Transcription of endogenous retroviruses is inhibited by de novo DNA methylation during gametogenesis, and DNA methylation reaches a low point in E13.5 primordial germ cells (PGCs) of both sexes. Liu et al. find that germline-specific conditional knockout of the H3K9 methyltransferase SETDB1 yields a decrease of H3K9me3 and H3K27me3 marks and DNA methylation at H3K9me3-enriched retrotransposon families. Setdb1 deficiency is associated with a reduced number of male E13.5 PGCs and postnatal hypogonadism in both sexes. Therefore, SETDB1 is an essential guardian against proviral expression prior to the onset of de novo DNA methylation in the germline.
Keywords: endogenous retroviruses, germ cell development, Setdb1, H3K9me3, H3K27me3, DNA methylation
Abstract
Transcription of endogenous retroviruses (ERVs) is inhibited by de novo DNA methylation during gametogenesis, a process initiated after birth in oocytes and at approximately embryonic day 15.5 (E15.5) in prospermatogonia. Earlier in germline development, the genome, including most retrotransposons, is progressively demethylated. Young ERVK and ERV1 elements, however, retain intermediate methylation levels. As DNA methylation reaches a low point in E13.5 primordial germ cells (PGCs) of both sexes, we determined whether retrotransposons are marked by H3K9me3 and H3K27me3 using a recently developed low-input ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) method. Although these repressive histone modifications are found predominantly on distinct genomic regions in E13.5 PGCs, they concurrently mark partially methylated long terminal repeats (LTRs) and LINE1 elements. Germline-specific conditional knockout of the H3K9 methyltransferase SETDB1 yields a decrease of both marks and DNA methylation at H3K9me3-enriched retrotransposon families. Strikingly, Setdb1 knockout E13.5 PGCs show concomitant derepression of many marked ERVs, including intracisternal A particle (IAP), ETn, and ERVK10C elements, and ERV-proximal genes, a subset in a sex-dependent manner. Furthermore, Setdb1 deficiency is associated with a reduced number of male E13.5 PGCs and postnatal hypogonadism in both sexes. Taken together, these observations reveal that SETDB1 is an essential guardian against proviral expression prior to the onset of de novo DNA methylation in the germline.
Retrotransposons constitute nearly 40% of the mouse genome (Mouse Genome Sequencing Consortium 2002; Rowe et al. 2010). A subset of these intracellular parasites, including long terminal repeat (LTR) and LINE1 elements, maintain the ability to retrotranspose, thus representing an ongoing threat to genome integrity (Maksakova et al. 2006; Jern and Coffin 2008). Numerous transcriptional and post-transcriptional silencing pathways have evolved to suppress expression of retrotransposons (Wolf and Goff 2008). For example, loss of DNA methylation in somatic tissues of embryos deficient for the maintenance DNA methyltransferase DNMT1 leads to derepression of intracisternal A particle (IAP) endogenous retroviruses (ERVs) and embryonic lethality (Walsh et al. 1998). Concomitant with a loss of DNA methylation (Kato et al. 2007), IAP ERVs are also derepressed in perinatal testes of mice deficient for DNMT3L, a cofactor involved in de novo DNA methylation (Bourc’his and Bestor 2004). Moreover, conditional knockout of the de novo DNA methyltransferase DNMT3a in germ cells leads to reactivation of IAP elements, imprinting defects, and male sterility (Kaneda et al. 2004). Intriguingly, however, induction of IAP expression was not observed in Dnmt1 knockout primordial germ cells (PGCs) (Walsh et al. 1998), suggesting that an alternative silencing pathway may function prior to the gonocyte stage.
In order to erase parental imprints and initiate gametogenesis, both male and female PGCs undergo widespread epigenetic reprogramming. DNA methylation levels progressively decrease in proliferating pre- and post-migratory PGCs, reaching a low point at approximately embryonic day 13.5 (E13.5) (Hajkova et al. 2002; Lees-Murdock et al. 2003; Seisenberger et al. 2012; Kobayashi et al. 2013). At this stage, both de novo and maintenance DNA methylation are compromised due to reduced levels of expression of Dnmt3a and sequestration of DNMT3B and the DNMT1 cofactor NP95 in the cytoplasm (Hajkova et al. 2002; Seisenberger et al. 2012). While most retrotransposons are demethylated during this period, relatively high levels of DNA methylation persist at a subset of unique genomic regions and repetitive elements, including at IAP and other ERV1 (class I) and ERVK (class II) families (Hajkova et al. 2002; Lane et al. 2003; Seisenberger et al. 2012; Kobayashi et al. 2013). Beginning at approximately E15.5 in male gonocytes and after birth in oocytes, the genome is broadly remethylated by DNMT3A and DNMT3B (Kato et al. 2007). De novo DNA methylation of retroelements in prenatal male gonocytes is also dependent on DNMT3L (Kato et al. 2007) and the nuclear Argonaute protein MIWI2 (Kuramochi-Miyagawa et al. 2008), which act together in a specialized piRNA-dependent pathway to target transcriptionally active LTR and LINE1 elements in the nucleus (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Indeed, piRNAs complementary to both classes of retroelements are highly expressed at this stage (Kuramochi-Miyagawa et al. 2008), and knockout of subunits of the piRNA biogenesis pathway, including MIWI2, leads to male sterility and reactivation of IAP and/or LINE1 elements in postnatal male germ cells (Ishizu et al. 2012).
However, whether alternative pathways act to maintain retroelements in a silent state prior to the wave of MIWI2/DNMT3L-dependent de novo methylation remains to be determined. We showed previously that genomic regions retaining DNA methylation in naïve embryonic stem cells (ESCs) or ESCs deficient in DNMT3a and DNMT3b, including IAP elements and imprinted differentially methylated regions (DMRs), are enriched for H3K9me3 (Blaschke et al. 2013; Leung et al. 2014). Based on our previous findings that SETDB1-dependent deposition of H3K9me3 is required for silencing of ERV1 and ERVK families independent of DNA methylation in ESCs (Matsui et al. 2010; Karimi et al. 2011), we hypothesized that this methyltransferase may play a role in transcriptional repression of ERVs in PGCs.
Using our new ultralow-input NChIP-seq (native chromatin immunoprecipitation [NChIP] combined with deep sequencing) assay (J Brind’Amour, in prep.), we show that a subset of young ERV families, including IAP and ETn, as well as young LINE1 families are indeed marked by H3K9me3 in E13.5 PGCs. We also found that these elements are marked by H3K27me3, and enrichment of both marks at these potentially active retroelements is positively correlated with DNA methylation levels. Germline ablation of Setdb1 in PGCs yields decreased levels of H3K9me3 and H3K27me3 as well as aberrant DNA methylation at marked ERVs and LINE1 elements. However, widespread reactivation was observed exclusively for ERVs, with specific families showing sex-dependent expression levels. Gonads isolated from male E13.5 Setdb1 knockout embryos also showed a significant reduction in the number of PGCs relative to somatic cells, and germline defects extending into adulthood were observed in both males and females. Taken together, these observations reveal that SETDB1 plays an essential role in the establishment and/or maintenance of maximal H3K9me3, H3K27me3, and DNA methylation at young LTR and LINE1 elements and is required for proviral silencing prior to the onset of de novo DNA methylation in the prenatal germline.
Results
E13.5 gonadal somatic cells (soma) and PGCs express distinct retroelements
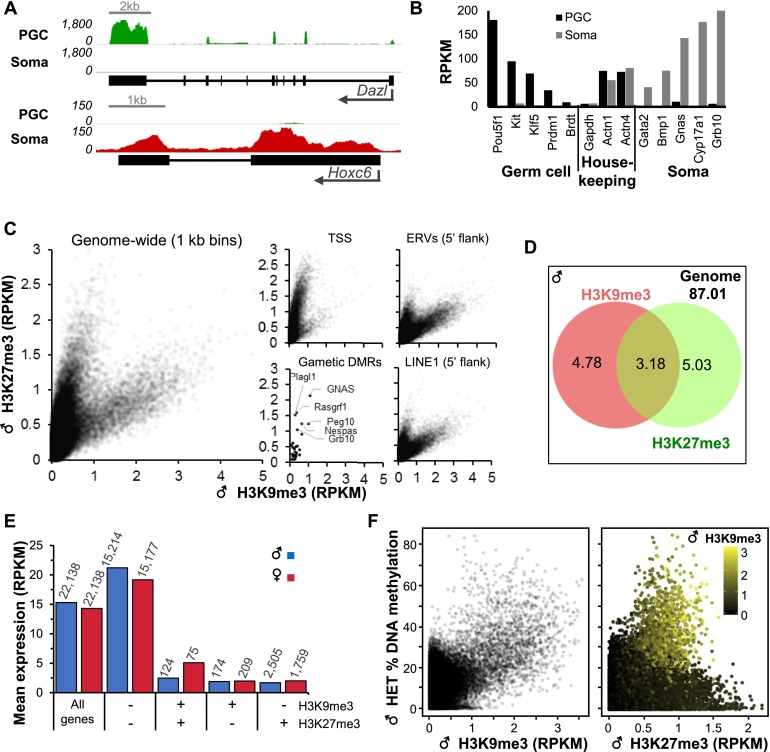
We first analyzed the expression of genes and repetitive elements in soma versus PGCs at E13.5, the stage in germ cell development when DNA methylation levels are at their lowest (Seisenberger et al. 2012; Hackett et al. 2013; Kobayashi et al. 2013). PGCs and soma from E13.5 gonads were purified by fluorescence-activated cell sorting (FACS) using the germ cell surface marker SSEA-1 (Supplemental Fig. 1A; Durcova-Hills et al. 1999). RNA sequencing (RNA-seq) analyses of total RNA from 103 PGCs or soma isolated from a single male embryo revealed that expression of male germ cell markers such as Dazl is restricted to PGCs, while expression of genes expressed in somatic cells, such as Hoxc6, is restricted to somatic/SSEA-1− cells (Jameson et al. 2012), confirming the purity of the sorted populations (Fig. 1A,B). Furthermore, gene ontology analysis revealed that genes associated with processes involved in male germline development, such as spermatogenesis, stem cell maintenance, and meiosis, are enriched in the SSEA-1+ population, as expected (Supplemental Fig. 1B; Supplemental Table 1). Notably, analysis of the expression of ERVs revealed high levels of expression of VL30 in both PGCs and soma but cell type-dependent differences in the expression of other families (Supplemental Fig. 1C). ETn elements, for example, are expressed at a higher level in PGCs, while the converse is true of several ERV1 and ERVK families, including RLTR4 (MLV) and IAPey3. Taken together, these data reveal clear differences in the regulation of retroelements in gonadal germ versus somatic cells at this developmental stage.
Figure 1.
Enrichment of H3K9me3, H3K27me3, and gene expression in E13.5 PGCs. (A) Genome browser screenshots of Dazl and Hoxc6 loci showing RNA-seq data for PGCs and soma. (B) Expression (RPKM) of germ cell-specific, somatic, and housekeeping genes in PGCs and soma. (C) Coverage of H3K9me3 and H3K27me3 genome-wide (1-kb bins; 50,000 random regions displayed), at transcription start sites (TSSs) (±1 kb) and gametic DMRs, and in the 5′ 1-kb region flanking annotated ERVs or LINEs. (D) Percentage of the mappable genome enriched for H3K9me3 and/or H3K27me3 in male E13.5 PGCs. Enriched regions were identified using MACS2 (P < 0.05). (E) Relationship between mean gene expression level and H3K9me3 and/or H3K27me3 enrichment at TSSs. The number of genes in each category is indicated above. (F) Genome-wide relationship (1-kb bins; 50,000 random regions displayed) between H3K9me3 enrichment, H3K27me3 enrichment, and DNA methylation in male E13.5 PGCs.
H3K9me3 and H3K27me3 mark distinct and overlapping genomic regions in E13.5 PGCs
To determine the role of SETDB1 in PGCs, we crossed the Tnap-Cre mouse line (Lomelí et al. 2000), which expresses Cre in germ cells from approximately E9.5 to late gestation, with a Setdb1 conditional knockout mouse line (Tan et al. 2012). E13.5 embryos were recovered and genotyped, and soma and PGCs were purified as described above (Supplemental Fig. 2; see the Materials and Methods). Using our recently developed low-input NChIP-seq protocol (J Brind’Amour, in prep.), we generated genome-wide profiles of H3K9me3 and H3K27me3 from 1000 PGCs of both sexes isolated from heterozygous (Setdb1flox/−; Tnap+/+) and knockout (Setdb1△/−; TnapCre/+) littermates, focusing initially on the former. Both marks show very similar distributions in male and female heterozygous PGCs, with genome-wide correlations (1-kb bins) of 0.89 and 0.9, respectively (Supplemental Fig. 3A). These results are consistent with previous microscopy-based studies of both marks in E13.5 PGCs (Lomelí et al. 2000; Hajkova et al. 2008; Abe et al. 2011). Intriguingly, most annotated genic transcription start sites (TSSs) are not marked or are enriched exclusively with H3K27me3 (Fig. 1C; Supplemental Fig. 3B). Consistent with erasure of parental DNA methylation imprints by E13.5 (Seisenberger et al. 2012), gametic DMRs show uniformly low levels of H3K9me3, as reported previously (Henckel et al. 2012). Interestingly, several gametic DMRs are enriched for H3K27me3 at this developmental stage (Fig. 1C; Supplemental Fig. 3B). In contrast, the flanks of a subset of ERVs and LINE1 elements are enriched for H3K9me3 and H3K27me3, with ∼3% and ∼2% of the genome enriched for both marks in male and female PGCs, respectively (Fig. 1D; Supplemental Fig. 3C). Consistent with the known functions of these covalent modifications, genes with unmarked TSSs are expressed at higher than average levels, while genes enriched for either or both silencing marks are expressed at low or undetectable levels (Fig. 1E; Supplemental Fig. 4A).
To determine the relationship between DNA methylation and these marks, we conducted whole-genome DNA methylation analysis on male heterozygous and knockout E13.5 PGCs using post-bisulfite adaptor tagging (PBAT) (Supplemental Table 2; Miura et al. 2012), focusing initially on the heterozygous data set. Strikingly, hypermethylated regions (>20% mean DNA methylation) are generally enriched for H3K9me3 and intermediate levels of H3K27me3 (Fig. 1F). In total, 2.9% of genomic bins are marked by H3K9me3 (reads per kilobase per million mapped reads [RPKM], >0.9) and show elevated DNA methylation. The apparent coexistence of H3K9me3 and intermediate levels of H3K27me3 at hypermethylated regions is also evident when compared with previously published PBAT data from E13.5 PGCs (Supplemental Fig, 4B,C; Kobayashi et al. 2013). While these observations likely reflect the coexistence of all three marks at the same genomic loci, mutually exclusive enrichment of H3K9me3 and H3K27me3 at these regions in distinct subpopulations of PGCs cannot be ruled out.
A subset of ERVs are marked by H3K9me3, H3K27me3, and DNA methylation in E13.5 PGCs
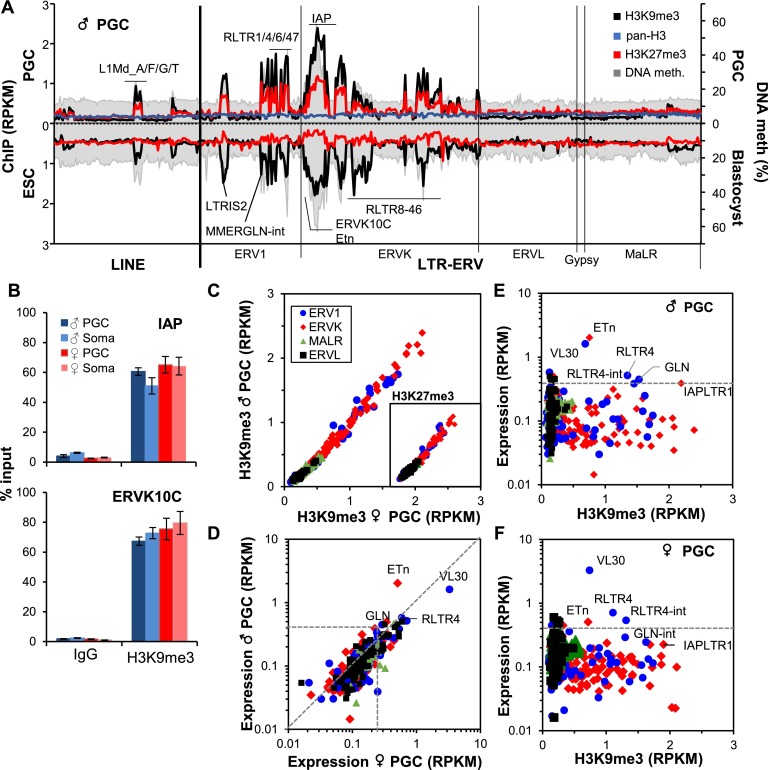
Having shown that the regions flanking ERV and LINE elements are frequently enriched for H3K9me3 and H3K27me3, we next analyzed the distribution of both marks within annotated repetitive elements. In male and female heterozygous PGCs, both marks are enriched at a number of ERV1 and ERVK families, including IAP (such as IAPez-int and its cognate LTRs, IAPLTR1 and IAPLTR1a) and ERVK10C (RLTR10C) elements (Fig. 2A; Supplemental Fig. 4D). These marks are also enriched, albeit to a lesser extent, at specific LINE1 elements, including the potentially active L1Md families (Dudley 1987), which show a 5′ bias in enrichment (Supplemental Fig. 4E). In contrast, H3 is evenly distributed across all repetitive elements (Fig. 2A). High H3K9me3 enrichment at IAP and ERVK10C families in male and female PGCs was confirmed by quantitative PCR (qPCR) (Fig. 2B). Surprisingly, H3K9me3 was also detected at these ERVs in soma, indicating that deposition at repetitive elements is not restricted to germ cells at this developmental stage.
Figure 2.
H3K9me3, H3K27me3, and DNA methylation at ERVs in E13.5 PGCs. (A) Enrichment of H3K9me3, H3K27me3, and H3 at Repbase annotated LINE and ERV families present at >100 copies in the BL6 genome is shown along with the mean percentage of DNA methylation (PBAT data for male heterozygous (HET) E13.5 PGCs (above the X-axis) or ESCs (shown below the X-axis) along with the mean percentage of DNA methylation in blastocysts (Kobayashi et al. 2012). LINE and ERV1, ERVK, ERVL, Gypsy, and MaLR classes of ERVs are presented in alphabetical order along the X-axis according to their Repbase nomenclature. Unique and multialigned reads were included in the analysis. (B) qPCR validation of H3K9me3 enrichment at IAP and ERVK10C ERVs in matched PGCs and soma isolated from male and female gonads. Error bars show standard deviation of technical replicates. (C) Enrichment of H3K9me3 and H3K27me3 (shown in inset) in male versus female E13.5 PGCs at all 385 Repbase annotated ERV families present at >100 copies. Unique and multialigned reads were included in the analysis. (D) Expression of ERVs in male versus female E13.5 PGCs. Relationship between ERV expression and H3K9me3 enrichment in male (E) and female (F) PGCs. Unique and multialigned reads were included in the analysis.
In contrast to mouse ESCs (mESCs) (Fig. 2A), enrichment levels of both marks are highly correlated at all ERV families in both male and female E13.5 PGCs (Fig. 2C), indicating that these retroelements harbor a unique combination of repressive histone marks in PGCs. H3K27me3 enrichment at ERVs was also recently reported in E13.5 PGCs using a distinct H3K27me3-specific antibody (Tan et al. 2012; Ng et al. 2013). Analysis of reads mapping uniquely to specific IAPez or Etn elements reveals a strong correlation between the levels of H3K9me3 and H3K27me3 in male and female PGCs, confirming that they mark the same individual retroelements (Supplemental Fig. 5A). Of note, ERV families with the highest level of enrichment of these repressive marks also show the lowest mappability (Supplemental Fig. 5B), indicating that they colonized the genome relatively recently. Thus, ERVs that are most likely to be competent for retrotransposition are preferentially targeted for deposition of H3K9me3 and H3K27me3 in PGCs.
As discussed above, specific repeats, IAP elements in particular, retain relatively high DNA methylation levels in E13.5 PGCs (Seisenberger et al. 2012; Kobayashi et al. 2013). As for genomic regions in general, ERV families enriched for H3K9me3 show relatively high levels of DNA methylation in E13.5 PGCs (Fig. 2A; Supplemental Fig. 4C). Analysis of recently published PBAT data (Kobayashi et al. 2012) confirms that these H3K9me3-marked retroelements are relatively hypermethylated in blastocysts (Fig. 2A). Elevated levels of both H3K9me3 and DNA methylation extend into the unique genomic regions flanking IAPez elements in PGCs, with both marks progressively diminishing to background levels ∼3 kb distal to the genomic DNA–ERV boundary (Supplemental Fig. 5C). Similar H3K9me3 and DNA methylation profiles are found in the flanks of IAP elements in ESCs and blastocysts, respectively. In contrast, elevated DNA methylation levels were not detected in the regions flanking MERVL elements, which are unmarked in each of these cell types.
While most enriched ERV1 and ERVK families are expressed at relatively low levels in PGCs (Fig. 2D), a subset, including ETn and RLTR4, is apparently expressed despite the presence of these repressive marks (Fig. 2E,F). Analysis of reads aligning uniquely to specific annotated elements reveals that while a majority of individual ERVs in both families show a low level of expression and high H3K9me3 enrichment, a subset of the mappable elements is expressed at a high level and shows relatively low levels of H3K9me3 (Supplemental Fig. 5D). Thus, while the majority of individual elements within each ERV family is repressed, likely by H3K9me3 alone or in concert with H3K27me3 and/or DNA methylation, rare members apparently evade such targeting and are constitutively expressed at this stage.
H3K9me3 and H3K27me3 are reduced at ERVs in Setdb1-deficient E13.5 PGCs
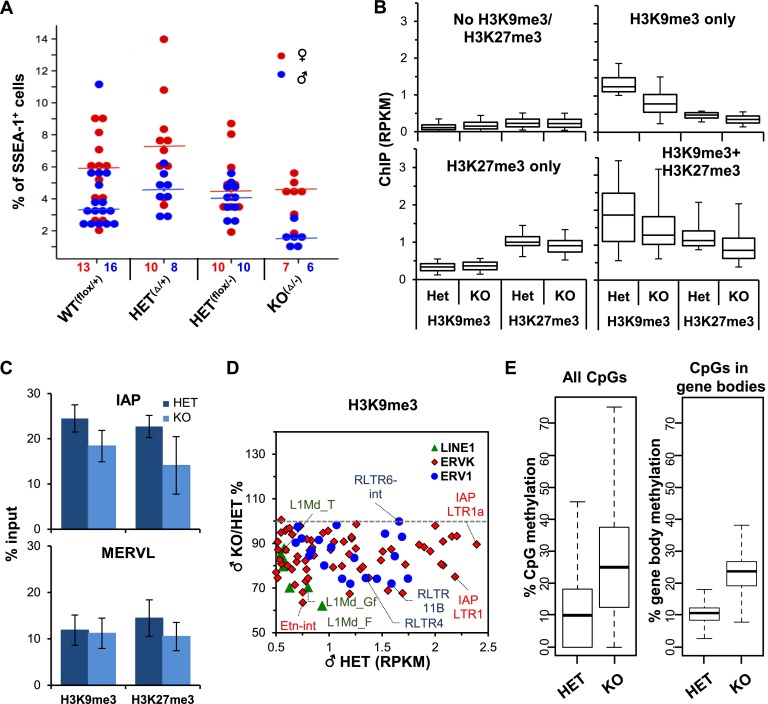
To determine the role of SETDB1 in H3K9 trimethylation, DNA methylation, and silencing of retroelements, we analyzed the data sets generated from the Setdb1 knockout (Setdb1△/−;TnapCre/+) littermates of the heterozygous (Setdb1flox/−;Tnap+/+) embryos described above. ChIP-seq, PBAT, and RNA-seq experiments on heterozygous and knockout pairs were all conducted in parallel. Strikingly, an approximately twofold to threefold decrease in the number of germ cells relative to soma was consistently observed in E13.5 male knockout gonads (Fig. 3A; Supplemental Fig. 6A). Nevertheless, sufficient numbers of PGCs were isolated from single embryos for genome-wide analyses. Based on relative read coverage of RNA-seq data generated from knockout and heterozygous littermates over Setdb1 exons 15 and 16, the deletion efficiency was estimated to be 60%–70% (Supplemental Fig. 6B), consistent with that originally reported for the Tnap-Cre line at E13.5 (Lomelí et al. 2000).
Figure 3.
Influence of Setdb1 knockout (KO) on PGC number and chromatin marks. (A) Percentage of PGCs in male and female gonads at E13.5, as determined by flow cytometry. The number of embryos analyzed for each genotype is presented below. Wilcoxon rank sum test P-value for WT(flox/+) versus KO(Δ/−): 0.0005 and 0.210; HET(Δ/+) versus KO(Δ/−): 0.001 and 0.013; and HET(flox/−) versus KO(Δ/−): 0.001 and 0.740 in male and female PGCs, respectively. (B) Box and whisker plots of global (1-kb bins) H3K9me3 and H3K27me3 in Setdb1 knockout versus heterozygous PGCs (box, 25th and 75th percentile; whisker, fifth and 95th percentile) in unmarked regions or regions enriched for H3K9me3 only, H3K27me3 only, or both marks. (C) qPCR validation of H3K9me3 enrichment at IAP and MERVL ERVs in matched Setdb1 heterozygous and knockout male PGCs. Error bars show standard deviation of three biological replicates. (D) Change in H3K9me3 at LINE1, ERVK, and ERV1 families in E13.5 male Setdb1 knockout relative to heterozygous PGCs versus the level of enrichment of H3K9me3 in heterozygous PGCs. No unmarked ERV showed an increase in H3K9me3 level above the background threshold (0.5 RPKM) in Setdb1 knockout PGCs (not shown). Unique and multialigned reads were included in the analysis. (E) Global changes of DNA methylation in E13.5 PGCs isolated from Setdb1 knockout versus heterozygous littermates. Box and whisker plots show the median percentage of CpG methylation (box, 25th and 75th percentile; whisker, 10th and 90th percentile) at all annotated CpGs in the genome (left panel) or at all CpGs within gene bodies of ENSEMBL annotated genes (right panel).
H3K9me3 levels were broadly reduced in Setdb1 knockout PGCs at both H3K9me3 and H3K9me3/H3K27me3 coenriched regions to 62% and 69%, respectively, of the level measured in their heterozygous littermates (Fig. 3B). Interestingly, while H3K27me3 levels were only modestly reduced at regions enriched solely for H3K27me3 (to ∼91% of heterozygous levels), H3K27me3 levels were reduced to 76% of normal levels at coenriched regions (Fig. 3B; Supplemental Fig. 7A). Reduced levels of H3K9me3 and H3K27me3 at IAP elements were confirmed by qPCR on biological replicates (Fig. 3C; Supplemental Fig. 7B). Consistent with the rest of the genome, while most H3K9me3-marked ERVK and ERV1 subfamilies, including ETn, IAPLTR1, and RLTR4, show a decrease in H3K9me3 (Fig. 3D; Supplemental Fig. 7C) and H3K27me3 (Supplemental Fig. 7D,E) in knockout PGCs, significant levels of these marks remain. Incomplete deletion (Supplemental Fig. 6B) may explain the residual levels of H3K9me3. Alternatively, other H3K9 methyltransferases, such as SUV39H1 and/or SUV39H2, could be responsible for the residual H3K9me3 present in Setdb1 knockout germ cells (Bulut-Karslioglu et al. 2014). No change in expression of the remaining known H3K9 or H3K27 histone methyltransferases was observed in the Setdb1 knockout (Supplemental Fig. 7E), indicating that deposition of H3K27me3 at coenriched regions, likely by EZH1 and/or EZH2, is at least partially dependent on SETDB1 or H3K9me3.
H3K9me3-depleted IAP LTRs show reduced DNA methylation in Setdb1 knockout PGCs
As genomic regions retaining relatively high levels of DNA methylation were found to be enriched for H3K9me3 (Fig. 1F), we next determined whether Setdb1 deletion perturbs DNA methylation at these regions in E13.5 PGCs. Surprisingly, analysis of PBAT data from male Setdb1 knockout PGCs revealed that at a global level, DNA methylation levels were ∼2.5-fold higher in Setbd1 knockout than heterozygous PGCs, with median CpG methylation values of 25.0% versus 9.0%, respectively (Fig. 3E, first panel; Supplemental Fig. 8A). Similar methylation levels were observed for all CpGs within the bodies of annotated genes (Fig. 3E, second panel). As we did not observe an increase in the expression of any of the DNMTs or a decrease in the expression of the dioxygenases TET1 or TET2 in Setdb1 knockout PGCs (Supplemental Fig. 8B), indirect effects are the most likely explanation. While we considered the possibility that development is delayed in Setdb1 knockout germ cells, we did not consistently observe higher expression of genes normally expressed at a higher level at E12.5 than E13.5 or lower levels of expression of genes normally expressed at a higher level at E13.5 than E12.5 (Jameson et al. 2012) in Setdb1 knockout relative to heterozygous PGCs (Supplemental Fig. 8C).
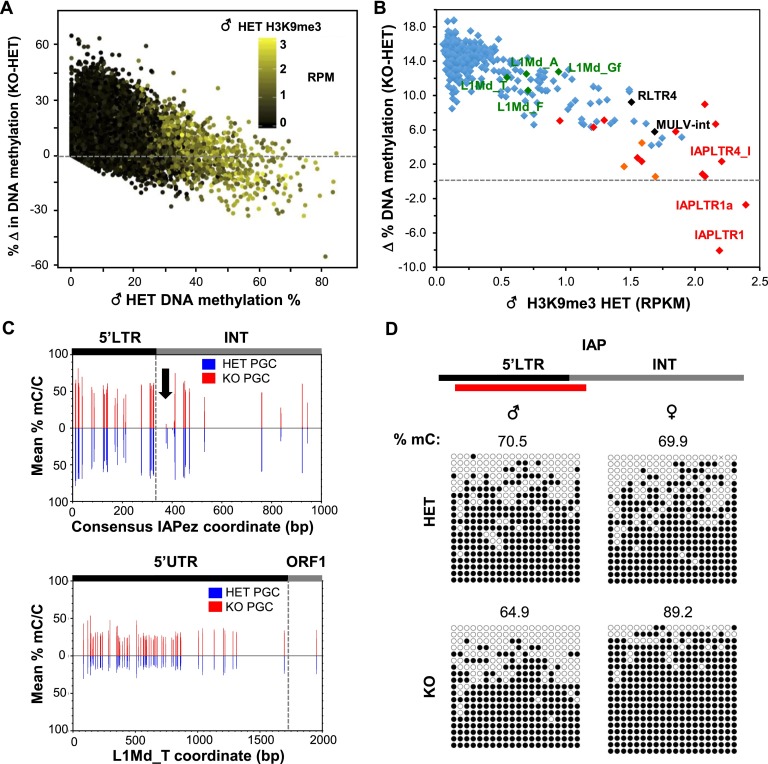
Intriguingly, analysis of the relationship between DNA methylation and both repressive histone modifications reveals that regions marked with H3K9me3 show a lower overall increase in DNA methylation than regions marked with H3K27me3 alone or regions lacking both marks (Supplemental Fig. 8D). Indeed, in nonrepetitive genomic regions, changes in DNA methylation levels are generally inversely correlated with H3K9me3 enrichment levels in heterozygous PGCs (Fig. 4A). Notably, hypermethylated regions with high levels of H3K9me3 and intermediate levels of H3K27me3 show decreased DNA methylation levels in Setdb1 knockout PGCs (Fig. 4A; Supplemental Fig. 8E). Similarly, the degree of DNA methylation gain at ERVs shows an inverse correlation with H3K9me3 enrichment levels (Fig. 4B), with the CpG-rich IAPLTR1 and IAPLTR1a of the IAPez family actually showing reduced mean DNA methylation levels of ∼8% and ∼3%, respectively. This decrease is likely an underestimate of the true decrease in DNA methylation, given the significant percentage of PGCs in which the deletion has not occurred.
Figure 4.
DNA methylation is reduced in Setdb1-deficient PGCs in regions highly enriched for H3K9me3. (A) Relationship between percentage of methylation in heterozygous (HET) E13.5 PGCs and the change (Δ) in the percentage of methylation in heterozygous PGCs relative to PGCs isolated from Setdb1 knockout (KO) littermates. PBAT data from 50,000 random regions (1-kb bins) are shown. (B) Relationship between H3K9me3 coverage at LINE1 elements and ERVs (>100 copies) in heterozygous E13.5 PGCs and the change (Δ) in the percentage of of DNA methylation at such ERVs relative to PGCs isolated from their Setdb1 knockout littermates. IAP and related ERK10C elements are labeled in red and orange, respectively. Unique and multialigned reads were included in the analysis. (C) The mean percentage of DNA methylation (mC/C) at all CpGs with five times or more coverage across the consensus LTR and proximal internal (INT) regions of IAPLTR1 elements or in the 5′ UTR of L1md_T elements (homologous to L1md_T chr10: 11,355,585–11,361,471) is shown for heterozygous and Setdb1 knockout E13.5 PGCs. (D) Sanger bisulfite sequencing of the 5′ LTR region of IAPLTR1a elements in male and female Setdb1 heterozygous and knockout PGCs. The region amplified is shown above (red bar).
Analysis of the distribution of DNA methylation across the consensus IAPLTR1 and proximal internal regions reveals a near-complete loss of methylation across a short stretch of CpGs just 3′ of the 5′ LTR (Fig. 4C). In contrast, a broad but modest increase in methylation was observed in the 5′ untranslated region (UTR) and ORF1 regions of L1Md_T elements (Fig. 4C), consistent with the overall methylation dynamics observed for LINE1 families. Decreased methylation in IAPLTR1a was confirmed by Sanger bisulphite sequencing in male E13.5 PGCs but, interestingly, was not observed in female E13.5 PGCs in the region amplified (Fig. 4D). Hypermethylation of IAP and other H3K9me3-marked ERVs in PGCs (Seisenberger et al. 2012) and preimplantation embryos (Smith et al. 2012) is consistent with the hypothesis that H3K9me3 plays a role in protecting marked genomic regions against active or passive demethylation (Leung et al. 2014).
A subset of ERVs is reactivated upon Setdb1 depletion in E13.5 PGCs
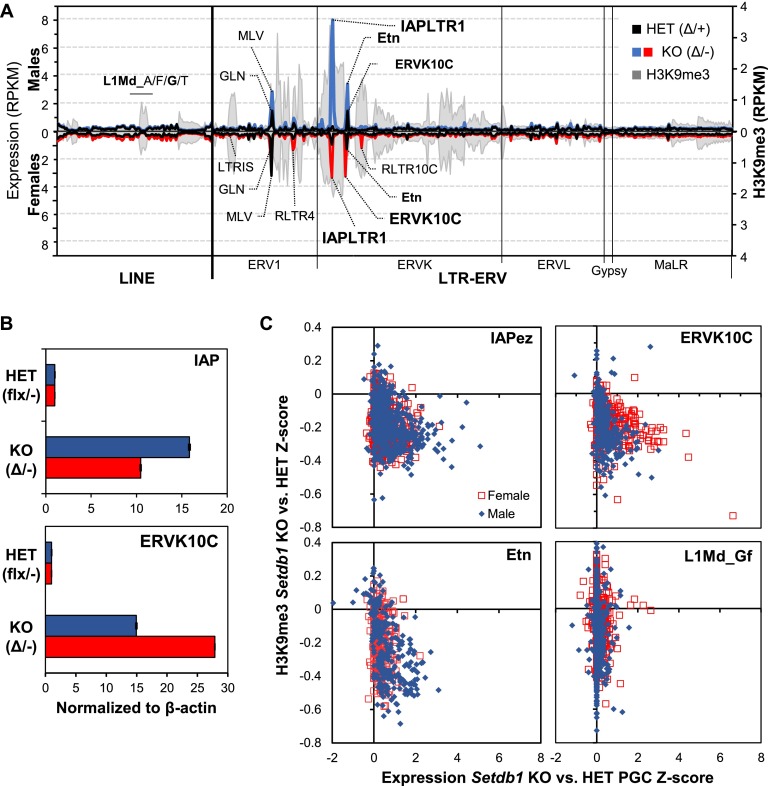
Comparison of RNA-seq data generated from PGCs isolated from E13.5 heterozygous with Setdb1 knockout littermates reveals that a number of ERV1 and ERVK families are dramatically up-regulated in both male and female knockout PGCs (Fig. 5A; Supplemental Table 3). Strikingly, for a subset of ERVs, the level of up-regulation is sex-dependent (Supplemental Fig. 9A). IAPez elements showed a 27.5-fold and a 15.5-fold increase in expression in male and female PGCs, respectively. While ETn elements also showed a higher level of derepression in males, the converse was true for ERVK10C elements. These trends were validated by qPCR (Fig. 5B) and confirmed by RNA-seq using PGCs isolated from an independent litter (Supplemental Fig. 9B). Notably, a subset of H3K9me3-marked ERVs, including LTRIS families, is not derepressed in Setdb1 knockout PGCs, implicating alternative repressive pathways and/or a nuclear milieu that does not support expression of these elements.
Figure 5.
Reactivation of ERVs upon Setdb1 depletion in mouse E13.5 PGCs. (A) Expression of Repbase annotated ERV and LINE1 families (>100 copies in the B6 genome) in male and female heterozygous (HET) and knockout (KO) E13.5 PGCs is shown along with male H3K9me3 enrichment levels. LINE and ERV1, ERVK, ERVL, Gypsy, and MaLR classes of ERVs are presented in alphabetical order along the X-axis according to their Repbase nomenclature. Unique and multialigned reads were included in the analysis. (B) qPCR validation of sex-specific differences in the expression of IAP and ERVK10C ERVs in heterozygous versus Setdb1 knockout E13.5 PGCs isolated from male and female littermates. (C) Expression versus H3K9me3 coverage Z-scores of individual “intact” IAP, ETn, and ERVK10C ERVs. Only uniquely aligned reads were considered in this analysis.
To determine the relationship between expression of reactivated ERV families and H3K9me3 enrichment at the level of individual elements, we compared H3K9me3, H3K27me3, and RNA-seq read coverage at intact elements, considering only uniquely aligned reads. Numerous IAPez, ERVK10C, and ETN elements show decreased H3K9me3, concomitant with increased expression (Fig. 5C), whereas marked MLV and unmarked ERVL elements show modest or no up-regulation, respectively (Supplemental Fig. 10A). Notably, LINE1 families enriched for H3K9me3 (Fig. 2A; Supplemental Fig. 4C), including L1Md, exhibited very little reactivation despite a clear decrease in H3K9me3 (Figs. 3C, 5C). Similar trends were observed for H3K27me3 (Supplemental Fig. 10B). Taken together, these results indicate that H3K9me3 at a subset of ERVs and LINE1 elements is dependent on Setdb1 and that, while these retroelements are also marked by H3K27me3 in PGCs, deletion of Setdb1 is sufficient to disrupt silencing of a subset of co-marked ERVs but not of LINE1 elements, implicating the activity of alternative transcriptional or post-transcriptional silencing pathways for the latter.
Transcription of a subset of genes derepressed in Setdb1 knockout PGCs initiates in LTRs
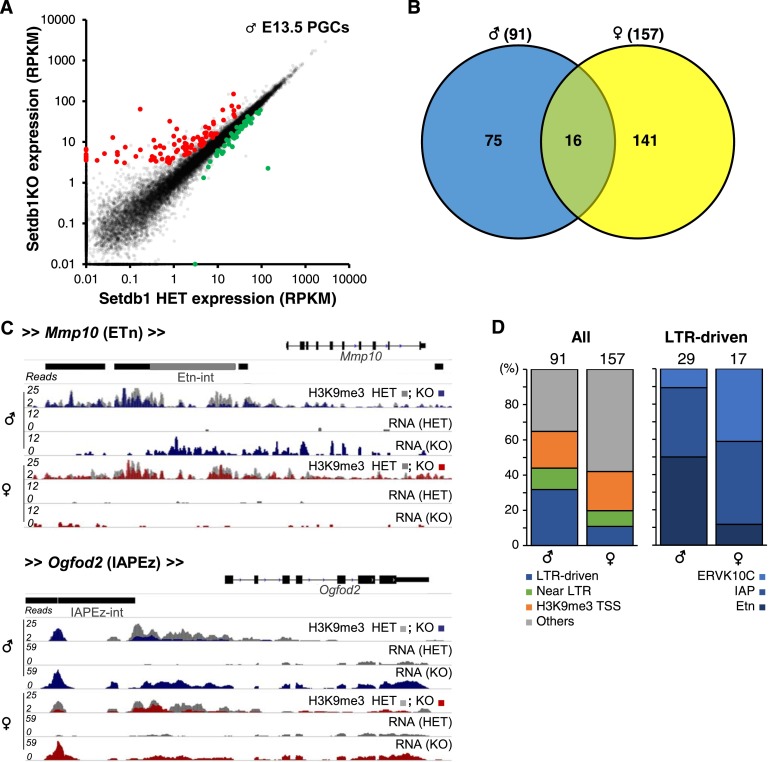
As discussed above, relatively few genic promoters are marked by H3K9me3. To determine how deletion of Setdb1 influences gene expression, we calculated RNA-seq coverage over all genic exons for wild-type, heterozygous, and knockout littermates. As expected, most genes that significantly change in expression in both male and female Setdb1 knockout PGCs were up-regulated (Fig. 6A; Supplemental Fig. 11A). Of the 91 and 157 genes up-regulated in male and female, respectively, only 16 are common to both using the stringent thresholds applied (Fig. 6B; Supplemental Table 4). Gene ontology analysis revealed that up-regulated genes were associated with nucleic acid binding and ion binding in males and extracellular matrix and definitive hematopoiesis in females (Supplemental Fig. 11B). Surprisingly, <25% of up-regulated genes have an H3K9me3 peak overlapping with their annotated TSS. Many of the other up-regulated genes in both sexes are either in close proximity to an LTR element or initiate transcription within an upstream LTR (Fig. 6C,D). For the latter cases, elevated gene expression is likely the result of transcription initiating in an ERV upstream (<25 kb) of the canonical TSS and extending through the 5′ end of the gene (Fig. 6C; Supplemental Fig. 11C). A minority of such genes (six out of 29 in males and three out of 17 in females) show evidence of “chimeric” transcription, where splicing occurs between the upstream ERV and a canonical genic exon. Interestingly, the LTRs driving aberrant genic transcripts also show clear sex-dependent differences. Almost 40% of the transcripts up-regulated in male PGCs originate from a nearby LTR (Fig. 6C,D), with ∼50% originating in ETn/MusD elements, and the remaining 40% and 10% originating in IAP and ERVK10C elements, respectively. In contrast, <15% of the genes up-regulated in females are LTR-driven, the majority of which originate from IAP or ERVK10C LTRs. These results are consistent with the sex-dependent differences observed in the expression of these ERVs (Supplemental Fig. 9A). Thus, as we previously reported for mESCs (Karimi et al. 2011), derepression of latent ERVs concomitantly influences the expression of nearby genes in PGCs.
Figure 6.
Genes up-regulated in Setdb1 knockout (KO) E13.5 PGCs. (A) Expression levels of annotated ENSEMBL genes in male heterozygous (HET) versus Setdb1 knockout E13.5 PGCs are shown. Genes up-regulated (91) in Setdb1 knockout PGCs (Z-scores: Setdb1 knockout/wild type and Setdb1 knockout/heterozygous, >1; RPKM: Setdb1 knockout/wild type and Setdb1 knockout/HET, >1.5-fold) are highlighted in red. Genes down-regulated (23) (Z-scores: Setdb1 knockout/wild type and Setdb1 knockout/heterozygous, <-1; RPKM: Setdb1 knockout/wild type and Setdb1 knockout/heterozygous, <0.75-fold) are highlighted in green. Only uniquely aligned reads were considered in this analysis. (B) Venn diagram of the overlap between genes up-regulated in Setdb1 knockout PGCs isolated from male and female littermates. (C) Genome browser views showing H3K9me3 and RNA-seq coverage of the Mmp10 and Ogfod2 gene loci in E13.5 PGCs. Note the coverage over the ETn ERV upstream of the Mmp10 gene and the IAP ERV upstream of the Ogfod2 gene. (D, left panel) Percentage of up-regulated genes in male and female Setdb1 knockout PGCs that are expressed from an LTR element (LTR-driven) within 20 kb of an LTR (Near LTR) or marked by H3K9me3 in the TSS (±1 kb) region. (Right panel) Percentage of LTR-driven up-regulated genes that are driven by ETn, IAP, or ERVK10C LTRs.
Setdb1 deletion early in germline development leads to gametogenesis defects in postnatal and adult mice
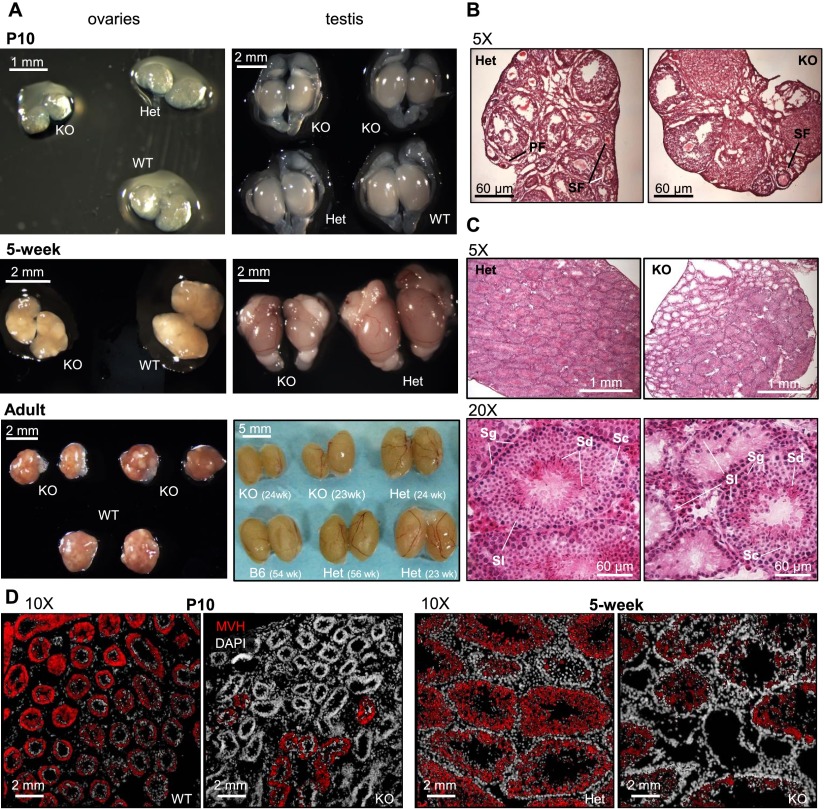
As Setdb1 deletion early in male germline development leads to a significant decrease in the number of PGCs relative to soma at E13.5, we assessed whether germ cell defects persist postnatally. Although Setdb1△/−; TnapCre/+ mice displayed normal size and morphology, postnatal day 10 (P10) testes, which normally harbor premeiotic germ cells, are clearly reduced in size relative to their heterozygous and wild-type littermates (Fig. 7A). Curiously, P10 ovaries, which normally harbor oocytes arrested in meiosis I, also show a decrease in size despite the fact that reduced numbers of female Setdb1 knockout PGCs were not observed. Male and female gonadal hypotrophy persisted through puberty and into adulthood (Fig. 7A), indicating that this phenotype is due to a germ cell defect rather than a developmental delay. Sections from ovaries isolated from 5-wk-old Setdb1 knockout females revealed the presence of both primordial and growing follicles, although in much smaller numbers than in ovaries isolated from heterozygous littermates (Fig. 7B). Sections of testes isolated from 10-d-old and 5-wk-old Setdb1 knockout mice revealed the presence of seminiferous tubules that were devoid of germ cells alongside normal seminiferous tubules displaying all layers of male germ cell development (Fig. 7C,D), consistent with incomplete Cre-mediated recombination. Although both male and female germline Setdb1 knockout animals appeared to be fertile, we did not observe transmission of Setdb1Δ/KO germ cells in litters born from Setdb1 flox/ΔTnap+/cre × B6 wild-type matings. Thus, offspring are apparently derived from germ cells in which Cre recombination has not taken place. Taken together, these observations reveal that deletion of Setdb1 in proliferating PGCs prior to the onset of de novo DNA methylation leads to prolonged defects in gametogenesis.
Figure 7.
Gonadal defects in male and female germline Setdb1-deficient mice. (A) Decrease in male and female gonadal size in germline Setdb1 knockout (KO) mice relative to heterozygous (HET) and/or wild-type (WT) littermates in postnatal mice at P10, 5 wk, and 23/24 wk (adult). Adult testes are also relative to ∼1-yr-old heterozygous (paternal, Setdb1flox/+ Tnapcre/+) and C57BL/6 testes. (B) Hematoxilin and eosin sections in 5-wk postnatal ovaries illustrating gonadal defects in germline Setdb1-deficient females. (PF) Primary follicle; (SF) secondary follicle; (TF) tertiary follicle. (C) Hematoxilin and eosin sections in 5-wk postnatal testis illustrating defects in gonadal morphology in germline Setdb1-deficient males. (Sl) Sertoli cells; (Sg) spermatogonia; (Sc) spermatocytes; (Sd) spermatids. (D) Immunofluorescence on sections from postnatal testes at day 10 (P10) and 5 wk. (White) DAPI (DNA); (red) mouse vasa homolog (MVH; germ cell marker).
Discussion
Genome-wide analysis of the distribution of H3K9me3 and H3K27me3 at nonrepetitive genomic regions revealed that these marks are generally mutually exclusive in E13.5 PGCs, consistent with previous analyses in ESCs (Mikkelsen et al. 2007; Karimi et al. 2011) and mouse embryonic fibroblasts (MEFs) (Pauler et al. 2009). Co-occurrence of high levels of H3K9me3 with intermediate levels of H3K27me3 at specific ERV and LINE1 elements was unexpected given that they are marked exclusively with H3K9me3 in ESCs (Fig. 2A; Mikkelsen et al. 2007). H3K27me3 levels significantly increase in migrating PGCs, perhaps to compensate for the progressive loss of DNA methylation and/or H3K9me2 that takes place during this developmental window (Seki et al. 2007; Hajkova et al. 2008). Co-occurrence of these marks at gene promoters was observed previously in trophectoderm cells (Alder et al. 2010), which, like PGCs, are globally hypomethylated (Oda et al. 2013). Thus, deposition of H3K27me3 at regions that do not generally harbor H3K9me3 may be a common feature of tissues with hypomethylated genomes.
Whether H3K27me3 is required for silencing of ERVs in PGCs remains to be determined. Since H3K27me3 and H3K9me3 levels were concomitantly reduced at ERVs in Setdb1 knockout PGCs, deposition of the former may depend on the presence of the latter in such regions. Several members of the Cbx protein family interact with the core Polycomb-repressive complex 1 (PRC1), which promotes deposition of H3K27me3 by PRC2/EZH2, and a subset of these chromodomain proteins (CBX4, for example) has relatively high affinity for H3K9me3 (Vermeulen et al. 2010; Kaustov et al. 2011). Similarly, the chromodomain protein CDYL, which binds with high affinity to H3K9me3 (Vermeulen et al. 2010) and directly recruits EZH2 (Zhang et al. 2011), is highly expressed in PGCs. Further experiments are required to determine the role of such H3K9me3 “readers” in targeting of H3K27me3 in PGCs.
Unexpectedly, Setdb1 knockout PGCs showed a global ∼2.5-fold increase in DNA methylation relative to control PGCs. As this increase in DNA methylation includes regions devoid of H3K9me3, an indirect effect such as premature initiation of the global wave of de novo methylation that normally occurs in male PGCs at E15.5 is the most likely explanation. Intriguingly, an inverse correlation was observed between DNA methylation gain in the Setdb1 knockout and the level of H3K9me3 enrichment in control PGCs. CpG-rich IAPLTR1 and IAPLTR1a, which show the highest level of H3K9me3 of all LTRs, show a decrease in DNA methylation in the absence of Setdb1, coincident with induction of IAPez expression. Given that the de novo DNA methyltransferases Dnmt3a and Dnmt3b are expressed at relatively low levels and Dnmt3l is not expressed at this stage (Kurimoto et al. 2008; Seisenberger et al. 2012), these results indicate that H3K9me3 likely protects methylated genomic regions against active and/or passive DNA demethylation. As Tet1 and Tet2 are expressed in both PGCs and preimplantation embryos (Seisenberger et al. 2012), H3K9me3 may specifically inhibit the activity of one or both of these putative DNA demethylation factors. Indeed, we showed recently that a subset of IAP ERVs also show reduced DNA methylation and increased binding of TET1 in Setdb1 knockout ESCs (Leung et al. 2014).
As the activity of DNMT1 is stimulated by the binding of its cofactor, NP95, to H3K9me3 (Rothbart et al. 2012, 2013), SETDB1-dependent deposition of this mark may also influence the efficiency of maintenance DNA methylation. Notably, NP95 expression is down-regulated in PGCs (Kurimoto et al. 2008), and the remaining protein is detected predominantly in the cytoplasm (Seisenberger et al. 2012), likely compromising maintenance methylation at a global level. Based on these observations, we propose that H3K9me3 inhibits TET-dependent DNA demethylation and independently promotes maintenance DNA methylation at marked regions in PGCs, minimizing the likelihood of mobilization of young ERVs at later stages in development. Indeed, our results are consistent with previous reports that IAP ERVs are expressed at very low levels in E13 PGCs (Dupressoir and Heidmann 1996). In contrast, IAPez elements were up-regulated in Setdb1-deficient PGCs, revealing that, as in ESCs, H3K9me3 likely plays a critical role in safeguarding the genome against IAP retrotransposition.
Coincident with derepression of numerous ERV families, many genes are aberrantly expressed in Setdb1 knockout PGCs as a consequence of transcriptional activation of nearby related LTRs, indicating that SETDB1 plays an important role in not only safeguarding against expression of intact ERVs but also minimizing the expression of LTR-initiated transcripts that extend into genes. Interestingly, however, numerous chimaeric transcripts detected in Setdb1 knockout PGCs were not up-regulated in Setdb1 knockout ESCs (Karimi et al. 2011) and vice versa, indicating that additional factors influence the expression of LTR elements.
We show that a subset of constitutively expressed ERVs, including ERVK10C and ETn elements, are clearly expressed at higher levels in female than in male E13.5 PGCs or vice versa. Consistent with the observation that germ cells began to adopt lineage-specific/sex-specific transcriptional states by E12.5 (Jameson et al. 2012), our data indicate that the intracellular complement of positive and/or negative transcriptional regulators differ in a sex-dependent manner. Notably, a significantly greater number of ERVs, including ETn elements, showed higher levels of derepression in male than in female Setdb1 knockout PGCs, suggesting that the male germline may be more susceptible to retrotransposition. Indeed, systematic analyses of the genomic landscape of transposable elements across 18 strains suggest that the majority of retrotranposons in mice are introduced through the male germline (Nellåker et al. 2012). The Y chromosome itself may act as a reservoir for specific ERVs, including IAP elements, which are amplified in nonpseudoautosomal regions (Reuss et al. 1996).
As Setdb1-deficient ESCs are also inviable (Dodge et al. 2004) and are characterized by activation of the same ERV families (Matsui et al. 2010; Karimi et al. 2011), it is possible that widespread activation of ERVs triggers cell death in PGCs via retrotransposition-dependent or retrotransposition-independent pathways, with activation of ERVs on the Y chromosome rendering male PGCs more susceptible. Consistent with this model, deletion of Setdb1 in MEFs does not lead to derepression of ERVs or decreased cell viability. Furthermore, deletion of KAP1/TIF1β, which interacts directly with SETDB1, also leads to derepression of ERVs in the early embryo (Rowe et al. 2010) and embryonic lethality (Cammas et al. 2000) as well as testicular degeneration when deleted specifically in the germ cell lineage (Weber et al. 2002). Given that perinatal induction of ERVs in Dnmt3l-null embryos at P2 does not affect gonocyte viability (Bourc’his and Bestor 2004), it is possible that proliferating PGCs are more sensitive to activation of ERVs than germ cells at later developmental stages. Alternatively, ERVs may be derepressed to a greater extent in Setdb1 knockout PGCs than Dnmt3l knockout perinatal gonocytes. Despite a significant depletion of male Setdb1 knockout PGCs at E13.5, we did not observe increased expression of genes involved in cell cycle arrest or apoptosis, although we cannot rule out that such events occur earlier in knockout PGCs. Interestingly, work published by Zeng and colleagues (An et al. 2014) while this paper was under review demonstrates that SETDB1 depletion in postnatal spermatogonial stem cells results in increased levels of apoptosis and reduced gonadal size in a transplantation model. Taken together, these data suggest that SETDB1 is required for germ cell viability not only during epigenetic remodeling in PGCs but also at later stages of germ cell development.
Global remethylation in males is initiated in gonocytes at approximately E15.5 and extends through the perinatal stage. Efficient de novo DNA methylation of a subset of ERV and LINE1 retroelements during this developmental window is dependent on the cofactor DNMT3L and piRNA pathway components, including MIWI2 (Kim et al. 2009). While IAP elements are expressed in Dnmt3l−/− P2 gonocytes, premeiotic Dnmt3l−/− germ cells develop normally in the perinatal stage (Bourc’his and Bestor 2004) but show reduced numbers of spermatocytes (which do not express Dnmt3l or Miwi2) and undergo meiotic failure. Derepression of LINE1 and, to a lesser extent, IAP elements has been observed in spermatogonia and/or spermatocytes deficient in the piRNA proteins mentioned above, but, like the Dnmt3l mutant, developmental defects are not manifest until the pachytene stage of meiotic prophase I in these mutants. This common postnatal germline phenotype indicates that while the DNA methylation defect is initiated in prenatal gonocytes, the loss of DNA methylation does not affect viability until the spermatogonial stages or later. Consistent with this model, induction of IAP expression was not observed in Dnmt1 knockout PGCs (Walsh et al. 1998).
Intriguingly, depletion of Eggless/dSETDB1, the ortholog of SETDB1 in Drosophila melanogaster required for H3K9me3 deposition in the ovary and early oogenesis (Clough et al. 2007), leads to derepression of LTR retrotransposons and reduced transcription of piRNA clusters in the ovary (Rangan et al. 2011), indicating that H3K9me3 acts upstream of the piRNA pathway in flies. Conversely, depletion of Piwi or its binding partner, Gtsf1, leads to a rapid loss of H3K9me3 and derepression of specific LTR elements, indicating that the Drosophila piRNA pathway impinges on this histone mark (Donertas et al. 2013). Similarly, the AGO1 silencing complex RITS interacts with nascent target RNAs to guide H3K9me3 at repetitive regions in Schizosaccharomyces pombe (Castel and Martienssen 2013). However, in Setdb1 knockout male and female E13.5 PGCs, expression levels of members of the piRNA pathway were not significantly altered relative to their heterozygous and wild-type littermates, and Miwi2 is not yet expressed at this stage, indicating that this nuclear piRNA pathway is not required for SETDB1-dependent transcriptional silencing in early mammalian germ cells.
Based on these observations, we propose that SETDB1-dependent deposition of H3K9me3 at ERVs occurs very early in germline development or prior to the emergence of germ cells and persists until the early spermatocyte stage. Programmed loss of H3K9me3 at this stage would explain why germ cells deficient in perinatal DNA methylation, including mutants in Dnmt3l or the nuclear piRNA pathway, are susceptible to derepression of ERVs only later in spermatogenesis. Whether H3K9me3 plays a role in piRNA-dependent de novo DNA methylation of marked retroelements and/or persistence of DNA methylation at these later stages remains to be determined. Addressing these questions would require detailed analyses of the distribution of H3K9me3 and DNA methylation in prenatal and/or perinatal gonocytes. Unfortunately, the rapid loss of germ cells following Setdb1 deletion in early PGCs necessitates the establishment of an alternative conditional deletion system. Regardless, our results clearly show that SETDB1 functions as an important guardian against transcriptional activation of young ERVs in PGCs and may simultaneously protect marked elements against the waves of global demethylation that take place early in germline development.
Addendum
While this paper was under review, Aravin and colleagues (Pezic et al. 2014) showed that deletion of Miwi2 led to a decrease in H3K9me3 levels and derepression specifically of young L1Md elements in P10 male germ cells. Taken together with the data presented here, these results indicate that the piRNA pathway may be required to maintain SETDB1-deposited H3K9me3 at young LINE1 elements or for deposition of H3K9me3 at previously unmarked elements at later stages of premeiotic spermatogenesis.
Materials and methods
Breeding and mating
Germline-specific Setdb1 knockout animals were generated using the Setdb1tm1.1Yshk (Tan et al. 2012) and Tnap-Cre (129-Alpltm1(cre)Nagy/J) (Lomelí et al. 2000) lines. Briefly, Setdb1flox/flox females were mated with a Setdb1flox/+ Tnapcre/+ male to generate the genotypes described in Supplemental Figure 5A. For timed matings, the day of the vaginal plug was considered E0.5, and females were sacrificed at E13.5 for embryo collection. Animal experimentation followed the guidelines from the Canadian Council on Animal Care (CCAC) under University of British Columbia animal care license numbers A13-0115 and A12-0208.
Isolation of PGCs
PGCs were isolated from the gonadal ridges of embryos 13.5 d post-coitum, as described previously (Nagy 2003). A more detailed procedure is presented in the Supplemental Material.
Genotyping
Tissue from tails of the E13.5 embryos was isolated and extracted using the HotShot method, as described previously (Truett et al. 2000). The sex of each embryo was determined using primers to Zfy1 and Xist, and the genotypes were determined using multiplex primers (primer sequences are presented in Supplemental Table 5). Setdb1 knockout primers were used to confirm the deletion of the flox allele.
ChIP
Chromatin was fragmented with micrococcal nuclease, precleared, and immunoprecipitated with H3K9me3 (Active Motif, no. 39161), H3K27me3 (Diagenode, pAb-069-050), and pan-H3 (Sigma, I8140). For ChIP-seq, 10% of the raw ChIP material was used for qPCR validation, and the remaining 90% was used for library construction. A detailed procedure is presented in the Supplemental Material.
RNA extraction and double-stranded cDNA preparation
RNA was extracted from 103 cells using TRIzol, and ribosomal RNA was depleted using riboMinus. First strand cDNA synthesis was carried using SuperScript III followed by second strand cDNA synthesis using Klenow polymerase. A 10% fraction was used for qPCR validation, and the rest was fragmented for library construction. A detailed procedure is presented in the Supplemental Material.
qPCR
qPCR was carried out with EvaGreen Supermix (Bio-Rad SsoFast EvaGreen Supermix) on an Opticon II qPCR machine (Bio-Rad). IAP, ERVK10C, and MERVL primers were used for validation of ChIP and expression. Primer sequences are available in Supplemental Table 5.
Library construction and sequencing
Libraries were constructed using a custom paired-end protocol (Illumina) (Falconer et al. 2012). Briefly, samples were end-repaired and A-tailed, and Illumina PE adapters were ligated. Libraries were amplified using indexed PE primers for eight to 10 PCR cycles. Amplified indexed libraries were pooled (four to six libraries per pool) and size-selected for paired-end sequencing. A detailed library construction procedure is presented in the Supplemental Material. Cluster generation and paired-end sequencing (100-base-pair [bp] reads) were performed on the Illumina cluster station and Illumina HiSeq 2000 sequencing platform. Sequence reads were mapped to mm9 (NCBI 37) using BWA (Li and Durbin 2009). Reads passing Illumina’s default chastity filter (Li et al. 2009) were used to generate library statistics.
ChIP and RNA-seq analysis
Detailed ChIP and RNA-seq analysis procedures are presented in the Supplemental Material. ChIP-seq and RNA-seq data are available at the Gene Expression Omnibus repository under the accession number GSE60377.
PBAT sequencing
Two-thousand to 3000 PGCs isolated from individual E13.5 heterozygous and Setdb1 knockout embryos were spiked with 0.1 ng of λ phage DNA, lysed, and subjected to bisulfite treatment as described previously (Shirane et al. 2013). Whole-genome bisulfite shotgun libraries were prepared using the PBAT method (Miura et al. 2012). Single-end sequencing generated 38.1 and 34.6 Gb of uniquely aligned sequencing data for male heterozygous and knockout PGC libraries. Alignment of PBAT data was performed using Bismark (Krueger and Andrews 2011). The bisulfite conversion rate as determined by analysis of λ DNA was 99.55% (converted C: 73,394,225; unconverted C: 332,718) for the heterozygous and 99.53% (converted C: 107,386,787, unconverted C: 504,201) for the knockout. PBAT data are available at the Gene Expression Omnibus repository under the accession number GSE60377.
Imaging
Gonads were dissected from P10, 5-wk-old, and adult germline Setdb1 knockout animals and littermates and imaged using a Leica MS5 stereomicroscope. Gonads isolated from 5-wk-old animals were fixed in 4% formaldehyde, incubated in sucrose, embedded in OCT (Tissue-Tek), and frozen at −80°C until sectioning. Ten-micrometer gonad sections were stained with hematoxilin and eosin according to standard procedures. Germ cells were detected in postnatal gonads using a mouse Vasa homolog (MVH/DDX4) antibody (Abcam, ab13840).
Acknowledgments
We thank the ubcFLOW cytometry facility for FACS analyses, Matthew Hudson for technical support, and Dixie Mager and Danny Leung for helpful discussions. We also thank Miho Miyake (Kyushu University) and Canada’s Michael Smith Genome Sciences Centre for Illumina sequencing. This work was supported by Canadian Institutes of Health Research (CIHR) grants 77805 and 92090 to M.C.L. and RMF-92093 and MOP-119357 to L.L. and Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) grant 25112010 to H.S. M.C.L. is a CIHR New Investigator, and M.M.K. was supported by a Michael Smith Foundation for Healthcare Research (MSFHR) post-doctoral fellowship. K.S. is a JSPS research fellow.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.244848.114.
References
- Abe M, Tsai SY, Jin S-G, Pfeifer GP, Szabó PE. 2011. Sex-specific dynamics of global chromatin changes in fetal mouse germ cells. PLoS ONE 6: e23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O’Neill L, Azuara V. 2010. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development 137: 2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Zhang X, Qin J, Wan Y, Hu Y, Liu T, Li J, Dong W, Du E, Pan C, et al. 2014. The histone methyltransferase ESET is required for the survival of spermatogonial stem/progenitor cells in mice. Cell Death Dis 5: e1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, La Rosa-Velázquez De IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B, et al. 2014. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell 55: 277–290 [DOI] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. 2000. Mice lacking the transcriptional corepressor TIF1β are defective in early postimplantation development. Development 127: 2955–2963 [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E, Moon W, Wang S, Smith K, Hazelrigg T. 2007. Histone methylation is required for oogenesis in Drosophila. Development 134: 157–165 [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang Y-K, Beppu H, Lei H, Li E. 2004. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24: 2478–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donertas D, Sienski G, Brennecke J. 2013. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev 27: 1693–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley JP 1987. Discrete high molecular weight RNA transcribed from the long interspersed repetitive element L1Md. Nucleic Acids Res 15: 2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Heidmann T. 1996. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol Cell Biol 16: 4495–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcova-Hills G, Tokunaga T, Kurosaka S, Yamaguchi M, Takahashi S, Imai H. 1999. Immunomagnetic isolation of primordial germ cells and the establishment of embryonic germ cell lines in the mouse. Cloning 1: 217–224 [DOI] [PubMed] [Google Scholar]
- Falconer E, Hills M, Naumann U, Poon SSS, Chavez EA, Sanders AD, Zhao Y, Hirst M, Lansdorp PM. 2012. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Methods 9: 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henckel A, Chebli K, Kota SK, Arnaud P, Feil R. 2012. Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO J 31: 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC. 2012. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 26: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B. 2012. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet 8: e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42: 709–732 [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903 [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280 [DOI] [PubMed] [Google Scholar]
- Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, et al. 2011. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem 286: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8: e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, et al. 2013. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res 23: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M. 2008. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 22: 1617–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter JR, Reik W. 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, De Felici M, Walsh CP. 2003. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics 82: 230–237 [DOI] [PubMed] [Google Scholar]
- Leung D, Du T, Wagner U, Xie W, Lee AY, Goyal P, Li Y, Szulwach KE, Jin P, Lorincz MC, et al. 2014. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc Natl Acad Sci 111: 6690-6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomelí H, Ramos-Mejía V, Gertsenstein M, Lobe CG, Nagy A. 2000. Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis 26: 116–117 [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, Van De Lagemaat LN, Mager DL. 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F, Enomoto Y, Dairiki R, Ito T. 2012. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Nagy A. 2003. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Nellåker C, Keane TM, Yalcin B, Wong K, Agam A, Belgard TG, Flint J, Adams DJ, Frankel WN, Ponting CP. 2012. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol 13: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J-H, Kumar V, Muratani M, Kraus P, Yeo J-C, Yaw L-P, Xue K, Lufkin T, Prabhakar S, Ng H-H. 2013. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell 24: 324–333 [DOI] [PubMed] [Google Scholar]
- Oda M, Oxley D, Dean W, Reik W. 2013. Regulation of lineage specific DNA hypomethylation in mouse trophectoderm. PLoS ONE 8: e68846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. 2009. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezic D, Manakov SA, Sachidanandam R, Aravin AA. 2014. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 28: 1410–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. 2011. piRNA production requires heterochromatin formation in Drosophila. Curr Biol 21: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss FU, Frankel WN, Moriwaki K, Shiroishi T, Coffin JM. 1996. Genetics of intracisternal-A-particle-related envelope-encoding proviral elements in mice. J Virol 70: 6450–6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. 2012. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. 2013. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev 27: 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, Kono T, Sasaki H. 2013. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet 9: e1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S-L, Nishi M, Ohtsuka T, Matsui T, Takemoto K, Kamio-Miura A, Aburatani H, Shinkai Y, Kageyama R. 2012. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 139: 3806–3816 [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotech 29: 52–54 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Weber P, Cammas F, Gerard C, Metzger D, Chambon P, Losson R, Mark M. 2002. Germ cell expression of the transcriptional co-repressor TIF1β is required for the maintenance of spermatogenesis in the mouse. Development 129: 2329–2337 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2008. Host restriction factors blocking retroviral replication. Annu Rev Genet 42: 143–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang X, Gui B, Xie G, Zhang D, Shang Y, Liang J. 2011. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J Biol Chem 286: 42414–42425 [DOI] [PMC free article] [PubMed] [Google Scholar]