The SAGA coactivator complex contains distinct chromatin-modifying activities and is recruited by DNA-bound activators to regulate the expression of a subset of genes. Bonnet et al. discovered that SAGA acetylates the promoters and deubiquitinates the transcribed region of all expressed genes. SAGA also plays a critical role for RNA polymerase II recruitment at all expressed genes. This study uncovers a new function for SAGA as a bona fide cofactor for all RNA polymerase II transcription.
Keywords: SAGA, chromatin, transcription, deubiquitinase, acetyltransferase, RNA polymerase II
Abstract
The SAGA (Spt–Ada–Gcn5 acetyltransferase) coactivator complex contains distinct chromatin-modifying activities and is recruited by DNA-bound activators to regulate the expression of a subset of genes. Surprisingly, recent studies revealed little overlap between genome-wide SAGA-binding profiles and changes in gene expression upon depletion of subunits of the complex. As indicators of SAGA recruitment on chromatin, we monitored in yeast and human cells the genome-wide distribution of histone H3K9 acetylation and H2B ubiquitination, which are respectively deposited or removed by SAGA. Changes in these modifications after inactivation of the corresponding enzyme revealed that SAGA acetylates the promoters and deubiquitinates the transcribed region of all expressed genes. In agreement with this broad distribution, we show that SAGA plays a critical role for RNA polymerase II recruitment at all expressed genes. In addition, through quantification of newly synthesized RNA, we demonstrated that SAGA inactivation induced a strong decrease of mRNA synthesis at all tested genes. Analysis of the SAGA deubiquitination activity further revealed that SAGA acts on the whole transcribed genome in a very fast manner, indicating a highly dynamic association of the complex with chromatin. Thus, our study uncovers a new function for SAGA as a bone fide cofactor for all RNA polymerase II transcription.
Through their different activities, coactivator complexes convey the function of transcription activators bound to DNA elements to drive specific gene expression profiles. SAGA (Spt–Ada–Gcn5 acetyltransferase) is an evolutionarily conserved multifunctional coactivator complex organized in functional and structural modules (for review, see Rodríguez-Navarro 2009; Koutelou et al. 2010; Weake and Workman 2012). A number of biochemical and structural studies of SAGA architecture in Saccharomyces cerevisiae (y) consistently identified histone acetyltransferase (HAT), deubiquitinase (DUB), and TATA-binding protein (TBP) regulatory and structural modules (Wu et al. 2004; Lee et al. 2011). The HAT and DUB activities within SAGA are carried out by Gcn5 and Ubp8, respectively, which interact with other SAGA subunits to form a HAT module composed of Gcn5, Ada2, Ada3, and Sgf29 and a DUB module composed of Ubp8, Sgf73, Sgf11, and Sus1. These modules are conserved in humans (h) and are composed of homologous proteins (GCN5/PCAF, ADA2b, ADA3, and SGF29 in the HAT module; USP22, ATXN7, ATXN7L3, and ENY2 in the DUB module). The full assembly of the given modules is crucial to increase the substrate recognition and specificity of yGcn5/hGCN5 and enhance the catalytic activity of yUbp8/hUSP22 on nucleosomes (Gamper et al. 2009; Kohler et al. 2010; Samara et al. 2010; Bian et al. 2011; Lang et al. 2011). Interestingly, deletion of subunits of the DUB or the HAT module affected the stability of the corresponding module but not the overall integrity of the SAGA complex (Lee et al. 2011).
In S. cerevisiae, the Tra1 subunit of SAGA interacts with different activators, leading to its recruitment at a subset of promoters. At genes where it is recruited, the yeast SAGA complex was suggested to stimulate preinitiation complex formation through interactions with TBP and facilitate transcription initiation through Gcn5-mediated acetylation of their promoters (for review, see Weake and Workman 2012). However, it is still unclear how the multiple activities of SAGA are integrated to regulate gene expression. Indeed, analysis of gene expression changes in yeast strains deleted for all nonessential SAGA subunits revealed that the subunits of each module (HAT, DUB, and TBP-binding) tend to cluster together, although some differences were observed between S. cerevisiae and Schizosaccharomyces pombe (Helmlinger et al. 2011; Lenstra et al. 2011).
Genome-wide location studies of subunits of the SAGA complex in different organisms revealed only a few hundred binding sites for SAGA (Vermeulen et al. 2010; Bian et al. 2011; Krebs et al. 2011; Venters et al. 2011; Weake et al. 2011). In most cases, SAGA subunits were detected at a subset of promoters. Such specific targeting of ySAGA could explain the regulation of “SAGA-dominated genes,” which were identified as genes that are down-regulated following the deletion of Spt3, a ySAGA-specific subunit, but unmodified upon inactivation of Taf1, a yTFIID-specific subunit (Huisinga and Pugh 2004). These genes, which preferentially use SAGA rather than TFIID to assemble the transcription machinery, account for ∼10% of the yeast genes and correspond mainly to stress-induced genes with a TATA box in their promoters (Basehoar et al. 2004). However, recent studies in yeast revealed that only a fraction of bound genes is misregulated upon deletion of SAGA subunits, and, conversely, only a fraction of misregulated genes is bound by the corresponding subunits (Lenstra et al. 2011; Venters et al. 2011; Lenstra and Holstege 2012). Furthermore, a wider distribution of the complex is suggested by chromatin immunoprecipitation (ChIP) experiments on a few model genes in yeast that also detected SAGA in the coding region and by ChIP-coupled high-throughput sequencing (ChIP-seq) analyses in flies that revealed a colocalization of SAGA with RNA polymerase II (Pol II) in the body of a subset of genes (Govind et al. 2007; Johnsson et al. 2009; Weake et al. 2011). In addition to its gene-specific regulatory role, it has been proposed that SAGA acetylates histones throughout the genome in a global and untargeted manner (Vogelauer et al. 2000). In good agreement, inactivation of the SAGA HAT or DUB activities was reproducibly reported to affect the global levels of histone H3K9 acetylation (H3K9ac) and histone H2B monoubiquitination (H2Bub), two marks associated with active transcription at promoters and in gene transcribed regions, respectively (Henry et al. 2003; Bian et al. 2011; Jin et al. 2011).
To better understand the genome-wide function of SAGA, we investigated the distribution of this coactivator complex by tracking chromatin modifications deposited or removed by its two different enzymatic activities. The genome-wide comparison of H3K9ac or H2Bub densities in wild-type cells and cells with impaired SAGA HAT or DUB activity revealed that SAGA modifies the promoter and the transcribed region of all expressed genes in both S. cerevisiae and human cells. Our study reveals a highly dynamic and transient association of SAGA with the entire active chromatin that is required for Pol II recruitment and transcription of almost all expressed genes.
Results
SAGA deubiquitinates H2B on the transcribed region of all expressed genes in human cells
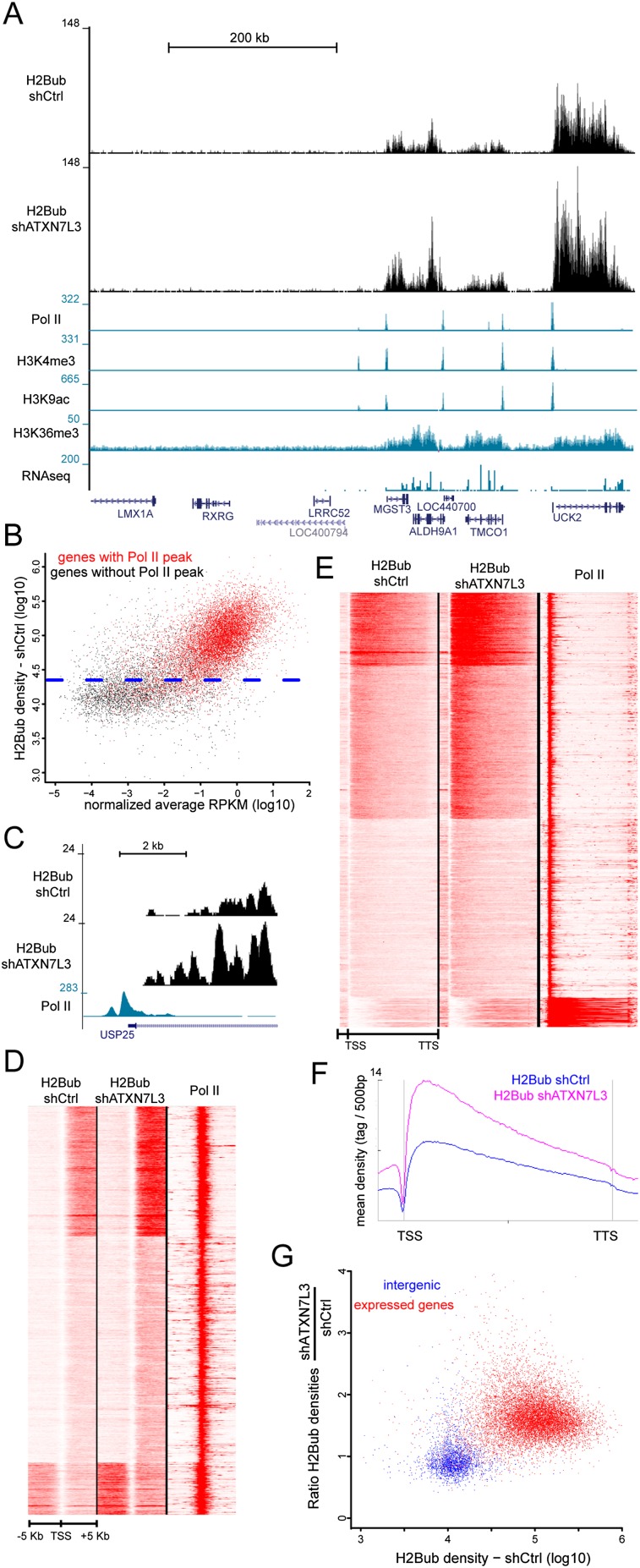
As an indicator of the genome-wide SAGA-binding profile, we aimed to localize histone marks regulated by the two enzymatic activities of the complex. We first analyzed the H2B DUB of the human SAGA complex. Former analyses in mammals revealed that H2Bub was extensively enriched in the transcribed region of expressed genes, in agreement with an association of the H2B ubiquitination enzymes with the elongating Pol II (Minsky et al. 2008; Shema et al. 2008; Vethantham et al. 2012). However, it remains to be determined whether SAGA can deubiquitinate all H2Bub-containing nucleosomes, which represent a large fraction of the genome, or whether its DUB activity would be restricted to the promoters of SAGA-regulated genes. To answer these questions, we first performed H2Bub ChIP-seq experiments on HeLa cells that revealed significant H2Bub signal exclusively on the transcribed region of different genes enriched for marks associated with active transcription (Fig. 1A). Scatter plot analysis of all human genes according to their mRNA levels and H2Bub densities in gene bodies (from transcription start site [TSS] to transcription termination site [TTS]) revealed two different gene populations with respect to H2B ubiquitination. Nonexpressed genes displayed background levels of H2Bub similar to that observed on intergenic regions, whereas significant levels of H2Bub were found at almost all expressed genes that were also characterized by the presence of a Pol II peak at their promoter regions (Fig. 1B; Supplemental Fig. S1B,C).
Figure 1.
SAGA acts in the transcribed region of all expressed genes in HeLa cells. (A) Genome browser tracks depicting H2Bub distribution at a representative region in control and ATXN7L3 knockdown HeLa cells. H2Bub localized in the transcribed region of expressed genes that are characterized by Pol II, H3K4me3, and H3K9ac peaks at their promoter; H3K36me3 signal on the gene body; and RNA sequencing (RNA-seq) reads on exons. (B) Gene expression levels (based on normalized average RPKM [reads per kilobase per million mapped reads]) were plotted versus the H2Bub densities on the corresponding gene bodies. The blue dotted line indicates the background density of H2Bub in control HeLa cells (95% of intergenic regions have lower H2Bub densities). Significant H2Bub and RNA-seq reads are found at most of the 10,934 genes that have a Pol II peak at their promoter (red dots) but not at the majority of the 8363 genes devoid of Pol II (black dots). When considering expressed genes, a very weak correlation (Pearson correlation coefficient, 0.20) could be observed between H2Bub levels and gene expression, indicating that H2B ubiquitination is not solely related to gene expression levels. (C) Absence of H2Bub at the promoter of an expressed gene in control and ATXN7L3 knockdown HeLa cells. (D,E) Heat maps showing the distribution of H2Bub in control HeLa cells (H2Bub shCtrl) and after the inactivation of SAGA DUB activity (H2Bub shATXN7L3) around the TSS (TSS −5 kb/+5 kb) (D) and on bodies of expressed genes (from TSS to TTS) (E). Nine-thousand-six-hundred-thirty genes with a Pol II peak at the TSS were considered. (F) Average profiles depicting H2Bub distribution on gene bodies. Upon inactivation of SAGA DUB activity, H2Bub is not increased at the promoter of expressed genes (C,D) but increases on the body of expressed genes (E,F). (G) Scatter plots representing H2Bub densities in control cells relative to SAGA DUB activity (ratio of H2Bub densities in shATXN7L3 and shCtrl cells). Two-thousand-eight-hundred-forty-nine intergenic regions (blue dots) were compared with 10,486 expressed genes (red dots).
To determine at which genomic locations SAGA would remove ubiquitin from H2B, we generated and compared H2Bub ChIP-seq profiles in control and ATXN7L3 knockdown HeLa cells after normalization of the two data sets by using large intergenic regions (see the Supplemental Material). We used our previously generated cell line expressing an ATXN7L3 shRNA in which the DUB module is dissociated from the SAGA complex, leading to an increase of global H2Bub levels (Supplemental Fig. S1A; Lang et al. 2011). After inactivation of the SAGA DUB, H2Bub densities in the whole transcribed regions of expressed genes were higher than that in control cells, but the overall H2Bub distribution was preserved. In contrast, comparable low background H2Bub levels were observed in nonexpressed genes in both ATXN7L3 knockdown and control HeLa cells (Fig. 1A). It has been previously suggested that the very low levels of H2Bub at gene promoters were due to the high DUB activity of SAGA recruited to promoters. However, the absence of any H2Bub signal in wild-type conditions and upon depletion of SAGA DUB activity at the promoter of a representative gene (USP25) indicates that H2B was not targeted by the ubiquitinating enzymes at this promoter (Fig. 1C). Similar observations were made on all expressed genes upon depletion of the SAGA DUB activity, suggesting that the H2B ubiquitinating enzymes are not active on these promoters (Fig. 1D). In contrast, we observed an increase of H2Bub levels in the bodies of expressed genes in ATXN7L3 knockdown as compared with control HeLa cells (Fig. 1E). Metagene analysis revealed a similar H2Bub profile in both cell lines, but the signal was increased by ∼1.7-fold throughout gene bodies upon SAGA DUB depletion (Fig. 1F).
As a quantification of SAGA DUB activity, we measured the H2Bub ratio in ATXN7L3 knockdown cells versus control cells in intergenic regions as well as in the transcribed region of all expressed genes. This analysis clearly differentiated intergenic regions from expressed genes, which had higher H2Bub densities and increased H2Bub ratios (Fig. 1G). Nonexpressed genes share the same profile as intergenic regions, with background levels of H2Bub and mean H2Bub ratios close to 1 (Supplemental Fig. S2A). To determine whether gene expression levels influence the SAGA DUB activity, we compared gene categories characterized by low, medium, and high expression (Supplemental Material). The three groups displayed strikingly similar H2Bub ratios that were clearly different from those of intergenic regions (Supplemental Fig. S2B). Furthermore, 50% of expressed genes had H2Bub ratios ranging from 1.5 to 2.0, indicating a homogenous DUB activity, although these genes had much more heterogenous H2Bub densities, varying by 10-fold (Fig. 1G; Supplemental Fig. S2C). Our results together demonstrate that the SAGA DUB activity is directed toward the transcribed region of almost all expressed genes and is directly proportional to H2Bub levels, suggesting that the modification itself is the main driving force for this activity.
SAGA can be recruited in gene bodies independently of Pol II to induce a very fast reversal of H2B ubiquitination
The above reported distribution of the SAGA DUB raises the question of the mechanisms for SAGA recruitment and activity in gene bodies. An obvious hypothesis posits that following its recruitment at promoters, SAGA would interact with the elongating Pol II and thus travel through the whole transcribed region. To test this hypothesis, we measured H2Bub bulk levels in HeLa cells at different time points following treatment with actinomycin D, an inhibitor of transcription elongation. Transcription inhibition suppresses the deposition of the mark as the H2B ubiquitination machinery is recruited in gene bodies by the elongating Pol II. Thus, changes in the levels of already deposited H2Bub upon actinomycin D treatment can be explained only by the activity of DUBs that can remove ubiquitin from H2B.
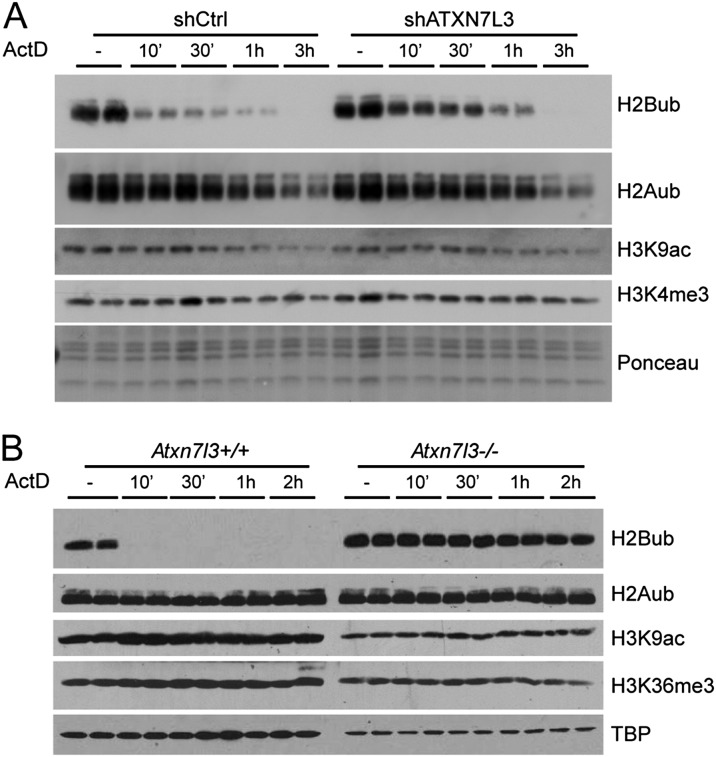
Indeed, we observed a very fast loss of H2Bub that was dramatically reduced after 10 min and undetectable after 3 h of treatment in wild-type HeLa cells, indicating an extremely dynamic action of DUBs (Fig. 2A). We could not detect such extremely rapid effects on other chromatin marks analyzed (H2Aub, H3K4me3, or H3K9ac), suggesting a unique and dynamic versatility of histone H2B ubiquitination. Importantly, the loss of H2Bub was significantly delayed in ATXN7L3 knockdown cells, indicating a role for SAGA DUB in H2Bub removal (Fig. 2A). To demonstrate unequivocally the role of the SAGA DUB in H2Bub turnover, we analyzed H2Bub kinetics in cells in which this activity was completely suppressed. We therefore generated mouse embryonic stem cells (mESCs) homozygous for an Atxn7l3-null allele (Atxn7l3−/−) (Supplemental Fig. S3). While H2Bub was almost undetectable after 10 min of actinomycin D treatment in wild-type mESCs, the H2Bub global levels stayed completely unchanged in Atxn7l3−/− mESCs at all of the analyzed time points (Fig. 2B). This result demonstrates unambiguously that the fast disappearance of H2Bub in control cells is exclusively carried out by the SAGA DUB activity, ruling out the role of other DUBs in this process. In addition, SAGA can act in the transcribed region of active genes independently of elongating Pol II. Importantly, our observations reveal an extremely transient and dynamic association of SAGA with active chromatin, as H2Bub can be erased genome-wide by the SAGA DUB activity in a few minutes.
Figure 2.
SAGA mediates a very fast reversal of H2B ubiquitination upon transcription inhibition. Control (left panels) and ATXN7L3 knockdown (right panels) HeLa cells (A) or Atxn7l3 knockout mESCs (B, right panels) were treated with actinomycin D for the indicated time points. Acidic extracts were analyzed by Western blotting using the indicated antibodies.
SAGA deubiquitinates the bodies of all expressed genes in yeast
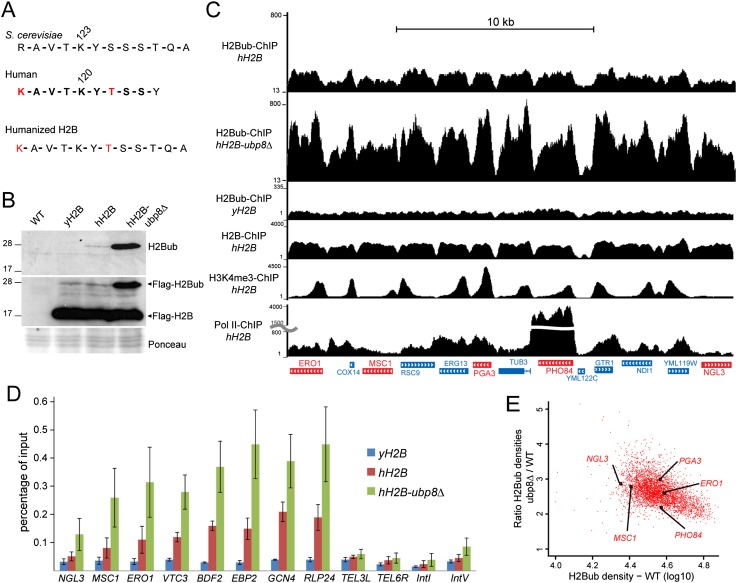
Next, we wanted to confirm in other organisms the wide distribution of the SAGA DUB activity that we observed in human cells. Former ChIP-on-chip experiments in S. cerevisiae revealed a colocalization of H2Bub with H3K79me3 in the coding region of only a subset of genes and suggested that Ubp8 would mainly remove ubiquitin from H2Bub in the 5′ region of genes at H3K4me3 peaks (Schulze et al. 2009, 2011). This would indicate different regulations for H2B ubiquitination between yeast and other eukaryotes, as indicated by differential H2Bub distribution (Kharchenko et al. 2011; Roudier et al. 2011; Vethantham et al. 2012). To compare the activity of the evolutionarily well-conserved DUB module of the yeast and the human SAGA complexes, we aimed to compare H2Bub profiles in wild-type and ubp8Δ strains. To get an accurate map of H2Bub in S. cerevisiae, we took advantage of the highly specific anti-H2Bub monoclonal antibody that was raised against a branched peptide corresponding to the ubiquitinated human histone H2B (Minsky et al. 2008) but did not recognize H2Bub in budding yeast. We thus humanized the yeast H2B through substitutions of R119 and S125 to K and T, respectively (Fig. 3A). This modified H2B was Flag-tagged and expressed in yeast depleted for endogenous H2B. Western blot analysis of yeast extracts with an anti-Flag antibody revealed that this humanized H2B (hH2B) was efficiently ubiquitinated to the same extent as a Flag-tagged yeast H2B (yH2B) (Fig. 3B). The anti-H2Bub antibody revealed a single band of 25 kDa in the hH2B, whose intensity was further increased upon UBP8 deletion. These results together demonstrate that the regulation of H2B ubiquitination in S. cerevisiae was not affected by the yeast H2B humanization.
Figure 3.
H2Bub ChIP-seq analyses in budding yeast expressing a humanized histone H2B reveal a global distribution of the SAGA DUB activity. (A,B) A humanized version of the yeast histone H2B is efficiently ubiquitinated and recognized by an anti-human H2Bub antibody. (A) S. cerevisiae and human H2B protein sequence comparison around their ubiquitination sites. The peptide used to raise the anti-H2Bub antibody (in bold) contains two residues (in red) that are not conserved in the yeast H2B. hH2B was obtained by mutating these two residues into their human counterpart. (B) H2B ubiquitination in htb1∆ htb2∆ yeast strains expressing Flag-tagged yH2B or hH2B and after further UBP8 deletion (hH2B-ubp8Δ) was analyzed by immunoblotting using an anti-H2Bub (top panel) or an anti-Flag (middle panel) antibody. (C) H2Bub ChIP-seq analyses in S. cerevisiae expressing hH2B (H2Bub-ChIP hH2B) and after further UBP8 deletion (H2Bub-ChIP hH2B-ubp8Δ) or the unmodified yH2B (H2Bub-ChIP yH2B). Genome browser tracks of a representative genomic region showing H2Bub enrichment in gene bodies. Representative genes shown in E are indicated in red. H2B, H3K4me3, and Pol II profiles in the hH2B background are shown in the lower tracks. (D) H2Bub ChIP was performed on yH2B, hH2B, and hH2B-ubp8∆ strains. H2Bub levels were quantified by real-time quantitative PCR (qPCR) on bodies of active genes and control intergenic regions (TEL3L, TEL6R, IntI, and IntV). The values (mean ± SD of three independent ChIP experiments) are expressed as percentage of input DNA signal. (E) Scatter plot showing a homogenous SAGA DUB activity on the 3916 expressed genes. The ratios of H2Bub densities on gene bodies between hH2B-ubp8Δ and hH2B strains were plotted against the H2Bub densities.
ChIP-seq experiments in hH2B-expressing yeast cells revealed that H2Bub was specifically enriched in gene bodies of almost all active genes and depleted from intergenic regions (Fig. 3C; Supplemental Fig. S4A). The signal specificity was demonstrated by the detection of background signal on the whole genome of cells expressing yH2B. Moreover, significant variation of H2Bub densities from gene to gene contrasted with a homogenous enrichment of H2B on all genes, indicating that H2B local densities cannot be the sole determinant of H2B ubiquitination (Fig. 3C; Supplemental Fig. S4B).
As the vast majority of the yeast genome can be ubiquitinated on H2B (see above) and thus could be a substrate for the SAGA DUB, we next determined the distribution of this activity. To this end, we analyzed H2B ubiquitination in yeast strains expressing hH2B and after further deletion of UBP8. H2Bub levels were increased by about threefold to fivefold in an ubp8Δ strain when compared with the parental strain, suggesting that SAGA has a global DUB activity on the yeast genome (Fig. 3B; Supplemental Fig. S5A). We next performed H2Bub ChIP-seq experiments in the same strains. To get an accurate normalization of the two data sets, we quantified H2Bub levels on a number of selected genes and intergenic regions by ChIP combined with quantitative PCR (ChIP-qPCR) (Fig. 3D). A mean 2.5-fold increase of H2Bub levels in gene bodies upon UBP8 deletion was calculated from three independent ChIP-qPCR experiments and was applied to normalize our two ChIP-seq data sets. On a representative region, the H2Bub levels were found increased in all gene bodies upon UBP8 deletion, in good agreement with our observation in human cells (Fig. 3C). Importantly, very similar results were observed when the ChIP-seq experiments were performed with the addition of S. pombe chromatin as spike-in control for normalization (Supplemental Fig. S5B,C). For all active gene bodies, we calculated the ratio of H2Bub densities between the ubp8Δ and parental strains and observed a twofold to threefold increase of the ratio between the two strains (Fig. 3E). This observation and the very high correlation between H2Bub densities between the two strains (Pearson correlation coefficient, 0.84) suggest a global and homogenous activity of Ubp8 in all regions that display significant H2Bub in wild-type cells. Similar to our observations in human cells, the yeast SAGA complex deubiquitinates H2B on the transcribed region of all expressed genes.
GCN5-containing complexes acetylate H3K9 at the promoter of all expressed genes in HeLa cells
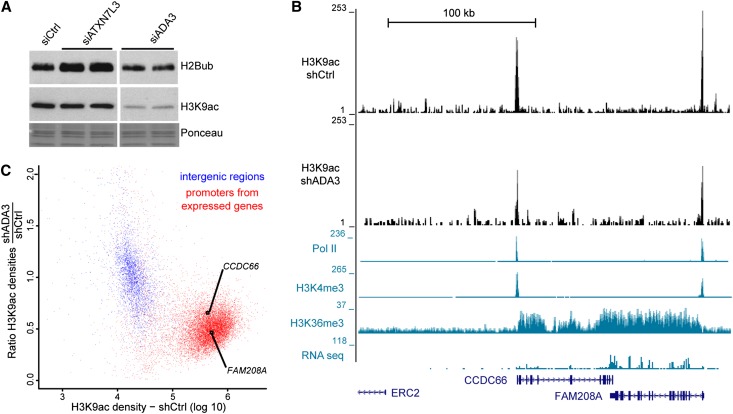
The observed localization of the SAGA DUB activity prompted us to determine whether its HAT activity also displays a broad genome-wide distribution. In human cells, GCN5 and its paralog, PCAF, are found in two different HAT complexes, SAGA and ATAC, in which they are regulated by interactions with other subunits of the HAT module, such as ADA3 (Lee and Workman 2007). We could observe a global decrease of H3K9ac levels upon ADA3 depletion in HeLa cells, corresponding to an inactivation of both GCN5-containing complexes (Fig. 4A). In the same conditions, H2Bub levels were unchanged. Conversely, H3K9ac was unchanged, but H2Bub levels were increased upon ATXN7L3 down-regulation, indicating that the two enzymatic activities of SAGA are independent (Fig. 4A).
Figure 4.
All active Pol II promoters are acetylated by GCN5-containing complexes in human cells. (A) Western blot analyses of changes in H2B ubiquitination and H3K9ac upon inactivation of the corresponding enzymes. Total histones were purified by acidic extractions from HeLa cells transfected with the specified siRNA and immunoblotted with the indicated antibodies. (B,C) Analysis of H3K9ac ChIP-seq profiles in control and ADA3 knockdown HeLa cells. (B) Genome browser tracks at a representative region reveal that H3K9ac peak intensity at the promoter of expressed genes in HeLa cells (H3K9ac-shCtrl) drops upon ADA3 knockdown (H3K9ac-shADA3). (C) GCN5 HAT activity (ratio of H3K9ac densities in shADA3 and shCtrl cells) was plotted relative to H3K9ac densities in 3189 intergenic regions (blue dots) and 11,300 regions surrounding the TSS of expressed genes (TSS −1.5 kb/+3.5 kb; red dots). Promoters of expressed genes shown in B are indicated.
To determine the genome-wide distribution of GCN5-containing complexes, we profiled and quantified the distribution of H3K9ac by ChIP-seq in cells expressing either control or ADA3 shRNA (Supplemental Fig. S6A,B). In control HeLa cells, H3K9ac peaks were detected exclusively at the promoters of expressed genes together with Pol II and H3K4me3 but were absent from silent genes (Fig. 4B). The control and ADA3 knockdown data sets were normalized by using intergenic regions where only background levels of H3K9ac could be detected (Supplemental Material). In ADA3-depleted cells, the overall profile of H3K9ac was conserved, but the peak heights were significantly reduced (Fig. 4B). As a measure of GCN5 HAT activity, we calculated the ratio of H3K9ac densities obtained in ADA3-expressing versus control shRNA-expressing cells both in intergenic regions and at the promoters of all expressed genes. This analysis clearly separated the two categories of genomic loci: (1) All intergenic regions had low levels of H3K9ac, and the H3K9ac ratios were centered on 1, and (2) the vast majority of promoters of active genes were enriched for H3K9ac and displayed decreased ratios of H3K9ac, indicating that GCN5-containing complexes significantly acetylate H3K9 at these locations (Fig. 4C).
GCN5 acetylates H3K9 at all active promoters in yeast
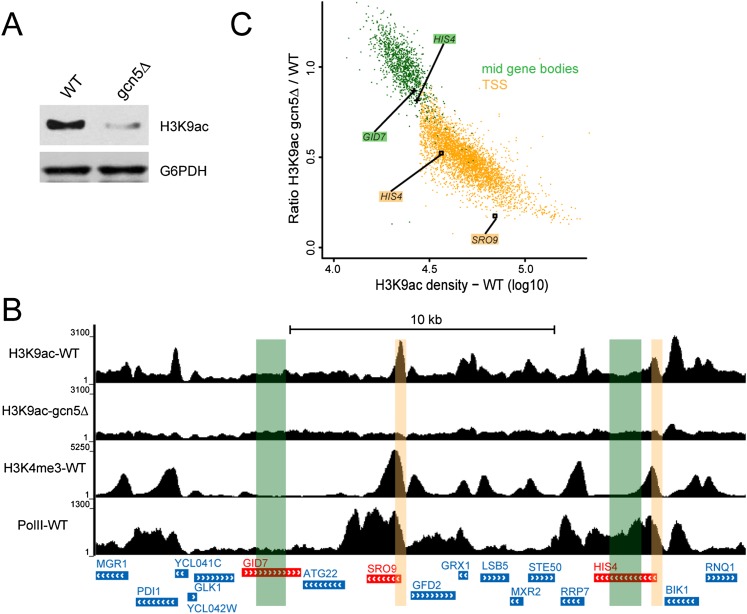
As GCN5 deletion in S. cerevisiae induced a decrease in global H3K9ac (Fig. 5A), we further investigated Gcn5 distribution in yeast by measuring H3K9ac levels in wild-type and gcn5Δ strains by ChIP-seq. In wild-type cells, H3K9ac peaks colocalized with H3K4me3 around the TSS of active genes. In agreement with our results in human cells, H3K9ac peaks were completely lost at active promoters in the gcn5Δ strain (Fig. 5B; Supplemental Fig. S6C). We measured the ratio of H3K9ac densities in gcn5Δ versus wild-type cells at all active promoters (as defined in Supplemental Fig. S6D). Mid-gene bodies (25%–75% of the coding region) of large genes (>2 kb) with only background levels of H3K9ac were used to normalize the two data sets. All active promoters could be easily differentiated from mid-gene bodies, as they were enriched for H3K9ac in wild-type cells and had a <1 ratio of H3K9ac densities (Fig. 5C; Supplemental Fig. S6E,F). Our results together indicate that, in both yeast and human cells, GCN5-containing complexes acetylate H3K9 specifically at the promoters of active genes and thus strongly suggest a wide distribution of the SAGA HAT activity.
Figure 5.
Gcn5 acetylates H3K9 at the promoter of all expressed genes in yeast. (A) Whole-cell extracts from wild-type (WT) and gcn5∆ yeasts were prepared in an 8 M urea buffer and probed as indicated. (B,C) Analysis of H3K9ac ChIP-seq profiles in wild-type and gcn5∆ yeast cells. (B) Genome browser tracks of a representative yeast genomic region. H3K9ac peaks detected in wild-type cells (H3K9ac-WT) are absent when GCN5 is deleted (H3K9ac-gcn5∆). Expressed genes are characterized by an H3K4me3 peak at their promoter (H3K4me3-WT) and a Pol II signal on the gene body (Pol II-WT). (C) Scatter plot representing Gcn5 acetylation activity (ratio of H3K9ac densities in gcn5∆ and wild type) versus H3K9ac density in 3916 regions centered on the first codon (±200 base pairs [bp]) of expressed yeast genes as defined by the presence of H3K4me3 and H3K9ac peaks at the TSS (yellow dots) or in 1184 mid-gene bodies of large genes (from 25% to 75% of gene bodies >2 kb; green dots). Promoters and control regions highlighted in B are indicated.
SAGA is required for RNA Pol II recruitment at all transcribed genes and for nascent mRNA synthesis
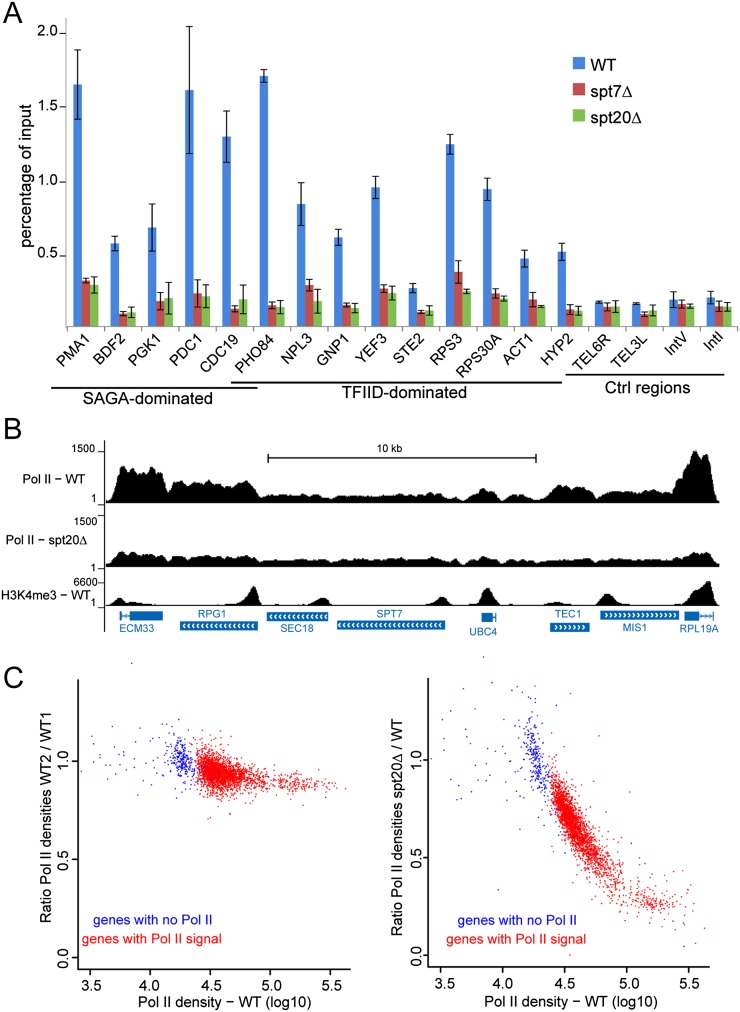
Based on our results that revealed an unexpected wide action of SAGA on the transcriptionally active genome, we asked whether this complex would have a more global function in transcription regulation than previously appreciated. We hypothesized that SAGA would behave as a general cofactor required for the transcription of all Pol II genes rather than being involved only in the regulation of stress-induced genes. Besides its enzymatic activities, other functions of SAGA could influence Pol II recruitment and transcription. Therefore, we analyzed Pol II recruitment upon deletion of SPT7 or SPT20, which are known to result in the disruption of the SAGA complex in yeast. Using an anti-Pol II antibody that recognizes both modified and unmodified forms of the C-terminal domain of Rpb1, the largest subunit of Pol II, background levels of Pol II were detected in control intergenic or telomeric regions in wild-type S. cerevisiae and were unmodified in spt7Δ or spt20Δ strains. In contrast, in all 14 genes tested that were previously defined as either SAGA- or TFIID-dominated (Huisinga and Pugh 2004), Pol II occupancy was strikingly reduced upon SPT7 or SPT20 deletion when compared with that observed in the parental strain (Fig. 6A). Comparable effects were observed for genes that were found to be down-regulated as well as other genes (either SAGA- or TFIID-dominated) whose expression was found to be unchanged upon deletion of these subunits (Lenstra et al. 2011). Two independent chromatin extractions gave similar results, and H2B or H3K4me3 ChIP on the same samples revealed comparable signals in wild-type, spt7Δ, and spt20Δ strains (Supplemental Fig. S7A). These results suggest that SAGA might be crucial for Pol II recruitment on a large number of active genes.
Figure 6.
SAGA is required for RNA Pol II recruitment at both SAGA- and TFIID-dominated genes. (A) Pol II ChIP-qPCR performed on chromatin extracted from wild-type (WT), spt7Δ, and spt20Δ yeast cells on SAGA- and TFIID-dominated genes or in control intergenic regions (mean ± SD, n = 2). (B,C) Pol II ChIP-seq analysis in S. cerevisiae wild type and spt20Δ. (B) Genome browser tracks of a representative genomic region show decreased Pol II occupancy in the spt20Δ strain. (C, left panel) Scatter plot showing the reproducibility of Pol II ChIP performed on two independent wild-type chromatin preparations. (Right panel) A global loss of Pol II recruitment in spt20Δ cells is observed on 2606 active genes with H3K9ac and H3K4me3 peaks at the TSS and significant Pol II signal (red dots). Data were normalized on 291 genes that have background levels of Pol II and lack H3K4me3 and H3K9ac (blue dots).
Next, we analyzed Pol II occupancy on all genes by performing ChIP-seq experiments that revealed a global decrease of the Pol II signal upon SPT20 deletion (Fig. 6B; Supplemental Fig. S7C). For most genes that displayed a significant Pol II occupancy in wild-type cells, the signal was strongly reduced in mutant cells. We identified ∼300 genes with background levels of Pol II signal and devoid of H3K4me3 and H3K9ac that were further used to normalize the two data sets (Supplemental Material). Next, we compared Pol II occupancy in wild-type and spt20Δ cells on ∼2600 active genes in which a clear Pol II signal was detected (Supplemental Fig. S7B). On all of these genes, we observed that the Pol II occupancy was strongly reduced upon SPT20 deletion (Fig. 6C). Very similar conclusions were made when normalization of the two data sets was performed by using Pol II densities measured on Pol III transcribed genes (Supplemental Fig. S7D). Therefore, our results indicate that SAGA was required for Pol II recruitment at all transcribed genes where a significant Pol II occupancy could be detected.
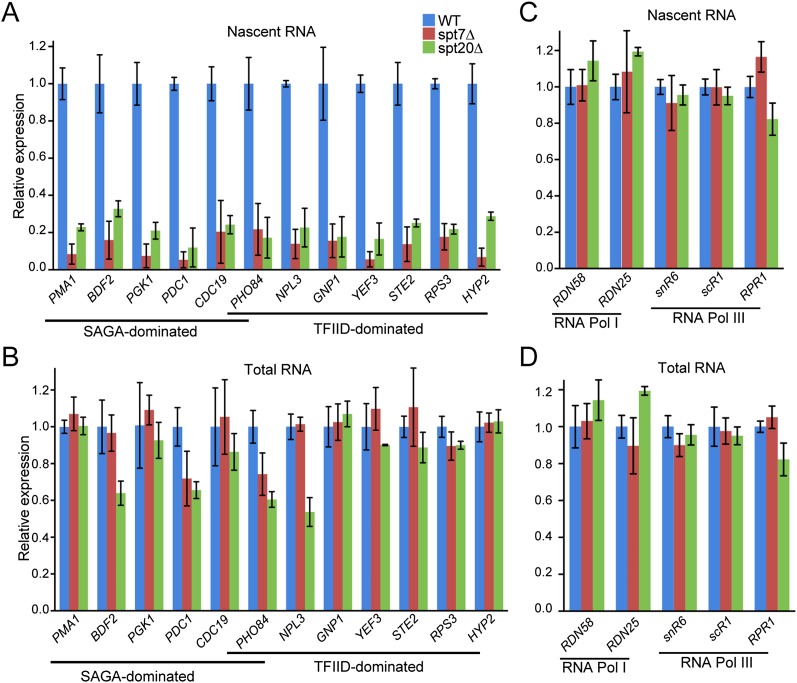
This result was unexpected, as previous measurements of mRNA expression changes in ySAGA mutant strains identified only a small subset of SAGA-dominated genes. However, measuring steady-state mRNA levels may not fully reflect the SAGA-dependent transcriptional effects, as cells can compensate for global transcription changes (Haimovich et al. 2013). To overcome this limitation, we analyzed how SAGA affects gene expression through the quantification of newly synthesized mRNA that measure mRNA synthesis uncoupled from mRNA degradation. Wild-type, spt20Δ, and spt7Δ S. cerevisiae strains cultured in YPD were metabolically labeled with 4-thiouracil for 6 min. Labeled cells were then mixed in a ratio of 3:1 with labeled S. pombe cells that provided an internal standard. After total RNA extraction, newly synthesized (labeled) RNAs were purified, and both fractions were analyzed by RT-qPCR. Surprisingly, we observed a threefold to 10-fold reduction in newly synthesized mRNA production from the 12 genes analyzed in spt20Δ or spt7Δ strains when compared with wild-type cells (Fig. 7A). In both strains, SAGA- or TFIID-dominated genes were down-regulated to the same extent. These changes in mRNA synthesis reflected the decreased Pol II occupancy that was observed on the same genes in spt20Δ or spt7Δ strains (Fig. 6A). In contrast, mRNA quantification performed on total RNA from the spt20Δ strain revealed unchanged levels for eight of these genes and a moderate decrease of less than twofold for four other genes, in good agreement with a previous study (Lenstra et al. 2011). Similarly, in the spt7Δ strain, mRNA steady-state levels from only two genes were found to be slightly decreased (Fig. 7B). The levels of RNA Pol I and RNA Pol III nascent transcripts used as controls were not affected upon SPT20 or SPT7 deletion (Fig. 7C,D). Altogether, our results indicate that the SAGA complex plays a role in Pol II recruitment and transcription on a vast majority of Pol II genes independently, whether they were defined as SAGA and/or TFIID dominated.
Figure 7.
SAGA is required for RNA Pol II transcription of either SAGA- or TFIID-dominated genes. Newly synthesized (A,C) and total (B,D) mRNA were extracted from wild-type (WT), spt7Δ, and spt20Δ yeast cells. mRNA from SAGA- and TFIID-dominated genes (A,B) or RNA Pol I and RNA Pol III genes (C,D) were quantified by real-time PCR. Results were normalized to S. pombe tubulin expression and are presented as fold variation in comparison with the wild type (mean ± SD, n = 3).
Discussion
In this study, we provide a series of results that coherently indicate that the SAGA complex behaves as a general cofactor that is required for Pol II transcription of almost all active genes. First, we analyzed the genome-wide location of the HAT and DUB activities of SAGA in both S. cerevisiae and human cells. These analyses revealed that SAGA is acting at all active genes, both at their promoters and in their whole transcribed regions. Such a distribution of the two SAGA enzymatic activities was highly unexpected, as it appears much broader than previously anticipated. Second, we showed that Pol II recruitment at all active genes was severely impaired upon deletion of SAGA structural subunits. Third, through the quantification of newly synthesized RNA, we demonstrated that SAGA inactivation induced a strong decrease of mRNA synthesis for all genes that were analyzed. In good agreement with this broad function, our analysis of the SAGA DUB activity upon transcription inhibition indicates that SAGA can act on a large fraction of the genome in a very fast manner. This underscores that SAGA association with chromatin occurs in a very transient and dynamic manner, indicating a new paradigm for the dynamic distribution and broad activity of this coactivator.
SAGA acts on a very large fraction of eukaryotic genomes
Our analyses of changes in H2B ubiquitination upon depletion of the SAGA DUB activity directly reflect SAGA localization at all expressed genes. Indeed, ChIP-seq analyses in the ubp8Δ yeast strain or ATXN7L3-depleted HeLa cells revealed H2Bub changes that can only be explained by a loss of the SAGA DUB activity. Actually, Ubp8 has to be incorporated within SAGA to be catalytically active and appears to be a unique constituent of SAGA (Ingvarsdottir et al. 2005; Lee et al. 2011). Similarly, USP22 was shown to be active in vitro only when interacting with all subunits of the DUB module, including ATXN7L3, and was found to be fully active in vivo only as part of the SAGA complex (Lang et al. 2011; Armour et al. 2013). Although it was recently shown that the proteasome regulatory particle can separate a DUB module from the yeast SAGA complex in vitro, such a free DUB module has no apparent effect on the regulation of H2Bub in vivo (Lim et al. 2013). In agreement with a role for SAGA in gene transcribed regions, different studies using ChIP on candidate genes previously suggested the recruitment of different SAGA subunits in gene bodies (Govind et al. 2007; Wyce et al. 2007), and genome-wide ChIP analyses in S. pombe and Drosophila embryos localized Gcn5 or Ada2b in the transcribed regions of a subset of expressed genes (Johnsson et al. 2009; Weake and Workman 2012). In contrast, our analyses of H2B ubiquitination upon SAGA DUB inactivation unambiguously demonstrate that SAGA is dynamically recruited in the transcribed region of all expressed genes. Although our experiments do not distinguish Gcn5 activity as part of the different Gcn5-containing complex, we demonstrate here that these complexes are recruited at the promoter of all expressed genes in budding yeast and human cells, further suggesting a broad genome-wide recruitment of the SAGA complex.
Former genome-wide location studies of the SAGA complex subunits in different organisms revealed only a few hundred SAGA-binding sites (Vermeulen et al. 2010; Krebs et al. 2011; Venters et al. 2011; Weake et al. 2011). These analyses rely on ChIP experiments of a subset of SAGA-specific subunits that likely capture the most stable associations with chromatin. In contrast, population-based approaches that average the signal across heterogenous cellular states would miss most of the locations where SAGA is only very transiently recruited. However, such short-lived associations can be revealed by tracking the chromatin changes mediated by the enzymatic activities of SAGA, as demonstrated by this study. In agreement with this conclusion, the analysis of the SAGA DUB activity upon transcription inhibition revealed that its association with the chromatin occurs in an extremely dynamic fashion.
SAGA is required for all Pol II transcription, in agreement with its broad genome-wide action at all transcribed regions
The observed distribution of the two enzymatic activities of SAGA contrasts with the current model that posits a recruitment of SAGA at a small subset of genes whose expression is regulated by the different activities of the complex. As indicated above, this is likely explained by the different modes of SAGA interaction with chromatin that are not equally detected, depending on the experimental methods. A broad distribution of SAGA on all active chromatin is in good agreement with our observation of a role for SAGA in Pol II recruitment and function on most active genes. Here, we show that the transcriptional role of SAGA is independent of its mode of recruitment. Indeed, upon SAGA depletion, Pol II recruitment and mRNA synthesis appear to be similarly affected at all genes whether ChIP of SAGA subunits detected a stable association of SAGA at the promoter or not. Our observation describing a general cofactor activity of SAGA at all Pol II genes appears to be in contradiction to the numerous expression studies reporting a low percentage of genes whose expression is deregulated in SAGA mutants (Huisinga and Pugh 2004; Lenstra et al. 2011). However, these gene expression analyses measured steady-state mRNA levels in wild-type and mutant strains. Nevertheless, several studies recently demonstrated that global effects on mRNA synthesis are accompanied by compensatory mechanisms such as parallel changes in mRNA degradation rates, resulting in mRNA level buffering (Helenius et al. 2011; Schulz et al. 2014). Such a mechanism has been previously demonstrated in a yeast strain expressing a point mutant of Rpb1 subunit of Pol II that causes decreased mRNA synthesis as expected but also decreases mRNA decay rates (Sun et al. 2012).
Two enzymatic activities of the same complex act on different functional genomic elements
Our study surprisingly revealed distinct genomic locations for the two enzymatic activities of the same complex. While H3K9ac by the HAT module of SAGA is restricted to the promoter of active genes, the DUB acts on gene bodies and is excluded from promoters in human cells. It is unclear how SAGA can be recruited at all active genes at both their promoters and transcribed regions. This observation also raises the question of a restriction of the HAT activity to promoter regions when the complex has a much wider distribution. Two alternative mechanisms can be proposed: (1) The specific localization of H3K9ac at the promoter could result from the cumulative actions of SAGA at both promoter and gene bodies and that of histone deacetylases such as Rpd3 acting in the transcribed region of these genes (Vogelauer et al. 2000; Govind et al. 2010; Weake et al. 2011). (2) Alternatively, at active promoters, the combination of transcription factors and histone marks known to interact with different domains of SAGA subunits could create a favorable environment for SAGA recruitment and/or retention and could position the HAT module for optimal substrate recognition (for review, see Weake and Workman 2012).
In contrast to GCN5, which lays a mark at the histone H3 tail, the SAGA DUB removes ubiquitin from H2Bub. Therefore, the DUB activity is restricted to locations where H2Bub is present, and this mark is clearly excluded from the promoter region in human cells. It is remarkable that H2B ubiquitination is very widely distributed in different genomes. Virtually all H2B molecules from any nucleosome, except those at telomeric or silenced regions, are potentially ubiquitinated in S. cerevisiae. In human cells, H2Bub is found in the transcribed region of all expressed genes that would correspond to as much as 20% of the whole genome. Our results demonstrate that SAGA can act on any H2Bub molecule genome-wide, raising the question of SAGA recruitment and dynamics on all targeted regions. Two different scenarios can be considered. In a first hypothesis, following its recruitment at active promoters, SAGA would then travel through the gene transcribed region either by binding to ubiquitinated substrates or via an interaction with elongating Pol II, as previously suggested (Wyce et al. 2007; Weake and Workman 2012). An alternative hypothesis could be that SAGA is not actively recruited on chromatin but would be diffusing rather freely in the nucleoplasm and would have a high affinity to any ubiquitinated H2B-containing region. In favor of such a model is our observation that, upon inhibition of transcription elongation, SAGA can completely remove ubiquitin from the whole active chromatin in a few minutes (Fig. 2). Furthermore, we showed that H2B ubiquitination per se is the main driving force for SAGA DUB, as its activity is directly proportional to the local H2Bub levels, which are poorly correlated with gene expression in human cells (Fig. 1; Supplemental Fig. S2).
In summary, our study reveals that SAGA is recruited at all active genes and will acetylate H3K9 at the promoters and deubiquitinate H2Bub in the gene bodies in both S. cerevisiae and human cells. This demonstrates an unexpected dynamic association of SAGA with the chromatin that can hardly be detected by conventional ChIP of subunits of the complex, suggesting a multifactorial recruitment of SAGA to chromatin that cannot be explained by previous models. Our work leads to the discovery of a new function for SAGA and indicates a role for SAGA as a general cofactor required for Pol II recruitment and transcription.
Materials and methods
Antibodies
Antibodies used in this study were as follows: H3K9ac (Abcam, ab4441), H2Bub (NRO3; Medimabs, MM-0029), H3K4me3 (Abcam, ab8580), H2B (Active Motif, 39237), H2Aub (Cell Signaling Technologies, XP rabbit mAb #8240), H3K36me3 (Abcam, ab9050), and Flag (Sigma, M2). RNA Pol II (PB-7G5) and TBP (3TF1-3G3) antibodies have been described earlier (Karmodiya et al. 2012).
siRNA transfection
Nontargeting Pool (catalog no. D-001810-10) as well as ON-TARGET plus SMART pool anti-ADA3 (catalog no. L-017508-00) siRNA were purchased from Dharmacon. Additionally, anti-ATXN7L3 (siRNA ID no. 271961) siRNA were purchased from Ambion. All siRNA were transfected in HeLa cells using Lipofectamine 2000 according to the manufacturer’s instructions.
shRNA cell line construction
HEK293T cells were cotransfected with pLKO.1-puro (vector containing the shRNA sequence), pLP1, pLP2, and pLP/VSVG plasmids with FuGENE (Promega). Forty-eight hours later, the medium containing lentiviruses was used to infect HeLa cells, and positive cells were selected with 2 µg/mL puromycine. The following MISSION shRNA constructs (Sigma) were used: shRNA anti-ADA3 (TRC no. TRCN0000015734) and shRNA nontarget control (product no. SHC002). shATXN7L3 and the corresponding shCtrl cells were described (Lang et al. 2011).
mESC derivation
The Atxn7l3 mouse strain used for this research project was created at the Institut Clinique de la Souris from the Atxn7l3tm1(KOMP)Wtsi ESC clone EPD0321_4_E11, obtained from the Knockout Mouse Project (KOMP) Repository (http://www.komp.org) and generated by the Wellcome Trust Sanger Institute (WTSI). Targeting vectors used were generated by the Wellcome Trust Sanger Institute and the Children’s Hospital Oakland Research Institute as part of the Knockout Mouse Project (3U01HG004080) (Skarnes et al. 2011). At 3.5 d post-conception, blastocysts obtained by crossing Atxn7l3+/− heterozygous mice were isolated and cultured on a monolayer of feeder cells in 2i medium (50% DMEM/F12, 50% neurobasal, 1× N2, 1× B27 with vitamin A, 10 μg/mL insulin, 1 mM glutamine, 25 μg/mL BSA type V, 0.1 mM β-mercaptoethanol, 40 μg/mL gentamycin, 3 μM CHIR 99021, 1 μM PD0325901). One week after blastocyst collection, the inner cell mass outgrowths were dissociated with 0.05% Trypsin-EDTA and plated into new feeder cell-containing plates. After three passages, cells were frozen (10% DMSO, 20% FCS ESCs tested, 70% 2i medium), and DNA was extracted from each clone for genotyping purposes. Genotypes were determined by PCR analysis using standard procedures. Three primers were used for this purpose: 5′-CAAGAAGCAGCATGCTTGGTCAAG-3′ (forward), 5′-CCAACAGCTTCCCCACAACGG-3′ (reverse 1, specific for the mutant allele), and 5′-CAGGAAGAAGTAGCCACACTTAACAGC-3′ (reverse 2, specific for the wild-type allele). Expected sizes of the amplicons were 277 base pairs (bp) for the mutant allele and 215 bp for the wild-type allele. Mouse care and procedures were in accordance with institutional and national guidelines.
Newly synthesized RNA analysis
Wild-type, spt7Δ, and spt20Δ S. cerevisiae cells were grown in YPD medium to an OD600 ≈ 0.8. Newly synthesized RNAs were labeled for 6 min by adding 4-thiouracil (Sigma-Aldrich) until a final concentration of 5 mM. In parallel, wild-type S. pombe cells were similarly grown in YES medium and labeled to be used as a spike-in across all samples. Cells were immediately pelleted and flash-frozen in liquid N2 and stored at −80°C until further use. S. cerevisiae and S. pombe cells were mixed in a ratio of 3:1. Total RNAs were extracted using RiboPure yeast kit (Ambion, Life Technologies) according to the description provided by the manufacturer. Subsequently, RNA biotinylation was carried out on 200 μg of total RNA using 200 μL of 1 mg/mL EZ-link HPDP-Biotin (Pierce) in 100 μL of biotinylation buffer (100 mM Tris-HCl at pH 7.5, 10 mM EDTA) and 600 μL of DEPC-treated RNase-free water (Sigma-Aldrich) for 3 h at room temperature, protected from light. After chloroform extraction and isopropanol precipitation, purified RNAs were suspended in 100 μL of DEPC-treated RNase-free water (Sigma-Aldrich).
Newly synthesized biotinylated RNAs were bound to 100 μL of μMACS streptavidin microbeads (Miltenyi Biotec) for 90 min at room temperature with gentle shaking. Purification of labeled RNA was then carried out using μMACS streptavidin starting kit (Miltenyi Biotec). Columns were first equilibrated with 1 mL of washing buffer (100 mM Tris-HCl at pH 7.5, 10 mM EDTA, 1 M NaCl, 0.1% Tween20). Samples were passed through the columns twice and washed five times with increasing volumes of washing buffer (600, 700, 800, 900, and 1000 μL). Ultimately, labeled RNAs were eluted twice with 200 μL of 100 mM DTT. Following ethanol precipitation, RNAs were resuspended in 10 μL of DEPC-treated RNase-free water (Sigma-Aldrich).
cDNA synthesis was performed on 1 μg of total RNA or 10 μL of labeled RNA using random hexamers and AMV reverse transcriptase (Roche) according to the manufacturer’s instructions. Real-time qPCR were performed using SYBR Green I Master (Roche). A list of all of the primers used can be found in Supplemental Table S2. All samples were run in triplicate. After qPCR, all raw values were corrected for the expression of S. pombe tubulin. Finally, results were represented graphically as a relative comparison between the wild-type (set to 1), spt7Δ, and spt20Δ samples.
Yeast strains and protein extraction
Different yeast strains used in this study were derived from the previously described FY406 (Hirschhorn et al. 1995). Deletion mutants were generated as described previously (Janke et al. 2004) or by transformation with PCR products amplified from genomic DNA of the corresponding strain obtained from the Saccharomyces Genome Deletion (SGD) strain database. The plasmid expressing the humanized version of yeast HTB1 was generated by site-directed mutagenesis of pRS413-Flag-HTB1-HTA1. The genotypes of the strains used in this study are listed in Supplemental Table S3. Yeast whole-cell extracts were prepared as described (Gardner et al. 2005) with minor changes. Cells (3 × 108) from log-phase yeast cultures were harvested by centrifugation and lysed in 400 μL of SUME buffer (8 M urea, 1% SDS, 10 mM MOPS at pH 6.8, 10 mM EDTA, 0.01% bromophenol blue) by mechanical shearing.
ChIP in HeLa cells or S. cerevisiae, library preparation, sequencing, and bioinformatics analyses of the data are detailed in the Supplemental Material.
Accession numbers
The sequencing data have been deposited in the Gene Expression Omnibus database under the accession number GSE59370. All of the data sets that were used in this study are listed in the Supplemental Table S1.
Acknowledgments
We thank F. Winston for the gift of material. We thank B. Jost and the Institut de Génétique et de Biologie Moléculaire et Cellulaire microarray and deep sequencing platform for high-throughput sequencing, and Y. Tao and S. Le Gras for their help with bioinformatics analyses. We thank the Institut Clinique de la Souris for the generation of the Atxn7L3 knockout mouse line. We are grateful to J. Müller, M. Timmers, R. Schneider, and D. Umlauf for critically reading the manuscript. This study was supported by funds from the Agence Nationale de la Recherche (ANR-10-INTB-1201) to D.D. and the NSC (100-2911-I-001-510 and 100-2923-B-001-001-MY3) to C.F.K. This study was also supported by grant ANR-10-LABX-0030-INRT under the frame program labeled ANR-10-IDEX-0002-02.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.250225.114.
References
- Armour S, Bennett E, Braun C, Zhang X-Y, McMahon S, Gygi S, Harper J, Sinclair D. 2013. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol 33: 1487–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee K, Burke T, Tempel W, Barsyte D, Li J, Wu M, Zhou B, et al. 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 30: 2829–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper A, Kim J, Roeder R. 2009. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol 29: 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. 2005. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol 25: 6123–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C, Zhang F, Qiu H, Hofmeyer K, Hinnebusch A. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25: 31–42 [DOI] [PubMed] [Google Scholar]
- Govind C, Qiu H, Ginsburg D, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman J, Li B, Hinnebusch A. 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Choder M, Singer RH, Trcek T. 2013. The fate of the messenger is pre-determined: a new model for regulation of gene expression. Biochim Biophys Acta 1829: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius K, Yang Y, Tselykh TV, Pessa HK, Frilander MJ, Mäkelä TP. 2011. Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover. Nucleic Acids Res 39: 5025–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villén J, Swaney D, Gygi S, Bähler J, Winston F. 2011. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J 30: 2843–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Bortvin AL, Ricupero-Hovasse SL, Winston F. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol 15: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol 25: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson A, Durand-Dubief M, Xue-Franzén Y, Rönnerblad M, Ekwall K, Wright A. 2009. HAT–HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep 10: 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Hirsch C, Dent S. 2010. Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs A, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. 2011. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell 44: 410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31: 3734–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Workman J. 2007. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol 8: 284–295 [DOI] [PubMed] [Google Scholar]
- Lee K, Sardiu M, Swanson S, Gilmore J, Torok M, Grant P, Florens L, Workman J, Washburn M. 2011. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol 7: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra T, Holstege F. 2012. The discrepancy between chromatin factor location and effect. Nucleus 3: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42: 536–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kwak J, Kim M, Lee D. 2013. Separation of a functional deubiquitylating module from the SAGA complex by the proteasome regulatory particle. Nat Commun 4: 2641. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10: 483–488 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro S 2009. Insights into SAGA function during gene expression. EMBO Rep 10: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. 2011. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D, Pirkl N, Lehmann E, Cramer P. 2014. Rpb4 functions mainly in mRNA synthesis by RNA polymerase II. J Biol Chem 289: 17446–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell 35: 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. 2011. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev 25: 2242–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Larivière L, Maier K, Seizl M, Tresch A, Cramer P. 2012. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res 22: 1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters B, Wachi S, Mavrich T, Andersen B, Jena P, Sinnamon A, Jain P, Rolleri N, Jiang C, Hemeryck-Walsh C, et al. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl H, Matarese F, Marks H, Denissov S, Butter F, Lee K, Olsen J, Hyman A, Stunnenberg H, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Vethantham V, Yang Y, Bowman C, Asp P, Lee JH, Skalnik DG, Dynlacht BD. 2012. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol Cell Biol 32: 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. 2000. Global histone acetylation and deacetylation in yeast. Nature 408: 495–498 [DOI] [PubMed] [Google Scholar]
- Weake V, Workman J. 2012. SAGA function in tissue-specific gene expression. Trends Cell Biol 22: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL. 2011. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev 25: 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P-YJ, Ruhlmann C, Winston F, Schultz P. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15: 199–208 [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 27: 275–288 [DOI] [PubMed] [Google Scholar]