLUDING REMARKS
Cloud4Psi represents an excellent Software as a Service solution, which is still rare in the domain of bioinformatics. The systemimplements the Microsoft Azure role-based and queue-based model, which also provides many operational benefits. Taking into account that the range of resources provided by the Microsoft Azure cloud is theoretically unlimited, Cloud4Psi is a highly scalable and high-performance solution for protein similarity searching and function identification.
ACKNOWLEDGEMENTS
We would like to thank Microsoft Research for providing us with free access to the computational resources of the Windows Azure cloud under the Windows Azure for Research Award program.
Further development of the system will be carried out by the Cloud4Proteins non-profit scientific group (http://zti.polsl.pl/dmrozek/science/cloud4proteins.htm).
Funding: This work was supported by the European Union from the European Social Fund (grant agreement number: UDA-POKL.04.01.01-00-106/09).
Conflict of Interest: none declared.
REFERENCES
- Angiuoli SV, et al. CloVR: A virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics. 2011;12:356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliven S, Prlic A. Circular permutation in proteins. PLoS Comput. Biol. 2012;8:e1002445. doi: 10.1371/journal.pcbi.1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CL, Hua GJ. Cloud computing for protein-ligand binding site comparison. Biomed. Res. Int. 2013;2013:170356. doi: 10.1155/2013/170356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krampis K, et al. Cloud BioLinux: pre-configured and on-demand bioinformatics computing for the genomics community. BMC Bioinformatics. 2012;13:42. doi: 10.1186/1471-2105-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S, et al. Hydra: a scalable proteomic search engine which utilizes the Hadoop distributed computing framework. BMC Bioinformatics. 2012;13:324. doi: 10.1186/1471-2105-13-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Microsoft. 2014. Windows Azure cloud services specification: virtual machine and cloud service sizes for Windows Azure. http://msdn.microsoft.com/en-us/library/windowsazure/dn197896.aspx (6 May 2014, date last accessed) [Google Scholar]
- Prlic A, et al. Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics. 2010;26:2983–2985. doi: 10.1093/bioinformatics/btq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic A, et al. BioJava: an open-source framework for bioinformatics in 2012. Bioinformatics. 2012;28:2693–2695. doi: 10.1093/bioinformatics/bts494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC. CloudBurst: highly sensitive read mapping with MapReduce. Bioinformatics. 2009;25:1363–1369. doi: 10.1093/bioinformatics/btp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov I, Bourne P. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19:246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]