Abstract
Efficient expression of transgenes in vivo is of critical importance in studying gene function and developing treatments for diseases. Over the past years, hydrodynamic gene delivery (HGD) has emerged as a simple, fast, safe and effective method for delivering transgenes into rodents. This technique relies on the force generated by the rapid injection of a large volume of physiological solution to increase the permeability of cell membranes of perfused organs and thus deliver DNA into cells. One of the main advantages of HGD is the ability to introduce transgenes into mammalian cells using naked plasmid DNA (pDNA). Introducing an exogenous gene using a plasmid is minimally laborious, highly efficient and, contrary to viral carriers, remarkably safe. HGD was initially used to deliver genes into mice, it is now used to deliver a wide range of substances, including oligonucleotides, artificial chromosomes, RNA, proteins and small molecules into mice, rats and, to a limited degree, other animals. This protocol describes HGD in mice and focuses on three key aspects of the method that are critical to performing the procedure successfully: correct insertion of the needle into the vein, the volume of injection and the speed of delivery. Examples are given to show the application of this method to the transient expression of two genes that encode secreted, primate-specific proteins, apolipoprotein L-I (APOL-I) and haptoglobin-related protein (HPR).
Keywords: Genetics, Issue 87, hydrodynamic gene delivery, hydrodynamics-based transfection, mouse, gene therapy, plasmid DNA, transient gene expression, tail vein injection
Introduction
Since its first description by Liu et al. and Zhang et al., hydrodynamic gene delivery (HGD) has become an invaluable tool for studying gene function in rodent model systems1,2. The technique involves the rapid injection (5-7 sec) of a large volume (8-12% body weight) of solution into the tail vein of mice to facilitate the uptake of plasmid DNA by the cells of target organs1,2. These conditions lead to robust gene expression in the liver and less gene expression in the kidney, spleen, lung and heart.
Injection of 10 μg pCMV-LacZ plasmid can transfect as much as 40% of hepatocytes, making HGD the most efficient, non-viral, in vivo gene delivery method to date1. Unlike viral carriers, pDNA is easy to prepare, does not elicit an immune response in the rodent host3 and does not pose a health risk by recombining with endogenous viruses. In addition, since the DNA molecules delivered by HGD do not need packaging, this method is suitable for the delivery of bacterial artificial chromosomes (BAC) as large as 150 kb4. Other types of molecules that have been delivered by a hydrodynamic method include RNA5-10, morpholinos11, proteins12,13 and other small molecules12,14. The advantages and disadvantages of HGD over other delivery methods have been discussed in excellent reviews in the literature15-20 and a number of authors have provided a detailed description of the procedure21-23.
Introducing transgenes into mice by HGD is safe and effective1-3,24 and the method has been used in rats with comparable success25. With certain modifications, proof-of-concept experiments have been carried out in chickens26, rabbits27 and pigs28, although, the in vivo application of this technique in larger animals remains a challenge. When using this method, another common limitation is that many of the available mammalian expression vectors lack the components to achieve a persistent, high level of gene expression. Using a pCMV-Luc plasmid, gene expression in the target organs is evident as early as ten minutes after HGD, however, the initial, high expression level drops sharply in the first week after injection1. Long-term transgene expression is possible depending on the promoter and intron used in plasmid design3,24 however, maintenance of high-level gene expression often requires repeated injections. For this reason, HGD might be less suitable to study chronic diseases that are a result of long-term exposure to damaging proteins or protein products. With these limitations, HGD is an exceptionally powerful tool for studying the potential role of a gene and the effect of it mutants in vivo, as well as the therapeutic effects and regulation of proteins and for establishing animal models of disease (for review, see15). For example, HGD may be used to assign function to domains and amino acids of proteins by individually introducing various gene constructs into mice that have the respective genes knocked out. Furthermore, this technique may be used in any mouse strain.
This protocol describes HGD in mice with a focus on the technical aspects necessary to achieve successful transfection: correct needle insertion into the vein, injection volume and speed of delivery. The application of this method is demonstrated in a mouse model of African trypanosomiasis, a fatal disease of humans and livestock29,30. While several species of trypanosomes cause disease in livestock, most cannot cause disease in humans due to innate immune complexes in blood called trypanosome lytic factors (TLFs)29,31,32. These pore-forming, high-density lipoproteins (HDL) contain two unique, primate-specific proteins: HPR, the ligand, which facilitates the uptake of TLFs into trypanosomes, and APOL-I, the lytic component31,33-38. Trypanosoma brucei rhodesiense is able to infect humans due to expression of a serum resistance-associated protein (SRA) that binds to and neutralizes human APOL-I34,39. Baboon TLF is not neutralized by SRA due to its divergent APOL-I protein40. As reported previously, using a mammalian expression vector (pRG977), transgenic expression of baboon TLF components in mice confers protection against human-infective trypanosomes40. The representative data presented here demonstrate how hydrodynamic gene delivery may be applied to study the therapeutic effects of a protein.
Protocol
All experiments described here were approved by the Institutional Animal Care and Use Committee of Hunter College, City University of New York.
1. Preparation of endotoxin-free plasmid DNA
Pick a single colony of bacteria containing the gene of interest in a mammalian expression vector from a freshly streaked selective plate.
Follow the recommendations found in the handbook of a commercially available endotoxin-free plasmid purification kit for growing and harvesting bacteria.
Purify the endotoxin-free plasmid DNA from bacterial cells by following the protocol of a commercially available endotoxin-free plasmid purification kit.
Use endotoxin-free plastic ware and handle DNA with care to ensure that endotoxin is not re-introduced into the DNA sample after the removal step.
- Measure the concentration of endotoxin-free plasmid DNA by determining its absorbance at 260 nm using a micro-volume UV spectrophotometer as described below or a standard, cuvette-based spectrophotometer.
- Clean the lower and upper optical surfaces of the micro spectrophotometer sample retention system as follows. Pipette 1 μl of clean deionized water onto the lower optical system. Close the lever arm and tap it a few times to bathe the upper optical system, then open lever and wipe clean with a tissue.
- Open the software of the micro spectrophotometer and select the nucleic acids module.
- Place 1 μl clean deionized water on the lower optical system, lower the lever arm and select “initialize” from the program software. Once initialization is complete, clean both optical surfaces with a tissue..
- Perform blank measurement by loading 1 μl of endotoxin-free TE buffer (10 mM Tris-Cl, pH 8.0; 1 mM EDTA) and selecting “blank” from the program software. Once blank measurement is done, clean both optical surfaces with a tissue.
- Perform sample measurement by loading 1 μl of endotoxin-free DNA sample and selecting “measure” from the program software. Record the DNA concentration. Assess DNA purity by recording the absorbance ratio 260/280 nm that appears on the screen. Since the use of pure DNA (260/280 ratio of 1.8) for HGD is highly desirable, avoid using a DNA sample with a 260/280 ratio below 1.7. Once measurement is done, clean both optical surfaces with a tissue.
2. Weighing of mice
- Mark mice in a manner appropriate for the strain. Note: marking of the tail is not recommended for this procedure.
- Mark mouse species that have white fur (e.g. Swiss Webster) on their backs with a black marker. Reapply marks over time as necessary.
- Mark mouse species that have black fur (e.g. C57BL/6) by ear punching several days in advance.
Weigh mice individually by gently placing them in an animal weighing pan or bucket placed on a digital laboratory balance. Record the weight of each mouse used in the experiment, on the day of the experiment.
3. Preparation of syringes
- Preparation of the injection mix
- Calculate the amount of DNA needed by assuming 5 - 50 μg endotoxin-free plasmid DNA for the injection of each mouse.
- Determine the optimal amount of plasmid DNA experimentally.
- When determining the amount of DNA required, assume injection of one additional mouse per group, as proper loading of the syringes will require some extra injection mix. To help calculations, consider the following example: inject 4 experimental and 4 control mice, each weighing 25 g, with 50 μg plasmid DNA per mouse. To determine the amount of DNA needed in this case, calculate with 5 mice per group: 5 x 50 μg = 250 μg DNA needed for each group.
- Determine the amount of saline (0.9% sodium chloride) needed by assuming 10% of body weight for the injection of each mouse.
- According to the above example, inject each 25 g mouse with 50 μg of plasmid DNA in 2.5 ml saline (2.5 g liquid).
- When determining the amount of saline required for an entire group of mice, assume injection of an additional mouse per group to allow for proper loading of the syringes. Considering the experiment given, calculate the amount of saline needed for 5 mice in each group: 5 x 2.5 ml =12.5 ml saline for each group (or 25 ml total). Note: If mice within the same group are not the same weight (more than 10% difference in body weight), prepare injection mixes separately.
- Use preservative- and endotoxin-free, sterile saline that has been approved for human use, in 10 ml, multiple use vials.
- Using a 20-gauge needle (0.9 mm x 25 mm) attached to a luer-lock tip of a 20 ml syringe, withdraw the liquid from saline vials and empty the syringes into a 50 ml conical tube. To help accurate pipetting of saline during the preparation of the DNA-saline injection mixes, collect more saline than needed for the injection of all the mice in the experiment. For the experiment above, withdraw saline from 3, 10 ml vials (a total of approximately 30 ml) even if the calculated amount of saline needed for the experiment is only 25 ml.
- Pipette the calculated amount of DNA into a different 50 ml conical tube. Take care not to touch the side of the tube with the pipette body to avoid introduction of endotoxin. According to the example, pipette 250 μg of control and 250 μg of experimental plasmid DNA into separate conical tubes.
- Add the required amount of saline to the DNA using a serological pipette. Considering the sample experiment, pipette 12.5 ml saline to each of the master mixes (experimental and control).
- Allow all solutions to reach room temperature prior to injection.
- Preparation of syringes
- For each mouse, fill a sterile, 3 ml, luer-lock syringe with the DNA-saline mix using a sterile, 20-gauge needle (0.9 mm x 25 mm). Take care to avoid air bubbles. Avoid using higher volume syringes as the flow rate of injection cannot be controlled very well, and it is difficult to manipulate larger syringes in one hand. Eject any excess DNA-saline mix back into the conical tube as this may be re-used.
- Switch the needle to a sterile, 27-gauge needle. Fill the needle with liquid completely without introducing air bubbles. Adjust the volume of the DNA-saline mix as calculated based on the weight of the mouse.
- Do not allow the uncapped needle to touch any non-sterile surface. If necessary, needles may be re-capped using the one-handed technique: place the cap on a flat surface, insert the needle without holding onto the cap with the other hand and press the capped needle against a firm object to secure the cap onto the needle. The syringes are now ready for tail-vein injections.
4. Hydrodynamic DNA delivery
Note that anesthetizing mice may reduce viability. Avoid performing HGD in a room that is too cold, as this will also reduce viability. If the room has an ambient temperature of 20 °C or if using anesthesia, place the mice on a heating pad set at 37 °C post procedure to avoid loss of any animals. If using anesthesia, ensure that the mouth and snout of the mouse are unobstructed while on the heat pad and observe the mouse until it fully recovers. Do not withhold food or water from the mice prior to HGD.
- Warm the mice to dilate blood vessels.
- Place the mouse cage (with bedding included) on a heat pad for 5 min so that the temperature of the bedding is approximately 38-39 °C. The temperature should be low enough to keep the mice on the heat pad for an extended period.
- Alternatively, place mouse into a dorsal access restrainer with minimal restraint. Warm the tail of the mouse for 1 min by gently holding it with a piece of gauze wetted in warm water. Allow mouse to re-position, if necessary. Wipe the tail with a tissue to dry.
- Alternatively, warm mouse by placing the cage under a heat lamp for no more than 2 min. If using a heat lamp, exercise caution to avoid overheating of the mouse.
- Large-volume tail vein injection
- Immobilize a dorsal access restrainer on a flat surface by laboratory tape.
- Holding by the tail, place the mouse gently into the restrainer and insert the plug. Use as little restraint as necessary to keep the tail immobilized. Ensure that the mouse is breathing freely.
- If the mouse is in severe distress at any point during the procedure, remove the mouse from the restrainer immediately and allow it to rest.
- Position the mouse so one of the lateral caudal veins is visible. Locate the lateral caudal veins by looking straight down at the back of the mouse: the red line running in the middle of the tail is an artery, while the blue lines on either side of the tail are the lateral caudal veins.
- Wipe the tail of the mouse thoroughly with an alcohol swab.
- Using the non-injecting hand, hold the tail tightly between the index and middle fingers so that the tail passes under the index finger, over the middle and third fingers and under the little finger. Allow the thumb to remain free to move. (Figure 1A)
- Using the other hand, wipe the area to be injected again with an alcohol swab. Allow area to dry.
- Pick up the loaded syringe with the injecting-hand and hold it by the barrel between the thumb and the other four digits. Do not hold onto the plunger.
- Position the syringe parallel to the tail with the needle pointing toward the body of the mouse and the bevel (slanting edge) of the needle facing up.
- Insert the needle into the tail vein. Proceed with the injection only if the vein is correctly located, which is evident from the needle sliding in with no resistance. Note that the depth of needle insertion is a matter of personal preference however, full insertion is not recommended due to an increased risk of shearing of the vein. Avoid moving the needle.
- Using the thumb of the tail-holding hand, close down on the hub of the needle and press hub against the tail to hold needle in position. Do not apply extreme pressure and do not hold onto needle shaft, as these will impede the flow of liquid into the vein. Hold the tail with the index finger loosely at this point so as not impede injection (Figure 1A).
- Re-position the syringe-holding hand so that the index finger and middle finger are holding onto the flange and the thumb is on the end of the plunger (Figure 1A).
- Press down on plunger in one, continuous motion and inject the full volume of liquid in 6-8 sec.
- Remove needle from vein and stop the bleeding by applying gentle pressure to the tail with a tissue.
- Remove mouse from the restrainer and place mouse into a recovery cage placed on top of a heating pad (bedding should be 37-38 °C). If the injection room is cold, this step is critical for mouse survival. Hold onto the tail of the mouse until bleeding stops completely.
- Observe the mouse for 1 hr following hydrodynamic DNA delivery. An initial period of panting and immobility is normal due to the temporary arrhythmia caused by HGD but make sure that the mouse shows signs of recovery in approximately 5 min. If the breathing of the mouse becomes exceedingly shallow, gently massage the abdomen of the mouse to facilitate breathing. Note that rate of recovery may be slightly influenced by the mouse strain used.
- Return mouse to housing cage and ensure that mouse has an abundant supply of food and water.
- Large-volume tail vein injection using an alternative hand position
- Place mouse into the restrainer and prepare for injection by following steps 4.3.1 through 4.3.5, as described previously.
- Rest the non-injecting hand on a flat surface with four fingers bent at a ninety-degree angle. The fingers (all except thumb) should rest on top of each other, creating a platform. Hold the tail of the mouse between the thumb and the pointing finger (Figure 1B).
- Using the other hand, wipe the area to be injected again with an alcohol swab. Allow area to dry.
- Grab syringe with the injecting-hand and hold it by the flange and the end of the plunger, ready for injection.
- Complete the procedure by following this protocol from step 4.3.9 through step 4.3.17, as described previously.
Representative Results
Correct positioning of the needle into the tail vein of the mouse (as illustrated in Figure 1) is a prerequisite for successfully delivering a transgene by hydrodynamics-based transfection. Often the most challenging part of the technique, however, is the retention of the needle within the tail vein without movement so that the entire volume of the injection can be delivered within a 6-8 s period. Minor errors during the injection process may result in vastly reduced transfection efficiency and protein expression. Therefore, it is imperative to monitor the success of the HGD for each experiment. If the transgenic protein of interest is difficult to detect by western blot due to the lack of sensitive antibodies, as in the case of baboon APOL-I, injection efficiency may be monitored by co-injection of a plasmid carrying a gene for a protein that is easily detectable. In the experiment shown in Figure 2, mice were injected with 50 μg of a plasmid carrying one of three differentially 6XHIS tagged baboon APOL-I genes and 50 μg of another plasmid (same expression vector design) that was carrying baboon HPR (bHPR, untagged). The principal goal of this experiment was to assess whether the addition of a 6XHIS tag at the chosen sequence locations interferes with correct processing and secretion of the baboon APOL-I protein. If however, the addition of the tag disrupts the correct folding and, consequently, secretion of the protein, it becomes difficult to determine whether loss of protein expression occurred because of impaired processing or unsuccessful HGD. Furthermore, due to the lack of sufficiently sensitive antibodies, direct detection of the baboon APOL-I in transfected mouse plasma is difficult even in case of a successful gene delivery (as determined by the protection of mice against a human-infectious trypanosome challenge). In this experiment, these issues were addressed by the addition of an equivalent amount of baboon HPR plasmid to the APOL-I HGD mixture to serve as a surrogate injection control. As demonstrated in the figure, baboon HPR was highly expressed in the plasma of almost all of the transfected mice indicating that the HGD was successful. Minor drops in expression levels, as seen in one of the two mice in bAPOL-I 6XHIS group 1 In Figure 2A, are generally attributable to a very small reduction (1-2 s) in injection speed. In Figure 2B, the first plasma sample lane 1 (in bAPOL-I 6XHIS group 3) shows a complete lack of protein expression indicating a failed HGD. In most cases, this is due to incomplete injection of the full volume of the delivery vehicle. Since this experiment confirms the success of HGD in all cases but one, the effect of 6XHIS tagging on bAPOL-I secretion can now be evaluated by an anti-6XHIS immunoblot (Figure 2C and D). As seen in Figure 2D, the only group of mice that had detectable levels of 6xHIS-tagged protein in their plasma was group 3. The protein expression pattern within this group confirmed that injection of one of the mice in this group was unsuccessful. The complete lack of detectable 6xHIS proteins in groups 1 and 2 (Figure 2C) underlines the importance of including a tracer protein in the injection mix to monitor the success of the HGD. Without considering the plasma levels of bHPR (Figure 2A and B), data from Figure 2C and D could easily be misinterpreted as a failure of HGD in groups 1 and 2.
Successful HGD of the baboon TLF1 components bAPOL-I and bHPR, gives mice resistance to human-infectious Trypanosoma brucei brucei-SRA parasites (T.b.b.-SRA, a laboratory equivalent of T. b. rhodesiense)40. This resistance is due to the transgenic expression of bAPOL-I, as injection of bHPR alone provides no protection against human-infectious parasites. To evaluate whether 6XHIS-tagged variants of bAPOL-I are still able to function as lytic proteins, mice that received both bAPOL-I and bHPR by HGD were challenged with 5000 T.b.b-SRA parasites two days after gene delivery. As seen in Figure 3, mice in 6XHIS group 2 succumbed to infection very early, whereas most mice in groups 1 and 3 survived as long as the untagged bAPOL-I positive control. (Baboon APOL-I confers partial protection in this mouse strain.) Since transgene expression within group 2 was universally high, the lack of protection in these mice may be correctly interpreted as loss of bAPOL-I lytic function due to the 6XHIS tag at that position. Care must be taken, however, that the premature (day 7) death of a mouse within group 3 is not misinterpreted, as this mouse was not successfully injected with the transgenes (see Figure 2B, lane 1). In light of combined data from Figures 2 and 3, we conclude that bAPOL-I may be tagged in position 3 (in 6xHIS group 3) without adverse effects, while tagging of this protein in position 2 (in 6xHIS group 2) disrupts its lytic function. In 6xHIS group 1, while the data are suggestive of protection the number of mice was insufficient to obtain significant data. Power analyses should be performed to determine number of mice required for each experiment. While the apparent lack of detectable protein in mouse plasma from group 1 may seem contradictory to the protection pattern seen in Figure 3, note that a negative anti 6xHIS western blot in this group may be the result of a lack of detection due to protein processing rather than a lack of gene expression. In group 1, the 6xHIS tag was added close after the cleavage site of the predicted signal peptide. The baboon APOL-I amino acid sequence contains an additional predicted proteolytic cleavage site approximately 20 amino acids from the cleavage site of the signal peptide40. Therefore, it is clear that bAPOL-I in these mice is secreted and functional yet lost the 6xHIS tag necessary for its detection. Thus illustrating the importance of HIS tag placement.
APOL-I kills African trypanosomes by pore formation in the parasite lysosome membrane36,37. Natural variants of human APOL-I, found in people with recent African ancestry, have been strongly associated with kidney disease in African Americans, although, the mechanism of kidney damage is yet unclear41,42. Since HGD results in highest gene expression in the liver, we wanted to investigate if human APOL-I or its synthetic sequence variant causes damage in this organ. To this end, mice were injected with 50 μg of WT human APOL-I or a single amino acid synthetic mutant designated as K4.To assess liver damage, we measured aspartate transaminase (AST) levels in mouse plasma collected on day 2 post-injection. Consistent with its lytic function, WT human APOL-I causes an elevation of serum AST levels (Figure 4A) as compared to saline-injected mice. In contrast, injection of mutant K4, which differs in only one amino acid, caused very little AST release. Differences in liver damage are unlikely to be attributable to levels of protein expression, as K4 protein levels are equivalent or higher than those of the WT human APOL-I (Figure 4B). Examination of livers from HGD mice collected 5 days post-injection shows that human WT APOL-I causes limited tissue necrosis with moderate macrophage infiltration (Figure 5B) as compared to saline-injected mice that show normal liver histology (Figure 5A, purple areas). Confirming the data obtained by measuring plasma AST levels, K4 injected in the same plasmid background as the WT shows normal liver histology (Figure 5C).
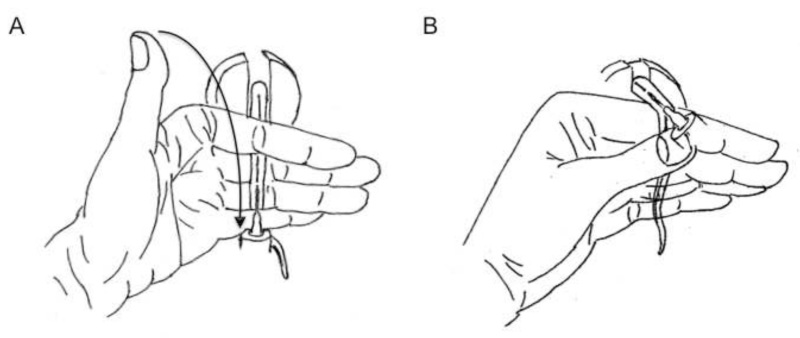 Figure 1. Illustration of hand positions for HGD. A.) Position 1. Recommended hand position to achieve maximal injection velocity and to avoid needle movement after the injection has been initiated. The arrow indicates the suggested movement of the thumb after insertion of the needle into the vein. Thumb should immobilize the needle by gently holding the hub against the tail of the mouse. The hand holding the syringe must be repositioned to be able to press down on the plunger prior to injection. B.) Position 2. This position is recommended if the tail vein injection must be carried out closer to the base of the tail due to repeated injections. Once the needle is inserted into the vein, this hand position needs no further adjustments.
Figure 1. Illustration of hand positions for HGD. A.) Position 1. Recommended hand position to achieve maximal injection velocity and to avoid needle movement after the injection has been initiated. The arrow indicates the suggested movement of the thumb after insertion of the needle into the vein. Thumb should immobilize the needle by gently holding the hub against the tail of the mouse. The hand holding the syringe must be repositioned to be able to press down on the plunger prior to injection. B.) Position 2. This position is recommended if the tail vein injection must be carried out closer to the base of the tail due to repeated injections. Once the needle is inserted into the vein, this hand position needs no further adjustments.
 Figure 2. Western blot analysis of plasma collected from five week-old, female, Swiss Webster (SW) mice injected with a mix of two separate plasmids: 50 μg baboon HPR and 50 μg of differentially 6XHIS tagged baboon APOL-I genes. Mice were injected with saline vehicle as negative control. To confirm the success of HGD, this experiment shows the circulating plasma levels of the baboon HPR protein, as baboon APOL-I is difficult to detect. Mouse plasma samples were collected 2 days post HGD and protein expression levels were analyzed by western blot (plasma diluted 1:20) on 10% Tris-Glycine polyacrylamide gels under denaturing, non-reducing conditions. HPR was detected using a rabbit polyclonal antibody against human haptoglobin (HP), which also recognizes baboon HPR. Tagged bAPOL-I was detected using a rabbit polyclonal antibody against a 6xHIS tag. Molecular weights of the control 6XHIS proteins included are 40 and 50 kDa. A.) Panel shows an example of western blot analysis of a secreted protein (HPR) when all mice in a group were injected successfully. Note the uniformly high protein expression in the second HIS-tagged group (bAPOL-I 6XHIS Group 2), where injection was highly successful. In the first group (bAPOL-I 6XHIS Group 1), protein expression from one of the mice is prominent but somewhat reduced, likely due to reduced speed of the injection. B.) Panel shows an example of protein expression levels when one of the injections (in bAPOL-I 6XHIS Group 3) was unsuccessful. C.) Panel demonstrates the importance of including a tracer protein (bHPR) to monitor the success of HGD when the detection of the protein of interest (bAPOL-I) is uncertain. Despite the confirmed success of HGD in bAPOL-I 6XHIS Group 1 and 2, no 6xHIS-tagged protein is detected in either group. D.) Panel shows that 6xHIS-tagged bAPOL-I is detectable in all three mice in Group 3 that were successfully injected.
Figure 2. Western blot analysis of plasma collected from five week-old, female, Swiss Webster (SW) mice injected with a mix of two separate plasmids: 50 μg baboon HPR and 50 μg of differentially 6XHIS tagged baboon APOL-I genes. Mice were injected with saline vehicle as negative control. To confirm the success of HGD, this experiment shows the circulating plasma levels of the baboon HPR protein, as baboon APOL-I is difficult to detect. Mouse plasma samples were collected 2 days post HGD and protein expression levels were analyzed by western blot (plasma diluted 1:20) on 10% Tris-Glycine polyacrylamide gels under denaturing, non-reducing conditions. HPR was detected using a rabbit polyclonal antibody against human haptoglobin (HP), which also recognizes baboon HPR. Tagged bAPOL-I was detected using a rabbit polyclonal antibody against a 6xHIS tag. Molecular weights of the control 6XHIS proteins included are 40 and 50 kDa. A.) Panel shows an example of western blot analysis of a secreted protein (HPR) when all mice in a group were injected successfully. Note the uniformly high protein expression in the second HIS-tagged group (bAPOL-I 6XHIS Group 2), where injection was highly successful. In the first group (bAPOL-I 6XHIS Group 1), protein expression from one of the mice is prominent but somewhat reduced, likely due to reduced speed of the injection. B.) Panel shows an example of protein expression levels when one of the injections (in bAPOL-I 6XHIS Group 3) was unsuccessful. C.) Panel demonstrates the importance of including a tracer protein (bHPR) to monitor the success of HGD when the detection of the protein of interest (bAPOL-I) is uncertain. Despite the confirmed success of HGD in bAPOL-I 6XHIS Group 1 and 2, no 6xHIS-tagged protein is detected in either group. D.) Panel shows that 6xHIS-tagged bAPOL-I is detectable in all three mice in Group 3 that were successfully injected.
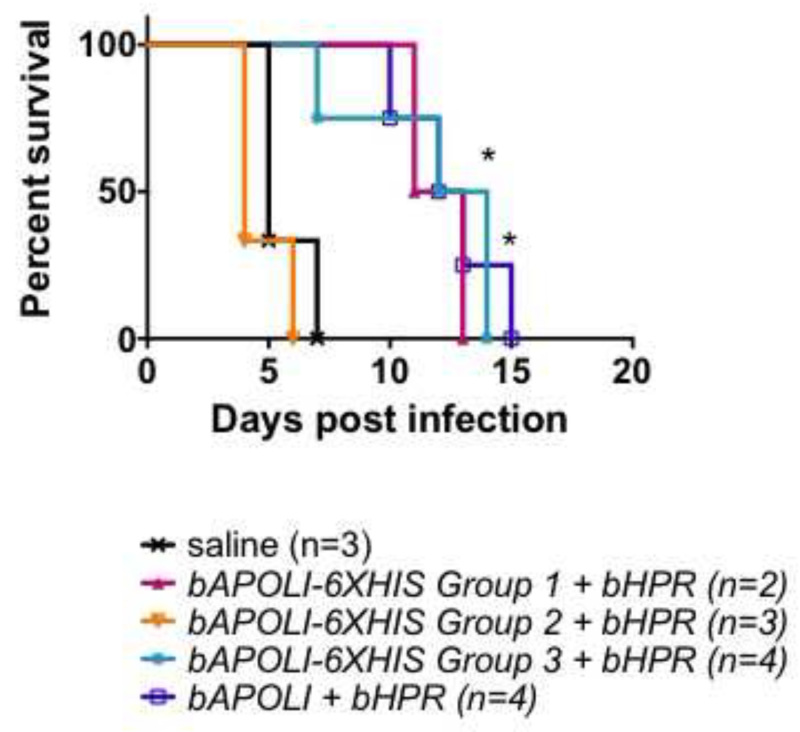 Figure 3. Survival of mice expressing baboon APOL-I 6XHIS variants and baboon HPR. Five week-old, female, Swiss Webster mice received 50 μg of each of two separate plasmids by HGD in the same injection mix. Mice used in this experiment correspond to the plasma analyzed in Figure 2. On day 2 post-injections, mice were infected with 5000 human-infective T.b.b-SRA parasites and parasitemia was monitored. When parasitemia reached 109 parasites/ml, mice were euthanized. Survival was significantly different from saline control where indicated (*- P < 0.05, log-rank test). Mouse survival upon parasite challenge is an example of how the success of HGD influences data obtained from subsequent experiments. When baboon APOL-I is injected successfully, the gene product is secreted into mouse blood where it is taken up by and kills human-infective trypanosomes. The survival curve shows that mice injected with the 6xHIS tag version 3 of baboon APOL-I were significantly protected against Tbb-SRA. The only premature death in this group corresponds to the mouse that was not successfully injected with the plasmid carrying the transgene (see Figure 2). This mouse should be excluded from the survival analysis as its survival upon parasite challenge is not attributable to differences of the transgene received but to the failure of gene delivery.
Figure 3. Survival of mice expressing baboon APOL-I 6XHIS variants and baboon HPR. Five week-old, female, Swiss Webster mice received 50 μg of each of two separate plasmids by HGD in the same injection mix. Mice used in this experiment correspond to the plasma analyzed in Figure 2. On day 2 post-injections, mice were infected with 5000 human-infective T.b.b-SRA parasites and parasitemia was monitored. When parasitemia reached 109 parasites/ml, mice were euthanized. Survival was significantly different from saline control where indicated (*- P < 0.05, log-rank test). Mouse survival upon parasite challenge is an example of how the success of HGD influences data obtained from subsequent experiments. When baboon APOL-I is injected successfully, the gene product is secreted into mouse blood where it is taken up by and kills human-infective trypanosomes. The survival curve shows that mice injected with the 6xHIS tag version 3 of baboon APOL-I were significantly protected against Tbb-SRA. The only premature death in this group corresponds to the mouse that was not successfully injected with the plasmid carrying the transgene (see Figure 2). This mouse should be excluded from the survival analysis as its survival upon parasite challenge is not attributable to differences of the transgene received but to the failure of gene delivery.
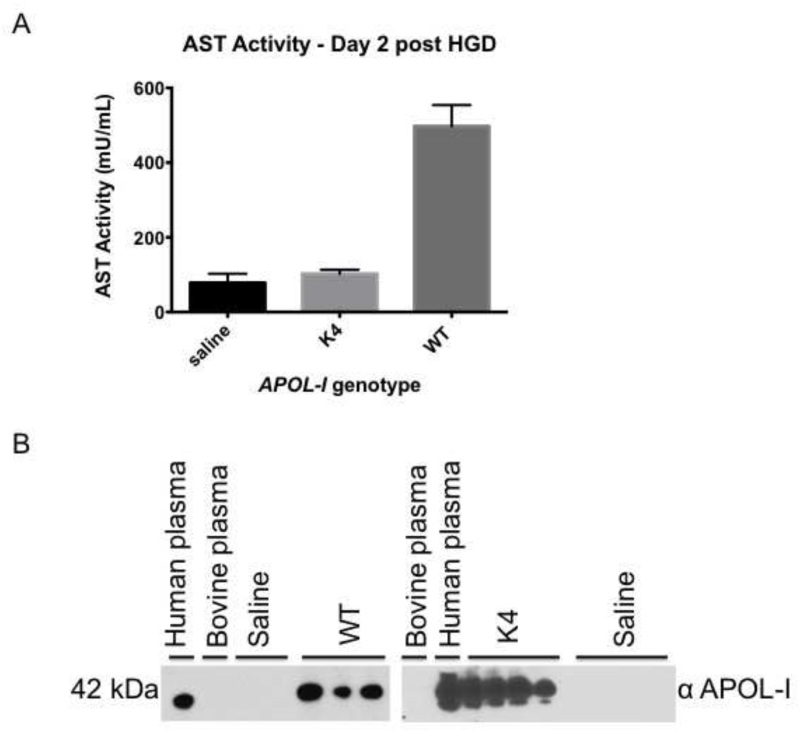 Figure 4. A.) Plasma AST levels from SW mice that received 50 μg of human WT or synthetic mutant (designated as K4) transgenes of APOL-I. The plasmid carrier (pRG977) was identical for all gene variants. Mouse plasma samples were collected two days after HGD. AST levels were measured using a commercially available colorimetric kit. Note that elevations in AST levels are associated with sequence variations in the APOL-I gene and not the trauma of HGD itself. Data are from one representative experiment assayed in triplicate using the following number of plasma samples: saline (n=6), K4 (n=4) and WT (n=3). The error bars represent standard deviation. B.) Western blot analysis of plasma collected on day 2 post HGD from mice that were assessed for AST levels in panel A. Protein expression levels were analyzed by immunoblot (plasma diluted 1:40) using 10% Tris-Glycine polyacrylamide gels under denaturing, non-reducing conditions. APOL-I was detected using a rabbit polyclonal antibody against the N terminus of human APOL-I.
Figure 4. A.) Plasma AST levels from SW mice that received 50 μg of human WT or synthetic mutant (designated as K4) transgenes of APOL-I. The plasmid carrier (pRG977) was identical for all gene variants. Mouse plasma samples were collected two days after HGD. AST levels were measured using a commercially available colorimetric kit. Note that elevations in AST levels are associated with sequence variations in the APOL-I gene and not the trauma of HGD itself. Data are from one representative experiment assayed in triplicate using the following number of plasma samples: saline (n=6), K4 (n=4) and WT (n=3). The error bars represent standard deviation. B.) Western blot analysis of plasma collected on day 2 post HGD from mice that were assessed for AST levels in panel A. Protein expression levels were analyzed by immunoblot (plasma diluted 1:40) using 10% Tris-Glycine polyacrylamide gels under denaturing, non-reducing conditions. APOL-I was detected using a rabbit polyclonal antibody against the N terminus of human APOL-I.
 Figure 5. A single amino acid substitution in the APOL-I sequence results in different levels of tissue pathology. The figure shows livers of SW mice that received hydrodynamic injections of saline vehicle (A), 50 μg human WT APOL-I (B), or 50 μg of a synthetic mutant variant of APOL-I, K4 (C). Livers were removed on day 5 post-injection and processed for hematoxylin and eosin staining. Representative examples are shown of the same mice assayed for AST levels in Figure 4. Panels (D-F) show additional examples of damaged areas in WT mice. Necrosis foci in panels (B), (D-F) are marked with black rectangles and are enlarged. White arrows in these panels point to areas of macrophage infiltration. For comparison, normal tissue from a representative area of the liver of a saline vehicle-injected mouse is marked by a light blue rectangle and is also enlarged. Panels (B) and (D) are representative images from the same WT mouse, while panels (E) and (F) are from two different WT mice.
Figure 5. A single amino acid substitution in the APOL-I sequence results in different levels of tissue pathology. The figure shows livers of SW mice that received hydrodynamic injections of saline vehicle (A), 50 μg human WT APOL-I (B), or 50 μg of a synthetic mutant variant of APOL-I, K4 (C). Livers were removed on day 5 post-injection and processed for hematoxylin and eosin staining. Representative examples are shown of the same mice assayed for AST levels in Figure 4. Panels (D-F) show additional examples of damaged areas in WT mice. Necrosis foci in panels (B), (D-F) are marked with black rectangles and are enlarged. White arrows in these panels point to areas of macrophage infiltration. For comparison, normal tissue from a representative area of the liver of a saline vehicle-injected mouse is marked by a light blue rectangle and is also enlarged. Panels (B) and (D) are representative images from the same WT mouse, while panels (E) and (F) are from two different WT mice.
Discussion
When performed correctly, HGD is a remarkably safe and effective means of transgene delivery. The critical steps to successful HGD are: 1) delivering the right amount of DNA in a large volume of saline vehicle 2) into the tail vein of the mouse 3) in less than 8 sec.
Although the process of injection itself undeniably requires some manual dexterity, the success of this six-second procedure often lies in careful preparation of the experiment. The amount of pDNA necessary to achieve maximal gene expression may vary greatly depending on the plasmid used and the gene to be expressed therefore, the optimal amount of DNA to be injected should be determined experimentally for each application. For mice, the recommended DNA range in general is 5-50 μg, beyond which gene expression might reach a plateau. In our hands, injection of 50 μg pRG977 carrying either the APOL-I or HPR gene leads to high levels of circulating protein levels in the first two days post HGD. Blood levels of both proteins decrease significantly over the next couple of days until protein levels fall below detectable levels around day 10 (38 and unpublished data). The optimal amount of DNA should be delivered in a physiological solution equivalent to 8-12% of the body weight of the mouse1,2. While most researchers, including us, use saline as the injection vehicle, Ringer’s solution has been used for HGD with comparable success 2. Note, that while it is not necessary to perform HGD in a sterile environment, it is imperative to avoid endotoxin contamination of the injection solution throughout the procedure. To this end, the saline solution used for injections should be of superior purity and the DNA to be injected should be prepared using a commercially available kit that removes endotoxin. Another important preparatory step is the careful weighing of mice. Since the volume of injection is of one of the critical aspects of successful HGD, it is important to make sure that the mouse is neither under-dosed nor over-dosed with saline. The former error results in suboptimal transfection, while the latter in the possible death of the animal. To further ensure the safety of the animals, we recommend loading the syringes with small-gauge needle to avoid introducing small air bubbles that are hard to get rid of. HGD should be performed with a 27- gauge needle as described in the protocol.
Correct injection is greatly facilitated if the caudal veins are clearly visible. Dilating blood vessels by gentle heating of the mouse may aid visualization of the veins if a restrainer with a tail-illuminator is not available. Placing the mouse cage on a heat pad for a few minutes or holding the tail with a warm, wet cloth work well in our hands. If using a heat-lamp, one must exercise extreme caution not to overheat the mice. We find that using a heat-lamp causes the mice unnecessary stress and discomfort and therefore, should be avoided. Wiping the tail with 70% ethanol further helps visualization by increasing the contrast between the tail vein and the skin. This step is also critical to avoid bacterial contamination during injection. Once the mouse is prepared for injection, the tail vein may be injected using either one of the hand positions described in this protocol. Note, that while position 2 appears easier and more straightforward, it may be more difficult to inject the full volume of liquid without moving the syringe. In contrast, establishing good needle position using the first method is more cumbersome however, once the needle hub is securely held against the tail, injection seldom fails. The first method is also recommended for maximum speed, although successful transfection is possible using either hand position.
The recommended speed of HGD injection in mice is 6-8 sec, although many authors suggest that faster injections yield even better results1,12. Slow injection results in markedly reduced gene expression. However, the most common reason for a complete lack of gene expression is failure to deliver the full volume of injection. This occurs when the needle is moved, after the injection was initiated. Correctly positioned, the needle enters the vein with no resistance. To check that the needle is inserted into the vein correctly, one may inject a very small amount (100 μl) liquid into the vein before full injection. If pressing down on the plunger meets with extreme resistance, the needle is in the wrong position and the liquid is entering the tail tissue. If the needle leaves the vein mid-injection, correction of the unsuccessful HGD may be attempted after the mouse was allowed to rest for a couple of hours. Re-injection after insufficient rest causes distress to the mouse and may result in loss of hydrodynamic pressure by opening up the first wound on the tail of the mouse. If repeated injections are necessary, the mouse may be injected with the full volume of saline (and amount of DNA) either closer to the base of the tail in the same vein or in the contralateral vein. Following HGD, the mouse should be observed in a cage placed on a heat pad for at least one hour. HGD causes a sharp increase in intravascular pressure resulting in an acute irregularity of heart function, liver expansion, and a disruption of the membrane pores of liver cells, sinusoids and fenestrae12,14,43. Despite the doubling of their blood volume in a few seconds, mice tolerate HGD remarkably well. The heart rate returns to normal in 2 min and the intravascular pressure, which decreases rapidly soon after injection, approaches basal levels in 3 min12,43. The disrupted hepatocytes reseal in less than 2 min and the expanded liver size returns to normal size in 30 min followed by the recovery of sinusoid function in approximately 24 hrs43. While many researchers use anesthesia successfully, in our hands, isofluorane anesthesia exacerbates the respiratory distress of mice due to HGD. We find that anesthetized mice take longer to recover and occasionally require resuscitation for survival. If injecting a large group of mice using anesthesia, it may be necessary to have an additional person in the room dedicated to oversee the well being of the recovering animals. Without anesthesia, even after repeated injections, mouse survival is 100% and recovery time is less than 5 minutes.
The protocol described here may be used to deliver a wide range of molecules to the mouse liver. Using the secreted protein APOL-I as an example, we showed how HGD could be used to establish a mouse model and study the function of a protective protein. When comparing the function of various gene products, the success of the HGD should be assessed whenever possible. For secreted proteins, it is easy to monitor expression levels by analyzing mouse plasma using western blot (Figures 2 and 4B.) It is particularly important to ensure the success of HGD when assaying natural or synthetic variants of a therapeutic protein, as transfection efficiency can severely influence disease outcome (Figures 2 and 3). Examples are given to show how this method can be expanded to analyze liver damage caused by APOLI, which is a pore-forming protein (Figures 4 and 5). Sequence variants of APOL-I in the same plasmid background result in markedly different damage profiles. This suggests that tissue damage is associated with protein function and not the expression vector or the trauma of the injection itself. It should be noted that, in our hands, AST levels are universally high on day 1 post-injection (data not shown). On day 2, however, AST levels of mice injected with saline return to normal and differences among gene variants are readily discernible (Figure 4). By day 3 post-injection, AST measured in all treatments return to their basal level (data not shown). Since necrotic tissue and inflammation in the liver are evident longer than a transient increase in AST, liver damage due to sustained gene expression may be assessed more reliably, albeit qualitatively, by staining of the livers collected on day 5 post-injection. The examples given show that variants of APOL-I that cause an increased release of AST 2 days post-injection also result in more sustained liver damage as assessed by staining with hematoxylin and eosin.
Hydrodynamic gene delivery allows the rapid analysis of gene expression in vivo. It requires the simple construction of the gene under study into a eukaryotic expression vector and the ability to perform the injection as outlined above. Generation of transgenic mice using other forms of gene delivery, such as recombinant lentiviral vectors derived from HIV-1 or recombinant adeno-associated viral vectors, require much more specialized expertise. However, their advantage is sustained gene expression, although viral gene expression often causes an inflammatory response due to the host sensing a viral infection and responding 15-19. Furthermore, the viral vectors have a limited packaging ability and therefore, are limited in gene size, whereas BACs of 150kb have been successfully used in HGD 4. After mastering this technique, the user can transfect any nucleic acid (cDNA or gDNA or RNA) in vivo and follow the production of protein and the biological consequences. The protein can be intracellular, membrane bound or extracellular; it can be tagged or modified at the nucleic acid and thus amino acid level. Most importantly, this is a very rapid and straightforward technique.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We thank Xia Liu (Regeneron Pharmaceuticals) for initially teaching us the HGD technique and Dr. Russell Thomson (Albert Einstein College of Medicine) for his continual guidance in various aspects of the technology. This work was funded by Hunter College, CUNY start up funds and NSF Bread award IOS-1249166. We thank the NYULMC Histopathology Core NYUCI Center Support Grant, NIH/NCI 5 P30CA16087-31 for histology on mouse livers.
References
- Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- Miao CH, Thompson AR, Loeb K, Ye X. Long-term and therapeutic-level hepatic gene expression of human factor IX after naked plasmid transfer in vivo. Mol Ther. 2001;3:947–957. doi: 10.1006/mthe.2001.0333. [DOI] [PubMed] [Google Scholar]
- Magin-Lachmann C, et al. In vitro and in vivo delivery of intact BAC DNA -- comparison of different methods. J Gene Med. 2004;6:195–209. doi: 10.1002/jgm.481. [DOI] [PubMed] [Google Scholar]
- Chang J, Sigal LJ, Lerro A, Taylor J. Replication of the human hepatitis delta virus genome Is initiated in mouse hepatocytes following intravenous injection of naked DNA or RNA sequences. J Virol. 2001;75:3469–3473. doi: 10.1128/JVI.75.7.3469-3473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi H, et al. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, et al. Vector-based in vivo RNA interference: dose- and time-dependent suppression of transgene expression. J Pharmacol Exp Ther. 2004;308:688–693. doi: 10.1124/jpet.103.059931. [DOI] [PubMed] [Google Scholar]
- Layzer JM, et al. In vivo activity of nuclease-resistant siRNAs. Rna. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, et al. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Karimi M, Contag CH, Kay MA. A potent and specific morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology. 2003;38:503–508. doi: 10.1053/jhep.2003.50330. [DOI] [PubMed] [Google Scholar]
- Zhang G, et al. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11:675–682. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Kuramoto T, Yamaoka K, Hashida M, Takakura Y. Hepatic uptake and gene expression mechanisms following intravenous administration of plasmid DNA by conventional and hydrodynamics-based procedures. J Pharmacol Exp Ther. 2001;297:853–860. [PubMed] [Google Scholar]
- Kobayashi N, Nishikawa M, Hirata K, Takakura Y. Hydrodynamics-based procedure involves transient hyperpermeability in the hepatic cellular membrane: implication of a nonspecific process in efficient intracellular gene delivery. J Gene Med. 2004;6:584–592. doi: 10.1002/jgm.541. [DOI] [PubMed] [Google Scholar]
- Bonamassa B, Hai L, Liu D. Hydrodynamic gene delivery and its applications in pharmaceutical research. Pharm Res. 2011;28:694–701. doi: 10.1007/s11095-010-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dosari MS, Gao X. Nonviral gene delivery: principle, limitations, and recent progress. Aaps J. 2009;11:671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Kim KS, Liu D. Nonviral gene delivery: what we know and what is next. Aaps J. 2007;9 doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Nishikawa M, Takakura Y. The hydrodynamics-based procedure for controlling the pharmacokinetics of gene medicines at whole body, organ and cellular levels. Adv Drug Deliv Rev. 2005;57:713–731. doi: 10.1016/j.addr.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Al-Dosari MS, Knapp JE, Liu D. Hydrodynamic delivery. Adv Genet. 2005;54:65–82. doi: 10.1016/S0065-2660(05)54004-5. [DOI] [PubMed] [Google Scholar]
- Bell JB, et al. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio FG, de la Rosa J, Freije JM. Luminescence-based in vivo monitoring of NF-kappaB activity through a gene delivery approach. Cell Commun Signal. 2013;11 doi: 10.1186/1478-811X-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp JE, Liu D. Hydrodynamic delivery of DNA. Methods Mol Biol. 2004;245:245–250. doi: 10.1385/1-59259-649-5:245. [DOI] [PubMed] [Google Scholar]
- Yang J, Chen S, Huang L, Michalopoulos GK, Liu Y. Sustained expression of naked plasmid DNA encoding hepatocyte growth factor in mice promotes liver and overall body growth. Hepatology. 2001;33:848–859. doi: 10.1053/jhep.2001.23438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, et al. High-level expression of naked DNA delivered to rat liver via tail vein injection. J Gene Med. 2002;4:333–341. doi: 10.1002/jgm.281. [DOI] [PubMed] [Google Scholar]
- Hen G, et al. Expression of foreign genes in chicks by hydrodynamics-based naked plasmid transfer in vivo. Domestic animal endocrinology. 2006;30:135–143. doi: 10.1016/j.domaniend.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Eastman SJ, et al. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum Gene Ther. 2002;13:2065–2077. doi: 10.1089/10430340260395910. [DOI] [PubMed] [Google Scholar]
- Yoshino H, Hashizume K, Kobayashi E. Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume. Gene Ther. 2006;13:1696–1702. doi: 10.1038/sj.gt.3302833. [DOI] [PubMed] [Google Scholar]
- Vanden Bossche P. Some general aspects of the distribution and epidemiology of bovine trypanosomosis in southern Africa. Int J Parasitol. 2001;31:592–598. doi: 10.1016/s0020-7519(01)00146-1. [DOI] [PubMed] [Google Scholar]
- Fevre EM, Picozzi K, Jannin J, Welburn SC, Maudlin I. Human African trypanosomiasis: Epidemiology and control. Adv Parasitol. 2006;61:167–221. doi: 10.1016/S0065-308X(05)61005-6. [DOI] [PubMed] [Google Scholar]
- Lugli EB, Pouliot M, Portela Mdel P, Loomis MR, Raper J. Characterization of primate trypanosome lytic factors. Mol Biochem Parasitol. 2004;138:9–20. doi: 10.1016/j.molbiopara.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Pays E, et al. The trypanolytic factor of human serum. Nat Rev Microbiol. 2006;4:477–486. doi: 10.1038/nrmicro1428. [DOI] [PubMed] [Google Scholar]
- Raper J, Fung R, Ghiso J, Nussenzweig V, Tomlinson S. Characterization of a novel trypanosome lytic factor from human serum. Infect Immun. 1999;67:1910–1916. doi: 10.1128/iai.67.4.1910-1916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- Nielsen MJ, et al. Haptoglobin-related protein is a high-affinity hemoglobin-binding plasma protein. Blood. 2006;108:2846–2849. doi: 10.1182/blood-2006-05-022327. [DOI] [PubMed] [Google Scholar]
- Molina-Portela Mdel P, Lugli P, Recio-Pinto E, Raper J. Trypanosome lytic factor, a subclass of high-density lipoprotein, forms cation-selective pores in membranes. Mol Biochem Parasitol. 2005;144:218–226. doi: 10.1016/j.molbiopara.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Perez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- Molina-Portela MP, Samanovic M, Raper J. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med. 2008;205:1721–1728. doi: 10.1084/jem.20071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oli MW, Cotlin LF, Shiflett AM, Hajduk SL. Serum resistance-associated protein blocks lysosomal targeting of trypanosome lytic factor in Trypanosoma brucei. Eukaryot Cell. 2006;5:132–139. doi: 10.1128/EC.5.1.132-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Molina-Portela P, Mott H, Carrington M, Raper J. Hydrodynamic gene delivery of baboon trypanosome lytic factor eliminates both animal and human-infective African trypanosomes. Proc Natl Acad Sci U S A. 2009;106:19509–19514. doi: 10.1073/pnas.0905669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur S, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Gao X, Stolz DB, Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129–137. doi: 10.1038/sj.gt.3302865. [DOI] [PubMed] [Google Scholar]


