Abstract
Background
ESMO consensus recommends EGFR mutation testing in never/former light smokers (<15 pack-years) or patients with non-squamous NSCLC. The aim of this work was to determine the frequency and clinical predictors of EGFR mutations, and the role of specimen sampling tests, in Caucasian standard practice setting.
Methods
We screened 297 patients according to this consensus. Mutational analysis of EGFR was performed using the Therascreen EGFR RGQ PCR mutation kit. Clinical and pathological correlative data were collected.
Results
An EGFR activating mutation was found in 32 patients (11%), twelve exon 19 deletions, two exon 18 and eighteen exon 21 point mutations. Most were in females, but half were in smokers. Negative TTF-1 staining had a very strong negative predictive value (all except one patient had TTF-1 positive adenocarcinoma). Both biopsies as well as cytology specimens (mainly EBUS-TBNA) did well: 24 mutations in 213 biopsy samples (11.2%) and 8 in 84 cytology samples (9.5%), respectively. The Therascreen acted as a sensitive test in all types of samples: 7 activating mutations were found in samples rated to have <5% of tumour cells, and there were only 4 test failures in the whole series.
Conclusion
In this Caucasian standard practice NSCLC cohort, tested according to the ESMO consensus, activating EGFR mutation occurred in 11% of the patients. Half of these were in former/current smokers. With our sampling technique and use of the Therascreen kit, EBUS-TBNA cell blocks performed as good as biopsies.
Electronic supplementary material
The online version of this article (doi:10.1186/s40247-014-0009-0) contains supplementary material, which is available to authorized users.
Keywords: Predictive factors, Real-time quantitative polymerase chain reaction (rt-qPCR), EGFR mutation, Endobronchial ultrasound, Biopsy methods
Background
About 85% of all lung cancer patients have non-small cell lung cancer (NSCLC), and the majority presents with advanced stage disease at the time of diagnosis. The standard of care for these patients is platinum-based doublet chemotherapy [1]. This `any platinum doublet fits all’ strategy results in a median overall survival of 8 to 10 months and a 1 year survival rate of about 33%.
Different strategies to prolong survival have been developed. Besides customisation of chemotherapy according to histological subtype [2], and the use of maintenance therapy in order to achieve prolonged tumour control [3], the most important change during the last decade was the introduction of treatment guided by the tumour's genetic profile.
The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) heralded this evolution, when very pronounced and durable responses to these agents were noted early in this century [4]. A few years later, somatic mutations in the EGFR gene were discovered in these highly responsive tumours. They affect the tyrosine kinase domain of the EGFR - involving exons 18 until 21 - and lead to constitutive activation of the receptor, independent of ligand binding. The tumour becomes highly dependent on this pathway, and thus very sensitive to blockade of this pathway by TKIs [5]. Recent randomised trials demonstrated that the presence of an EGFR activating mutation was the best predictive factor for response and progression-free survival (PFS) to EGFR TKIs when compared to platinum doublet chemotherapy in the first-line therapy of advanced NSCLC [6],[7]. Given this strong benefit of EGFR-TKIs in patients with a tumour with an EGFR activating mutation (EGFR mut + tumour), molecular profiling became necessary in the assessment of stage IV NSCLC.
EGFR mutations are known to be associated with clinical characteristics such as never-smoking status, female gender, adenocarcinoma histology and South-East Asian ethnicity [8]. Adenocarcinoma is by far the most common NSCLC histology in South-East Asia, and about 40% of these tumours are EGFR mutant [9]. The EGFR mutation occurrence is much lower in Caucasian populations. As both gefitinib and erlotinib are now registered in Europe for the treatment of patients with an EGFR mut + tumour, selection criteria to identify which patients are most likely to have an EGFR mutation are needed. Based on the South-Asian experience and the occurrence of the different NSCLC histologies in Europe, a European guideline (ESMO consensus [1]) recommends EGFR mutation testing in never-/former light (<15 pack-years) smokers or patients with non-squamous NSCLC.
As a consequence and in contrast with 10 years ago, precise histological subtyping and EGFR testing is now mandatory. Many colleagues of the multidisciplinary team involved in the diagnostic flow of NSCLC - pulmonologists, surgeons and radiologists - need to optimise biopsy samples (size, content of tumour cells).
Until now, the most common diagnostic procedure for the diagnosis of NSCLC was bronchoscopy, which may however not always provide enough tissue for molecular analysis. We therefore evaluated the performance of different small tissue samples obtained during bronchoscopic procedures, such as bronchial biopsies and endobronchial ultrasound guided transbronchial needle aspirations (EBUS-TBNA), in the molecular diagnostic setting of NSCLC. Study aims were (1) the frequency of EGFR mutations (exon 18-21) in a large Caucasian cohort; (2) clinical and pathological predictors; (3) the sensitivity of the Therascreen kit; and (4) the performance of cytology versus biopsy samples for mutation analyses.
Methods
Study subjects
All consecutive EGFR mutation analysis reports in the period from September 2010 until December 2011 included were retrieved from the molecular genetics database. During this period, our local policy was to order EGFR mutation testing in all patients diagnosed with advanced NSCLC, either of adenocarcinomas or not-otherwise specified (NOS) histology, irrespective of smoking history, or in those rare patients with squamous cell carcinoma with a negative or light (<15 pack years) smoking history.
Methods
The clinical data of the patients were retrieved retrospectively from the clinical records. We categorised male and female patients in age, grouped as less than 60 years old, between 60 and 70 years and older than 70 years. Smoking history of the patients was obtained at baseline, and patients were categorised as never-smokers, former smokers (stop >1 year), or current smokers (stop < 1 year or still smoking). We also looked at the number of pack years.
Each patient had one sample in this analysis, either a cytological or histological one. Most of the histology samples were bronchial biopsies, with a few resection specimens, and most of the cytology samples were EBUS-TBNA samples. In each patient, at least 4-5 bronchial biopsies using a 2 mm forceps (Boston) were taken or at least 3 representative needle aspirations using a 22Ga EBUS-TBNA needle (Olympus) until a sufficient amount of microcores was obtained as judged by the operator. Samples underwent fixation in 6% neutral-buffered formalin, during 6-12 hours for bronchial biopsies and 8-18 hours for surgical specimens. EBUS-TBNA samples were collected in Cytorich Red fluid, formalin fixed for 6-12 hours and paraffin-embedded in a cell block.
Immunohistochemistry (IHC) for EGFR was carried out using a mixture of two mouse monoclonal antibodies, clone NCL-L-EGFR-384 (1:100, Novocastra) and clone E30 (1:50, Dako) in Dako's EnVison FLEX Antibody Diluent (Dako) on a Dako Autostainer Plus (Dako Denmark) according to the manufacturer's recommendations. EGFR expression was reported as positive in case of membranous brown staining in at least 10% of the cells. TTF-1 was also immunostained using a mouse monoclonal antibody (clone 8G7G3/1, ready-to-use, Dako), following the same procedure as described for EGFR. Tumours were considered TTF-1 positive in case of nuclear staining of neoplastic cells.
For EGFR mutation analysis, one H&E slide followed by 10 serial unstained sections (4-μM thick) and a final H&E were prepared from the paraffin block and both H&E sections were evaluated for the presence and amount of tumour cells by an experienced pathologist. After macro-dissection and deparaffinisation, the tissue was digested overnight with proteinase K. DNA extraction was done using the Maxwell16 FFPE tissue LEV DNA purification kit on the Maxwell16 instrument (Promega Corporation, Madison, WI) according to manufacturer's instructions. Mutational analysis of EGFR was performed using the Therascreen EGFR RGQ PCR kit (produced by Qiagen) on the LightCycler 480II instrument (Roche). The Therascreen assay is a sequence of two techniques, ARMS and Scorpion, to detect mutations in real-time PCRs with a reported detection limit of 1% mutant alleles in DNA from tumour tissue. This kit allows the detection of the nineteen deletions between 2235 and 2258 in exon 19, three insertions in exon 20 and the point mutations G719A/S/C (exon 18), S768I (exon 20), T790M (exon 20), L858R (exon 21) and L861Q (exon 21). The test has both EU as well as FDA approval.
Ethics
This research was approved as a retrospective, non-interventional, study by the local Institutional Review Board of the University Hospitals KU Leuven.
Results and discussion
Clinical characteristics
From September 2010 until December 2011, a total of 297 NSCLC samples were analysed (Table 1). The population consisted of 113 females (38%) and 184 males (62%). The median age was 68 years (SD 11), eighty patients were younger than 60 years, 99 were between 60 and 70 years old and 118 were older than 70 years. All patients were Caucasians. Smoking status was known in 248 patients, 40 (16%) were never smokers, 109 (44%) were former smokers (stop >1 year) and 99 (40%) were current smokers.
Table 1.
Clinical and pathological patient characteristics
| Patients (N) | EGFR mut + (N) | EGFR mut + (%) | |
|---|---|---|---|
| Total group | 297 | 32 | 11 |
| Gender | |||
| Female | 113 | 26 | 23 |
| Male | 184 | 6 | 3 |
| Age | |||
| <60 | 80 | 7 | 9 |
| 60-70 | 99 | 10 | 10 |
| >70 | 118 | 15 | 13 |
| Smoking | |||
| Never | 40 | 16 | 40 |
| Former | 109 | 8 | 7 |
| Current | 99 | 7 | 7 |
| Missing | 49 | 1 | - |
| Specimen | |||
| Cytology | 84 | 8 | 10 |
| Biopsy | 213 | 24 | 11 |
| Tumour histology | |||
| Adeno | 274 | 31 | 11 |
| NSCLC-NOS | 9 | 1 | 11 |
| Squamous | 14 | 0 | 0 |
| EGFR IHC | |||
| Positive | 254 | 29 | 12 |
| Negative | 9 | 0 | 0 |
| Missing | 34 | 3 | - |
| TTF-1 IHC | |||
| Positive | 171 | 29 | 17 |
| Negative | 45 | 1 | 2 |
| Missing | 81 | 2 | - |
EGFR mut+: presence of activating EGFR mutation; N: number; NSCLC-NOS: non-small cell lung cancer not otherwise specified; IHC: immunohistochemistry; TTF-1: thyroid transcription factor 1.
Pathology
We examined 84 (28%) cytology samples and 213 (72%) biopsy samples. Out of the 84 cytology specimens, 80 were EBUS-TBNA cellblocks, and 4 were pleural cytology. Biopsy samples were 114 bronchial biopsies, 48 archived resection specimens, 12 lymph nodes (7 mediastinoscopy, 5 supraclavicular), 17 pleural biopsies, 11 trans-thoracic lung tumour punctures, and 11 metastatic sites (2 bone, 2 brain, 2 liver, 4 soft tissue, 1 adrenal gland).
Two hundred and seventy-four patients had adenocarcinoma (92%), fourteen had squamous cell carcinoma (5%) and nine had NSCLC-NOS (3%). EGFR immunohistochemistry was known for 263 patients, and was negative (i.e. <10% membranous staining) in only 9 patients. TTF-1 status was known in 216 patients, and was positive in 171 patients (79%).
The percentage of tumour cells was described in 293 samples. Fifty-seven samples had less than 10% tumour cells (19%), 152 samples had between 10 and 50% tumour cells (52%) and 84 samples had more than 50% tumour cells (29%). There was no significant difference in percentage of tumour cells between the cytology specimens and the biopsy samples. Out of 84 cytology specimens, 22 had less than 10% tumour cells (26%) versus 35 samples in 213 biopsy specimen (16%). Out of 84 cytology specimens, 41 had between 10 and 50% tumour cells (49%) versus 111 samples in 213 biopsy specimens (52%). Out of 84 cytology specimens, 21 one had more than 50% tumour cells (25%) versus 63 samples in 213 biopsy specimens (30%) (Figure 1).
Figure 1.
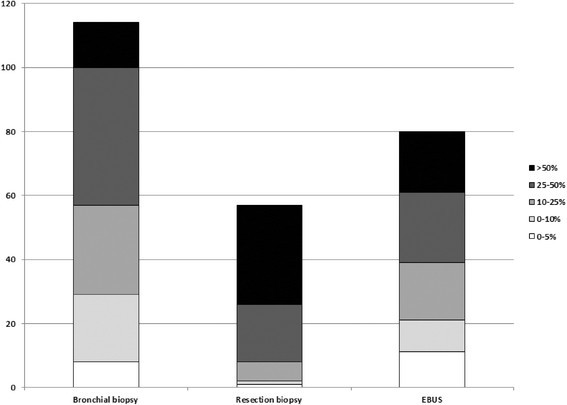
Cumulative numbers of tumour cell content in different samples. EBUS: needle aspiration in lymph node guided by endobronchial ultrasound guided procedure.
EGFR mutation test results
Activating EGFR mutations (EGFR mut+) were found in 32 patients (11%, Table 1): twelve patients had exon 19 deletions (38%), two patients had G719A/S/C point mutations in exon 18 (6%) and eighteen patients had exon 21 point mutations (56%), of which fifteen L858R mutations and three L861Q mutations. Inhibitory exon 20 insertion mutations were detected in five patients (2%).
Most mutations were found within the elderly population. Thirteen percent mutations in patients older than 70 years, 10% mutations in patients age between 60 and 70 years and 9% activating mutations in patients younger than 60 years.
EGFR mutation positive patients were mostly female (81%), 26 females versus 6 males. All but one were adenocarcinomas (97%), no activating mutations were found in squamous cell carcinomas. EGFR-mut + were clearly more present in never smokers (16/39, 41%) than in current (7/97, 7%) or former smokers (8/108, 7%).
All but one EGFR-mut + patients had immunohistochemistry (IHC) >50% for EGFR. This patient had 25% IHC + for EGFR. We found no mutations in samples with low (≤10%) EGFR-IHC. All but one EGFR-mut + patients had TTF-1 positivity.
We found 8 EGFR-mut + in 84 cytology specimens (10%) and 24 EGFR-mut + in 213 biopsy samples (11%).
Despite the fact that 21 out of 289 samples (7%) had low tumour cell percentage (0-5%), we found 7 activating mutations in these (33%). The Therascreen kit failed 4 times (1%). One failure was a bone biopsy, two had low (5%) tumour percentage and one was a bronchial biopsy with 25% of tumour cells. This confirms that the Therascreen kit is a very sensitive test, with low test failures, mostly when the tumour cell count was rated <5% by the pathologist.
Discussion
In the time interval September 2010 - December 2011, we selected advanced NSCLC patients for EGFR mutation screening in exon 18-21 by a PCR based test according to the ESMO criteria [1]. In our 297 patients, the testing was successful in all but four, and we detected an activating mutation in 11%. Not surprisingly, activating mutations were more frequently reported in women, never smokers, and pure adenocarcinoma.
Based on this series and review of other European findings, we believe that 9 to 12% may well be the real frequency of activating EGFR mutations in advanced NSCLC in Europe (Tables 2 and 3). Our frequency finding is in line with earlier Spanish work (12% [10], and very recent data from routine practice in e.g. the UK (9.7% [11], 10% [12]), Lithuania (9.7% [13]), and the Netherlands (8.4% [14]).
Table 2.
Demographics in European series on EGFR mutations testing
| Series | Year | Origin | N | Stages | Further description |
|---|---|---|---|---|---|
| Cortes-Funes et al. [10] | 2005 | Spain | 83 | Advanced | Failing 1st line/TKI therapy |
| Marchetti et al. [15] | 2005 | Italy | 860 | Stage I-II-III | Resection specimen |
| Rosell et al. [19] | 2009 | Spain | 2105 | Advanced | - |
| Penzel et al. [21] | 2011 | Germany | 1047 | All | - |
| Helland et al. [16] | 2011 | Norway | 240 | Stage I-II-III | Resection specimen |
| Vincenten et al. [20] | 2012 | Netherlands | 810 | All | - |
| Smits et al. [14] | 2012 | Netherlands | 778 | Advanced | - |
| Leary et al. [12] | 2012 | England | 144 | All | - |
| Pennycuick et al. [11] | 2012 | England | 215 | All | Non-squamous NSCLC |
| Vaguliene et al. [13] | 2012 | Lithuania | 103 | Advanced | Non-squamous NSCLC |
| Cadranel et al. [22] | 2012 | France | 522 | All | Failing 1st line/TKI therapy |
| This series | 2012 | Belgium | 297 | Advanced | ESMO guideline selected |
N: number; TKI: tyrosine kinase inhibitor; NSCLC: non-small cell lung cancer; ESMO: European Society of Medical Oncology.
Table 3.
Results in European series on EGFR mutation testing
| Series | Adeno (%) | Female (%) | Never-smokers (%) | EGFR mut + (%) | SQCC (N) |
|---|---|---|---|---|---|
| Cortes-Funes et al. [10] | 51 | 30 | 24 | 12 | 0 |
| Marchetti et al. [15] | 44 | 13 | 13 | 4.5 | 0 |
| Rosell et al. [19] | 85 | 39 | 29 | 16.6 | 0 |
| Penzel et al. [21] | NA | NA | NA | 14.7 | 0 |
| Helland et al. [16] | 59 | 48 | 6 | 6.6 | 2 |
| Vincenten et al. [20] | 65 | 49 | NA | 14 | 0 |
| Smits et al. [14] | 80 | 46 | 10 | 8.4 | 0 |
| Leary et al. [12] | 67 | 59 | NA | 10 | 0 |
| Pennycuick et al. [11] | NA | 57 | NA | 9.7 | 0 |
| Vaguliene et al. [13] | 63 | 25 | 21 | 9.7 | 0 |
| Cadranel et al. [22] | 65 | 32 | 18 | 14 | 0 |
| This series | 92 | 38 | 16 | 11 | 0 |
Some series have reported remarkably lower frequencies. These were investigations on resection specimens, an earlier one from Italy with a 4.5% occurrence [15], and a recent one from Norway (6.6% [16]). In another of our series on resection specimens, the frequency was lower as well at 7.0% (personal communication C. Dooms). This could be due to truly lower occurrence in early resectable stages, although this seems less likely. It has been postulated that mutations in the tyrosine kinase domain of EGFR are early events in lung carcinogenesis, already present in 50% of precancerous conditions such as atypical adenomatous hyperplasia, as well as in normal lung tissue surrounding a tumour [17]. More probable, several technical factors may play a role, as resection specimens usually don't suffer from tissue quantity but may suffer from tumour DNA quality. Indeed, the thoroughness of formalin fixation (which may be less good in large resection specimens) and the duration of storage (with DNA degradation over time) may affect the quality of extracted DNA and render the sample less suitable for mutation analysis. Williams et al. illustrated that as much as one artificial mutation per 500 bases may be recorded in the analysis of formalin-fixed material, because of formalin damaging or cross-linking cytosine nucleotides [18].
Several European studies also found higher frequencies than our study, but (unconscious) selection for testing based on clinical factors may have played a role in these findings. As a reminder and for comparison, in the hitherto largest - predominantly European - study in advanced NSCLC [2], there were 30% female patients and 14% never-smokers. In the more recent Spanish findings in the context of a randomised controlled trial, occurrence of exon 19 or 21 mutations was 16.6% [19]. In this series, there were 39% females, 29% never-smokers, and 75% never- and former smokers combined. In a recent Dutch series [20], the mutation rate was 14%, again with 49% females. In the very large German series, no information on gender or smoking is provided, but as the screening program was set up for the introduction of 1st line gefitinib in the treatment of advanced NSCLC, some pre-selection on clinical factors may again have been in place [21]. The series from France had a very different aim than the studies above [22], in particular examining the feasibility of EGFR and KRAS testing in the quite different context of use of erlotinib for 2nd or 3rd line treatment for all NSCLC histologies. In this group consisting of 86% never- or former smokers, the EGFR mutation rate was 14%.
We found no EGFR mutations in our never- or light-smokers with squamous cell carcinoma. This is consistent with previously published data. All European series found no mutations in squamous cell carcinoma, except for the Norwegian study, where two activating mutations were reported in surgical resection specimens. In a recent interesting pathology review study, all specimens with diagnosis of EGFR/KRAS-mutant squamous cell cancer were reassessed with modern techniques to differentiate squamous from adenocarcinoma differentiation [23]. Detailed morphologic and immunohistochemical study resulted in reclassification of 10 cases as adenosquamous cancer and 5 as poorly differentiated adenocarcinoma morphologically mimicking squamous cell cancer.
To optimize genetic testing in NSCLC, it is important to maximize the amount of tumour cells within the tissue sample. We investigated the tumour cell percentage on cytology and biopsy samples, and its effect on PCR-based detection of EGFR mutations. There was no significant difference between our bronchial biopsy samples and cell blocks of the cytology samples obtained by EBUS-TBNA. Our findings are in line with another recent multicentre series that examined if EBUS-TBNA cytology specimens are suitable for phenotyping and genotyping of NSCLC. A brief report from the Netherlands in 2010 suggested that specimens from FNA of the left adrenal gland were suitable for EGFR mutation analysis in 77% of the cases [24]. In a comparative study of biopsy and fine needle aspiration samples, the latter yielded lower DNA amounts but a comparable success rate in EGFR mutation analysis [25]. In a recent large series, EGFR mutation analysis was possible on cell block samples of 107/119 (90%) patients in whom it was requested [26].
ESMO guidelines advocate an EGFR TKI as the preferred first-line treatment in NSCLC patients whose tumour harbours an activating EGFR mutation [1]. A remaining practical controversy is how waiting time for EGFR mutation analysis should be balanced with delay of start of treatment with platinum doublet chemotherapy, especially as the overall survival with both strategies has hitherto not been shown to be significantly different [27]. Clinical features are related to but not sufficiently reliable to estimate the presence of EGFR mutations. Consequently, a rapid test with high negative predictive value (NPV) for EGFR mutations is welcome. TTF-1 IHC seems to fill this gap. Indeed, in series where this relationship is reported (Table 4), the NPV varies between 96 and 100%, although the NPV may be overestimated, as all series have a rather low prevalence of EGFR mutation. As many advanced NSCLC samples nowadays undergo IHC staining for the differentiation between adenocarcinoma or squamous cell carcinoma [28], TTF-1 staining is included or a little extra with great potential for the clinician: if the TTF-1 IHC is negative, doublet chemotherapy can be started. If an EGFR mutation is eventually detected, the EGFR-TKI can be given thereafter. Moreover, there is a good biological rationale for this high NPV, as TTF-1 expression is found in the terminal respiratory unit (TRU) cells, where peripheral tumours with a non-mucinous lepidic or papillary predominant pattern arise. Both types are known to harbour more frequently activating EGFR mutations [29].
Table 4.
Correlation between TTF-1 immunohistochemistry and presence of EGFR activating mutation
| Series | EGFR mut + (N) | EGFR mut- (N) | |
|---|---|---|---|
| Vincenten et al. [20] | NPV | ||
| TTF-1 + | 105 | 403 | 96% |
| TTF-1 - | 9 | 228 | |
| Leary et al. [12] | NPV | ||
| TTF-1 + | 15 | 40 | 100% |
| TTF-1 - | 0 | 15 | |
| This series | NPV | ||
| TTF-1 + | 29 | 140 | 98% |
| TTF-1 - | 1 | 43 | |
| Pooled results | NPV | ||
| TTF- 1 + | 149 | 583 | 97% |
| TTF-1 - | 10 | 286 |
EGFR mut+: presence of activating EGFR mutation; N: number; TTF-1: thyroid transcription factor 1; NPV: negative predictive value.
Conclusion
Both biopsies and cell blocks of EBUS-TBNA are suitable samples for EGFR mutation analysis when a sensitive method such as the Therascreen EGFR RGQ PCR mutation kit is used. We document an 11% frequency of EGFR activating mutations in our Caucasian standard practice NSCLC cohort, selected according to the ESMO consensus guideline.
Acknowledgments
This work was supported by the Jean-Francois Peterbroeck Chair for Translational Lung Cancer Research (patient donation for research), and the Astra Zeneca Chair in Personalised Lung Cancer Care (research funding) at the University of Leuven.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Competing interests
J. Vansteenkiste is holder of the Astra Zeneca Chair in Personalised Lung Cancer Care at the Leuven University (research funding).
Authors' contributions
IH: Acquisition of data; Analysis and interpretation of data; Writing, review and/or revision of the manuscript. CD: Development of methodology; Acquisition of data; Analysis and interpretation of data; Writing, review and/or revision of the manuscript. EV: Execution of pathological assessment of samples and macrodissection for genetic testing; Writing, review and/or revision of the manuscript. PV: Development of methodology; Execution of molecular genetic studies; Writing, review and/or revision of the manuscript. LV: Development of methodology; Execution of molecular genetic studies; Writing, review and/or revision of the manuscript. TR: Development of methodology; Execution of molecular genetic studies. SV: Development of methodology; Execution of molecular genetic studies; Analysis and interpretation of data; Writing, review and/or revision of the manuscript. KN: Acquisition of data; Writing, review and/or revision of the manuscript. IW: Acquisition of data; Writing, review and/or revision of the manuscript. JV: Conception and design; Acquisition of data; Analysis and interpretation of data; Writing, review and/or revision of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Inge Hantson, Email: ingehantson@hotmail.com.
Christophe Dooms, Email: christophe.dooms@uzleuven.be.
Eric Verbeken, Email: eric.verbeken@uzleuven.be.
Peter Vandenberghe, Email: peter.vandenberghe@uzleuven.be.
Liesbet Vliegen, Email: liesbet.vliegen@med.kuleuven.be.
Tania Roskams, Email: tania.roskams@uzleuven.be.
Sara Vander Borght, Email: sara.vanderborght@uzleuven.be.
Kris Nackaerts, Email: kristiaan.nackaerts@uzleuven.be.
Isabelle Wauters, Email: isabelle.wauters@uzleuven.be.
Johan Vansteenkiste, Email: johan.vansteenkiste@uzleuven.be.
References
- 1.Felip E, Gridelli C, Baas P, Rosell R, Stahel R. Metastatic non-small cell lung cancer: consensus on pathology and molecular tests, first-line, second-line, and third-line therapy: 1st ESMO Consensus Conference in Lung Cancer; Lugano 2010. Ann Oncol. 2011;22:1973–1980. doi: 10.1093/annonc/mdr150. [DOI] [PubMed] [Google Scholar]
- 2.Scagliotti G, Parikh P, Von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarthy SR, Zukin M, Lee JS, Mellemgaard A, Park B, Patil S, Rolski J, Goksel T, De Marinis F, Simms L, Sugarman K, Gandara D. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced stage non-small cell lung cancer. J Clin Oncol. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 3.Sibille A, Wauters I, Vansteenkiste J. Maintenance therapy for advanced non-small cell lung cancer: ready for clinical practice? Expert Rev Anticancer Ther. 2012;12:529–539. doi: 10.1586/era.12.22. [DOI] [PubMed] [Google Scholar]
- 4.Fukuoka M, Yano S, Giaccone G, Tamura T, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudo S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, McLeod A, Wolf M, Feyereislova A, Dong RP, Baselga J. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small cell lung cancer. J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the Epidermal Growth Factor Receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, De Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Munoz-Langa J, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CC, Hsu HY, Liu HP, Chang JW, Chen YT, Hsieh WY, Hsieh JJ, Hsieh MS, Chen YR, Huang SF. Reversed mutation rates of KRAS and EGFR genes in adenocarcinoma of the lung in Taiwan and their implications. Cancer. 2008;113:3199–3208. doi: 10.1002/cncr.23925. [DOI] [PubMed] [Google Scholar]
- 10.Cortes-Funes H, Gomez C, Rosell R, Valero P, Garcia-Giron C, Velasco A, Izquierdo A, Diz P, Camps C, Castellanos D, Alberola V, Cardenal F, Gonzalez-Larriba JL, Vieitez JM, Maeztu I, Sanchez JJ, Queralt C, Mayo C, Mendez P, Moran T, Taron M. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small cell lung cancer patients. Ann Oncol. 2005;16:1081–1086. doi: 10.1093/annonc/mdi221. [DOI] [PubMed] [Google Scholar]
- 11.Pennycuick A, Simpson T, Crawley D, Lal R, Santis G, Cane P, Tobal K, Spicer J. Routine EGFR and KRAS mutation analysis using COLD-PCR in non-small cell lung cancer. Int J Clin Pract. 2012;66:748–752. doi: 10.1111/j.1742-1241.2012.02961.x. [DOI] [PubMed] [Google Scholar]
- 12.Leary AF, Castro DG, Nicholson AG, Ashley S, Wotherspoon A, O'Brien ME, Popat S. Establishing an EGFR mutation screening service for non-small cell lung cancer - sample quality criteria and candidate histological predictors. Eur J Cancer. 2012;48:61–67. doi: 10.1016/j.ejca.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Vaguliene N, Zemaitis M, Sarauskas V, Vitkauskiene A, Miliauskas S. The role of mutation status of the epidermal growth factor receptor gene in advanced non-small cell lung cancer. Medicina (Kaunas) 2012;48:175–181. [PubMed] [Google Scholar]
- 14.Smits AJ, Kummer JA, Hinrichs JW, Herder GJ, Scheidel-Jacobse KC, Jiwa NM, Ruijter TE, Nooijen PT, Looijen-Salamon MG, Ligtenberg MJ, Thunnissen FB, Heideman DA, De Weger RA, Vink A. EGFR and KRAS mutations in lung carcinomas in the Dutch population: increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell Oncol (Dordr) 2012;35:189–196. doi: 10.1007/s13402-012-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non-small cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 16.Helland A, Skaug HM, Kleinberg L, Iversen ML, Rud AK, Fleischer T, Sagerup C, Solberg S, Jorgensen L, Ariansen S, Brustugun OT. EGFR gene alterations in a Norwegian cohort of lung cancer patients selected for surgery. J Thorac Oncol. 2011;6:947–950. doi: 10.1097/JTO.0b013e31820db209. [DOI] [PubMed] [Google Scholar]
- 17.Sakuma Y, Matsukuma S, Yoshihara M, Nakamura Y, Nakayama H, Kameda Y, Tsuchiya E, Miyagi Y. Epidermal growth factor receptor gene mutations in atypical adenomatous hyperplasias of the lung. Mod Pathol. 2007;20:967–973. doi: 10.1038/modpathol.3800929. [DOI] [PubMed] [Google Scholar]
- 18.Williams C, Ponten F, Moberg C, Soderkvist P, Uhlen M, Ponten J, Sitbon G, Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sanchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 20.Vincenten J, Smit EF, Vos W, Grunberg K, Postmus PE, Heideman DA, Snijders PJ, Meijer G, Kuik J, Witte BI, Thunnissen E. Negative NKX2-1 (TTF-1) as temporary surrogate marker for treatment selection during EGFR-mutation analysis in patients with non-small cell lung cancer. J Thorac Oncol. 2012;7:1522–1527. doi: 10.1097/JTO.0b013e3182635a91. [DOI] [PubMed] [Google Scholar]
- 21.Penzel R, Sers C, Chen Y, Lehmann-Mühlenhoff U, Merkelbach-Bruse S, Jung A, Kirchner T, Büttner R, Kreipe HH, Petersen I, Dietel M, Schirmacher P. EGFR mutation detection in NSCLC - assessment of diagnostic application and recommendations of the German panel for mutation testing in NSCLC. Virchows Arch. 2011;458:95–98. doi: 10.1007/s00428-010-1000-y. [DOI] [PubMed] [Google Scholar]
- 22.Cadranel J, Mauguen A, Faller M, Zalcman G, Buisine MP, Westeel V, Longchampt E, Wislez M, Coudert B, Daniel C, Chetaille B, Michiels S, Blons H, Solassol J, De Fraipont F, Foucher P, Urban T, Lacroix L, Poulot V, Quoix E, Antoine M, Danton G, Morin F, Chouaid C, Pignon JP. Impact of systematic EGFR and KRAS mutation evaluation on progression-free survival and overall survival in patients with advanced non-small cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project-part 2) J Thorac Oncol. 2012;7:1490–1502. doi: 10.1097/JTO.0b013e318265b2b5. [DOI] [PubMed] [Google Scholar]
- 23.Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, Travis WD, Zakowski MF, Kris MG, Ladanyi M. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–1176. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuurbiers OC, Looijen-Salamon MG, Ligtenberg MJ, Van der Heijden HF. A brief retrospective report on the feasibility of epidermal growth factor receptor and KRAS mutation analysis in transesophageal ultrasound- and endobronchial ultrasound-guided fine needle cytological aspirates. J Thorac Oncol. 2010;5:1664–1667. doi: 10.1097/JTO.0b013e3181f0bd93. [DOI] [PubMed] [Google Scholar]
- 25.Arcila ME, Oxnard GR, Nafa K, Woolhouse I, Harrison RN, Jeebun V, Munavvar M, Ng BJ, Rassl DM, Falzon M, Kocjan G, Rintoul RC, Nicholson AG, Janes SM. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navani N, Brown JM, Nankivell M, Woolhouse I, Harrison RN, Jeebun V, Munavvar M, Ng BJ, Rassl DM, Falzon M, Kocjan G, Rintoul RC, Nicholson AG, Janes SM. Suitability of endobronchial ultrasound-guided transbronchial needle aspiration specimens for subtyping and genotyping of non-small cell lung cancer: a multicenter study of 774 patients. Am J Respir Crit Care Med. 2012;185:1316–1322. doi: 10.1164/rccm.201202-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran T, Sequist LV. Timing of epidermal growth factor receptor tyrosine kinase inhibitor therapy in patients with lung cancer with EGFR mutations. J Clin Oncol. 2012;30:3330–3336. doi: 10.1200/JCO.2012.43.1858. [DOI] [PubMed] [Google Scholar]
- 28.Warth A, Muley T, Herpel E, Meister M, Herth FJ, Schirmacher P, Weichert W, Hoffmann H, Schnabel PA. Large-scale comparative analyses of immunomarkers for diagnostic subtyping of non-small cell lung cancer biopsies. Histopathology. 2012;61:1017–1025. doi: 10.1111/j.1365-2559.2012.04308.x. [DOI] [PubMed] [Google Scholar]
- 29.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]


