Polycomb-repressive complex 2 (PRC2) localizes to the promoter regions of repressed genes giving rise to H3K27me2/3 but also localizes to nascent transcripts from active genes that are devoid of H3K27me2/3 in embryonic stem cells. Kaneko et al. show that RNA interaction inhibits SET domain-containing proteins, such as PRC2. CRISPR-mediated truncation of a PRC2-interacting nascent RNA rescued PRC2-mediated deposition of H3K27me2/3. This study supports a model in which PRC2 can only mark for repression those genes silenced by transcriptional repressors.
Keywords: chromatin, EZH2, JARID2, PRC2, ncRNA
Abstract
Polycomb-repressive complex 2 (PRC2) facilitates the maintenance and inheritance of chromatin domains repressive to transcription through catalysis of methylation of histone H3 at Lys27 (H3K27me2/3). However, through its EZH2 subunit, PRC2 also binds to nascent transcripts from active genes that are devoid of H3K27me2/3 in embryonic stem cells. Here, biochemical analyses indicated that RNA interaction inhibits SET domain-containing proteins, such as PRC2, nonspecifically in vitro. However, CRISPR-mediated truncation of a PRC2-interacting nascent RNA rescued PRC2-mediated deposition of H3K27me2/3. That PRC2 activity is inhibited by interactions with nascent transcripts supports a model in which PRC2 can only mark for repression those genes silenced by transcriptional repressors.
Based on early genetic studies in Drosophila (i.e., Schuettengruber et al. 2007; Simon and Kingston 2009), the Polycomb group (PcG) of proteins is key to maintaining gene repression already established by master regulators during lineage commitment. This feature of maintaining as opposed to establishing initial repression is an important consideration in understanding the mechanism underlying PcG protein recruitment to chromatin and the resultant reinstatement of appropriate PcG-patterned gene expression upon cell division when master regulators are no longer present. The means by which this already programmed gene repression is conveyed to newly replicated genomes entails epigenetic processes that include the activities of PcG proteins.
In Drosophila, the Polycomb-repressive elements (PREs) comprise domains recognized by sequence-specific DNA-binding proteins that facilitate PcG protein recruitment to chromatin (for review, see Simon and Kingston 2009). However, in mammals, very few such PREs have been identified, their role in PcG recruitment is controversial, and, in general, the recruitment of mammalian PcG proteins appears not to be mediated by PREs (for review, see Margueron and Reinberg 2011). Mammalian PcG proteins exist in two major complexes, Polycomb-repressive complex 1 (PRC1) and PRC2, that are functionally interrelated and maintain appropriately repressed chromatin domains. The PRC2 core complex comprises EZH2, EED, SUZ12, and RBBP4/7, where EZH2 catalyzes methylation of histone H3 at Lys27 (H3K27me2/3), a modification associated with transcriptionally silent facultative heterochromatin (for review, see Margueron and Reinberg 2011; Simon and Kingston 2013). PRC2 recognizes the product of its own catalysis, H3K27me2/3, through its EED subunit, leading to a stimulation of its histone lysine methyltransferase (HKMT) activity and providing a means for H3K27me2/3 propagation (Margueron et al. 2009). The core PRC2 complex associates with other factors that significantly enhance its HKMT activity, such as AEBP2 and JARID2 (Cao and Zhang 2004; Li et al. 2010, respectively). JARID2 is essential for early development, being expressed in undifferentiated cells, declining in expression as a function of differentiation, and undetectable in fully differentiated cells (Takeuchi et al. 1995; Peng et al. 2009; Shen et al. 2009; Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010, Son et al. 2013). Our previous studies demonstrated that JARID2 binds to the catalytic component of core PRC2, EZH2, and stimulates its HKMT activity through an allosteric mechanism (Li et al. 2010; Son et al. 2013). Moreover, as opposed to core PRC2, JARID2 exhibits nucleosome-binding activity, facilitating PRC2 access to nucleosomes (Son et al. 2013).
While we are beginning to understand how H3K27me2/3 domains are maintained over time and cell divisions, it remains unclear what triggers the placement of these histone modifications in the first place, when transcriptional repression is initially established. How is transcriptional repression monitored by PRC2, for example? Both EZH2 and JARID2 contain RNA-binding regions (RBRs) that recognize specific long noncoding RNA (lncRNAs) (Kaneko et al. 2010), such as MEG3, which enhance the association of EZH2 with JARID2 (Kaneko et al. 2014). Our studies with MEG3 and those of others with RepA point to an important role for lncRNAs in recruiting PRC2/JARID2 to the appropriate loci (Zhao et al. 2008; Kaneko et al. 2014). However, EZH2 does not recognize only lncRNAs. We and others found that, remarkably, PRC2 localizes to not only the promoter regions of repressed genes giving rise to H3K27me2/3, as expected, but also those of active genes in embryonic stem cells (ESCs), and its EZH2 component binds to the 5′ terminus of the nascent transcripts (ezRNAs) emanating from these active genes (Davidovich et al. 2013; Kaneko et al. 2013). Interestingly, promoters of ezRNA-producing genes contain reduced levels of H3K27me2/3 despite the fact that they are occupied by PRC2 (Davidovich et al. 2013; Kaneko et al. 2013). These results led us to hypothesize that PRC2 senses the transcriptional activity of genes through nascent RNA binding that in turn might preclude its HKMT activity. When transcription is silenced by, for example, master regulatory repressors, the absence of nascent transcripts would relieve PRC2 inhibition, allowing for H3K27me2/3 catalysis at the newly repressed loci. Here, we investigate this hypothesis as well as the input to this phenomenon from the functionally relevant PRC2 partner JARID2.
Results and Discussion
RNA interaction inhibits PRC2 holoenzyme (Holo-PRC2 HKMT) activity in vitro
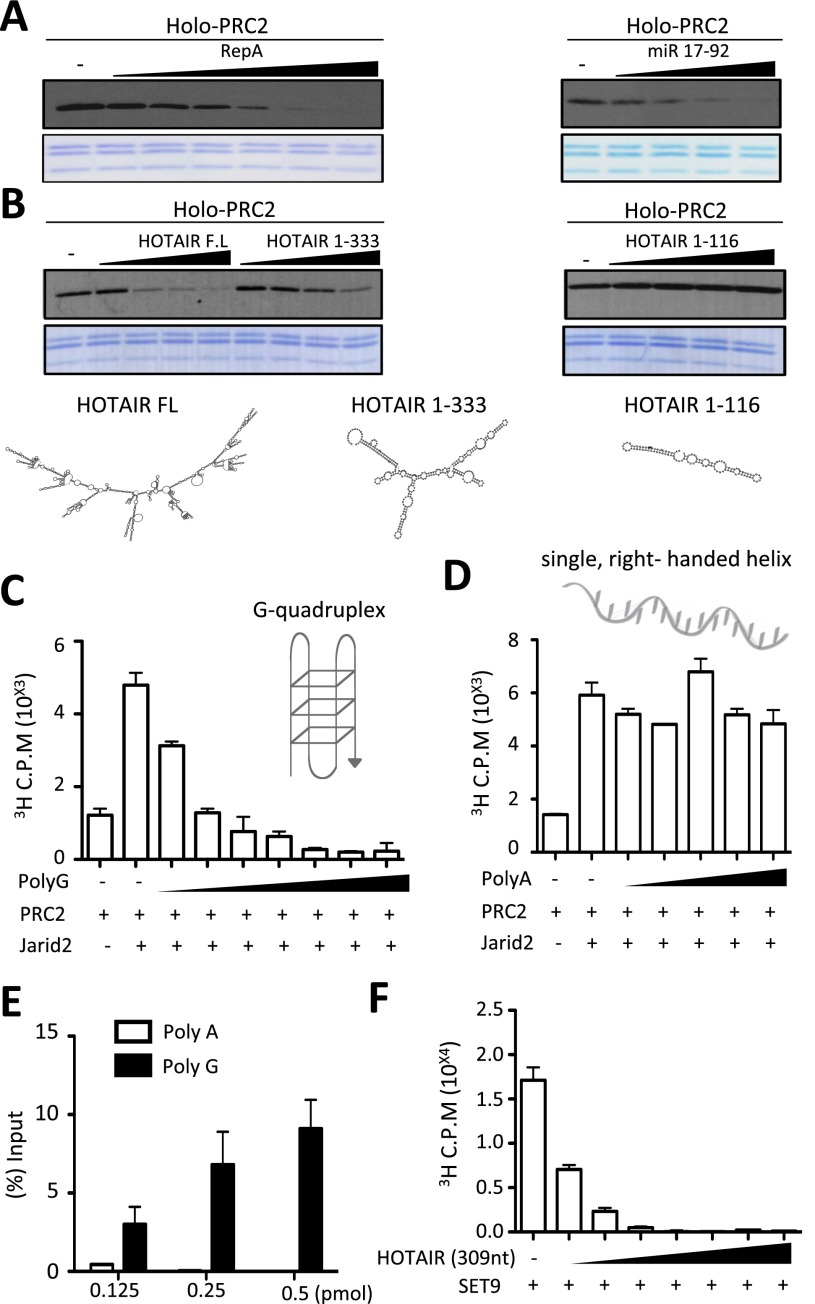
To test our hypothesis that RNA interaction might inhibit PRC2 activity, we first examined the levels of HKMT activity from recombinant Holo-PRC2 comprising core PRC2 components EZH2, EED, SUZ12, and RBBP4 and associated partners JARID2 and AEBP2 in vitro as a consequence of the presence of candidate RNAs that associate in vitro and in vivo with Holo-PRC2: RepA (Zhao et al. 2008) and pre-microRNA 17–92 (mir17–92), which forms two stem–loops (Kaneko et al. 2014). Holo-PRC2-mediated methylation of recombinant nucleosomal substrates decreased as a function of the presence of increasing amounts of either RepA or mir17–92 (Fig. 1A).
Figure 1.
Holo-PRC2 activity is inhibited by RNA in vitro. (A) HKMT assay performed with increasing amounts of RepA (0.2–6.4 pmol) (left panel) or miR17–92 (0.2–1.6 pmol) (right panel) on 4.56 pmol of recombinant nucleosomes using 1 pmol of PRC2 with 2.5 pmol of JARID2 (residues 119–574) and 4 pmol of AEBP2 for 1 h. Results were analyzed by autoradiography using [3H]ENHANCE spray after SDS-PAGE. (B, top) HKMT assay as in A using HOTAIR FL (0.16–1.25 pmol), HOTAIR1–333 (1–8 pmol), or HOTAIR1–116 (2.87–23 pmol). (Bottom) RNA structure prediction using the ViennaRNA package. (C,D) HKMT assay performed with increasing amounts of either Poly(G) (1–64 pmol) (C) or Poly(A) (4–64 pmol) (D) on 2.28 pmol of native nucleosomes using 1 pmol of PRC2 with 1.5 pmol of JARID2 (residues 119–574). The assay was incubated for 15 min with 12.5 μM SAM (radiolabeled:cold: 1:45) and analyzed by scintillation counting after SDS-PAGE. (E) Core PRC2 and RNA-binding assays. (F) HKMT assay using 1 pmol of SET9 and recombinant octamers with increasing amounts of HOTAIR1–309 (1–64 pmol).
Previous reports indicated that the presence of putative secondary structural motifs within lncRNAs might impact their interaction with PRC2 (Zhao et al. 2008; Tsai et al. 2010). Indeed, the predicted structure of full-length (FL) HOTAIR RNA that interacts with Holo-PRC2 exhibits an extensive secondary structure (Fig. 1B, bottom schematic) that is also evident in its truncated form, comprising 1–333 nucleotides (nt). Increasing amounts of either FL or 1- to 333-nt versions of HOTAIR RNA also led to decreased levels of Holo-PRC2-mediated HKMT activity (Fig. 1B, left panel). Core PRC2 activity was also inhibited by HOTAIR 1–333 in a dose-dependent manner (Supplemental Fig. S1). In contrast, a shorter version of HOTAIR comprising its first 116 nt with a much less complex predicted structure was ineffectual (Fig. 1B, right panel).
These results suggested that the length and/or structure of the interacting RNA are determinant in Holo-PRC2 inhibition. To distinguish these possibilities, we next compared ribonucleotide polymers of comparable lengths (data not shown) but starkly different structures: Poly(G) and Poly(A). Poly(G) RNA forms a three-dimensional structure known as a G quadruplex (Millevoi et al. 2012), whereas, in the neutral pH conditions of this assay, Poly(A) RNA does not fold and remains in an extended right-handed helix (Seol et al. 2007). As previously shown (Li et al. 2010; Son et al. 2013), core PRC2 HKMT activity is greatly stimulated in the presence of its associated partner, JARID2 (Fig. 1C,D). The highly disparate structural features of Poly(G) and Poly(A) were reflected in their contrasting effects on JARID2/PRC2-mediated HKMT activity, with increasing amounts of the quadruplex structure associated with Poly(G) being inhibitory and that of the unfolded, right-handed helix associated with Poly(A) being ineffectual (Fig. 1C,D, respectively). In keeping with this finding, PRC2 was able to bind Poly(G) but not Poly(A) (Fig. 1E).
Although the structure of Poly(G) RNA is likely very different from the stem–loops formed by bona fide EZH2-interacting RNAs such as HOTAIR and RepA, nonetheless, we speculate that some of the three-dimensional features of Poly(G), such as the simultaneous presence of double-stranded segments and single-stranded loops, are sufficient to mimic binding to structured RNA in vitro and therefore result in PRC2 inhibition. Thus, our results strongly implicate not only RNA length, as previously shown (Davidovich et al. 2013), but also sequence composition and secondary structure as a defining feature in Holo-PRC2 binding and suggest that such binding leads to inhibition of Holo-PRC2-mediated catalysis of H3K27me2/3 in vitro.
Lack of specificity in RNA-mediated inhibition of SET activity in vitro
The HKMT activity inhibited by RNA interaction, as shown above, resides within the SET domain of the core PRC2 component EZH2. However, SET domain-containing proteins have in general been shown to interact with ssDNA and ssRNA (Krajewski et al. 2005). One such SET domain-containing protein is SET9. Unlike EZH2, SET9 contains a split SET domain and is capable of monomethylating different proteins, including histone H3, rather than nucleosomes, in vitro; therefore, its activity is independent of DNA and RNA. Nonetheless, similar to its inhibitory effect on Holo-PRC2 as well as core PRC2 (Supplemental Fig. S1), HOTAIR RNA also inhibited SET9 HMT activity using histone octamers as substrates in vitro (Fig. 1F). Similar results were obtained with PR-Set7 (data not shown), leading us to conclude that RNA-mediated inhibition of SET domain-containing proteins, including Holo-PRC2, was not specific, at least under these in vitro conditions.
In spite of this lack of specificity, two important features regarding the nature and consequences of the interaction of Holo-PRC2 with RNA became evident from this and previous analyses. First, the length of RNA is determinant, consistent with findings from the Cech laboratory (Davidovich et al. 2013) that demonstrated an RNA length-dependent interaction with Holo-PRC2. Although this was also apparent in the case of HOTAIR RNA as shown above (Fig. 1B) and Ntn1 nascent RNAs (Fig. 2E; see below), we cannot exclude the possibility that different length RNA might have different branched structures. Second, RNA structure is also a key determinant, as evidenced by the observation that equally sized polymers of adenosine versus guanosine monophosphates give rise to striking differences in Holo-PRC2 binding and inhibition (Fig. 1C,D).
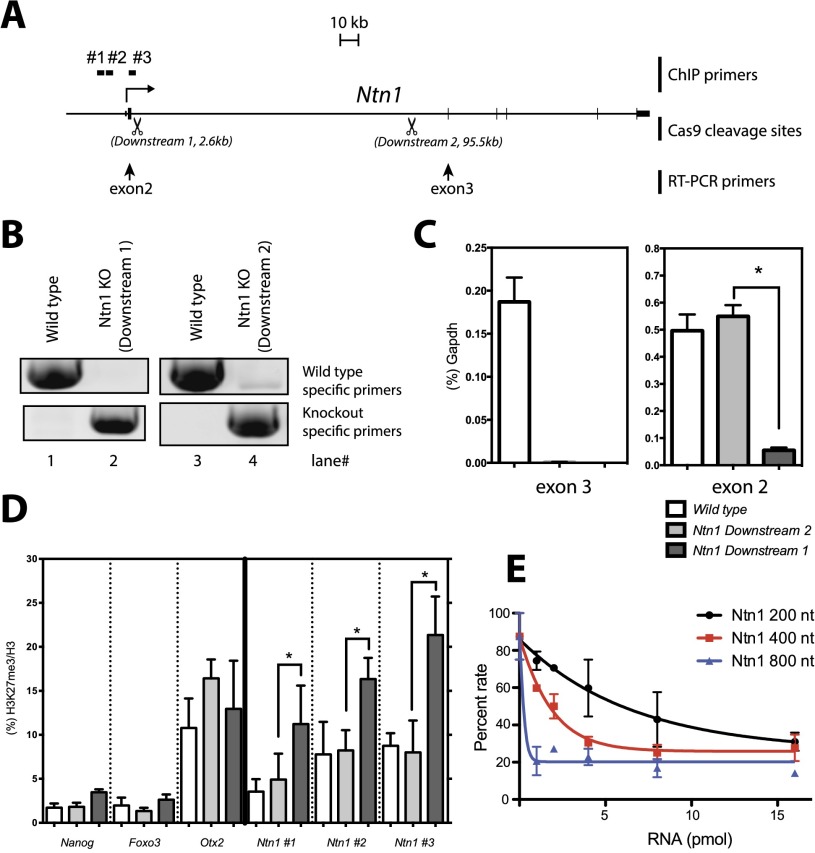
Figure 2.
Transcription termination proximal to the TSS results in increased H3K27me3 at the Ntn1 promoter. (A) Schematic of the Ntn1 gene locus. CRISPR-Cas9 cleavage sites (Downstream 1: 2.6 kb downstream from the TSS; Downstream 2: 95.5 kb downstream from the TSS) are shown by scissors. Position of the RT–PCR primer sets used in C are shown by arrows. Positions of ChIP primer sets (#1, #2, #3) used in D are also indicated. (B) PCR genotyping. (C) RT-qPCR normalized to Gapdh levels on wild-type (white bars) and Ntn1 Downstream 2 (gray bars) and Downstream 1 (black bars) mESCs. Bars represents the mean of three biological replicates +SEM. (*) P < 0.05 by Mann-Whitney U-test. (D) ChIP-qPCR for H3K27me3 normalized by H3 on wild-type (white bars) and Ntn1 downstream knockout (KO) (gray bars) and upstream knockout (black bars) mESCs for the genes indicated at the bottom. All ChIP primer sets are targeted to respective promoter regions. Bars represents the mean of three biological replicates +SEM. (*) P < 0.05 by Mann-Whitney U-test. (E) HKMT assay as in Figure 1 using 1–16 pmol of each of the various sized runoff transcripts derived from Ntn1. Values are given as a percentage of enzymatic activity obtained in the absence of RNA.
Holo-PRC2 HKMT activity is inhibited by nascent RNAs in vivo
Given the lack of specificity in RNA-mediated inhibition of HKMT activity observed in vitro, we sought an in vivo setting to investigate further the relevance of RNA interaction to PRC2 function. To this end, in mouse ESCs (mESCs), we chose one of the cohorts of genes previously identified as actively producing nascent transcripts to which the EZH2 component of PRC2 binds (ezRNAs) yet containing reduced amounts of H3K27me2/3 at their respective promoter regions (Kaneko et al. 2013). We used the CRISPR-Cas9 methodology (Supplemental Fig. S2) to generate mESCs containing derivatives of the chosen candidate Ntn1 gene in which an effective polyadenylation site was inserted independently at two positions: 2.6 kb downstream (Downstream 1) and 95.5 kb downstream (Downstream 2) from the transcriptional start site (TSS) of Ntn1 (Fig. 2A,B). To minimize potential adverse effects of puromycin, we drug-selected mESC clones for 48 h, and these clones were subsequently maintained without puromycin (see the Materials and Methods).
Comparison of RT–PCR results using primers specific to exons 2 and 3 and mRNA isolated from wild-type, Downstream 1, and Downstream 2 mESC lines demonstrated that Downstream 1 is deficient in production of both exon 2- and exon 3-containing mRNAs from the Ntn1 gene, while Downstream 2 is deficient in only exon 3-containing mRNAs when normalized to Gapdh mRNA levels (Fig. 2C). Although the effect of the Downstream 1 insertion on the immediately upstream exon 2 was unexpected, it resulted in an mESC line with much decreased levels of nascent Ntnt1 ezRNAs, which we were able to use for the subsequent experiments.
Importantly, mRNA levels from several pluripotent genes (Oct4, Nanog, and Sox2) were consistent between these three mESC lines, suggesting that the genetic manipulations of the Ntn1 locus did not affect pluripotency in mESCs (Supplemental Fig. S3). In addition, chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) performed with antibody against H3K27me2/3 in the case of wild-type versus Downstream 1 and Downstream 2 cell lines did not reveal any differences of note in the levels of H3K27me2/3 at the promoter region of actively transcribing pluripotent genes such as Nanog and Foxo3 or of the repressed developmental gene Otx2 (Fig. 2D). However, while the levels of H3K27me2/3 at the respective Ntn1 promoter regions of wild type and Downstream 2 were similar, the levels of H3K27me2/3 were markedly elevated in the case of Downstream 1 (Fig. 2D). Indeed, the length of the Ntn1 transcript was determinant to its inhibitory effect on PRC2 HKMT activity in vitro, as evidenced by results obtained using in vitro transcribed fragments of the Ntn1 pre-mRNA of different lengths (Fig. 2E). Given that PRC2-mediated catalysis of H3K27me2/3 at the Ntn1 promoter was dependent on the length and perhaps the structure of the nascent Ntn1 transcript permitted in the case of Downstream 1 versus Downstream 2, we conclude that Holo-PRC2 binding to nascent RNA does inhibit its HKMT activity at the wild-type promoter in vivo. Thus, the absence of H3K27me2/3 at actively transcribing promoters that nonetheless exhibit PRC2 bound to their respective nascent transcripts (Kaneko et al. 2013) is a consequence of interacting RNA-mediated inhibition of Holo-PRC2 catalysis.
This in vivo observation is consistent with the inhibition of Holo-PRC2 and core PRC2 activity that we obtained in vitro despite its lack of specificity in those settings. A recent report also investigated the role of RNA interaction with respect to PRC2 activity, concluding that RNA interaction inhibits PRC2 activity in vitro (Cifuentes-Rojas et al. 2014). In addition, that report presented evidence that JARID2 relieved RNA-mediated PRC2 repression (Cifuentes-Rojas et al. 2014). While, in principle, this and our report agree that RNA inhibits core PRC2 activity, the lack of specificity in this inhibition observed in vitro led us to find the data inconclusive. Moreover, in pursuing this question in vivo, we found that in undifferentiated mESCs that contain JARID2, RNA interaction inhibits Holo-PRC2 activity. We believe that these discrepancies are due to the methodology used for reconstituting PRC2 in vitro. We and others have observed that while the integrity of PRC2 is not impinged when all relevant subunits are reconstituted through their coexpression via baculovirus, the reconstitution of PRC2 from subunits expressed in isolation might result in PRC2 aggregation/misfolding. Indeed, such a discrepancy in the reconstitution of PRC2 led to a previous controversy as to whether JARID2 itself stimulates or inhibits PRC2 activity (Peng et al. 2009; Shen et al. 2009). It is now recognized that JARID2 stimulates PRC2 activity (Li et al. 2010; Mejetta et al. 2011; Zhang et al. 2011; Son et al. 2013; Cifuentes-Rojas et al. 2014; Kalb et al. 2014; this study).
Similar to EZH2, JARID2 binds nascent transcripts in vivo
Our previous studies demonstrated that JARID2 contains an RBR necessary for its interaction with the lncRNA MEG3 in vitro and in vivo and that MEG3 interaction promotes JARID2 interaction with the core PRC2 catalytic subunit EZH2 as well as PRC2 recruitment to chromatin (Kaneko et al. 2014).
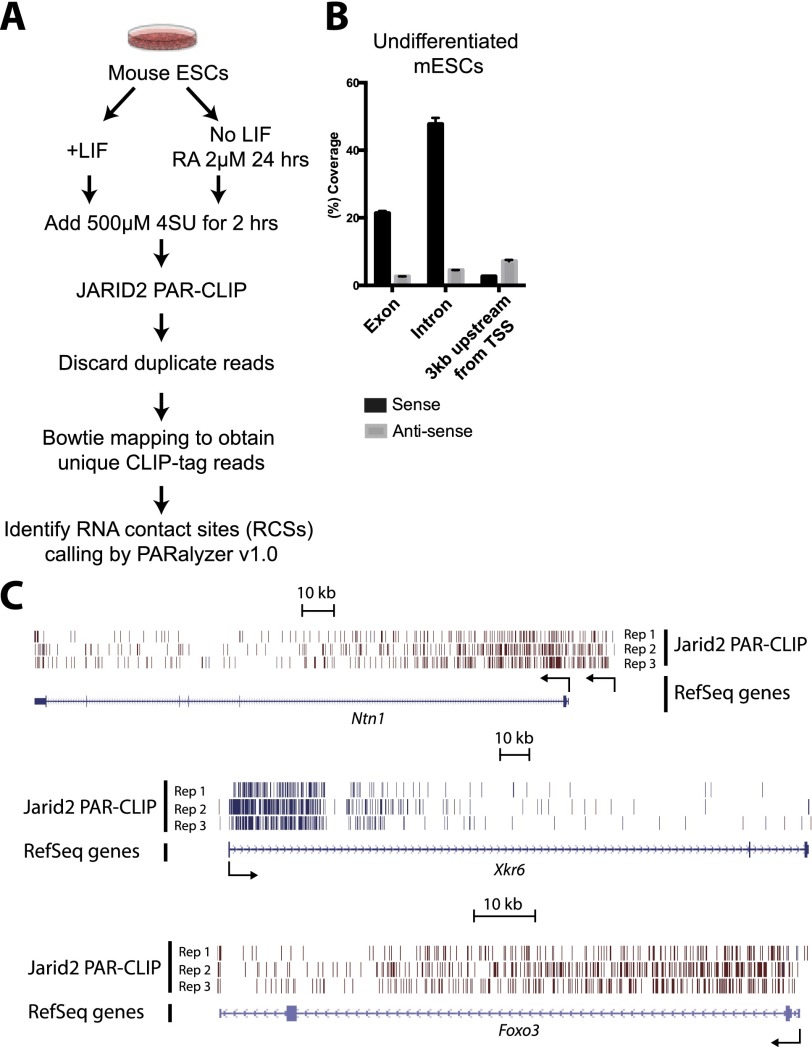
Given the facts that JARID2 has an affinity for RNA and that it exists in close association with PRC2 in undifferentiated mESCs, we hypothesized that it might also engage some of the same nascent transcripts that interact with EZH2 in vivo (Kaneko et al. 2013), such as, for example, those emanating from the Ntn1 gene. In a manner similar to our studies of EZH2 binding to nascent transcripts (ezRNAs) (Kaneko et al. 2013), we used the modified photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) methodology (Kaneko et al. 2013) using, in this case, extensively validated antibody to JARID2 (Li et al. 2010; Kaneko et al. 2014). After immunoprecipitation and SDS-PAGE, radiolabeled signal was erased by RNase treatment, demonstrating JARID2–RNA cross-linking (Supplemental Fig. S4A). Since JARID2 levels steadily decline during ESC differentiation, we compared PAR-CLIP results from undifferentiated mESCs grown in LIF versus partially differentiated mESCs derived in the absence of LIF but in the presence of retinoic acid (RA) such that JARID2 is still present (protocol shown schematically in Fig. 3A; summarized in Supplemental Table S2; Li et al. 2010).
Figure 3.
JARID2 contacts nascent RNAs. (A) Procedure for obtaining JARID2 PAR-CLIP data as a function of mESC differentiation. (B) Histogram of mapped CLIP tags reads to the whole genome. Three genome categories for read assignments, which are exon, intron, and promoter regions (from the TSS to −3 kb), are shown in sense (black bar) and antisense (gray bar) directions. (C) Schematic of Ntn1, Xkr6, and Foxo3 gene loci. Unique JARID2 PAR-CLIP-tags are shown in bars with blue (plus strand) and dark red (minus strand). (Rep1–3) Biological replicates. Note that there are upstream CLIP tags that originated from Ntn1 alternative transcript ENSMUST00000108674. TSSs are shown by arrows.
We identified RNA contact sites (RCSs) using PARalyzer (Supplemental Tables S3,S4; Corcoran et al. 2011), and observed that they mapped to the bodies of protein-coding genes, as evidenced by extensive coverage of intronic regions by PAR-CLIP reads (Fig. 3B), which would be absent if the interacting RNA species were mature and spliced mRNA. We observed a similar trend in differentiating mESCs (data not shown). In fact, the same three genes that we previously identified as giving rise to ezRNAs with the strongest binding to EZH2, which include Ntn1, also accumulated large numbers of reads from the JARID2 PAR-CLIP (jarRNAs) (Fig. 3C). Indeed, there was extensive overlap (∼75%) between ezRNAs and jarRNAs, suggesting that EZH2 and JARID2 might make simultaneous contacts with these transcripts, as we previously observed for the lncRNA MEG3 (Kaneko et al. 2014).
Despite the fact that 4-SU cross-linking requires hydrogen-bonding distances between the RNA and the protein (Ascano et al. 2012), it has been recently shown that considerable amounts of background RNA can be retrieved with this technique, and such background PAR-CLIP tags are enriched in Poly(G) stretches (Friedersdorf and Keene 2014). We analyzed the sequence composition of the reads from our JARID2 PAR-CLIP and found no evidence of Poly(G) stretches (Supplemental Fig. S4B), suggesting that background contamination could not explain our results. Similar results were obtained when reanalyzing our previous EZH2 PAR-CLIP data set (Supplemental Fig. S4C; Kaneko et al. 2013).
Thus, we conclude that JARID2, like EZH2, establishes direct contacts with nascent RNAs from genes whose promoters are occupied by intermediate or low levels of PRC2 and might therefore participate in the ezRNA-mediated inhibition of PRC2, consistent with our biochemical results presented above (Fig. 1).
JARID2-interacting RNAs change during differentiation
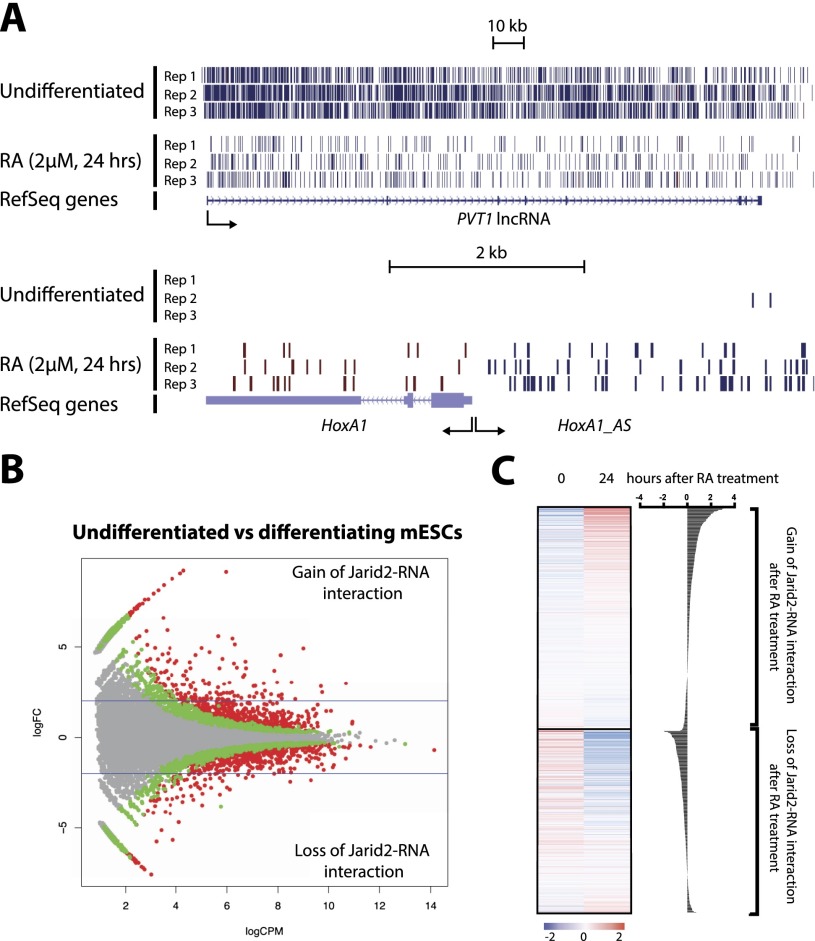
Next, we sought to determine whether jarRNAs are a static population or change dynamically with the transcriptional program as ESCs differentiate. We observed that PVT1, a lncRNA expressed predominantly in undifferentiated ESCs, gave rise to a large number of JARID2 PAR-CLIP tags and that, upon differentiation, the number of tags recovered from this RNA was diminished considerably (Fig. 4A). Conversely, the HOXA1 antisense lncRNA (HOXA1_AS) was associated with JARID2 almost exclusively after RA-mediated differentiation (Fig. 4A), suggesting that the pool of jarRNAs in ESCs is dynamic and modulated by changes in their transcriptional and epigenetic state (Supplemental Fig. S5).
Figure 4.
Dynamics of JARID2–nascent RNA interaction correlates with gene expression during mESC differentiation. (A) JARID2 CLIP tag mapping to two representative lncRNAs before and after RA-mediated differentiation in mESCs. (B) MA plots for JARID2–RNA interaction on gene bodies, comparing undifferentiated versus differentiating mESCs. The graph indicates fold change (logFC; Y-axis) versus abundance (logCPM; X-axis) of mapped CLIP tags. Differentially binding genes (DBGs) with an FDR < 0.05 (red dots) and 0.25 < FDR < 0.05 (green dots) are displayed. (C, left) Heat maps for mRNA levels in undifferentiated versus differentiating mESCs, showing up-regulated (gain of JARID2–RNA interaction; FDR > 0.05, logFC > 1) and down-regulated (loss of JARID2–RNA interaction; FDR > 0.05, logFC < −1) DBGs (see Supplemental Table S5). (Right) Differential expression is also displayed with individual bars. Data were obtained from GSE43221 (Kashyap et al. 2013).
Comparing PAR-CLIP reads genome-wide and using a false discovery rate (FDR) of 5%, we identified 661 differentially bound RNAs (DBRs) specific to the pluripotent state and 631 DBRs specific to the differentiating cells (Fig. 4B, red dots; Supplemental Table S5). Using more comprehensive parameters (25% FDR), the number of DBRs grew to 1524 and 1395 for pluripotent and differentiating cells, respectively, indicating that a large fraction of genes modulated during differentiation have the ability to generate jarRNAs.
In line with our model for an RNA-sensing mechanism regulating PRC2, we found that genes giving rise to jarRNAs in pluripotent cells but not differentiated cells became silent, and, vice versa, newly activated genes gave rise to the majority of the new RA-specific jarRNAs (Fig. 4C). This is consistent with the idea that contacts of PRC2 subunits with nascent RNA result in reduced enzymatic activity and in turn allow for the maintenance of a chromatin structure more permissive to transcription.
Taking into account the role of PcGs in maintaining already established repression, we propose that Holo-PRC2 marks the gene with its modification only when transcriptional repression has already been established by DNA sequence-specific repressors; for example, during differentiation. The subsequent shutoff of nascent transcript production relieves Holo-PRC2 inhibition such that these promoters can now be marked and maintained as repressed through deposition of H3K27me2/3. The inherent lack of specificity in nascent RNA binding by EZH2 and JARID2 in vivo may enable Holo-PRC2 to be available at transcribing genes at large and on hold for marking the gene once repression is established.
Materials and methods
Recombinant proteins, synthesis of ncRNAs, in vitro binding assay, and cell culture are described in the Supplemental Material.
HKMT assay
Standard HKMT assays were performed as described (Son et al. 2013). For most reactions scored by scintillation counting, if not indicated otherwise, the conditions were as follows: 0.67 pmol of PRC2, 12.5 μM SAM (3H SAM/cold SAM at a 1:45 ratio), and 2.28 pmol of recombinant nucleosomes with 1.5 pmol of JARID2 and 4 pmol of AEBP2. The reaction mixture was incubated in HKMT buffer (50 mM Tris-HCl at pH 8.5, 5 mM MgCl2, 4 mM DTT) for 15 min at 30°C.
CRISPR-Cas9-mediated gene knockout in mESCs
Experimental procedures were essentially described previously (Cong et al. 2013; Mali et al. 2013). Wild-type Cas9 plasmid was obtained from Addgene (plasmid #41815). Primers for generating 5′ and 3′ arms and template DNA for guide RNAs (gBlocks, Integrated DNA Technologies) are summarized in Supplemental Table S1. Cassette sequences are available on request. Donor plasmids were prepared using Gibson Assembly (New England Biolabs). For knockout of Ntn1 in embryonic day 14 (E14) mESCs, we transiently transfected wild-type Cas9, donor plasmids, and PCR-amplified template DNAs for guide RNA expression by using Lipofectamine 2000 (Life Technologies). In 48 h post-transfection, selection was done using 2 μg/mL puromycin for 48 h, and clones were screened by RT-qPCR and genomic PCRs using the primers summarized in Supplemental Table S1. Ntn1 knockout E14 mESCs were maintained without puromycin.
ChIP-qPCR
Experimental procedures and antibodies were described previously (Kaneko et al. 2013, 2014). Primers are summarized in Supplemental Table S1.
PAR-CLIP
Experimental procedures and antibodies for PAR-CLIP and bioinformatic analysis were described previously (Kaneko et al. 2013, 2014).
Accession numbers
CLIP sequences reported in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus as SuperSeries GSE60516.
Acknowledgments
We thank the Genome Technology Center at New York University for help with sequencing, and Dr. Lynne Vales for comments and help with manuscript preparation. This work was supported by New York State Stem Cell Science (NYSTEM; C028105), National Institutes of Health (GM64844), and Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.247940.114.
References
- Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. 2012. Identification of RNA–protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA 3: 159–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol Cell 15: 57–67 [DOI] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. 2014. Regulatory interactions between RNA and Polycomb repressive complex 2. Mol Cell. doi: 10.1016/j.molcel.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 12: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. 2013. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20: 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedersdorf MB, Keene JD. 2014. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol 15: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21: 569–571 [DOI] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. 2010. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24: 2615–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. 2013. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. 2014. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 53: 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Laursen KB, Brenet F, Viale AJ, Scandura JM, Gudas LJ. 2013. RARγ is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells. J Cell Sci 126: 999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski WA, Nakamura T, Mazo A, Canaani E. 2005. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol 25: 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA polymerase II to developmental regulators. Nat Cell Biol 12: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejetta S, Morey L, Pascual G, Kuebler B, Mysliwiec MR, Lee Y, Shiekhattar R, Di Croce L, Benitah SA. 2011. Jarid2 regulates mouse epidermal stem cell activation and differentiation. EMBO J 30: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Moine H, Vagner S. 2012. G-quadruplexes in RNA biology. Wiley Interdisp Rev RNA 3: 495–507 [DOI] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. 2007. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys Rev Lett 98: 158103. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Shen SS, Margueron R, Reinberg D. 2013. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev 27: 2663–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. 1995. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev 9: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, et al. 2011. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]