The mammalian circadian clock is based on a transcription–translation feedback loop in which CLOCK and BMAL1 proteins act as transcriptional activators of Cryptochrome and Period genes, which encode proteins that repress CLOCK–BMAL1 with a periodicity of ∼ 24 h. Ye et al. show that CRY binds to CLOCK–BMAL1 at the promoter and inhibits CLOCK–BMAL1-dependent transcription without dissociating the complex. PER alone has no effect on CLOCK–BMAL1-activated transcription, but in the presence of CRY, nuclear entry of PER inhibits transcription by displacing CLOCK–BMAL1 from the promoter.
Keywords: circadian, Cryptochrome, Period, transcription, DNA binding
Abstract
The mammalian circadian clock is based on a transcription–translation feedback loop (TTFL) in which CLOCK and BMAL1 proteins act as transcriptional activators of Cryptochrome and Period genes, which encode proteins that repress CLOCK–BMAL1 with a periodicity of ∼24 h. In this model, the mechanistic roles of CRY and PER are unclear. Here, we used a controlled targeting system to introduce CRY1 or PER2 into the nuclei of mouse cells with defined circadian genotypes to characterize the functions of CRY and PER. Our data show that CRY is the primary repressor in the TTFL: It binds to CLOCK–BMAL1 at the promoter and inhibits CLOCK–BMAL1-dependent transcription without dissociating the complex (“blocking”-type repression). PER alone has no effect on CLOCK–BMAL1-activated transcription. However, in the presence of CRY, nuclear entry of PER inhibits transcription by displacing CLOCK–BMAL1 from the promoter (“displacement”-type repression). In light of these findings, we propose a new model for the mammalian circadian clock in which the negative arm of the TTFL proceeds by two different mechanisms during the circadian cycle.
The circadian rhythm is the cyclic change in biochemical, physiological, and behavioral functions of organisms with a periodicity of ∼24 h. In mammalian organisms, this cell-autonomous and self-sustained rhythm is generated by a transcription–translation feedback loop (TTFL) (Reppert and Weaver 2002; Hardin and Panda 2013; Partch et al. 2014). According to the commonly accepted model (“canonical model”), the core clock circuitry is composed of four genes/proteins and their paralogs (Clock [Npas2], Bmal1, Cry [Cry1 and Cry2], and Per [Per1 and Per2]), which generate rhythmicity in the following manner: CLOCK and BMAL1 transcriptional activators bind to E-box sequences in the promoters of Cry and Per genes and activate their transcription; CRY and PER proteins then accumulate in the cytoplasm and, after a time delay, enter the nucleus as a heterodimer and inhibit their own transcription (Kume et al. 1999; Vitaterna et al. 1999; Zheng et al. 1999, 2001; Shearman et al. 2000) as well as the transcription of other output genes controlled by CLOCK–BMAL1 (Fig. 1A; Hughes et al. 2009). The core clock circuitry is stabilized by secondary Ror/Rev-Erb (Nr1d1, Nr1d2) loops that are controlled by the core loop and in turn regulate the activation and repression of Bmal1 and Cry1 transcription, respectively (Preitner et al. 2002; Etchegaray et al. 2003; Liu et al. 2008; Ukai-Tadenuma et al. 2011; Bugge et al. 2012; Cho et al. 2012). Further robustness of the rhythm and stability of the clock are ensured by post-translational modifications and proteolysis of the clock proteins (Lee et al. 2001; Masri and Sassone-Corsi 2010; Partch et al. 2014).
Figure 1.
Experimental systems for analysis of the repressive phase of the mammalian clock TTFL. (A) Canonical model of the mammalian circadian clock. In this highly simplified model, only the core TTFL is shown: The CLOCK–BMAL1 heterodimer binds to the E-boxes in Per and Cry genes and activates their transcription. The CRY and PER proteins dimerize in the cytoplasm and, after a time lag, enter the nucleus, bind to CLOCK–BMAL1, and inhibit their own transcription. The cycle starts over after CRY and PER levels decrease by proteolysis. (B) Targeted nuclear delivery system for analysis of CRY1 and PER2 functions. CRY1–ER* or PER2–ER* are expressed in Cry1/2−/− or Per1/2−/− cells, respectively. The fusion proteins are retained in the cytoplasm in complex with heat-shock protein (HSP). Upon addition of 4-hydroxytamoxifen (4-OHT), HSP dissociates, and the fusion proteins enter the nucleus. (C,D) 4-OHT-induced nuclear entry of CRY1–ER* and PER2–ER* analyzed by immunofluorescence microscopy (C) and immunoblotting analysis of nuclear extracts (D). Red arrows point to nuclei. A nonspecific band (N.S.) and tubulin protein are shown for loading controls.
The canonical model is largely based on genetic data with mutant mice, reporter gene assays, and protein–protein interaction analysis (Kume et al. 1999; Vitaterna et al. 1999; Shearman et al. 2000; Bae et al. 2001; Zheng et al. 2001; Ishikawa et al. 2002; Tamai et al. 2007). Although the model has provided a framework for molecular clock research, the mechanism of action of the core clock proteins—in particular the roles of PERs and CRYs, which make up the negative arm of the loop—has remained ill-defined. A recent comprehensive biochemical study with the four core clock proteins and in vivo chromatin immunoprecipitation (ChIP) and transcription analyses revealed an unexpected fact that is inconsistent with the canonical model: CRY binds to the CLOCK–BMAL1–E-box complex in vitro and in vivo independent of PER and inhibits transcription (Ye et al. 2011). A subsequent genome-wide ChIP study (Koike et al. 2012) and a small molecule inhibitor/computational modeling study (St John et al. 2014) supported this finding; namely, that CRY is the dominant repressor in the TTFL.
In contrast to these findings that define a role of CRY in the feedback loop, the role of PER in repression has remained unclear. PER heterodimerizes with CRY, protects it from ubiquitylation and proteolysis (Czarna et al. 2013; Hirano et al. 2013; Xing et al. 2013; Yoo et al. 2013), and promotes nuclear entry of CRY, thus contributing to repression (Lee et al. 2001). However, its physical participation in the repressive complex has been controversial (Zheng et al. 1999; Miki et al. 2012). The biochemical study (Ye et al. 2011) revealed that PER (PER1 or PER2) does not bind to the CLOCK–BMAL1–E-box complex and, importantly, provided the first evidence for a ternary CLOCK–BMAL1–CRY complex in vitro and in vivo that is incompatible with PER binding (Partch et al. 2014). Most surprisingly, in vitro, it was found that PER causes the dissociation of CRY from the CLOCK–BMAL1–E-box complex (Ye et al. 2011), suggesting that it may actually interfere with E-box repression by CRY. However, genome-wide ChIP experiments have indicated that PER actively participates in repression by binding to E-box promoters in a multiprotein complex that includes PER and CRY in addition to other transcription factors that interact with PERs (Brown et al. 2005; Duong et al. 2011; Koike et al. 2012; Padmanabhan et al. 2012; Duong and Weitz 2014). Furthermore, it was reported that the associations of CRY and the PER multiprotein complex with E-boxes are temporally separated, and thus it was proposed that each mode of binding confers unique regulatory properties (Koike et al. 2012; Gustafson and Partch 2014; Partch et al. 2014).
To reconcile these seemingly conflicting findings and define more precisely the roles of CRYs and PERs in the core clock mechanism, we generated a mouse fibroblast cell line lacking CRYs and PERs and derivative lines that express CRY or PER that can be targeted to the nucleus in a controllable manner. Using this system, we were able to analyze the effects of CRY alone, PER alone, and CRY plus PER on the binding of CLOCK–BMAL1 to chromatin and on transcription of genes exclusively controlled by CLOCK–BMAL1. We found that CRY alone binds to CLOCK–BMAL1 on chromatin and inhibits the transcriptional activation without affecting the binding of CLOCK–BMAL1 to chromatin. In contrast, PER alone had no effect on the binding of CLOCK–BMAL1 to cognate promoters or on CLOCK–BMAL1-activated transcription. Unexpectedly, however, in cells expressing CRY, nuclear entry of PER resulted in removal of CLOCK–BMAL1 from chromatin and inhibition of CLOCK–BMAL1-mediated transcription. We propose a new model for the core mammalian molecular clock that incorporates these new findings and previously described properties of CRY and PER proteins.
Results
A system for controllable delivery of CRY and PER into the nucleus
Although our previous study demonstrated that CRY is the dominant repressor in the core circadian clock TTFL, the role of PER, other than stabilizing CRY and facilitating its nuclear entry, remained ill-defined. In particular, the fact that PER specifically interacts with CLOCK–BMAL1 off DNA (Ye et al. 2011) suggested the reasonable possibility that PER, in the absence of CRY, could function as a repressor/coregulator of CLOCK–BMAL1. To address this specific question, we used the estrogen receptor (ER)-mediated controllable nuclear entry system (Littlewood et al. 1995) to study the effects of CRY1 and PER2 on CLOCK–BMAL1 activity. In this system (Fig. 1B), the target protein is fused to a mutant form of the ER ligand-binding domain (ER*), which binds the estrogen agonist 4-hydroxytamoxifen (4-OHT) but not estrogen, and hence its subcellular localization can be controlled by 4-OHT addition to the medium.
We made CRY1–ER* and PER2–ER* constructs and stably expressed them in desired Cry- or Per-null mutant mouse skin fibroblasts. Figure 1C shows the subcellular distribution of the two proteins by immunofluorescence microscopy. Both proteins are largely cytoplasmic before 4-OHT treatment and become concentrated in the nucleus after addition of the drug. Immunoblotting data in Figure 1D support the immunofluorescence data by demonstrating the movement of these proteins into the nucleus by 4-OHT in as little as 1 h. Of note, although the recombinant proteins were expressed in amounts greatly exceeding their endogenous binding partners (data not shown), Cry1/2−/− cells still contain considerable amounts of PER2, and Per1/2−/− cells contain considerable amounts of CRY1 (Fig. 1D), and this fact must be taken into account in interpreting data regarding the effects of nuclear entry of CRY1–ER* and PER2–ER* on CLOCK–BMAL1-mediated gene expression.
Regulation of CLOCK–BMAL1 by controlled nuclear entry of CRY1 or PER2
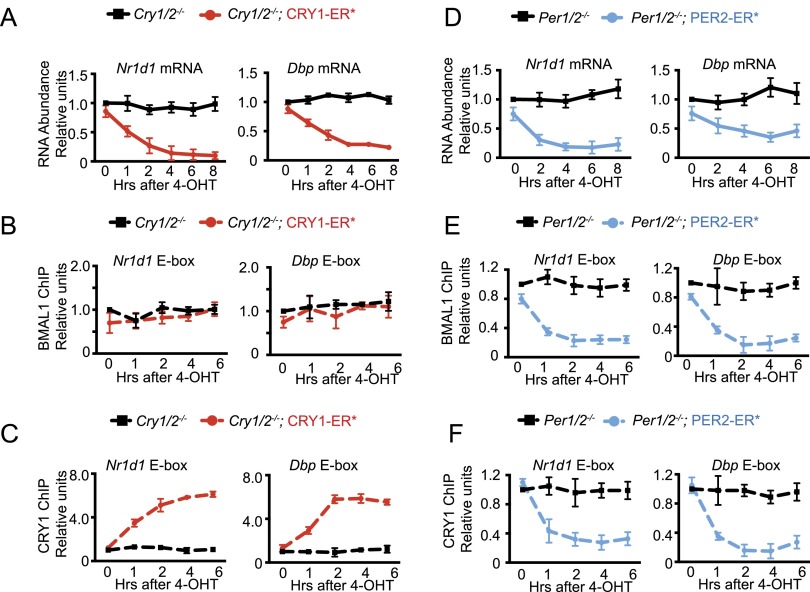
The Nr1d1 (Rev-Erbα) and Dbp genes are controlled by binding of CLOCK–BMAL1 to E-box cis-regulatory elements in their promoters with negligible contribution from other transcription factors (Rey et al. 2011). Therefore, we used these genes to define the roles of CRY and PER in E-box repression. We treated our Cry1/2−/−; CRY1–ER* and Per1/2−/−; PER2–ER* mouse skin fibroblasts with 4-OHT and monitored transcription of Nr1d1 and Dbp and binding of core clock proteins to their promoters. The results in Figure 2, A–F, show that although both CRY1 (Fig. 2A) and PER2 (Fig. 2D) repress the transcription of the sentinel genes, they appear to do so by different mechanisms: CRY1 binds to CLOCK–BMAL1 on DNA (Fig. 2C) and inhibits transcription (Fig. 2A) without affecting the binding of CLOCK–BMAL1 to chromatin, as determined by ChIP analysis (Fig. 2B; Supplemental Fig. S1A). Strikingly, in contrast to CRY1, even though PER2 inhibits the transcription of the target genes to the same extent as CRY1 (Fig. 2D), it appears to do so by dissociating CLOCK–BMAL1 and CRY1 from the promoters (Fig. 2E,F; Supplemental Fig. S1B) without changing their protein levels (Supplemental Fig. S1C). However, in this system, although the recombinant proteins were expressed in excess, both the Cry1/2−/−; CRY1–ER* and the Per1/2−/−; PER2–ER* cells also contain endogenous PER and CRY proteins, respectively (Fig. 1D). Thus, it was conceivable that the “repression” by CRY1 and PER2 in this system was actually mediated by the CRY–PER heterodimer, and the different modes of repression were due to the differential abundance of CRY1 and PER2 in the two cell lines. To test this model, it was necessary to analyze the effects of CRY and PER on CLOCK–BMAL1 activity in cells that contained either CRY alone or PER alone.
Figure 2.
Effect of CRY1 or PER2 nuclear entry on E-box binding and transcription in Cry1/2−/− or Per1/2−/− mutants. Nuclear entry of CRY1–ER* inhibits transcription (A) without disrupting the CLOCK–BMAL1–E-box complex (B) and is accompanied by increased binding of CRY1 to the E-box (C). Nuclear entry of PER2–ER* inhibits transcription (D) by displacing CLOCK–BMAL1 (E) and CRY1 (F) from the E-box.
Quadruple clock mutant cell line and specific functions of CRY and PER in the circadian TTFL
To define the functions of CRY and PER more precisely, we generated a mouse embryonic fibroblast (MEF) cell line lacking both CRY1/2 and both PER1/2 proteins. The cell line was generated from Per1/2−/− MEFs by using TALEN nucleases targeting the Cry1 and Cry2 genes. Figure 3A shows that in this quadruple mutant cell line (Cry1/2−/−; Per1/2−/−), there are no CRY1/2 or PER1/2 proteins, as determined by immunoblotting. To analyze the effects of CRY and PER proteins in isolation, the quadruple mutant cell line was stably transfected with either CRY1–ER* or PER2–ER*. As shown in Figure 3B, the fusion proteins have a mostly cytoplasmic location in untreated cells and enter the nucleus upon 4-OHT treatment, as expected.
Figure 3.
Effect of CRY1 or PER2 nuclear entry on E-box binding and transcription in the Cry1/2−/−; Per1/2-/ mutant. (A) Repressor protein expression in wild-type and mutant mouse cell lines analyzed by immunoblotting. (B) 4-OHT-induced nuclear entry of CRY1–ER* and PER2–ER* in the quadruple mutant cell line analyzed by immunofluorescence microscopy. (C–F) Effects of CRY1 or PER2 nuclear entry on CLOCK–BMAL1 function: CRY1–ER* nuclear entry inhibits transcription (C) without displacing CLOCK–BMAL1 from the promoter (D). In the absence of CRYs, PER2–ER* nuclear entry does not affect E-box transcription (E) or CLOCK–BMAL1 binding to the E-box (F). (EtOH) Ethanol.
Next, we used this system to test the effects of CRY1 and PER2 in isolation on CLOCK–BMAL1 binding to E-boxes and on CLOCK–BMAL1-activated transcription, and the results are shown in Figure 3, C–F. The effects of CRY1–ER* on both endpoints (Fig. 3C,D) are essentially identical to those obtained in the presence of PER2 (see Fig. 2A,B): CRY1–ER* inhibits CLOCK–BMAL1-activated transcription (Fig. 3C) without affecting the binding of CLOCK–BMAL1 to the cognate promoter (Fig. 3D). However, PER2, in stark contrast to the finding in Per1/2−/−; PER2–ER* cells (see Fig. 2D,E), is recruited to the E-box sites (Supplemental Fig. S2A) in the quadruple mutant (which lacks CRYs) but has no effect on either CLOCK–BMAL1 binding to target promoters (Fig. 3F) or the CLOCK–BMAL1-activated transcription from these promoters (Fig. 3E). When CRY1 protein was expressed in the Cry1/2−/−; Per1/2−/−; PER2-ER* cells, the CLOCK–BMAL1-releasing activity of PER2 was restored (Supplemental Fig. S2B,C). Taken together, the data in Figures 2 and 3 lead us to conclude that CRY is a bona fide repressor of CLOCK–BMAL1 and that it represses transcription by two mechanisms. First, by binding to CLOCK–BMAL1–E-box complexes, CRY blocks CLOCK–BMAL1/basal transcription machinery interactions independent of PERs (“blocking”-type repression). Second, when PER proteins are at sufficiently high concentration (and have potentially undergone appropriate post-translational modification), CRY promotes the dissociation of CLOCK–BMAL1 from cognate promoters together with PER (“displacement”-type repression).
The results obtained from cells expressing CRY1–ER* protein agree with the in vitro data, in which CRY1 binds stably to the CLOCK–BMAL1–E-box ternary complex in the absence of PER (Ye et al. 2011). However, dissociation of the transactivator complex from promoters by PER2 in the presence of CRY (Fig. 2E; Supplemental Fig. S1B) differs from the in vitro data, in which PER2 removes only CRY1 from the CRY1–CLOCK–BMAL1–E-box complex (Ye et al. 2011). It is possible that the in vivo system provides additional factors to allow PER2 to remove not only CRY but also the entire complex (Brown et al. 2005; Duong et al. 2011; Padmanabhan et al. 2012; Duong and Weitz 2014).
Mapping the PER2 domains required to disrupt the CRY–CLOCK–BMAL1–E-box complex
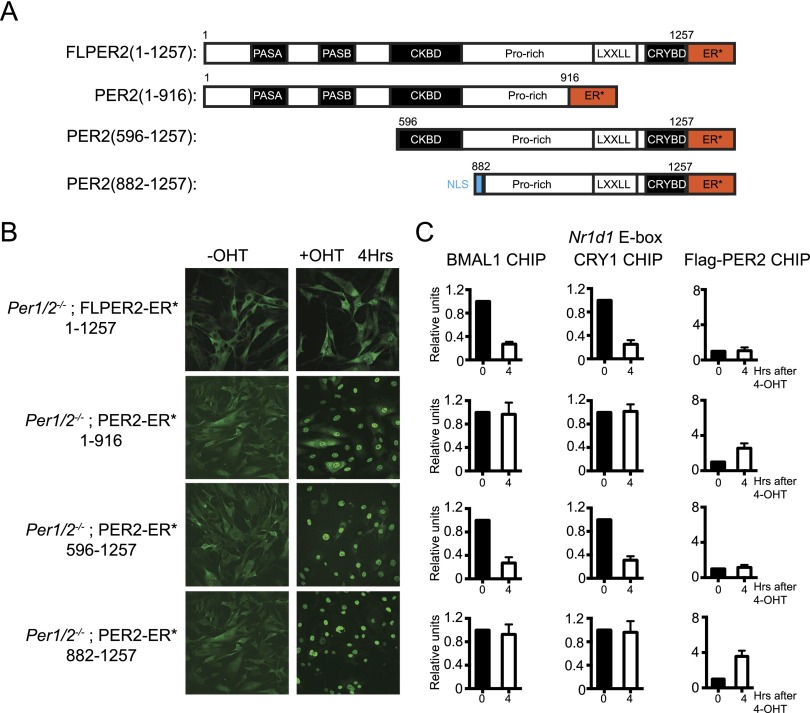
PER2 has distinct sequence/structural motifs that have been shown or predicted to play specific roles in PER function (Fig. 4A): PAS domains, which have been shown to interact with PAS domains of CLOCK and BMAL1 (Chen et al. 2009); a casein kinase-binding domain (CKBD), which interacts with casein kinases (Ck1δ and CK1ɛ) and also contains highly conserved sequences that may be important for protein–protein interaction (Vielhaber et al. 2000; Lee et al. 2004); a proline-rich region; and an LXXLL nuclear receptor interaction motif (Albrecht et al. 2007; Schmutz et al. 2010) followed by a 100-amino-acid stretch known to be the primary CRY-interacting domain (Eide et al. 2002; Yagita et al. 2002; Chen et al. 2009; Schmalen et al. 2014). In view of these aspects of PER2 structure, we wished to define the PER2 domains responsible for disrupting the binding of CRY1 and CLOCK–BMAL1 to E-boxes in vivo. To this end, we constructed three PER–ER* deletion mutants (Fig. 4A) and stably expressed them in the Per1/2−/− cells lines. As apparent in Figure 4B, 4-OHT induced nuclear accumulation of these PER2–ER* derivatives as it did the full-length (FLPER2) protein.
Figure 4.
Mapping of PER2 domains necessary for disrupting the CRY–CLOCK–BMAL1–promoter complex. (A) PER2-ER* constructs used in Per1/2−/− cells. The numbers associated with each mutant represent the amino acids of full-length PER (1257 amino acids) contained in each mutant protein. An SV40 nuclear localization signal is fused to the PER2(882–1257) to assure its nuclear translocation. (B) Nuclear entry of the fusion proteins following 4 h of treatment with 4-OHT analyzed by immunofluorescence. (C) Analyses of BMAL1, CRY1, and PER2 chromatin binding by ChIP. Full-length PER2 and PER2(596–1257) disrupt CRY1–CLOCK–BMAL1 binding to chromatin without measurable PER2 binding. Neither the N-terminal half [PER2(1–916)] nor the C-terminal half [PER2(882–1257)] of the protein have an effect on CRY1–CLOCK–BMAL1 binding to chromatin, but both do weakly associate with the promoter.
Next, we performed ChIP to measure binding of BMAL1, CRY1, and PER2 to the Nr1d1 E-box before and after 4-OHT-induced nuclear entry of the PER2 derivatives (Fig. 4C). The results show that, as expected, FLPER2(1–1257) disrupts binding of BMAL1 and CRY1 to the promoter. The same activity is seen with PER2(596–1257), which indicates that the PER2 PAS domains are not required for the PER2 corepressor activity. The PER2(1–916) and the PER2(882–1257) proteins do not dissociate the clock complex but, of special interest, are bound to the E-box, indicating that they both contain sites that interact stably with the CRY–CLOCK–BMAL1–E-box complex, although this interaction in the case of the full-length protein appears to be transient, resulting in complex dissociation.
From these results, the following conclusions can be made. First, although the PAS domains of PER2 were presumed to be responsible for recruitment of the PER–CRY complex to CLOCK–BMAL1 to initiate E-box repression (Chen et al. 2009), they are in fact dispensable for PER2 to dissociate the CLOCK–BMAL1–promoter complex. Second, deletion of the CRY-interacting domain eliminated the ability of PER to function because it can no longer bind CRY (Schmalen et al. 2014). Third, deletion of the CKBD from PER2 abolishes its repressive activity [cf. PER2(596–1257) vs. PER2(882–1257)], suggesting that kinases or other PER-interacting partners might be recruited to this region and required for PER2 to displace CLOCK–BMAL1 from chromatin.
Discussion
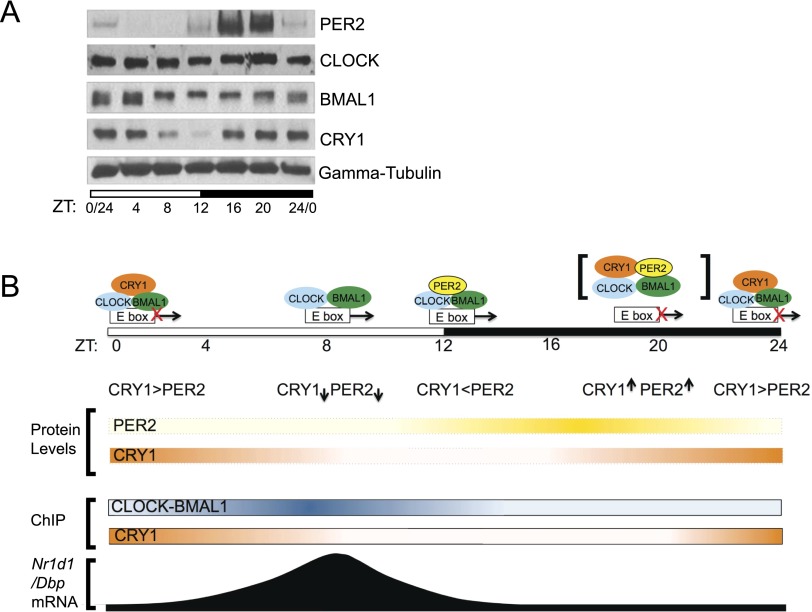
In addition to the information gathered from the reporter gene assay (Kume et al. 1999) and analysis of gene expression in clock mutant mice (Vitaterna et al. 1999; Zheng et al. 1999, 2001; Shearman et al. 2000), the nuclear delivery system that we used here provides novel insights into the repressive phase of the molecular clock (Fig. 5). As seen in the Western blot in Figure 5A, CRY1 and PER2 levels in mouse liver nuclei peak at different circadian times as controlled by both rhythmic transcription and ubiquitylation and proteolytic degradation (Lee et al. 2001; Ukai-Tadenuma et al. 2011). Taking into account this pattern of CRY1 and PER2 levels over a circadian cycle and the findings reported in this study as well as previously published rhythmic CLOCK, BMAL1, and CRY1 chromatin-binding data (Ripperger and Schibler 2006; Stratmann et al. 2010, 2012; Rey et al. 2011; Koike et al. 2012), we propose the following qualitative model for E-box repression over the course of the cycle in mouse livers (Fig. 5B): At the beginning of the circadian cycle (Zeitgeber time 0 [ZT0]), the CRY1 level is high, and CRY1 binds to the CLOCK–BMAL1–E-box complex and inhibits transcription (blocking-type repression). Over the course of the light phase, CRY1 is degraded, and CLOCK–BMAL1-mediated transcription is derepressed. In the evening, PER2 accumulates and may bind to the CLOCK–BMAL1–E-box, but the CRY1 level is low, and E-box transcription continues. At night, both CRY1 and PER2 levels are high, resulting in CRY- and PER-dependent dissociation of CLOCK–BMAL1 from the promoter and transcription repression (displacement-type repression; this phase of repression may involve a transient [PER2–CRY1–CLOCK–BMAL1–E-box] complex, but we were not able to detect PER2 at Nr1d1 or Dbp promoters by the standard ChIP method using mouse livers). Late at night and in the early morning hours, PER2 is degraded, but CRY1 continues to accumulate in the nucleus and maintains the repressive phase of the cycle (blocking-type repression). This sets the stage for the activator phase by allowing the formation of the CRY1–CLOCK–BMAL1–E-box complex, which is primed to transition to the activator phase upon proteolytic degradation of CRY1. We note that while the model presented here (Fig. 5B) refers to events occurring in the liver, the model likely applies to the SCN as well, in which the phase is advanced by ∼4 h (Lopez-Molina et al. 1997).
Figure 5.
Model for the primary TTFL of the mammalian clock in the mouse liver. (A) Expression patterns of the four core clock proteins in mouse liver nuclei over a circadian period revealing different peak expression times of CRY1 and PER2. (ZT) Zeitgeber time; (ZT0) light on; (ZT12) light off for mice under a 12-h light:12-h dark cycle. (B) New model for the mammalian circadian clock. The figure shows a semiquantitative heat map representation of CRY1 and PER2 protein expression as well as the ChIP data for CLOCK–BMAL1 and CRY1 over a circadian cycle and its consequences with regard to interactions of core clock proteins with the E-box and the effects of these interactions on transcription of genes (Nr1d1 and Dbp) regulated exclusively by the core TTFL.
We believe that this model clarifies some of the seemingly conflicting findings regarding the roles of CRYs and PERs in the repressive arm of the mammalian circadian clock. Thus, while CRY is an essential repressor in blocking-type repression, PER also plays multiple roles in the repression: First, it binds to CRY and prevents its ubiquitylation in the cytoplasm and nucleus by different E3 ligases (Hirano et al. 2013; Xing et al. 2013; Yoo et al. 2013). Second, it aids the nuclear import of CRY (Lee et al. 2001). Finally, when present at high concentration, it causes CRY-dependent displacement-type repression by removing CLOCK–BMAL1 from the E-box.
The model in Figure 5B is a rather idealized model for transcriptional control of the clock that is further refined into a smooth sinusoidal pattern by the contribution of CRY2 and PER1 proteins, which have slightly different expression kinetics than CRY1 and PER2, which are known to have a more dominant role in the negative arm of the circadian TTFL (Partch et al. 2014). We also note that the proposed repression and transcriptional expression pattern is for genes such as Dbp and Nr1d1, which are exclusively controlled by the core clock proteins. For other genes, such as Cry1, which is controlled by both the primary TTFL and the consolidating Ror/Rev-Erb loop (Preitner et al. 2002; Liu et al. 2008; Ukai-Tadenuma et al. 2011; Cho et al. 2012), the repression pattern will be different because of the kinetics of REVERB and ROR accumulation and differential contributions of the various transcriptional activators and repressors to the control of expression.
Finally, we wish to point out the remarkable similarities at the mechanistic level between the mammalian and Drosophila core clock repression mechanisms even though the role of CRY has highly diverged between the two systems. In Drosophila, CRY is the primary circadian photoreceptor (Stanewsky et al. 1998), and PER has subsumed the role of the mammalian CRY as the primary repressor and carries out inhibition by two mechanisms (Menet et al. 2010): At night (ZT16–ZT24), dPER is abundant and inhibits CLK–CYC by binding to the complex on circadian promoters (“on-DNA” repression). During the early light phase (ZT24–ZT6), it inhibits by sequestering CLK in a 1:1 PER–CLK complex (“off-DNA” repression). In mammals, as we show in this study, CRY does one type of repression (blocking type), and PER–CRY does the other (displacement type). However, in the displacement-type repression, in contrast to Drosophila, we did not detect a stoichiometric off-DNA complex of CLOCK–BMAL1–CRY–PER (data not shown), and the precise mechanism of this mode of repression remains to be determined by future investigations.
Materials and methods
Animals
Mice were handled in accordance with the guidelines of the National Institutes of Health, and procedures were approved by the Institute Animal Care and Use Committee of the University of North Carolina, School of Medicine. Per1/2−/− double-knockout mice were generated by crossing Per1−/− and Per2−/− mice (Bae et al. 2001) obtained from Jackson Laboratory. Wild-type mouse strain C57BL/6 was from Jackson Laboratory. All mice were maintained on an LD 12:12 schedule.
Plasmids
Plasmids used in this study can be obtained from Addgene with detailed sequence information. Flag-mCRY1-pBABEpuro and Flag-mPer2-pBABEpuro were made by inserting a PCR product containing mCRY1 or mPer2 into pBABEpuro. To generate Flag-mCry1ER-pBABEpuro and Flag-mPer2ER-pBABEpuro, the estrogen ligand-binding domain was amplified from HA-FOXO3a WT-ER (Tran et al. 2002) by PCR and inserted into Flag-mCRY1-pBABEpuro and Flag-mPer2-pBABEpuro. Deletion mutants of PER2–ER were similarly generated by PCR according to the two-step cloning strategies described above. To generate PER2(882–1257)–ER*pBABEpuro, a SV40 large T antigen nuclear localization sequence (DPKKKRKV) was added to the N terminus. Note that all of the ER* constructs described in this study contain a Flag tag at the N terminus.
Cells and antibodies
The Cry1/2−/− mouse skin fibroblasts have been described previously (Ye et al. 2011). The Per1/2−/− embryonic fibroblasts were prepared from Per1/2−/− double-knockout mice (Bae et al. 2001). Fibroblasts expressing mCRY1–ER* and mPER2–ER* were made by retrovirus infection. Briefly, retroviral constructs were cotransfected together with pVSVG and pCIHPZ plasmid DNAs into HEK293T to produce retrovirus particles. Next, fibroblasts were infected with the retrovirus, and transfectants were selected in medium containing 4 μg/mL puromycin for 2 wk. Single colonies were isolated and cultured for subsequent analysis.
Anti-mCRY1 (IgM-type monoclonal) antibodies were described previously (Ye et al. 2011). Anti-CLOCK (Bethyl Laboratories), anti-BMAL1 (Bethyl Laboratories), anti-PER2 (Alpha Diagnostic International), and anti-Flag (Sigma) antibodies were obtained from commercial sources.
Disruption of Cry1 and Cry2 with TALEN nuclease
TALENs designed to disrupt mCry1 and mCry2 were generated with the assistance of resources available at https://tale-nt.cac.cornell.edu (Cermak et al. 2011; Doyle et al. 2012). Components of the Golden Gate TALEN and TAL effector kit 2.0, obtained from Addgene, were used for assembly of TALENs in the pC-GoldyTALEN destination vector (Bedell et al. 2012; Carlson et al. 2012), which was obtained separately from Addgene.
Briefly, Per1/2−/− double-knockout MEFs were used to generate Cry1/2−/−, Per1/2−/− cells. To begin, MEFs at approximately passage 5 were cotransfected with the Cry2-directed TALEN pair in Supplemental Table S1 (C2X1Ahd TALENs 1 and 2); a Cry1-directed TALEN pair (data not shown); plasmid pBSSVD2005 (Addgene plasmid 21826), which permits cells to become immortalized; and a GFP-expressing construct (pEGFP). One clone, called “M32,” had wild-type Cry1 alleles, and both Cry2 alleles had a single base-pair deletion. The deleted base pair is number 56 of the coding sequence (an A) and causes codons 19–39 to be out of frame and codon 40 to be a stop codon.
Next, the M32 knockout cell line (Cry1+/+; Cry2−/−; Per1/2−/−) was cotransfected with the Cry1 TALEN pair directed toward exon 1 of the Cry1 gene in Supplemental Table S1 (C1X1 TALENs 1 and 2) and pBABEpuro plasmid DNA. Transfected cells were selected with puromycin for 3 d. Numerous clones were isolated, and one of the mutants, “M32P17,” was used as Cry1/2−/−; Per1/2−/− cells in this study. M32P17 has two mutations (a 4-base-pair [bp] deletion and a 5-bp deletion in exon 1) that knock out the Cry1 gene and resulted from the action of the C1X1 TALEN pair. The 4-bp deletion is from nucleotide 64 to nucleotide 67 (AAGG) of the coding sequence, makes codons 22–67 out of frame, and makes codon 68 a stop codon. The 5-bp deletion is from nucleotide 64 to nucleotide 68 (AAGGA), makes codons 22–57 out of frame, and makes codon 58 a stop codon.
All clones were isolated and screened by Western blot with anti-CRY1 or anti-CRY2 antibodies. DNA from target regions of clones lacking CRY expression as determined by Western blot was amplified by PCR and subcloned, and subclones were sequenced.
Fluorescence microscopy
For Flag-CRY1–ER* and Flag-PER2–ER* immunofluorescence staining, fibroblasts were cultured on poly-D-lysine Cellware 12-mm round coverslips (BD Bioscience) placed in 35-mm dishes. After treatment with 4-OHT, cells were fixed with 2% formaldehyde in PBS, permeabilized by PBT (0.1% Triton X-100 in PBS), blocked with antibody-blocking buffer (5% BSA in PBT) for 1 h, and incubated with anti-Flag antibody for 45 min. After washing three times with PBT, the cells were incubated with Alexa Fluor 488 (goat anti-mouse IgG; Invitrogen) secondary antibody for 30 min. Next, the coverslip was washed three times and mounted in Prolong Gold anti-fade reagent. Images were captured using a LeicaSP2 confocal microscope.
ChIP
Formaldehyde was added to adherent cell cultures to a final concentration of 1% (v/v). After 10 min of mixing at room temperature, glycine was added to a final concentration of 0.1 M and incubated for another 10 min. Cells were washed and collected in cold PBS. Collected cells were lysed on ice in 1 mL of ice-cold cell lysis buffer I (50 mM Tris-HCl at pH 8.0, 85 mM KCl, 0.5% NP40) containing protease inhibitor cocktail (Roche) for 10 min. Nuclei were recovered by centrifugation and resuspended in 0.8 mL of RIPA ChIP lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, protease inhibitor cocktail) and incubated for 10 min. Lysates were sonicated 24 rounds for 10 sec using a Misonix Sonicator 3000 with microtip on ice water and then centrifuged at 14,000 rpm for 10 min to pellet debris. Chromatin samples were precleared for 1 h at 4°C with 20 μL of protein A beads (Millipore) followed by incubation with antibody overnight at 4°C. Immune complexes were recovered for 40 min at 4°C using 30 μL of protein A/ssDNA agarose beads. Complexes were washed twice with RIPA buffer, three times with LiCl wash buffer (100 mM Tris-HCl at pH 8, 500 mM LiCl, 1% NP40, 1% NaDoc, protease inhibitor cocktail), and twice with TE buffer. Immunoprecipitation reactions and input chromatin were eluted by elution buffer (1% SDS, 0.1 M NaHCO3) and digested with proteinase K. Cross-links were reversed overnight at 65°C in elution buffer plus 150 mM NaCl. DNA was purified by Qiagen PCR purification kit and subjected to PCR.
Quantitative real-time PCR and data analysis
Real-time PCR assays were performed by using an ABI 7300 system (Applied Biosystems) and iTaq SYBR Green/ROX qPCR Master Mix (Bio-Rad). Quantitation of ChIP was performed with the primer sets described previously (Annayev et al. 2014). Each sample was analyzed in triplicate. To compare ChIP data from different chromatin samples, signals obtained from each chromatin sample were first divided by signals obtained from the corresponding input sample. Next, results were normalized as follows: In Figure 2, B and C, results obtained with 0h of 4-OHT treatment of Cry1/2−/− cells were given a value of 1. In Figure 2, E and F, results with Per1/2−/− cells after 0 h of 4-OHT were given a value of 1. In Figure 3, D and F, results with 0 h of EtOH were given a value of 1.
Quantitation of Nr1d1 and Dbp mRNA levels was performed with the primer sets as described (Annayev et al. 2014). Each sample was analyzed in triplicate, and cycle thresholds of individual genes were first normalized to corresponding Gapdh mRNA expression values obtained with the primer set. Next, data sets were normalized in the same way as ChIP data sets, giving a value of 1 to 0 h of 4-OHT treatment of Cry1/2−/− cells in Figure 2A, 0 h of 4-OHT treatment of Per1/2−/− cells in Figure 2D, and 0 h of treatment with EtOH in Figure 3, C and E.
Acknowledgments
We thank Dr. Carrie L. Partch (University of California at Santa Cruz) for useful comments on the manuscript. This work was supported by National Institutes of Health grant GM31082 and funding to A.S. from the Science Research Council and Academia Sinica, Taiwan, China.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.249417.114.
References
- Albrecht U, Bordon A, Schmutz I, Ripperger J. 2007. The multiple facets of Per2. Cold Spring Harb Symp Quant Biol 72: 95–104 [DOI] [PubMed] [Google Scholar]
- Annayev Y, Adar S, Chiou YY, Lieb JD, Sancar A, Ye R. 2014. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J Biol Chem 289: 5013–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536 [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. 2012. In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308: 693–696 [DOI] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. 2012. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. 2012. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109: 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. 2009. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell 36: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarna A, Berndt A, Singh HR, Grudziecki A, Ladurner AG, Timinszky G, Kramer A, Wolf E. 2013. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153: 1394–1405 [DOI] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. 2012. TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40: W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HA, Weitz CJ. 2014. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 21: 126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, Weitz CJ. 2011. A molecular mechanism for circadian clock negative feedback. Science 332: 1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. 2002. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iɛ. J Biol Chem 277: 17248–17254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–182 [DOI] [PubMed] [Google Scholar]
- Gustafson CL, Partch CL. 2014. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Panda S. 2013. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol 23: 724–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152: 1106–1118 [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Hirayama J, Kobayashi Y, Todo T.. 2002. Zebrafish CRY represses transcription mediated by CLOCK–BMAL heterodimer without inhibiting its binding to DNA. Genes Cells 7: 1073–1086 [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855–867 [DOI] [PubMed] [Google Scholar]
- Lee C, Weaver DR, Reppert SM. 2004. Direct association between mouse PERIOD and CKIɛ is critical for a functioning circadian clock. Mol Cell Biol 24: 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23: 1686–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. 2008. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. 1997. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J 16: 6762–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Sassone-Corsi P. 2010. Plasticity and specificity of the circadian epigenome. Nat Neurosci 13: 1324–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. 2010. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev 24: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Xu Z, Chen-Goodspeed M, Liu M, Van Oort-Jansen A, Rea MA, Zhao Z, Lee CC, Chang KS. 2012. PML regulates PER2 nuclear localization and circadian function. EMBO J 31: 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ. 2012. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337: 599–602 [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. 2002. Coordination of circadian timing in mammals. Nature 418: 935–941 [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. 2011. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. 2006. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38: 369–374 [DOI] [PubMed] [Google Scholar]
- Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, Benda C, Kramer A, Wolf E. 2014. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157: 1203–1215 [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. 2010. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24: 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692 [DOI] [PubMed] [Google Scholar]
- St John PC, Hirota T, Kay SA, Doyle FJ 3rd. 2014. Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc Natl Acad Sci 111: 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Stadler F, Tamanini F, van der Horst GT, Ripperger JA. 2010. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev 24: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Suter DM, Molina N, Naef F, Schibler U. 2012. Circadian Dbp transcription relies on highly dynamic BMAL1–CLOCK interaction with E boxes and requires the proteasome. Mol Cell 48: 277–287 [DOI] [PubMed] [Google Scholar]
- Tamai TK, Young LC, Whitmore D. 2007. Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc Natl Acad Sci 104: 14712–14717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr, DiStefano PS, Chiang LW, Greenberg ME. 2002. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296: 530–534 [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. 2011. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144: 268–281 [DOI] [PubMed] [Google Scholar]
- Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. 2000. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase Iɛ. Mol Cell Biol 20: 4888–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. 1999. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci 96: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, Zheng N. 2013. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496: 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J 21: 1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. 2011. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem 286: 25891–25902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. 1999. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400: 169–173 [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–694 [DOI] [PubMed] [Google Scholar]