Using conditional mutagenesis to remove the core PRC2 subunits EED and SUZ12 during male germ cell development, Mu et al. identified a requirement for PRC2 in both mitotic and meiotic germ cells. Mutant spermatocytes exhibited ectopic expression of somatic lamins and an abnormal distribution of SUN1 proteins on the nuclear envelope. These defects were coincident with abnormal chromosome dynamics, affecting homologous chromosome pairing and synapsis.
Keywords: meiosis, spermatogenesis, spermatogonia, Polycomb group proteins, epigenetic repression, gene expression
Abstract
Polycomb-repressive complex 2 (PRC2) catalyzes the methylation of histone H3 Lys27 (H3K27) and functions as a critical epigenetic regulator of both stem cell pluripotency and somatic differentiation, but its role in male germ cell development is unknown. Using conditional mutagenesis to remove the core PRC2 subunits EED and SUZ12 during male germ cell development, we identified a requirement for PRC2 in both mitotic and meiotic germ cells. We observed a paucity of mutant spermatogonial stem cells (SSCs), which appears independent of repression of the known cell cycle inhibitors Ink4a/Ink4b/Arf. Moreover, mutant spermatocytes exhibited ectopic expression of somatic lamins and an abnormal distribution of SUN1 proteins on the nuclear envelope. These defects were coincident with abnormal chromosome dynamics, affecting homologous chromosome pairing and synapsis. We observed acquisition of H3K27me3 on stage-specific genes during meiotic progression, indicating a requirement for PRC2 in regulating the meiotic transcriptional program. Together, these data demonstrate that transcriptional repression of soma-specific genes by PRC2 facilitates homeostasis and differentiation during mammalian spermatogenesis.
Polycomb-repressive complex 2 (PRC2) consists of four core subunits—EED, SUZ12, EZH1/2, and RbBP4/7—and is responsible for catalyzing di- and trimethylation of Lys27 of histone H3 (H3K27). In embryonic stem cells (ESCs), PRC2 and H3K27me3 occupy inactive promoters of key developmental regulators (Bernstein et al. 2006; Boyer et al. 2006; Lee et al. 2006). In vivo, PRC2 regulates developmental transitions by promoting or blocking differentiation, fine-tuning cell fate acquisition, or preserving proper cell identity during the progression from proliferation to differentiation (Aldiri and Vetter 2012). In other contexts, PRC2 is required for the self-renewal of somatic stem and progenitor cells through repression of the cell cycle inhibitors p16INK4A and p19ARF (Aoki et al. 2010; Ezhkova et al. 2011; Juan et al. 2011). Transcriptional regulation by PRC2 during development is complicated by the coexistence of H3K27me3 with active gene marks such as H3K4me3 and the removal of H3K27me3 from promoters by cognate demethylases (Agger et al. 2007).
Mammalian spermatogenesis is a complex process by which mature gametes are produced through spermatogonial stem cell (SSC) proliferation, differentiation, and meiosis and haploid differentiation. It requires establishing genomic imprints, germ cell-specific transcription patterns, and a genome-wide transition from histone to protamine-based chromatin packaging. Because H3K27 methylation is present throughout germ cell development (Sasaki and Matsui 2008), it stands to reason that PRC2 may have a prominent role in the associated transition of cellular identity. However, the requirement for PRC2 and H3K27me2/3 during mammalian gametogenesis remains unknown. Because of the importance of PRC2 and H3K27me3 in somatic differentiation, we speculated that PRC2 has a similar essential role in the mammalian germline.
Male germ cell development is characterized by dynamic changes to gene expression patterns, including a transition from somatic to germ cell-specific genes, global repression of transposon activity, and meiotic sex chromosome inactivation (MSCI) (Sasaki and Matsui 2008). As a key transcriptional repressor, PRC2 may play a vital role in the reprogramming of the transcriptome during spermatogenesis. To determine the role of PRC2 in mammalian gametogenesis, we generated PRC2 mutant mice with germ cell-specific ablation of EED or SUZ12. Our results demonstrate that PRC2 is essential for male fertility, maintenance of SSCs, and the progression of meiosis. Loss of PRC2 resulted in ectopic expression of somatic lamins in spermatocytes, which may interfere with the LINC (linker of nucleoskeleton and cytoskeleton) complex to facilitate telomere-mediated chromosome movement for homologous chromosome pairing and synapsis. Moreover, genomic analysis of H3K27me3 distribution indicates the involvement of PRC2 in the precise execution of the meiotic transcriptional program.
Results
Germ cell-specific ablation of PRC2 results in male infertility
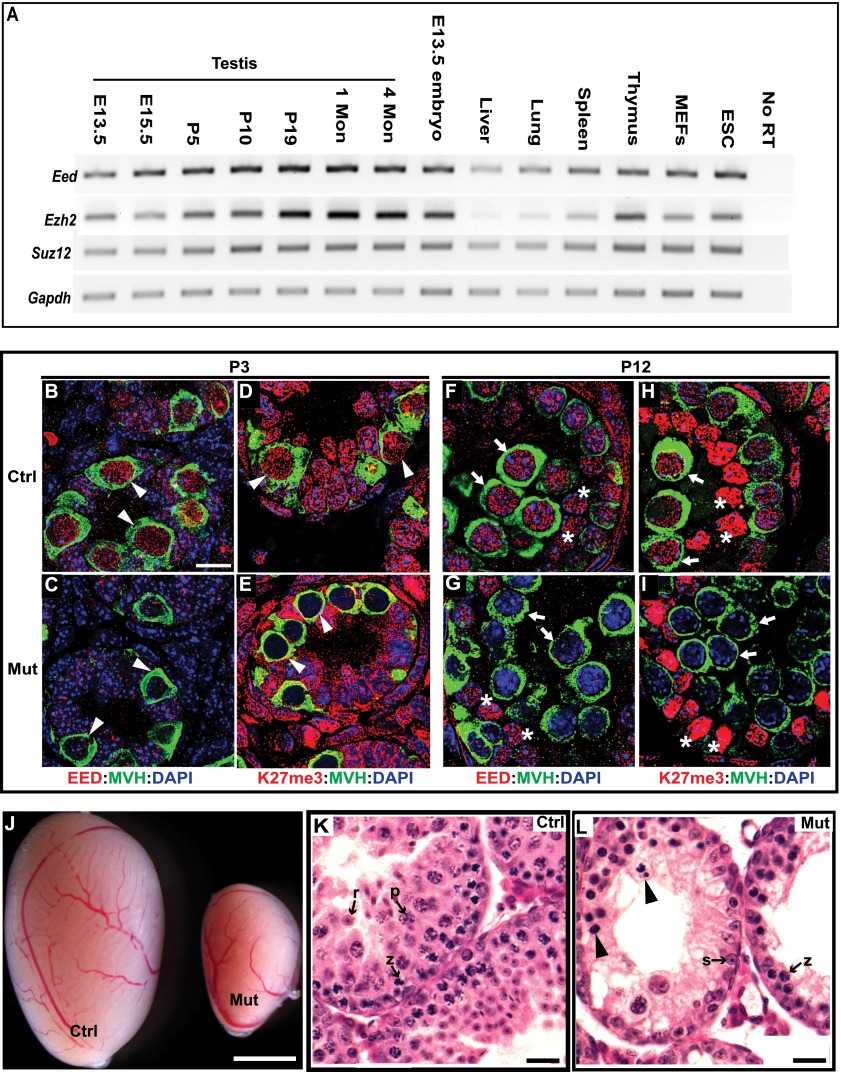
We measured mRNA levels of PRC2 core subunits, including EED, EZH2, and SUZ12, at developmental stages in testes by RT–PCR (Fig. 1A). The stages assessed included (1) embryonic day 13.5 (E13.5), when primordial germ cells migrate to and reside in newly formed gonads; (2) E15.5, when gonocytes undergo mitotic arrest; (3) postnatal day 5 (P5), when gonocytes commit to spermatogonia; (4) P10, when meiosis initiates; (5) P19, when spermatocytes are at prophase I; (6) 1 mo, when elongating spermatids are present; and (7) 4 mo, when all stages of spermatogenesis are represented. Compared with other mouse tissues, embryos, and primary cell lines, Eed and Ezh2 are transcribed at higher levels in the testes beginning at P19. In contrast, Suz12 expression is not significantly higher in testis.
Figure 1.
EED deficiency causes germ cell depletion. (A) RT–PCR analysis of PRC2 core components in mouse testis of different ages, tissues, embryos, and cell lines. Gapdh was used as a control. (B–I) Testis sections from P3 and P12 testes were coimmunostained with the antibodies indicated. The arrowheads, arrows, and asterisks point to spermatogonia, spermatocytes, and Sertoli cells, respectively. MVH staining marks germ cells. (J) Two-month-old testes. (K,L) Hematoxylin and eosin-stained sections of 4-wk-old testes. Arrowheads indicate pyknotic cells with condensed chromatin or fragmented nuclei. (z) A zygotene spermatocyte; (p) a pachytene spermatocyte; (r), a round spermatid; (s), a Sertoli cell. In all of the figures, Ctrl indicates control (MvhCre;Eedfl/+), and Mut indicates Eed mutant (MvhCre;Eedfl/Δ). Bars: B–I, 10 μm; K,L, 20 μm; J, 1 mm.
Since EED is required for stabilization of other PRC2 components and methylation of H3K27 (Montgomery et al. 2005), we analyzed animals with a genetic ablation of Eed, reasoning that this would cause the most severe phenotype. We created Eed conditional mutant mice using the European Conditional Mouse Mutagenesis (EUCOMM) Eed knockout ESC line (project 35891, Eedtm1a(EUCOMM)Wtsi), which has a gene trap in intron 6 and a floxed exon 7 (Supplemental Fig. S1A). Deletion of the floxed exon would result in a predicted null allele by removing an indispensable WD40 motif and shifting the reading frame. Southern blotting (Supplemental Fig. S1C) and PCR genotyping (Supplemental Fig. S1D) verified the correct targeting of this allele in both ESCs and mice.
To ablate EED in developing germ cells, we used the Mvh-Cre recombinase, which is first expressed in male germ cells around E15 and results in efficient deletion of floxed alleles by birth (Gallardo et al. 2007). Since histone turnover rate is low in nonproliferating cells (Commerford et al. 1982), we reasoned that H3K27me2/3 would be retained if excision occurred in early stages of meiosis after the completion of DNA synthesis. Therefore, deletion in the proliferating germ cell populations (via Mvh-Cre) would be a more effective approach to deplete germ cells of H3K27me2/3. Disruption of PRC2 function in Eed homozygous mutant testis (MvhCre;Eedfl/∆) was verified by the lack of EED protein and H3K27me3 in spermatogonia of P3 testis (Fig. 1B–E) and spermatocytes of P12 testis (Fig. 1F–I). EED protein was detected in <1% of germ cells (seven out of 1000 MVH-positive cells) in mutant P3 testes, indicating a high efficiency of MVH-mediated excision of floxed Eed alleles. Compared with the controls, the mutants showed similar levels of EED and H3K27me3 in Sertoli cells (Fig. 1G,I), which is indicative of the specificity of Mvh-Cre-directed recombination in germ cells.
Although Eed mutant males exhibited normal mating behavior, they were unable to sire any litters. At 1 mo of age, testes from mutants were much smaller than the controls (Fig. 1J). Histological analysis revealed a dramatic decrease in spermatocytes in the seminiferous tubules of mutant animals. A subset of mutant spermatocytes exhibited atypical nuclei: They were very condensed or fragmented, indicative of apoptosis (Fig. 1K,L). In contrast to the control testes, post-pachytene spermatocytes and spermatids were absent, and only a few prepachytene spermatocytes were observed in sections of mutant tubules (Fig. 1K,L), suggesting that PRC2 is required for meiotic progression.
PRC2 is required for efficient synapsis and double-strand break (DSB) repair in meiosis
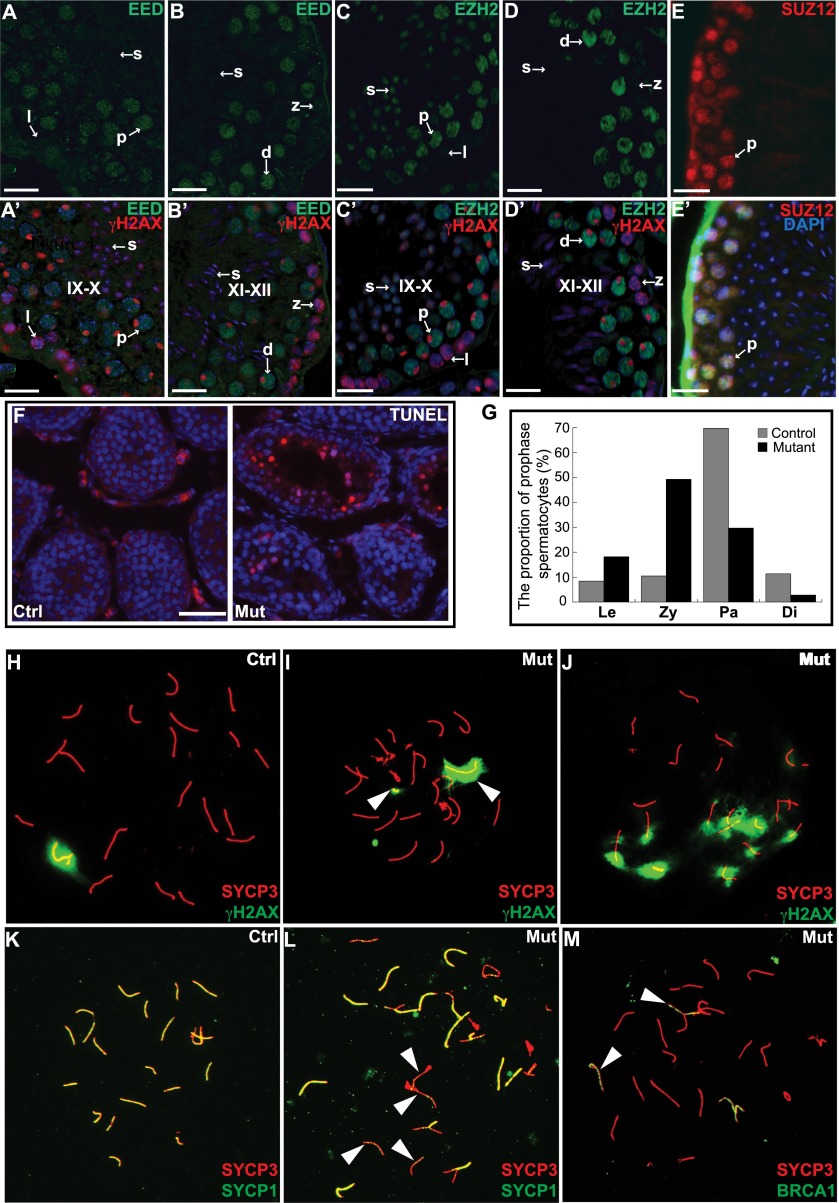
To identify the meiotic stage during which mutant spermatocytes become arrested, we examined the dynamics of PRC2 subunits during spermatogenesis by immunohistochemical analysis of their protein levels in wild-type testis tubule sections. EED (Fig. 2A,A′,B,B′), EZH2 (Fig. 2C,C′,D,D′), and SUZ12 (Fig. 2E,E′) were barely detectable in leptotene and zygotene spermatocytes but were highly expressed in pachynema and diplonema, suggesting that PRC2 may be active during the latter stages of prophase I. Furthermore, TUNEL staining showed increased numbers of apoptotic cells in Eed mutants when the first wave of primary spermatocytes advances to pachynema at day 13 (Fig. 2F). We quantified the cell populations of the first meiotic prophase I by counting surface-spread nuclei stained with SYCP3 and the phosphorylated histone variant H2AX (γH2AX). The spermatocytes were staged according to the standard as described in Supplemental Figure S2A. Among control spermatocytes, 69.8% were in pachynema at P13. In contrast, only 29.7% of mutant spermatocytes reached pachynema, with the majority (49.7%) arrested at zygonema (Fig. 2G). Thus, the onset of defects occurs at the zygotene-to-pachytene transition, which is coincident with the increase in protein levels of PRC2 components in wild-type spermatocytes.
Figure 2.
Disruption of PRC2 complex leads to meiotic defects. (A–E) Testis sections immunostained with antibodies against EED (A,B), EZH2 (C,D), and SUZ12 (E). (A′–D′) Merged images with γH2AX staining and DAPI. (E′) SUZ12 staining merged with DAPI. IX–X and XI–XII indicate the stages of seminiferous epithelial cycle. IX–X is defined by the presence of leptotene spermatocytes and elongated spermatid heads, and XI–XII is defined by the presence of zygotene spermatocytes and more elongated spermatids. (l) Leptotene spermatocytes; (z) zygonema; (p) pachynema; (d) diplonema. (F) TUNEL-stained frozen sections of 13-d-old testis. TUNEL-positive apoptotic cells are stained red. (G) The percentage of spermatocytes at each substage of prophase I analyzed from 1000 meiotic chromosome spreads. (Le) Leptotene; (Zy) zygotene; (Pa) pachytene; (Di) diplotene. (H–M) Surface-spread spermatocyte nuclei were immunostained for the indicated proteins. The arrowheads in I point to asynapsed X and Y chromosomes. The arrowheads in L point to asynapsed autosomes. The arrowheads in M point to asynapsed autosomes. Bars: A–E′, 20 μm; F, 50 μm.
To elucidate the cause of meiotic arrest, we analyzed meiotic nuclei with a variety of markers for DSBs, recombination, and synapsis. Meiosis in Eed mutant spermatocytes appeared to initiate normally as judged by the presence of γH2AX in leptonema (Supplemental Fig. S2B), reflecting the presence of meiotically induced DSBs. RAD51, a component of early recombination nodules, was also present as abundant foci in Eed mutant zygotene spermatocytes (Supplemental Fig. S2C), suggesting that repair of DSBs was initiated. However, defects in DSB repair and synapsis became apparent in pachynema. A relatively large percentage (30%; n = 100) of mutant spermatocytes exhibited nonassociated X and Y chromosomes, as indicated by the two separate γH2AX-positive spots (Fig. 2H,I). In controls, complete synapsis of autosomes can be judged by the colabeling of SYCP1 and SYCP3 along the full length of chromosomes (Fig. 2K). However, in the mutant spermatocytes, some chromosomes lacked SYCP1 staining (Fig. 2L), suggesting a failure in the maintenance or establishment of synapsis. Accordingly, mutant pachytene spermatocytes showed widespread localization of BRCA1, a known marker of chromosomes that fail to synapse (Fig. 2M; Supplemental Fig. S2D). In addition, broad γH2AX persisted into later stages of the first meiotic prophase (Fig. 2J). Thus, meiotic arrest of Eed mutant spermatocytes corresponds with failures of synapsis and DSB repair.
To verify the requirement for PRC2 in spermatogenesis, we also generated Suz12 conditional mutant mice carrying the floxed exons 2 and 3 of the Suz12 gene (Supplemental Fig. S1B). Disruption of the Suz12 conditional allele by Mvh-Cre resulted in the loss of H3K27me3 in spermatocytes (Supplemental Fig. S3A) and a similar phenotype, as exhibited in Eed mutants (Supplemental Fig. S3B–E). Together, these results demonstrate an essential role for PRC2 in male meiosis.
Altered chromosome dynamics in the absence of PRC2
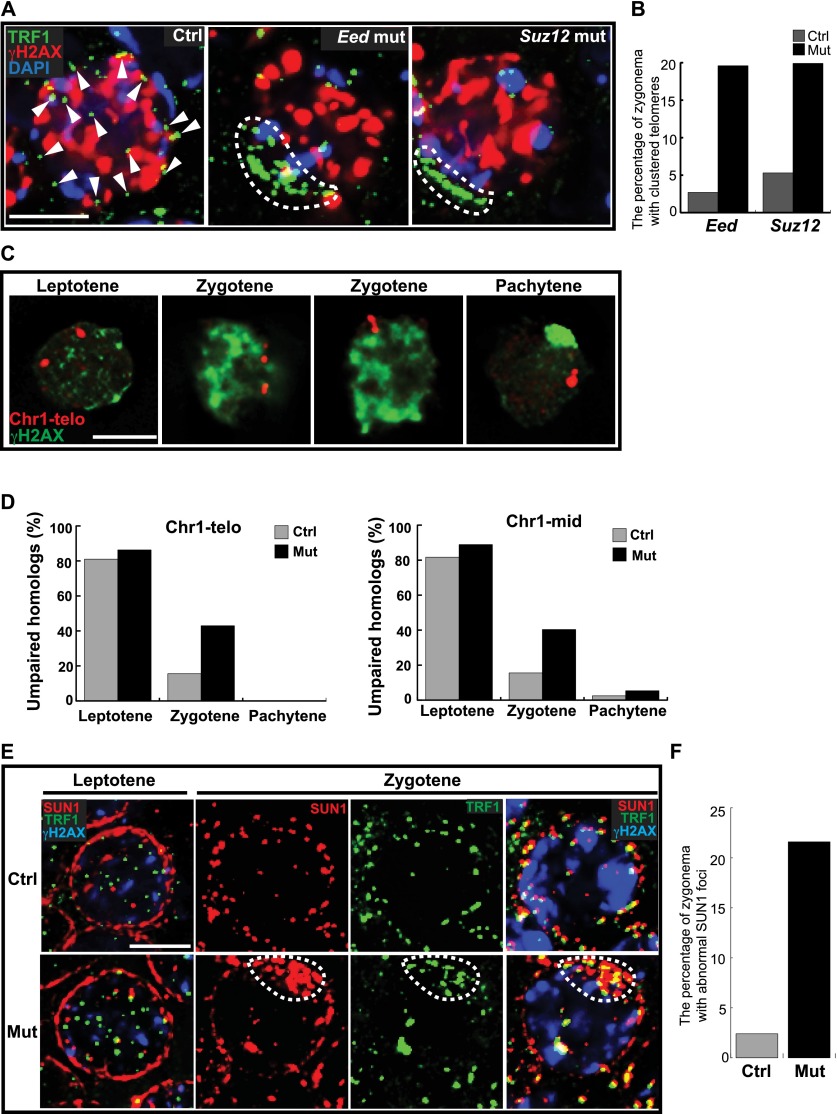
An early step in homologous pairing involves the dynamic movement of telomeres across the nuclear envelope (NE). At the onset of zygonema, the telomeres transiently cluster together before spreading throughout the NE (Liebe et al. 2006). We assessed the effect of PRC2 deficiency on telomere distribution through the localization of telomeric repeat-binding factor1 (TRF1). In controls, telomeres were typically spread around the NE during the zygotene stage (Fig. 3A). However, in Eed and Suz12 mutants, the telomeres were frequently clustered (19.6% and 19.9%, respectively) during the zygotene stage compared with stage-matched spermatocytes from littermate controls (2.7% and 5.4%, respectively) (Fig. 3A,B). Persistent or delayed clustering of telomeres in mutant spermatocytes would be predicted to impact chromosome pairing. Therefore, we examined homologous pairing by DNA fluorescence in situ hybridization (FISH) assays on structurally preserved spermatocytes using probes that detect specific sequences in the subtelomere region and the mid-arm region of chromosome 1 (Fig. 3C). There were more unpaired homologs measured in the zygotene stage Eed mutant spermatocytes compared with littermate controls (43% vs. 15.6% for the subtelomere region and 40.7% vs. 17.1% for the mid-arm region) (Fig. 3D). Therefore, the normal meiotic chromosome mobility that underlies homolog pairing and synapsis is disrupted in the absence of PRC2 function.
Figure 3.
PRC2 regulates telomere movement within the NE and homolog pairing during meiotic prophase I. (A) Zygotene spermatocytes from cryosections of P13 testis were immunostained with the antibodies indicated. The arrowheads indicate discrete telomeres. The dashed lines surround clustered telomeres. (B) Zygotene spermatocytes with clustered telomeres at the NE were quantified (n = 250). (C) Illustrative images of homolog pairing in spermatocytes analyzed by DNA FISH with a probe to detect the subtelomeric region of chromosome 1. γH2AX staining was used to identify specific stages. (D) The frequency of homolog pairing was examined in 400 spermatocytes of each stage (50 for mutant pachytene spermatocytes) using the probes to detect the mid-region and the subtelomeric region on chromosome 1. (E) Immunostaining analysis of dynamic patterns of SUN1 at the NE during meiotic prophase I progression. The dashed lines indicate clustered SUN1 foci. Bar, 5 μm. (F) Quantification of wild-type and mutant zygotene spermatocytes with clustered SUN1 foci at the NE.
Telomere-driven chromosome movement is mediated through the LINC complex, which is situated at the NE and interacts with microtubule networks during meiosis (Hiraoka and Dernburg 2009; Morimoto et al. 2012; Wynne et al. 2012). SUN1 is the inner nuclear membrane (INM) subunit of the LINC complex and is important for telomere attachment to the NE and homolog pairing/synapsis formation during meiosis (Ding et al. 2007; Boateng et al. 2013; Ishiguro et al. 2014). To investigate whether chromosome rearrangement defects in the mutants were attributable to LINC complex dysfunction, we examined the dynamics of SUN1 in prophase I spermatocytes. In controls, SUN1 moved from a diffuse distribution at the leptotene stage to discrete foci colocalized with telomeres by the zygotene stage. In contrast, mutant spermatocytes more frequently had clustered SUN1 foci located in the telomere clusters at zygonema (Fig. 3E,F).
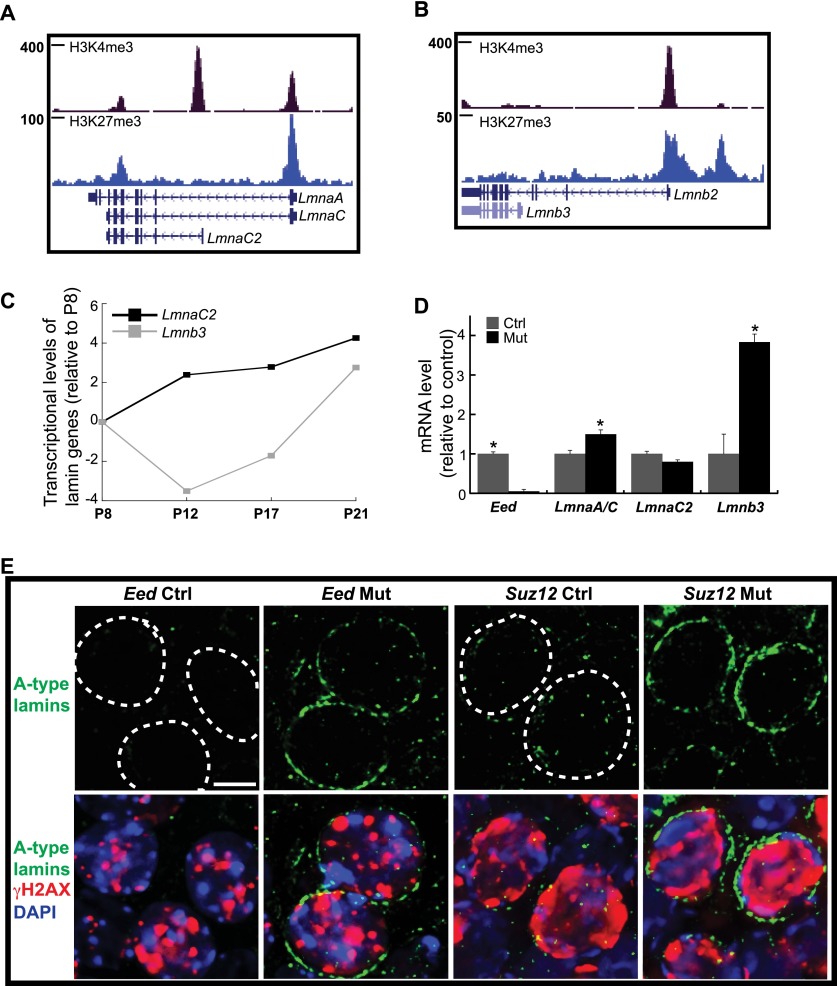
SUN1 interacts with NE lamin proteins. Mammalian meiotic cells are distinguished by the absence of lamins A, C, and B2, which are typically expressed in differentiated somatic cells (Burke and Stewart 2013; Haque et al. 2006; Link et al. 2013). A-type lamins are encoded by Lmna genes and are composed of three isoforms, including somatic lamins A and C and meiotic lamin C2. In particular, lamin A levels were negatively correlated with SUN1 mobility at the NE (Ostlund et al. 2009). We postulated that loss of PRC2 would result in an up-regulation of the somatic lamins in meiocytes, thereby restricting SUN1 movement during meiosis. A ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) data set for H3K27me3 in wild-type meiocytes (described below) showed that promoter regions of these lamin isoforms were enriched for H3K27me3 (Fig. 4A,B), suggesting regulation by PRC2. However, the promoters of germ cell-specific lamins C2 and B3 were devoid of H3K27me3 (Fig. 4A,B), and their expression was up-regulated with meiotic progression (Fig. 4C). In contrast to wild-type spermatocytes, we observed an increase in RNA levels of somatic lamins A and C (Fig. 4D) in Eed mutants. Moreover, immunofluorescence analysis of A-type lamins revealed an increase at the nuclear periphery of both Eed and Suz12 mutant zygotene spermatocytes (Fig. 4E). Finally, the transcript levels of the post-meiotic lamin B3 were higher in Eed mutant testes than controls (Fig. 4D).
Figure 4.
PRC2 depletion affects expression of somatic lamins in spermatocytes. (A,B) ChIP-seq analysis on purified P17 wild-type spermatocytes demonstrates that somatic lamin genes are marked by H3K27me3. (C) RT-qPCR analysis of germ cell-specific lamin gene transcription during spermatogenesis. Germ cells were isolated from 8-, 12-, 17-, and 21-d-old testes. The Y-axis is log2 scale. (D) qPCR analysis of lamin mRNA levels in P12 testis. (*) P < 0.05 (t-test). (E) The up-regulation of lamins A and C was detected in mutant spermatocytes by immunostaining with an antibody recognizing all A-type lamins. The dashed lines demarcate the NE. Bar, 5 μm.
PRC2 regulates expression of stage-specific meiosis genes
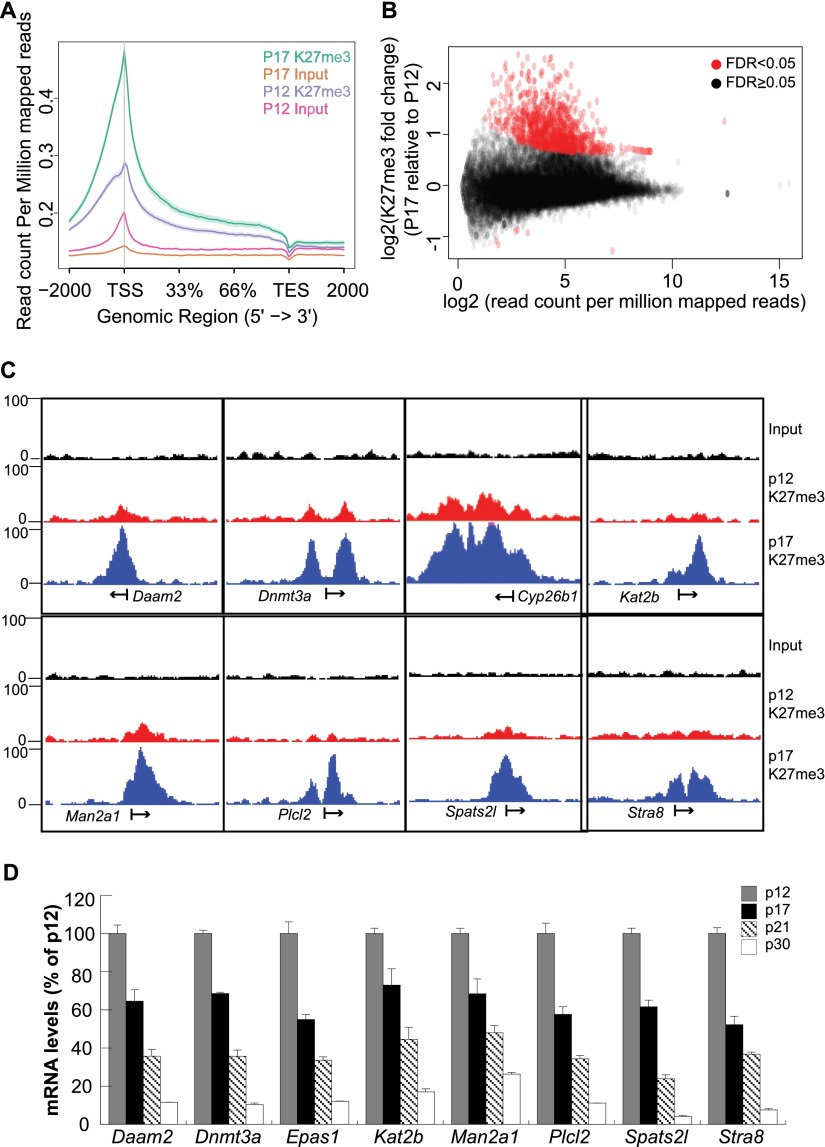
Since PRC2 levels were elevated in normal meiocytes during the zygonema-to-pachynema transition, we analyzed the genome-wide dynamics of EED and H3K27me3 via ChIP-seq during this time. Spermatocytes were isolated from wild-type P12 and P17 testis, where the cell population is primarily prepachytene and pachytene spermatocytes, respectively. The enrichment of H3K27me3 on the gene bodies was higher in P17 than in P12 spermatocytes, especially for the regions near the transcriptional start site (TSS) (Fig. 5A). We identified numerous genes with strong H3K27me3 signals at P17 but not at P12 (Fig. 5B). Among the 1316 genes that have significantly more H3K27me3 around the TSS at P17 than at P12, many are statistically enriched in cell membrane-associated biological pathways (Supplemental Fig. S4). Daam2, Dnmt3a, Cyp26b1, Kat2b, Man2a1, Plcl2, Spats2l, and Stra8—all of which had a dramatic increase in H3K27me3 around the TSS at P17 (Fig. 5C)—were further analyzed for their expression during meiosis. The transcriptional levels of all eight genes decreased as meiocytes progressed from leptonema/zygonema (P12) to pachynema (P17), diplonema (P21), and spermatids (P30) (Fig. 5D). Importantly, Stra8 (Anderson et al. 2008), Dnmt3a (Yuan et al. 2006), and Cyp26b1 (Koubova et al. 2006; Bowles and Koopman 2007) have been implicated in the initiation of the meiotic program.
Figure 5.
Dynamics of genomic H3K27me3 distribution in spermatocytes. (A) A metaplot showing the distribution of ChIP-seq reads as normalized read counts per million across the gene bodies in P12 and P17 wild-type spermatocytes. (B) H3K27me3 enrichment on gene bodies between P17 and P12 wild-type spermatocytes. Each dot represents a single gene. (C) Genome browser images of representative genes newly marked by H3K27me3 around the TSS during meiosis progression. (D) qPCR analysis profiles the transcriptional dynamics of the genes in C during spermatogenesis. Spermatocytes for these experiments were isolated from 12-, 17-, 21-, and 30-d-old testes.
Repetitive DNA silencing and MSCI occur in the absence of PRC2
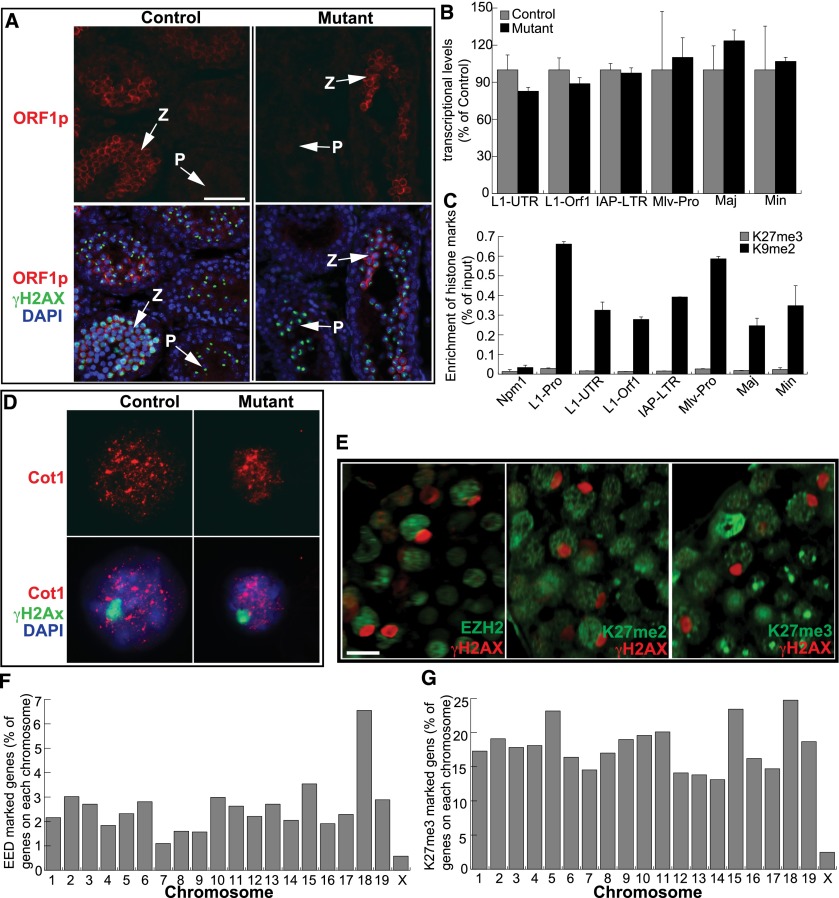
Failure to silence repetitive DNA sequences in germ cells impairs homologous chromosome pairing and synapsis (Sasaki and Matsui 2008). Since Polycomb complexes are required for stable repression of long terminal repeat retrotransposons in ESCs (Leeb et al. 2010), we speculated that PRC2 might participate in suppressing repetitive elements through H3K27 methylation in the male germline. Therefore, we compared the expression levels of transposon and satellite sequences between Eed mutants and controls. It has been reported that ORF1p mediates the transposition of transposon LINE1 and is detected in meiocytes (Branciforte and Martin 1994; Soper et al. 2008). In P13 control testes, ORF1p was enriched in the cytoplasm of zygotene spermatocytes but was no longer present at the entry into pachynema (Fig. 6A). The mutants exhibited similar ORF1p expression patterns during the zygotene-to-pachytene transition (Fig. 6A). Importantly, the transcription of LINE1, IAP, MLV, major satellites, and minor satellites was not elevated in Eed mutants (Fig. 6B). To confirm that PRC2 is not involved in silencing repeat elements, we determined whether H3K27me3 is associated with DNA repeat regions by ChIP-PCR assays on wild-type spermatocytes. Accordingly, we found that H3K27me3 was not enriched in LINE1, MLV, IAP, major satellites, and minor satellites compared with H3K9me2 (as a positive control) (Fig. 6C), suggesting that PRC2 does not have a role in silencing repetitive elements in spermatocytes.
Figure 6.
PRC2 is not required for silencing of repetitive elements and MSCI. (A) Immunostaining for ORF1p and γH2AX on 14-d-old testis. (z) Zygotene spermatocytes; (p) pachytene spermatocytes. Bar, 50 μm. (B) RT-qPCR analysis of repeat element transcription in P11 testis. (C) ChIP-PCR analysis of repressive histone marks on different classes of repetitive elements in spermatocytes from P12 males. The Npm1 promoter region served as a negative control. (D) Cot1 RNA FISH on pachytene spermatocytes isolated from 15-d-old testis. Sex chromosomes were visualized by γH2AX immunofluorescence. (E) Frozen testis sections of 4-mo-old mice were immunostained with the antibodies indicated. Bar, 10 μm. (F,G) ChIP-seq analysis of total EED and H3K27me3 enrichment for each chromosome.
Since PRC2 is essential for the maintenance of imprinted X inactivation in the extraembryonic tissues of female mice (Wang et al. 2001), PRC2 may also be involved in MSCI, which occurs during pachynema. To evaluate transcriptional activity in pachytene spermatocytes, we performed RNA FISH for Cot-1, the repetitive fraction of the genome, which is used as a surrogate to detect nascent transcription (Britten and Kohne 1968). We found that the sex chromosomes were devoid of Cot1 RNA FISH signals in both the controls and Eed mutants (Fig. 6D), suggesting that MSCI occurs in the absence of PRC2. Furthermore, EZH2, H3K27me2, and H3K27me3 were excluded from sex bodies in wild-type pachytene spermatocytes (Fig. 6E). Analysis of EED and H3K27me3 ChIP-seq data sets from wild-type spermatocytes showed that the X chromosome lacks both association with EED (Fig. 6F) and incorporation of H3K27me3 compared with autosomes (Fig. 6G). Therefore, PRC2 does not have a prominent role in the chromosome-wide transcriptional repression associated with MSCI.
MSCI may function as a meiotic checkpoint, where its failure results in arrest at the pachytene stage. Activation of either Zfy1 or Zfy2 on the Y chromosome in meiocytes is sufficient to cause meiotic arrest (Royo et al. 2010). We verified the down-regulation of these two genes in zygotene (P12) and pachytene (P17) spermatocytes (Supplemental Fig. S5A). Loss of EED did not cause the up-regulation of Zfy1 and Zfy2 in P12 testis (Supplemental Fig. S5B). Furthermore, the paucity of H3K27me3 at the promoter regions of these two genes confirmed that the repression of Zfy1 and Zfy2 occurs independently of PRC2 (Supplemental Fig. S5C).
PRC2 is responsible for maintaining SSCs
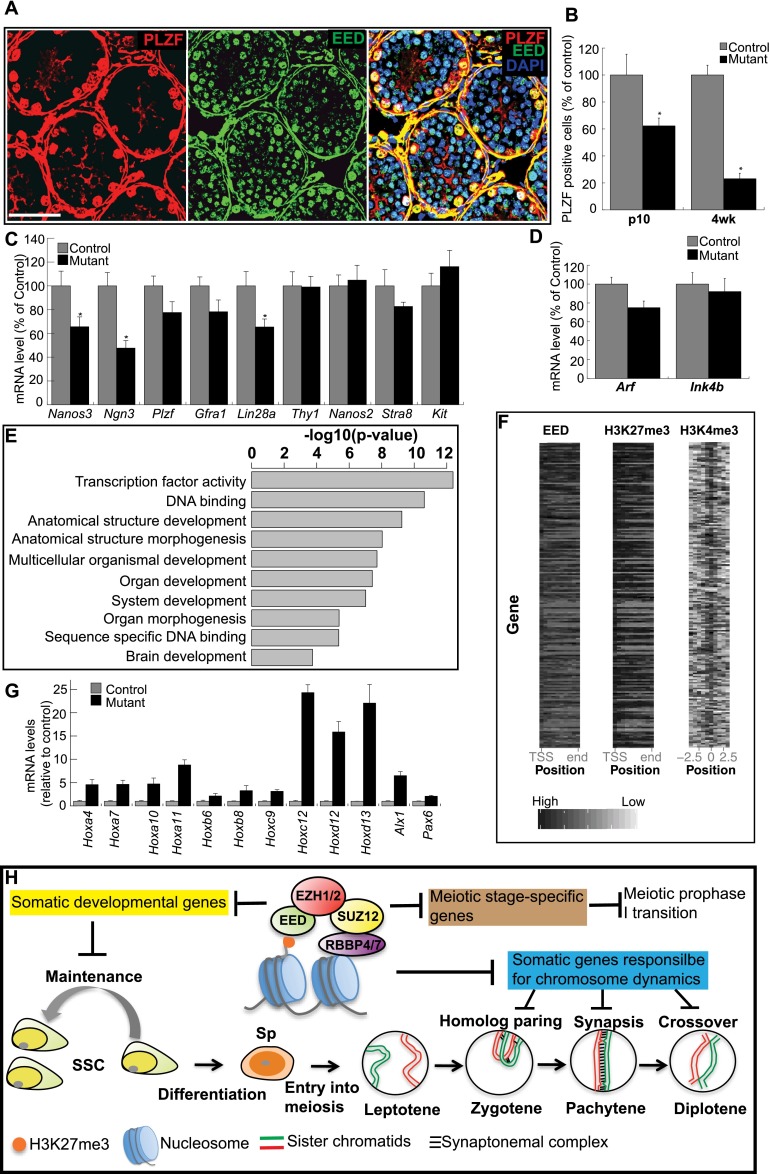
By 2 mo, most tubules in Eed mutant testis were entirely depleted of germ cells (Supplemental Fig. S6A), suggesting that PRC2 is required for the maintenance of the mitotic spermatogonial population. Spermatogonia are a heterogeneous cell population that includes SSCs, undifferentiated spermatogonia, and differentiated spermatogonia. EED is present in SSCs and undifferentiated spermatogonia, which can be identified with PLZF (Fig. 7A), suggesting a potential function in these cell populations. In Eed mutant testes, EED was undetectable in PLZF-positive mutant cells at P3, prior to spermatogonial differentiation (Supplemental Fig. S7). Furthermore, we found that the number of PLZF-positive cells decreased with age in Eed mutant testes. Relative to testes from wild-type animals, only 62.3% and 23.1%, respectively, of the mutant cells were PLZF-positive at P10 and 4 wk (Fig. 7B), indicating a loss of SSCs and undifferentiated spermatogonia in the absence of PRC2. However, this loss was not associated with a difference in proliferation (indicated by proliferating cell nuclear antigen [PCNA]) (Supplemental Fig. S7B) or apoptosis (data not shown) in P7 mutant testes. Therefore, the reduction of undifferentiated spermatogonia in Eed mutants is caused by either precocious differentiation into differentiated spermatogonia or an insufficient supply from their progenitors, SSCs.
Figure 7.
Undifferentiated spermatogonia in PRC2 mutants. (A) Cryosection of P10 testis immunostained for the indicated proteins. (B) The number of PLZF-positive cells was quantified (as a percentage of PLZF-positive cells in control testes) by counting five sections. (*) P < 0.01. (C) RT-qPCR analysis of spermatogonial markers in P7 testes. (D) RT-qPCR analysis of cell cycle regulators (p19/Arf and Ink4b) in P7. (E) gene ontology (GO) analysis of EED-enriched genes. The top 200 genes enriched for EED were included in this analysis. The top 10 most enriched biological processes based on their P-values are shown. (F) Heat maps depicting the binding patterns of EED, H3K27me3, and H3K4me3 within −2.5/+2.5 kb of the predicted TSS (H3K4me3) or across gene bodies (EED and H3K27me3). The genes are ranked based on EED read density. The ChIP-seq reads of EED and H3K27me3 were normalized to the length of each gene. (G) RT-qPCR analysis of the expression of key developmental genes in P7 testis. (H) Proposed model for regulatory roles of the PRC2 complex during spermatogenesis. PRC2 and H3K27me3 repress three groups of genes in germ cells to safeguard SSC maintenance and meiotic progression. In the absence of functional PRC2, (1) derepression of somatic developmental genes may alter SSC homeostasis, (2) ectopic expression of somatic genes interferes with the functions of germ cell-specific counterparts in the orchestration of homologous chromosome rearrangement, and (3) the failure to down-regulate meiotic stage-specific genes hinders transitions through prophase I. Bar, 50 μm.
To determine the underlying cause of the observed reduction in undifferentiated spermatogonia in Eed mutants, we used RT-qPCR to measure the expression levels of markers for differentiated spermatogonia (Stra8 and Kit) and for undifferentiated spermatogonia and SSCs (Nanos3, Ngn3, Plzf, Gfra1, Lin28a, Thy1, and Nanos2). To correct for differences in germ cell number between wild-type and Eed mutant testes, we normalized the expression levels of these genes to the levels of a germ cell marker, Mvh. There was no change in the levels of the markers of differentiated spermatogonia (Fig. 7C), indicating that the reduction of undifferentiated spermatogonia is not a result of precocious differentiation. In contrast, the expression levels for SSC and undifferentiated spermatogonia markers Nanos3, Ngn3, and Lin28a were significantly decreased in Eed mutants compared with controls. Collectively, these data suggest that the reduction of undifferentiated spermatogonia in mutant testes can be attributed to defects in the maintenance of SSCs.
One possible mechanism for PRC2 in maintaining SSC populations could be through transcriptional repression of the Ink4b–Arf–Ink4a locus (Bracken et al. 2007; Kotake et al. 2007). In stem and progenitor cells of the liver, skin, and muscle, lack of PRC2 function up-regulates the cell cycle inhibitors p16INK4A and p19ARF, leading to cell cycle arrest and senescence (Aoki et al. 2010; Ezhkova et al. 2011; Juan et al. 2011). However, in Eed mutant testes, Arf and Ink4b mRNA levels were not elevated compared with controls (Fig. 7D); Ink4a levels were too low to be detected by quantitative PCR (qPCR) in both the mutants and the controls. These observations suggest that the requirement for PRC2 in maintaining a self-renewing SSC population may not occur through PRC2-dependent regulation of the Ink4b–Arf–Ink4a locus.
In ESCs, H3K27me3 and PRC2 are enriched on genes encoding key developmental regulators, maintaining a poised state with low levels of transcription (Bernstein et al. 2006; Boyer et al. 2006; Lee et al. 2006). To determine whether PRC2 may function in an analogous manner in developing germ cells, we analyzed genome-wide EED, H3K27me3, and H3K4me3 in spermatocytes by ChIP-seq. Using gene ontology (GO) term enrichment analysis, we found that the enrichment of EED coincided with H3K27me3 and H3K4me3 at key developmental genes (Figs. 7E). While H3K4me3, H3K27me3, and EED were found to be enriched in the promoter regions, only EED and H3K27me3 were enriched throughout the gene body, covering both introns and exons (Fig. 7F). Notably, genes essential for somatic development, which we found associated with H3K27me3 in wild-type spermatocytes, were up-regulated in P7 Eed mutant testis (Fig. 7G). Consistent with germ cell-specific depletion of EED in mutant testis, RNA in situ hybridization revealed the up-regulation of Hoxa4 genes in spermatogonia but not in Sertoli cells (Supplemental Fig. S8). These results support a model in which PRC2 contributes to SSC maintenance by restricting expression of developmental genes.
Discussion
Transcriptional profiling has revealed the dynamic changes to gene expression in differentiating germ cells (Schlecht et al. 2004; Shima et al. 2004), yet regulators of this program remain largely unidentified. We observed dynamic changes in the genome-wide distribution of H3K27me3 in spermatocytes of postnatal testes, including the promoters of meiotic initiation genes, such as Stra8, Dnmt3a, and Cyp26b1. Increases in H3K27me3 coincided with decreased transcriptional levels during meiotic progression, suggesting that PRC2 may repress the expression of early meiotic genes during the meiosis phase transition. Consistent with our findings, Yokobayashi et al. (2013) reported that the expression of Stra8 is silenced with the accumulation of H3K27me3 on its promoter and is then activated with the erasure of this mark during female germ cell development. Based on these observations, we propose a model (Fig. 7H) in which PRC2 controls male germ cell development through repression of soma-specific genes and timely down-regulation of meiotic genes. Up-regulation of these genes due to PRC2 deficiency impairs meiotic chromosome dynamics and SSC maintenance. Together, these data demonstrate that PRC2 is a critical regulator of mammalian spermatogenesis, with essential roles in both the meiotic and mitotic compartments of the male germline.
Derepression of the somatic lamins in spermatocytes due to loss of PRC2 may have a particularly deleterious effect on meiotic progression. Ectopic expression of lamins is harmful to cell viability and proliferation (Stuurman et al. 1999; Dreesen et al. 2013) and NE integrity (Schutz et al. 2005; Jahn et al. 2010). During meiosis, the interaction between telomeres and specific LINC complexes is essential for the chromosome mobility that underlies homologous chromosome pairing and synapsis (Dernburg 2013). PRC2 mutants had more clustered telomeres and unpaired homologs, which is indicative of impaired chromosome rearrangement. Lamin C2 is a meiotic-specific lamin transcribed from an alternative promoter at the Lmna locus, where the somatic lamin A and C promoters are normally associated with high levels of H3K27me3 in wild-type spermatocytes. It is therefore likely that ectopic expression of lamins A and C in mutant spermatocytes interferes with the function of lamin C2 in telomere-mediated rearrangement of meiotic chromosomes (Link et al. 2013). These data are compatible with the model in which ectopic expression of somatic lamins in meiocytes impairs the hypermobility of the LINC complex, possibly through the interaction with SUN1. As a result, telomere-mediated chromosome movement is restricted, interfering with homolog pairing and synapsis formation.
The progressive loss of spermatogonia in Eed mutants suggests that PRC2 plays a unique role in maintaining the undifferentiated spermatogonial pool through regulating SSC self-renewal but independently of Ink4a/Arf-dependent repression. PRC2 is known to facilitate somatic stem cell and progenitor cell proliferation in skin (Ezhkova et al. 2009), liver (Aoki et al. 2010), skeletal muscle (Juan et al. 2011), and cardiac muscle (Delgado-Olguin et al. 2012). Homeostasis in all of these tissues is facilitated by PRC2-mediated suppression of the Ink4a and Arf tumor suppressors. In contrast, Arf is specifically expressed in spermatogonia of adult mice. The induction of Arf expression in spermatogonia does not interfere with mitotic cell divisions but instead contributes to the survival of meiotic male germ cells (Churchman et al. 2011). Spermatogonial proliferation is synchronized with cycles of seminiferous epithelium and also coincides with cellular differentiation, indicating that the cell cycle regulation of SSC is distinct from that of somatic cells. Thus, it appears that a separate, p16INK4A/p19ARF-independent but PRC2-dependent mechanism exists for maintaining SSC self-renewal.
We hypothesize that the ectopic expression of developmental genes in the absence of PRC2 compromises germ cell-specific transcription. In adult mice, the transcription factor NGN3 is involved in SSC self-renewal (Yoshida et al. 2006). However, in Eed mutants, Ngn3 is one of the most down-regulated genes among the markers of undifferentiated spermatogonia. The impairment of SSC maintenance and subsequent germ cell depletion in Eed mutants may be a result of deregulation of Ngn3. Because PRC2 functions as a transcriptional repressor, the down-regulation of key undifferentiated spermatogonia genes (Ngn3, Nanos3, and Lin28a) cannot be attributed directly to the loss of PRC2. Consistent with our hypothesis, removal of PRC2 permits ectopically expressed, neuron type-specific transcription factors to convert germ cells directly into specific neuron types in Caenorhabditis elegans (Patel et al. 2012). Therefore, PRC2 may control spermatogonial maintenance through repression of developmental genes.
Materials and methods
Generation of Eed and Suz12 mutant mice
The EUCOMM Eed knockout ESC line was microinjected into albino B6 blastocysts to generate chimeric mice. These chimeras were subsequently crossed with CD1 females, and heterozygous offspring bearing the targeted alleles were crossed with CAG-FLPe transgenic lines to excise the Frt-βGal-Neo-Frt cassette. Eed mutant and control littermates were obtained from crosses between homozygous females (Eed fl/fl) and Mvh-Cre/+; EedΔ/+ males. Eed-floxed mice and Mvh-Cre mice were maintained on a mixed background. To generate Suz12 mutant mice, we a used recombineering-based method (Liu et al. 2003) to build a Suz12 conditional allele, which contains loxP-flanked exons 2 and 3 and a Frt-flanked Neo cassette (Supplemental Fig. S1B). The Kpn1-linearized targeting vector DNA was electroporated into E14 cells followed by selection in the presence of 250 μg/mL G418 and 2 μM ganciclovir. Correctly targeted clones were identified by Southern blot analysis with 5′ and 3′ probes (Supplemental Fig. S1B,E) and microinjected into C57BL/6J blastocysts to generate chimeric mice. The Suz12 mutant animals were bred through the same breeding strategy as the Eed mutants and genotyped by PCR (Supplemental Fig. S1F). The University of North Carolina Institutional Animal Care and Use Committee approved all animal use protocols.
Histological analyses
Testes were fixed in Bouin’s overnight, embedded in paraffin, and sectioned at 6 μm. Following standard protocols, sections were deparaffinized, rehydrated, and then stained with hematoxylin and eosin for histology.
Immunocytochemistry
Immunolabeling of surface-spread spermatocytes was performed as described (Peters et al. 1997). Spermatocytes were isolated from testes of 14-d-old mice. Immunostaining of testis cryosections was prepared as previously described. The information of primary antibodies used in this study is provided in the Supplemental Material. Secondary antibodies conjugated with either Alexa Fluor 488 or 594 (Molecular Probes) were used at a dilution of 1:500.
Apoptosis assay
The apoptotic cells were detected by fluorescent TUNEL assay on testis cryosections using the in situ cell death detection kit (Roche, no. 11 684 795 001).
Cot1 RNA FISH and RNA in situ hybridization
The spermatocytes were collected from 15-d-old testes for Cot1 RNA FISH analysis. Details of the procedure were described previously (Mahadevaiah et al. 2009). RNA in situ hybridization was performed on P7 testis section as described (Chandler et al. 2007).
Combined immunofluorescence staining and DNA FISH
For DNA FISH assays on homolog pairing, structurally preserved nuclei were prepared from spermatocytes isolated from 14-d-old testes and then pretreated with NaSCN, pepsin, and RNase A as described (Scherthan 2009). Immunofluorescence staining followed by DNA FISH was performed as previously described (Fedoriw et al. 2012). The probes derived from RP24-99N6 and RP24-367A12 detected the subtelomeric region and mid-region of chromosome 1, respectively. Two homologs were defined as paired if the distance between their focus centers was ≤1.35 μm.
Isolation of spermatogenic cells
The methodology for isolation of spermatocytes was described previously (Chang et al. 2011). Testes were collected from P12, P17, P21, and P30 mice and digested with collagenase, trypsin, and DNase I. Percoll solution, and cell strainers were applied to separate somatic cells from spermatogenic cells.
RT–PCR and qPCR
Total RNA was extracted from whole testis or isolated spermatocytes using TRIzol reagent (Invitrogen) followed by an RNA cleanup step and on-column DNA digestion using the RNeasy mini prep kit (Qiagen) according to the manufacturers’ instructions. cDNA was synthesized using SuperScript III (Invitrogen). Real-time qPCR analysis was performed using SsoFast EvaGreen supermix (Bio-Rad) and CFX96 thermocycler (Bio-Rad).
ChIP-seq
ChIP-seq was performed as described (Calabrese et al. 2012). Five million isolated spermatocytes and 3 μg of antibody were used per immunoprecipitation. ChIP-seq libraries were prepared according to Illumina’s instructions and sequenced on Illumina’s Genome Analyzer IIx or HiSeq2000 instrument. Sequence reads were aligned to genomic sequence with Bowtie (Langmead et al. 2009). All mm9 genome annotations were obtained from the University of California at Santa Cruz genome browser. MACS identified ChIP-seq enrichment (Feng et al. 2012). Metaplots of H3K27me3 across gene bodies were drawn using ngs.plot (Shen et al. 2014). To compare H3K27me3 enrichment of the gene bodies, we used HTSeq-count (Anders et al. 2014) to determine the number of reads per gene using mm9 annotations. We then use edgeR to determine significant differences in read counts (Robinson et al. 2010). MSigDB provided GO analysis (Subramanian et al. 2005).
Acknowledgments
We thank all members of the Magnuson laboratory, especially J.M. Calabrese, for helpful comments on manuscript preparation and technical support for the ChIP-seq experiment. M. Alsheimer (University of Wüzburg, Wüzburg, Germany), A. Bortvin (Johns Hopkins University, Baltimore, MD), and A. Otte (Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, The Netherlands) kindly provided antibodies against SUN1, ORF1p, and EED, respectively. This work was supported by National Institutes of Health grants RO1 GM101974 and U42 OD010924.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.246124.114.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Aldiri I, Vetter ML. 2012. PRC2 during vertebrate organogenesis: a complex in transition. Dev Biol 367: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2014. HTSeq— a Python framework to work with high-throughput sequencing data. bioRxiv doi: 10.1101/002824 [DOI] [PMC free article] [PubMed]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. 2008. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci 105: 14976–14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki R, Chiba T, Miyagi S, Negishi M, Konuma T, Taniguchi H, Ogawa M, Yokosuka O, Iwama A. 2010. The polycomb group gene product Ezh2 regulates proliferation and differentiation of murine hepatic stem/progenitor cells. J Hepatol 52: 854–863 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. 2013. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell 24: 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Koopman P. 2007. Retinoic acid, meiosis and germ cell fate in mammals. Development 134: 3401–3411 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. 2007. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branciforte D, Martin SL. 1994. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol 14: 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE. 1968. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161: 529–540 [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. 2013. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol 14: 13–24 [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. 2007. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol Cell Biol 27: 2934–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Lee-Chang JS, Panneerdoss S, MacLean JA 2nd, Rao MK. 2011. Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. Biotechniques 51: 341–342, 344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Roig I, Jasin M, Keeney S, Sherr CJ. 2011. Expression of arf tumor suppressor in spermatogonia facilitates meiotic progression in male germ cells. PLoS Genet 7: e1002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commerford SL, Carsten AL, Cronkite EP. 1982. Histone turnover within nonproliferating cells. Proc Natl Acad Sci 79: 1163–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. 2012. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet 44: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF 2013. Pushing the (nuclear) envelope into meiosis. Genome Biol 14: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12: 863–872 [DOI] [PubMed] [Google Scholar]
- Dreesen O, Chojnowski A, Ong PF, Zhao TY, Common JE, Lunny D, Lane EB, Lee SJ, Vardy LA, Stewart CL, et al. 2013. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J Cell Biol 200: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw AM, Calabrese JM, Mu W, Yee D, Magnuson T. 2012. Differentiation-driven nucleolar association of the mouse imprinted Kcnq1 locus. G3 2: 1521- 1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. 2007. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45: 413–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. 2006. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 26: 3738–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. 2009. The SUN rises on meiotic chromosome dynamics. Dev Cell 17: 598–605 [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Shibuya H, Hernandez-Hernandez A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, et al. 2014. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev 28: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D, Schramm S, Benavente R, Alsheimer M. 2010. Dynamic properties of meiosis-specific lamin C2 and its impact on nuclear envelope integrity. Nucleus 1: 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. 2011. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev 25: 789–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes Dev 21: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. 2006. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci 103: 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe B, Petukhova G, Barchi M, Bellani M, Braselmann H, Nakano T, Pandita TK, Jasin M, Fornace A, Meistrich ML, et al. 2006. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp Cell Res 312: 3768–3781 [DOI] [PubMed] [Google Scholar]
- Link J, Jahn D, Schmitt J, Gob E, Baar J, Ortega S, Benavente R, Alsheimer M. 2013. The meiotic nuclear lamina regulates chromosome dynamics and promotes efficient homologous recombination in the mouse. PLoS Genet 9: e1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Costa Y, Turner JM. 2009. Using RNA FISH to study gene expression during mammalian meiosis. Methods Mol Biol 558: 433–444 [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. 2005. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 15: 942–947 [DOI] [PubMed] [Google Scholar]
- Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. 2012. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. 2009. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci 122: 4099–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T, Tursun B, Rahe DP, Hobert O. 2012. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Reports 2: 1178–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5: 66–68 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM. 2010. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20: 2117–2123 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. 2008. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140 [DOI] [PubMed] [Google Scholar]
- Scherthan H 2009. Analysis of telomere dynamics in mouse spermatogenesis. Methods Mol Biol 558: 383–399 [DOI] [PubMed] [Google Scholar]
- Schlecht U, Demougin P, Koch R, Hermida L, Wiederkehr C, Descombes P, Pineau C, Jegou B, Primig M. 2004. Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol Biol Cell 15: 1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz W, Benavente R, Alsheimer M. 2005. Dynamic properties of germ line-specific lamin B3: the role of the shortened rod domain. Eur J Cell Biol 84: 649–662 [DOI] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. 2014. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. 2004. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71: 319–330 [DOI] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15: 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Delbecque JP, Callaerts P, Aebi U. 1999. Ectopic overexpression of Drosophila lamin C is stage-specific lethal. Exp Cell Res 248: 350–357 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet 28: 371–375 [DOI] [PubMed] [Google Scholar]
- Wynne DJ, Rog O, Carlton PM, Dernburg AF. 2012. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196: 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. 2013. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 495: 236–240 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. 2006. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 133: 1495–1505 [DOI] [PubMed] [Google Scholar]
- Yuan W, Leisner TM, McFadden AW, Clark S, Hiller S, Maeda N, O’Brien DA, Parise LV. 2006. CIB1 is essential for mouse spermatogenesis. Mol Cell Biol 26: 8507–8514 [DOI] [PMC free article] [PubMed] [Google Scholar]