Abstract
Under global change, populations have four possible responses: ‘migrate, acclimate, adapt or die’ (Gienapp et al. 2008 Climate change and evolution: disentangling environmental and genetic response. Mol. Ecol. 17, 167–178. (doi:10.1111/j.1365-294X.2007.03413.x)). The challenge is to predict how much migration, acclimatization or adaptation populations are capable of. We have previously shown that populations from more variable environments are more plastic (Schaum et al. 2013 Variation in plastic responses of a globally distributed picoplankton species to ocean acidification. Nature 3, 298–230. (doi:10.1038/nclimate1774)), and here we use experimental evolution with a marine microbe to learn that plastic responses predict the extent of adaptation in the face of elevated partial pressure of CO2 (pCO2). Specifically, plastic populations evolve more, and plastic responses in traits other than growth can predict changes in growth in a marine microbe. The relationship between plasticity and evolution is strongest when populations evolve in fluctuating environments, which favour the evolution and maintenance of plasticity. Strikingly, plasticity predicts the extent, but not direction of phenotypic evolution. The plastic response to elevated pCO2 in green algae is to increase cell division rates, but the evolutionary response here is to decrease cell division rates over 400 generations until cells are dividing at the same rate their ancestors did in ambient CO2. Slow-growing cells have higher mitochondrial potential and withstand further environmental change better than faster growing cells. Based on this, we hypothesize that slow growth is adaptive under CO2 enrichment when associated with the production of higher quality daughter cells.
Keywords: phenotypic plasticity, adaptation, Ostreococcus tauri, climate change, oceanography
1. Introduction
Shifts in the environment drive both plastic and evolutionary responses in organisms, and theoretical studies have shown that plastic responses are good candidates for predicting evolutionary ones [1–3], but to our knowledge, no direct experimental tests of this exist. Here, we present an empirical study that tests and quantifies links between the two processes. Phenotypic plasticity is a single genotype's ability to produce variable phenotypes in response to environmental conditions [4] and views on the possible relationship between phenotypic plasticity and evolution fall into two main groups with mutually exclusive predictions. The first is that populations made up of plastic individuals are more likely to adapt to novel and changing environments [3,5–9]. This is based on population genetics models [10–12], where plasticity acts mainly by keeping population sizes high enough to maintain and/or produce variation, though it has never been tested in the absence of demographic effects. The second is that populations made up of plastic individuals are less likely to adapt [13–15]. This is possibly a result of qualitative descriptions for possible outcomes of climate change scenarios that phrase possibilities as either/or (‘migrate, acclimatize, adapt or die’), and is usually based on verbal arguments rather than mathematical models or simulations.
The misleading statement that populations respond to environmental change through either plasticity or evolution stems from two sources: first, in studies of natural populations, it is difficult to disentangle plastic and evolutionary responses, and when plastic responses cannot be ruled out, the implication is often that evolutionary responses are absent (e.g. [16,17]). While failing to rule out a plastic response does not imply that no evolutionary response is present or possible, it is harder to definitively detect an evolutionary response than a plastic one [18]. Evolutionary responses will consequently be reported less than plastic ones in proportion to the extent that they actually occur [19]. Second, a common thought experiment proposes that the environment changes such that fitness decreases, causing populations made up of individuals that can mitigate fitness loss by plasticity to be under weak or no selection. A population that is not under selection will not have to adapt. By contrast, populations with no or insufficient adaptive plastic responses will have to either migrate or adapt to avoid going extinct. This leads to the conclusion that adaptive plasticity and genetic adaptation should be negatively correlated [13,14], despite the growing body of theoretical work predicting the opposite [1–3,20]. While the relationship between adaptive plasticity and adaptation is uncontroversial within disciplines, this relationship is a source of conflicting predictions between disciplines, particularly evolutionary and marine biology, and must be empirically tested in order to estimate the extent to which plasticity data can be used to predict the evolutionary fate or adaptive potential of populations [21]. These conflicting predictions are especially important in the context of understanding how marine populations are likely to respond to global change. There are empirical studies on how large microbial populations respond to environmental perturbations in the short term through phenotypic plasticity in the absence of genetic change [22–26], or evolve in the long term using genetic change [27–32]. Despite this, neither group of studies has measured links between phenotypic plasticity and evolution.
The link between plasticity and evolution is ecologically relevant, and here we explore it in the context of marine microbes: as the world changes and oceans become less basic, more stratified and depleted in nutrients, large populations of marine microbes with short generation times will have ample scope for evolution [33]. In the context of climate change, marine biologists often base predictions on future oceans on short-term experiments [34,35], but the predictive power of plasticity data remains untested. Evolutionary biologists use models to predict, for example, how different rates of environmental change may require different levels of plasticity in order to keep fitness constant (e.g. [2]), but little is known about how the costs and benefits of plasticity affect the adaptive potential of large populations, where even a substantial drop in fitness is unlikely to lower population size to the point where natural selection cannot act effectively. Environmental fluctuations are also expected to increase in the future, which may select for increased plasticity, which could subsequently alter the speed or outcome of evolution and consequently, affect evolved phenotypes (e.g. [36,37]). The evolution of plasticity is expected when the frequency of fluctuations is on a scale of few generations relative to the organism's generation time [38]. While there are studies that characterize evolution in complex and fluctuating environments [3,39], this has yet to be applied to understanding how marine phytoplankton are likely to evolve under climate change scenarios.
Here, we use experimental evolution with a globally distributed marine picoplankton to measure how phenotypic plasticity affects evolution. We show that populations founded from more plastic ancestors evolve more, and that phenotypic plasticity in a fitness-related trait can be used as a predictor for the magnitude of an evolutionary response. We evolved 16 physiologically distinct lineages of the species-complex [40,41] Ostreococcus from single cells for 400 generations in constant and fluctuating environments at ambient (430 ppm CO2) and elevated partial pressure of CO2 (pCO2) levels (predicted for the year 2100: 1000 ppm CO2, based on the Intergovernmental Panel on Climate Change report 2007, [42]). We refer to the selection environments as follows: stable ambient (SA), fluctuating ambient (FA), stable high (SH) and fluctuating high (FH). The lineages varied initially in their plastic responses in oxygen evolution rates to CO2 enrichment [22]. We present four main findings. First, under CO2 enrichment, variations in plastic responses (change in oxygen evolution rates) before evolution predict evolutionary responses (change in growth rate). Second, plasticity evolves in fluctuating environments and degrades in constant ones. Third, natural selection in constant and fluctuating environments produces radically different phenotypes. Finally, lineages evolved under long-term carbon enrichment eventually grow more slowly than lineages under short-term carbon enrichment, thereby producing less but better quality daughter cells, which indicates that slow growth can be adaptive in our experimental set-up.
2. Material and methods
(a). Lineages and culturing conditions
Ostreococcus lineages were obtained from the Roscoff Culture Collection and the Plymouth Marine Laboratory, grown in Keller Medium [43] and made clonal by dilution and propagated as described in [22]. Lineages were grown in a closed system in semi-continuous batch cultures at low densities (maximum density of 104). In the selection experiment, algae were subjected to one of the following four selection regimes: selection for growth at 430 ppm CO2 (final average ± s.d. CO2: 444 ± 43 ppm CO2), selection for growth at 1000 ppm CO2 (final average ± s.d: 1031 ± 87 ppm CO2), selection for plasticity in an environment that fluctuated around a mean of 430 ppm CO2 (490 ± 97 ppm CO2), and selection for plasticity at high pCO2, in an environment where CO2 levels were fluctuating around a mean of 1000 ppm CO2 (1012 ± 244 ppm CO2). More precisely, in the fluctuating selection regime, pCO2 in the incubator was changed to a random value between 430 and 630 µatm CO2 once per week in the FA environment and was changed to a random value between 700 and 1300 µatm (also once per week) in the FH environment. This rate of environmental fluctuation maintains plasticity rather than multiple specialist lineages within populations (see Results). The slightly higher mean of pCO2 between FA and SA treatment (see the electronic supplementary material, table S1) has no overall effect (p = 0.42)—when we use mean and variation from mean as explaining variables in our ANOVAs, the differences between SA and FA lineages are indeed driven by differences in environmental variability. For assays, samples were pre-acclimatized and acclimatized for five to seven asexual generations to 430 ppm CO2 or 1000 ppm CO2 (see the electronic supplementary material, figure S1 for experimental design). The selection environments were established by setting the incubator to the appropriate pCO2 and by using air-stones to aerate the seawater to be used at each transfer for at least 24 h prior to transfer.
(b). Growth rate μ
At t = 0, 100 and 400 (generations), cell count was determined using flow cytometry (FACSCalibur and FACS CANTO) and growth rate (μ) calculated as described in [22].
Growth rate data were used to calculate evolutionary responses, i.e. heritable differences in growth rates between populations evolved for 400 generations.
Growth responses were calculated using the following formula:
 |
The formulae for direct and correlated responses after evolution can be found in the electronic supplementary material. Here, direct responses refer to traits of SH and FH evolved lineages measured at 1000 ppm CO2 relative to growth rates of SA and FH evolved lineages measured at 1000 ppm CO2. Correlated responses refer to traits of FH-evolved and SH-evolved lineages measured at 430 ppm CO2 (ancestral environment) relative to phenotypes of SA- and FA-evolved lineages measured at 430 ppm CO2. Unless stated otherwise, measurements were carried out after 400 generations of selection.
All populations were pre-acclimatized and acclimatized to their respective assay environment for five to seven generations each.
(c). Flow cytometry for determination of cell density and health
FACS flow cytometry was used to determine event number (cell density), orange fluorescence, mitochondrial potential in rhodamine 123 stained cells and green fluorescence in green fluorescent protein (GFP)-modified Ostreococcus strains. To determine rhodamine 123 fluorescence, 1 µl of a (0.2 µg ml−1) rhodamine solution was added to 200 µl of sample and left to incubate for 30 min prior. Rhodamine 123 fluorescence quantifies the strength of the proton gradient across mitochondrial membranes [44] and was detected as green fluorescence.
(d). Heat shock assay
To assess how well lineages would deal with stress, we first determined the lethal temperature threshold for our populations, and then chose a temperature that reduced viability of SA-evolved lineages by 50%. In previous trials, we had found that this is the case at a sudden 4°C increase and incubation at that temperature (22°C) for 1 h. Viability and growth rate were measured using flow cytometry over the next five to seven cell divisions.
(e). Oxygen evolution rates
Here, we use oxygen evolution rates as an example of a plastic trait other than fitness that can be related to evolution. This requires that the trait be correlated with growth, which is indeed the case in Ostreococcus [22]. Net oxygen evolution and consumption were measured and used to calculate plastic responses as described in [22].
(f). Carbonate chemistry
Seawater carbonate chemistry was calculated from pH and alkalinity using the CO2sys software [45]. Dissolved inorganic carbon (DIC) was measured colourimetrically. Total alkalinity was inferred from linear Gran-titration plots (see the electronic supplementary material, table S1). DIC and pH samples for all lineages were taken at the beginning and at the end of the selection experiment. Additionally, we measured DIC and pH or alkalinity prior to using the bubbled medium in a transfer.
(g). Green fluorescent protein (GFP) strains/competition assay
To test the hypothesis that slow growers are better competitors [46], we competed eight representative lineages against a GFP lineage (transformed oth95) [40]. For the competition experiment, 20 ml of medium were inoculated with 100 µl of wild-type lineages and GFP populations, and cell numbers for each were recorded every day for a 14 day period (two transfers). Each lineage's competitive ability was calculated relative to the GFP-modified lineages (as fold difference in growth) and plotted as a function of the lineage's growth rate in single culture.
(h). Clones/composition of evolved populations
In order to assess whether populations were composed of plastic lineages or of a mixture of non-plastic lineages, bio-replicates of a subset of seven representative lineages were made clonal by dilution, and growth rates were then determined for at least three clones per lineage.
(i). Data analysis
Data were analysed in the R environment, using linear mixed effects models in the nlme and lme4 packages. Data were tested for normality and heterogeneity prior to performing ANOVAs on the models with selection regime and assay environment as fixed effects. Depending on the test performed, there were up to three random effects per model that were all treated as un-nested.
3. Results
(a). Plasticity predicts the extent of evolution
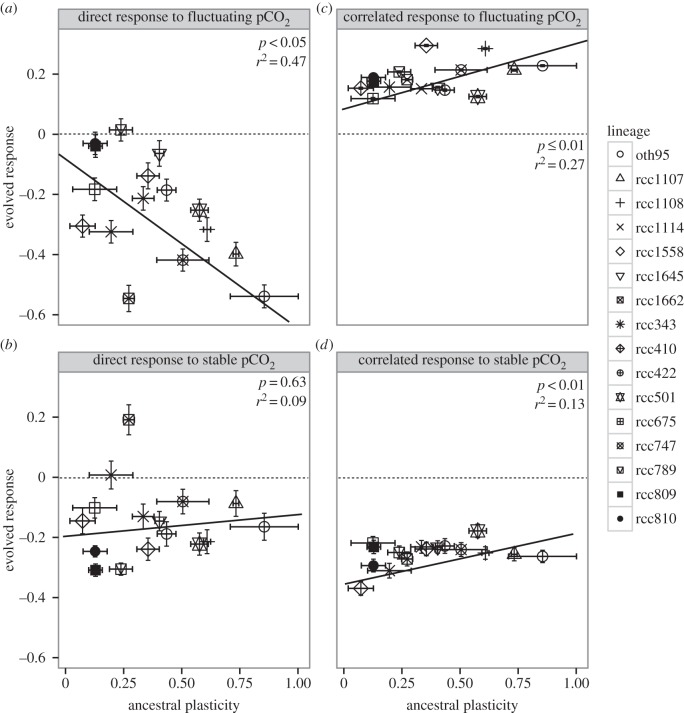
The main question that our experiment was designed to answer is how ancestral plasticity, measured as change in oxygen evolution rates to in response to elevated pCO2 prior to selection (at t = 0), relates to evolution (heritable changes in growth rate) within a lineage. The direct response to selection for growth at high pCO2 is calculated by comparing the growth rates in high pCO2 of populations evolved at high pCO2 (here, FH or SH) with the growth rates in high pCO2 of populations evolved in ambient CO2 (here, FA or SA). We find that Ostreococcus evolves in response to CO2 enrichment (figure 1; F3,132 = 155.66, p < 0.0001), and populations with more plastic ancestors evolve more in high pCO2 than populations founded from less plastic ancestors (F3,132 = 55.90, p < 0.05). In FH lineages, ancestral plasticity explains almost half of the direct response to selection (figure 1a; F1,120 = 167.66, p < 0.001) and 20% of the correlated response to selection (F1,120 = 238.77, p < 0.0001). By contrast, clade/species explains 5–15% of the variation in evolutionary responses in FH populations. Lineages selected in the stable SH environment also evolve (figure 1b; F1,120 = 122.27, p < 0.001 and figure 1d; F1,120 = 593.50 p < 0.0001 for direct and correlated responses, respectively), but the relationship between ancestral plasticity and evolution is weaker, and trends in the opposite direction, perhaps because plasticity is not under selection in constant environments.
Figure 1.
(a–d) Lineages with higher ancestral plasticity evolve more. Direct and correlated responses to selection plotted as a function of plasticity in oxygen evolution rates before evolution (ancestral plasticity). For all panels (a–d), different shapes represent mean values for each lineages ± 1 s.e. For each lineage n = 3. Dashed line indicates no response to selection. Panel (a) (selection in FH, assay at 1000 ppm CO2): ancestral plasticity in FH evolved lineages predicts up to 47% of the evolutionary response (F2,13 = 210.67, p < 0.001). FH populations evolve slow growth in response to high pCO2. Panel (b) (selection in SH, assay at 1000 ppm CO2): with no selection for plasticity, a linear relationship using ancestral plasticity as the only explaining variable is not statistically significant (p = 0.63). Still, most lineages evolve lower growth rates (range from −0.31 to −0.08, mean −0.15 ± 0.12). Panel (c) (selection in FH, assay at 430 ppm CO2): ancestral plasticity is a significant nonlinear predictor of the correlated response to selection (F2,13 = 563.38, p < 0.0001). Lineages from FH increased their growth rate at ambient pCO2 the most when their ancestral plasticity was high (increase in growth of 0.12–0.30, mean 0.19 ± 0.05). Panel (d) (selection in SH, assay at 430 ppm CO2): lineages selected in SH had a negative correlated response, and the relationship between ancestral plasticity and the correlated response to selection was significant (F2,13 = 22.28, p < 0.01), though best described by a nonlinear fit (p-values and r2 reported on the panels are for linear regression).
The correlated response to selection for growth at high pCO2 is calculated by comparing the growth rates in ambient pCO2 of populations evolved at high pCO2 (here, FH or SH) with the growth rates in ambient pCO2 of populations evolved in ambient pCO2 (here, FA or SA). The correlated response to selection in SH shows a strong nonlinear correlation between ancestral plasticity and evolution, where low-to-medium ancestral plasticity correlates with an increase in the evolutionary response, but where there is no further increase in the correlated evolutionary response for ancestral plasticity over 0.3. Plasticity degraded (figure 2) in SH evolved lineages, and these lineages display arrested growth (figure 3) in ambient pCO2.
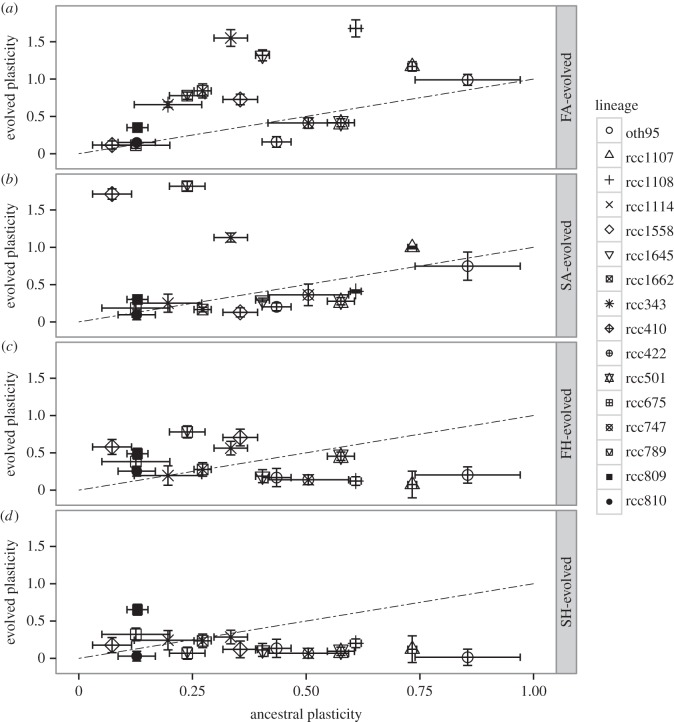
Figure 2.
Plasticity evolves in fluctuating environments. Plasticity in oxygen evolution rates in four selection regimes at the beginning at the selection experiment (x-axis) differs from plasticity in oxygen evolution rates after 400 generations of selection (y-axis). The dotted 1 : 1 line indicates no change in plasticity after 400 generations of evolution. Each symbol represents mean plasticity ± 1 s.e. for a lineage, with three replicate populations per lineage. Panel (a) (FA regime): plasticity changes significantly (F1,98 = 5.58, p < 0.05) and increases in 69% of lineages. Panel (b) (SA regime): in all but four lineages, plasticity does not change significantly. In the four lineages where plasticity does change, it increases (post hoc p < 0.001). Panel (c) (FH regime): 56% of all populations evolve higher plasticity (F1,98 = 6.01, p < 0.05) after selection at FH. Panel (d) (SH regime): plasticity increases in four out of 16 lineages, but decreases or remains unchanged in all other lineages (but see the electronic supplementary material, figure S3).
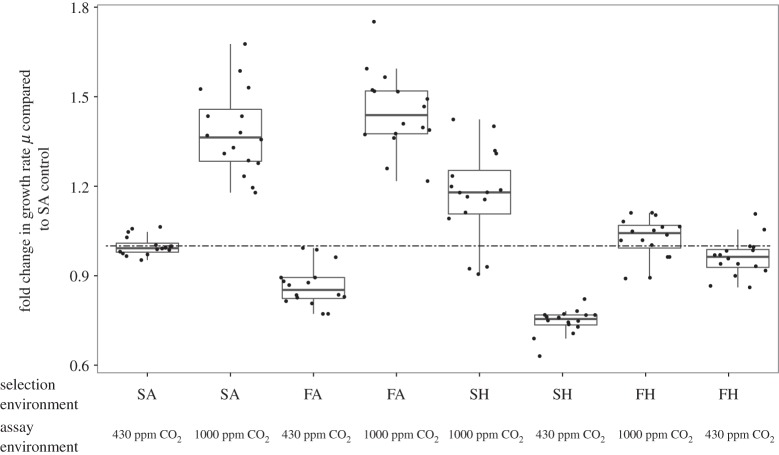
Figure 3.
Evolution at elevated pCO2 reverses short-term growth responses to changes in pCO2. Selection regime changes how lineages respond to CO2 enrichment following 400 generations of selection (F7,105 = 123.38, p < 0.001). Here, n = 48 for each selection and each assay environment. The dashed line indicates no change compared with SA populations. Populations selected in SA show the expected plastic response in growth to CO2 enrichment after 400 generations of selection, where mean growth rate at elevated pCO2 is higher than at 430 ppm CO2 (F7,105 = 114.88, p < 0.001, mean growth rate of SA populations assayed at 430 ppm: 0.67 ± 0.02 d−1, mean growth rate of SA population assayed at 1000 ppm CO2: 0.79 ± 0.03 d−1). When grown at elevated pCO2, the growth rate of lineages selected at FH or SH is lower than the short-term response of their respective controls (i.e. FA or SA) growing at high pCO2 (post-hoc FH p < 0.05 and SH p < 0.05). Populations selected at SH also have negative correlated responses to selection. Growth rate of SH lineages at 430 ppm CO2: 0.5 ± 0.01 d−1, after growth rates have recovered, while lineages selected in FH have an average growth rate of 0.69 ± 0.02 d−1.
(b). Plasticity evolves or is maintained in fluctuating environments
Fluctuating environments select for plasticity in our experiment. Growth for 400 generations in fluctuating environments yields populations composed of individuals that are more plastic than their ancestors (figure 2a,c; also see the electronic supplementary material, figure S2). Plasticity changes significantly in the FA selection environment, where selection was for plasticity alone (figure 2a, F1,98 = 5.58, p < 0.05). There, it increases in 11 of 16 lineages, remains unchanged in two, and is reduced in three out of all 16 lineages (average fold increase 1.69 ± 0.39, post hoc p < 0.05). In the FH environment, where populations were selected for both plasticity and growth at high pCO2, evolved plasticity after 400 generations of selection is on average higher than ancestral plasticity (F1,98 = 6.01, p < 0.05), with the average increase in plasticity being 1.67 ± 0.51-fold (post hoc p < 0.05).
Stable environments do not select for plasticity here, and plasticity does not change significantly after 400 generations of evolution in stable environments. In the SA environment (F1,98 = 0.16493, p = 0.2), it remains unchanged in 12 lineages and increases significantly in four. Plasticity decreases over time in 10 of 16 SH lineages (F1,98 = 10.6, p < 0.05). However, this apparent overall decrease in plasticity may be due to arrested growth in the ancestral environment in some SH lineages (no growth means we cannot quantify plasticity; see the electronic supplementary material, figure S3).
After 400 generations, we find a tendency in all selection regimes for plasticity to decrease in lineages with the highest ancestral plasticity (average 0.86-fold ± 0.44 change, also electronic supplementary material, figure S4) and to increase in most other lineages (average change 1.69-fold ± 0.39).
(c). Evolution reverses the plastic response to CO2 enrichment
Populations evolved in the ambient CO2 environments, SA and FA, respond to short-term increases in CO2 by increasing their growth rates (F7,105 = 114.88, p < 0.001; figure 3). Increasing growth is the usual short-term response of Ostreococcus to CO2 enrichment [22]. However, after about 100 generations (electronic supplementary material, figure S5), populations selected in high pCO2 environments (SH and FH) decrease their growth rates at high pCO2 relative to both their own ancestors and to populations selected at ambient pCO2. SH- and FH-selected populations eventually and completely reverse the plastic response to high pCO2 and return to growth rates at high pCO2 that are similar to SA-selected populations growing at 430 ppm CO2, showing the evolution of slow growth during an experiment where the culturing method should select for rapid growth.
After 400 generations of selection in elevated pCO2 environments, responses to ambient CO2 have changed. FH lineages grow better in their ancestral environment of 430 ppm CO2 (correlated response) than do SH lineages. When SH lineages are transferred back to 430 ppm CO2, they do not grow detectably for about two weeks. By contrast, FH lineages do not show arrested growth.
(d). Slow-growing cells are better at competing, withstanding heat shock and maintaining mitochondrial potential than fast-growing cells
FH-selected populations have lower growth rates than SH-selected populations at high pCO2, which would reflect a cost of plasticity of 14–18%, though we argue in the discussion that this interpretation is probably incorrect, or at least a misleading oversimplification, as slow growers are better competitors, better able to withstand heat shock, and have higher mitochondrial potential than lineages with chronically elevated growth rates.
In populations that had evolved to grow slow (FH) or been selected in environments that did not increase growth initially (SA, FA), lineages with low growth rates in monoculture had better competitive abilities than lineages with high growth rates in monoculture (F1,177 = 810.61, p < 0.0001; electronic supplementary material, figure S6). In SH-evolved populations however, lineages in monoculture with low growth rates were worse competitors in mixed culture—though SH lineages overall grew quickly and were also all poor competitors compared to lineages evolved in other environments. A similar pattern was observed in a study using Chlamydomonas [46]. If low growth rates in monoculture reflected a cost of plasticity, it would be expected that populations evolved in SH be better competitors than those evolved in FH. Instead, populations evolved in FH grow more slowly in monoculture but are in fact better competitors in mixed culture than populations evolved in SH (electronic supplementary material, figure S7).
We hypothesized that slow-growing cells were better competitors because they produced better quality daughter cells than did fast-growing cells. We tested this by measuring two indicators of cellular health. First, elevated levels of orange fluorescence have been shown to increase in stressed or moribund algae [47]. We find that levels of orange fluorescence at the end of the experiment are up to 20 times higher in cells from SH populations than in cells from SA populations or fluctuating populations, indicating that non-plastic populations are more stressed than plastic populations when grown under chronic high pCO2 conditions. Second, mitochondrial potential is higher in the slower growing lineages from the FH environment than the faster growing SH-selected lineages (F3,109 = 15.74, p < 0.05; see the electronic supplementary material, figure S8). In addition to appearing less healthy when alive, fast-growing cells are less likely to survive heat shock. FH-selected lineages have lower growth at high pCO2, but higher viability and growth rates after heat shock than lineages selected in SH, indicating that they are better able to withstand stress than fast-growing lineages (F1,236 = 9.52, p < 0.005 and F1,236 = 53.27, p < 0.0001, for viability and growth rates, respectively; electronic supplementary material, figure S9). This supports our hypothesis that a greater reduction in growth rate in populations evolved under chronic CO2 enrichment correlates with the production of better quality cells.
4. Discussion
(a). Plasticity predicts evolution
We have shown that, in a fluctuating environment, ancestral plasticity can explain almost half of the direct response to selection (figure 1), and that Ostreococcus lineages founded from more plastic ancestors evolve more in high pCO2 environments than lineages founded from less plastic ancestors. This makes ancestral plasticity a good predictor of eventual evolutionary responses and supports theoretical studies arguing that plasticity should facilitate evolution [1–3,20]. Most explanations on why adaptive plasticity facilitates evolution focuses on the effects of differences in population sizes between populations made up of plastic and non-plastic individuals [3,20,48]. Here, there is no systematic difference in population size between treatments (see the electronic supplementary material). Our results show that even in the absence of demographic effects that would affect the amount of genetic variation possible in the population, plasticity can facilitate evolution. This is consistent with plasticity affecting the phenotypic and fitness effects of mutations directly [1]. Interestingly, this suggests that individual plasticity in large microbial populations may be maintained partially as a by-product of more plastic types being more able to adapt, and thus being less likely to go extinct, than less plastic types, and may partly explain why microbes that can respond to environmental change genetically also maintain high levels of individual plasticity.
(b). Plasticity evolves or is maintained in fluctuating environments
Depending on its rate and predictability, environmental variation can select either for the evolution of plastic individuals [3], of generalists with invariable phenotypes over several environments [49,50], or of communities made up of many specialists [49,51,52]. Here, we found an overall increase of individual plasticity in lineages selected in fluctuating environments. We also observe that the lineages with the highest ancestral plasticity evolve slightly lower plasticity. This could be caused by lineages decreasing plasticity enough to limit its cost, while still remaining plastic enough to persist in a fluctuating environment, a strategy known as phenotypic buffering [53,54]. There may be some optimum amount of trait plasticity for the rate and magnitude of change in fluctuating environments used in this experiment, and populations may be converging on that—in the high pCO2 selection environments variance of evolved plasticity is almost half that of ancestral plasticity (see the electronic supplementary material, table S2). Here, an optimal level of plasticity is more likely than a reduction in plasticity that is due to costs associated with higher levels of plasticity.
Additionally, evolutionary history may limit the ability to evolve plasticity, since none of the ‘deep-sea’ lineages of Ostreococcus display any significant change in plasticity during the evolution experiment; these lineages were isolated from relatively constant environments and had lower ancestral plasticity than surface strains [22]. However, among the surface strains, variation in plasticity is the best predictor of variation in evolutionary responses.
We have discussed heritable changes as mutations, but these could be a combination of genetic and epigenetic contributions, if epigenetic changes are stable for at least 14 generations (the time used for acclimatization) or are encoded by genetic mutations.
(c). Evolution reverses the plastic response to CO2 enrichment
Ostreococcus evolves in response to selection at elevated pCO2. After 400 generations of selection at elevated pCO2, lineages selected in the SH environment fail to grow in their ancestral environment (SA). This pattern has also been reported in fresh water green algae [31] and coccolithophores [28,29]. By contrast, FH-selected populations do not show arrested growth at 430 ppm CO2. These results are consistent with the maintenance or the evolution of plasticity in fluctuating environments. Under climate change scenarios, there may be different modes of what selection acts on, favouring either phenotypic plasticity where phenotype changes in response to environmental fluctuations, phenotypic buffering [53,54] or a combination of both: as described in [22,55], in the short-term, CO2 enrichment may be beneficial to the most plastic lineages that are best at taking advantage of the new situation. More plastic lineages will be selected for in the short-term, and adaptive evolution will occur through lineage sorting of these lineages. In the long-term, the new environment may cause stress at the limit of tolerance levels [56], so lineages are now selected for maintaining cellular functions and metabolic capacities, rather than how well suited they are for outgrowing other lineages in the matter of a few generations. So long as a lower growth rate does not result in immediate competitive exclusion by being overgrown, the benefit of producing higher quality daughter cells less likely to die or fail to divide has the potential to outweigh the cost of producing fewer cells. We have shown that slowed growth evolves repeatedly under our high CO2 culture conditions. Further experiments are needed to ascertain how frequently slowed growth evolves under different enrichment scenarios.
Green algae (including Ostreococcus) and cyanobacteria usually respond to short-term increases in pCO2 by increasing their growth rates [22,31,57,58]. Unexpectedly, growth rates then slow back down after a few hundred generations of growth at elevated pCO2 in our experiment. This contrasts with selection experiments in other non-calcifying and non-silicifying algae, where plastic responses to constant high pCO2 are maintained during evolution for up to 1000 generations [30,31,59]. It is unusual (but not unprecedented—see [60]) for growth rates to decrease in a laboratory selection experiment using large populations of microbes, especially in semi-continuous cultures that do not reach carrying capacity, which selects for rapid growth [61]. We suggest that the slow growth rates measured here are adaptive in environments with chronically elevated pCO2. The evolution of slow growth has previously been described in [60]. There, however, slow growth was reported to be a consequence of, rather than an avoidance strategy for, damage in ageing cells. The phenomena described are similar but the causes are not. We argue that slower growth rates seen in FH are adaptive and reflect a benefit, not a cost, of plasticity and provide more detail of our reasoning below.
Our data indicate that slowing growth is associated with higher mitochondrial function. We see that fast-growing cells have lower mitochondrial potential than slow-growing cells, which is consistent with rapid growth causing more oxidative damage than slower growth [60,62]. This may be particularly important here, since Ostreococcus, has only a single mitochondrion [40]. We hypothesize that lower mitochondrial function may decrease fitness more in cells with only one mitochondrion than in cells with multiple mitochondria. In cells with multiple mitochondria, healthy mitochondria may have enough function to make up for the damaged ones, or, malfunctioning mitochondria may not be passed on to daughter cells, leading to no change in mitochondrial potential across generations [63,64]. In Chlamydomonas rheinhardtii, elevated pCO2 has been found to affect mitochondrial size and potential, with high pCO2-evolved cells shown to have smaller, more efficient mitochondria than in cells evolved in control (there, current ambient) levels of CO2 [65]. In organisms with one mitochondrion per cell, neither of these strategies can be applied.
The lower mitochondrial potential we find in the faster growing lineages may explain the difference in outcomes between selection experiments in Ostreococcus, where ancestral increases in growth under CO2 enrichment are reversed after several hundred generations of growth in high pCO2, and other algae, where growth remains high under similar conditions [30,31,59]. In addition to having cells with higher mitochondrial potentials, populations selected in fluctuating environments also survive heat shock better than populations selected in constant environments.
Taken together, these supporting results show that chronic growth at elevated pCO2 is stressful for Ostreococcus and that lower growth rates are associated with lowering that stress. This, along with the high relative fitness of slower growing lineages, supports our interpretation that slower cell division rates during evolution at high pCO2 are adaptive in Ostreococcus. In addition, plastic lineages decrease their growth rates most, indicating a benefit, not a cost, of plasticity.
5. Conclusion
Plastic responses can predict the magnitude of evolutionary responses, and this relationship between plasticity and evolution offers a pragmatic solution to predicting which phytoplankton populations are likely to evolve more under global change. Here, adaptation reverses plastic responses to CO2 enrichment and leads to the evolution of slow growth rates. In similar systems, short-term responses will overestimate changes in growth rate and trait values between contemporary and future phytoplankton populations, while underestimating genetic changes and the organisms' ability to adapt. More generally, our data suggest that since most laboratory evolution experiments so far have been carried out in constant environments even though marine environments can be highly variable, large microbial populations are more likely to adapt to ocean acidification than previously thought. This is especially relevant given predictions that both the magnitude and frequency of changes in pCO2 in oceans will increase in the future [66]. Our study predicts that first, large populations which are more plastic now will evolve more under global change, second, that most large populations will evolve to become more plastic in the future and third, that plastic responses which drastically increase growth rates can be reversed by natural selection because of the stress associated with maintaining rapid growth.
Supplementary Material
Acknowledgements
The selection experiment was conducted at the University of Edinburgh (UK). DIC was measured at the Scottish Association for Marine Science (SAMS, UK). We thank M. Allen and ASSEMBLE (Association of European Marine Biology Laboratories) Roscoff for providing the Ostreococcus lineages, SynthSys Edinburgh for providing the GFP strain; J. Raven, A. Millar, H. Kuehne, J. Raven, B. Kerr, I. Kronholm and D. Lawrence for discussion; and H. Stahl (SAMS) and M. Waterfall (UoE) for assistance with DIC measurements and flow-cytometry, respectively. E.S. designed and performed the experiments, analysed data and wrote the manuscript. S.C. designed the experiments, analysed data, wrote the manuscript and supervised laboratory work.
Data accessibility
Data will be available at https://datadryad.org/handle/10255/3/submit?workspaceID=93761.
Funding statement
The research was supported by a Royal Society (UK) University Research Fellowship and a European Research Council (ERC) under the European Community's Seventh Framework Program (FP7/2007–2013) to S.C. and a Scottish Universities Life Science Alliance scholarship to C.E.S.
Conflict of interests
The authors declare no conflict of interest.
References
- 1.Draghi JA, Whitlock MC. 2012. Phenotypic plasticity facilitates mutational variance, genetic variance, and evolvability along the major axis of environmental variation. Evolution 66, 2891–2902. ( 10.1111/j.1558-5646.2012.01649.x) [DOI] [PubMed] [Google Scholar]
- 2.Chevin L-M, Lande R, Mace GM. 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8, e1000357 ( 10.1371/journal.pbio.1000357.g002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lande R. 2009. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J. Evol. Biol. 22, 1435–1446. ( 10.1111/j.1420-9101.2009.01754.x) [DOI] [PubMed] [Google Scholar]
- 4.West-Eberhard MJ. 2003. Developmental plasticity and evolution. New York, NY: Oxford University Press. [Google Scholar]
- 5.Pichancourt J-B, van Klinken RD. 2012. Phenotypic plasticity influences the size, shape and dynamics of the geographic distribution of an invasive plant. PLoS ONE 7, e32323 ( 10.1371/journal.pone.0032323.g007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellers J, Stuefer JF. 2010. Frontiers in phenotypic plasticity research: new questions about mechanisms, induced responses and ecological impacts. Evol. Ecol. 24, 523–526. ( 10.1007/s10682-010-9375-4) [DOI] [Google Scholar]
- 7.Matesanz S, Gianoli E, Valladares F. 2010. Global change and the evolution of phenotypic plasticity in plants. Ann. NY Acad. Sci. 1206, 35–55. ( 10.1111/j.1749-6632.2010.05704.x) [DOI] [PubMed] [Google Scholar]
- 8.Nicotra AB, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. ( 10.1016/j.tplants.2010.09.008) [DOI] [PubMed] [Google Scholar]
- 9.Merilä J. 2012. Evolution in response to climate change: in pursuit of the missing evidence. Bioessays 34, 811–818. ( 10.1002/bies.201200054) [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Mestre I, Jovani R. 2013. A heuristic model on the role of plasticity in adaptive evolution: plasticity increases adaptation, population viability and genetic variation. Proc. R. Soc. B 280, 20131869 ( 10.1098/rspb.2013.1869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlichting CD, Pigliucci M. 1998. Phenotypic evolution. Sunderland, MA: Sinauer Associates Incorporated. [Google Scholar]
- 12.Snell-Rood EC, Van Dyken JD, Cruickshank T, Wade MJ, Moczek AP. 2010. Toward a population genetic framework of developmental evolution: the costs, limits, and consequences of phenotypic plasticity. Bioessays 32, 71–81. ( 10.1002/bies.200900132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin DA. 1988. Local differentiation and the breeding structure of plant populations. In Plant evolutionary biology (eds Gottieb LD, Subodh JK.), pp. 305–329. Dordrech, The Netherlands: Springer. [Google Scholar]
- 14.Falconer DS. 2008. Selection in different environments: effects on environmental sensitivity (reaction norm) and on mean performance. Genet. Res. 56, 57–70. ( 10.1017/S0016672300028883) [DOI] [Google Scholar]
- 15.Ghalambor CK, McKay JK, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407. ( 10.1111/j.1365-2435.2007.01283.x) [DOI] [Google Scholar]
- 16.Travisano M, Shaw RG. 2012. Lost in the map. Evolution 67, 305–314. ( 10.1111/j.1558-5646.2012.01802.x) [DOI] [PubMed] [Google Scholar]
- 17.Reusch TBH. 2013. Climate change in the oceans: evolutionary versus phenotypically plastic responses of marine animals and plants. Evol. Appl. 7, 104–122. ( 10.1111/eva.12109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen MM, Olivieri I, Waller DM, Nielsen EE, The GeM Working Group 2012. Monitoring adaptive genetic responses to environmental change. Mol. Ecol. 21, 1311–1329. ( 10.1111/j.1365-294X.2011.05463.x) [DOI] [PubMed] [Google Scholar]
- 19.Merilä J, Hendry AP. 2014. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol. Appl. 7, 1–14. ( 10.1111/eva.12137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fierst JL. 2011. A history of phenotypic plasticity accelerates adaptation to a new environment. J. Evol. Biol. 24, 1992–2001. ( 10.1111/j.1420-9101.2011.02333.x) [DOI] [PubMed] [Google Scholar]
- 21.Collins S, Rost B, Rynearson TA. 2013. Evolutionary potential of marine phytoplankton under ocean acidification. Evol. Appl. 7, 140–155. ( 10.1111/eva.12120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaum E, Rost B, Millar AJ, Collins S. 2013. Variation in plastic responses of a globally distributed picoplankton species to ocean acidification. Nature 3, 298–302. ( 10.1038/nclimate1774) [DOI] [Google Scholar]
- 23.Rokitta SD, Rost B. 2012. Effects of CO2 and their modulation by light in the life-cycle stages of the coccolithophore Emiliania huxleyi. Limnol. Oceanogr. 57, 607–618. ( 10.4319/lo.2012.57.2.0607) [DOI] [Google Scholar]
- 24.Langer G, Geisen M, Baumann K-H, Kläs J, Riebesell U, Thoms S, Young JR. 2006. Species-specific responses of calcifying algae to changing seawater carbonate chemistry. Geochem. Geophys. Geosyst. 7, 2637–2646. ( 10.1029/2005GC001227) [DOI] [Google Scholar]
- 25.Hutchins D, Fu F, Zhang Y, Warner M. 2007. CO2 control of Trichodesmium N2 fixation, photosynthesis, growth rates, and elemental ratios: implications for past, present, and future ocean biogeochemistry. Limnol. Oceanogr. 52, 1293–1304. ( 10.4319/lo.2007.52.4.1293) [DOI] [Google Scholar]
- 26.Kranz SA, Eichner M, Rost B. 2011. Interactions between CCM and N2 fixation in Trichodesmium. Photosynth. Res. 109, 73–84. ( 10.1007/s11120-010-9611-3) [DOI] [PubMed] [Google Scholar]
- 27.Tatters AO, Schnetzer A, Fu F, Lie AYA, Caron DA, Hutchins DA. 2013. Short- versus long-term responses to changing CO2 in a coastal dinoflagellate bloom: implications for interspecific competitive interactions and community structure. Evolution 7, 1879–1891. ( 10.1111/evo.12029) [DOI] [PubMed] [Google Scholar]
- 28.Lohbeck KT, Riebesell U, Reusch TBH. 2012. Adaptive evolution of a key phytoplankton species to ocean acidification. Nat. Geosci. 5, 346–351. ( 10.1038/ngeo1441) [DOI] [Google Scholar]
- 29.Lohbeck KT, Riebesell U, Collins S, Reusch TBH. 2012. Functional genetic divergence in high CO2 adapted Emiliania huxleyi populations. Evolution 67, 1892–1900. ( 10.1111/j.1558-5646.2012.01812.x) [DOI] [PubMed] [Google Scholar]
- 30.Crawfurd KJ, Raven JA, Wheeler GL, Baxter EJ, Joint I. 2011. The response of Thalassiosira pseudonana to long-term exposure to increased CO2 and decreased pH. PLoS ONE 6, e26695 ( 10.1371/journal.pone.0026695.s002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins S, Bell G. 2004. Phenotypic consequences of 1,000 generations of selection at elevated CO2 in a green alga. Nature 431, 566–569. ( 10.1038/nature02945) [DOI] [PubMed] [Google Scholar]
- 32.Jin P, Gao K, Beardall J. 2013. Evolutionary responses of a coccolithophorid Gephyrocapsa oceanica to ocean acidification. Evolution 67, 1869–1878. ( 10.1111/evo.12112) [DOI] [PubMed] [Google Scholar]
- 33.Collins S. 2011. Many possible worlds: expanding the ecological scenarios in experimental evolution. Evol. Biol. 38, 3–14. ( 10.1007/s11692-010-9106-3) [DOI] [Google Scholar]
- 34.Rost B, Zondervan I, Wolf-Gladrow D. 2008. Sensitivity of phytoplankton to future changes in ocean carbonate chemistry: current knowledge, contradictions and research directions. Mar. Ecol. Prog. Ser. 373, 227–237. ( 10.3354/meps07776) [DOI] [Google Scholar]
- 35.Riebesell U, Tortell PD. 2011. Effects of ocean acidification on pelagic organisms and ecosystems. In Ocean acidification (eds Gattuso JP, Hanson L.), pp. 99–121. Oxford, UK: Oxford University Press. [Google Scholar]
- 36.Cooper TF, Lenski RE. 2010. Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol. Biol. 10, 11 ( 10.1186/1471-2148-10-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puentes-Tellez PE, Hansen MA, Sorensen SJ, van Elsas JD. 2013. Adaptation and heterogeneity of Escherichia coli MC1000 growing in complex environments. Appl. Environ. Microbiol. 79, 1008–1017. ( 10.1128/AEM.02920-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Botero CA, Weissing FJ, Wright J, Rubenstein DR. Submitted. Evolutionary tipping points in the capacity to adapt to environmental change. Proc. Natl Acad. Sci. [DOI] [PMC free article] [PubMed]
- 39.Bonduriansky R, Crean AJ, Day T. 2011. The implications of nongenetic inheritance for evolution in changing environments. Evol. Appl. 5, 192–201. ( 10.1111/j.1752-4571.2011.00213.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courties C, VAQUER A, Troussellier M, Lautier J, Chretiennotdinet MJ, Neveux J, Machado C, Claustre H. 1994. Smallest eukaryotic organism. Nature 370, 255 ( 10.1038/370255a0) [DOI] [Google Scholar]
- 41.Subirana L, et al. 2013. Morphology, genome plasticity, and phylogeny in the genus. Ann. Anat. 164, 643–659. ( 10.1016/j.protis.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 42.IPCC. 2007. Intergovernmental Panel on Climate Change (2007) Working Group I report ‘The physical science basis’. See http://ipcc-wg1.ucar.edu/wg1/wg1-report.html .
- 43.Keller MD, Selvin RC, Claus W, Guillard R. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638. ( 10.1111/j.1529-8817.1987.tb04217.x) [DOI] [Google Scholar]
- 44.Baracca A, Sgarbi G, Solaini G, Lenaz G. 2003. Rhodamine 123 as a probe of mitochondrial membrane potential: evaluation of proton flux through F0 during ATP synthesis. Biochim. Biophys. Acta 1606, 137–146. ( 10.1016/S0005-2728(03)00110-5) [DOI] [PubMed] [Google Scholar]
- 45.Lewis E, Wallace D. 1998. CO2SYS: program developed for the CO2 system calculations. Carbon Dioxide Inf. Anal. Center. Oak Ridge National Laboratory, US Department of Energy, Oak Ridge, TN, USA.
- 46.Collins S. 2010. Competition limits adaptation and productivity in a photosynthetic alga at elevated CO2. Proc. R. Soc. B 278, 247–255. ( 10.1098/rspb.2010.1173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pouneva I. 1997. Evaluation of algal culture viability and physiological state by fluorescent microscopic methods. Bulg. J. Plant Physiol 23, 67–76. [Google Scholar]
- 48.Price TD, Qvarnstrom A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. B 270, 1433–1440. ( 10.1098/rspb.2003.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reboud X, Bell G. 1997. Experimental evolution in Chlamydomonas. III. Evolution of specialist and generalist types in environments that vary in space and time. Heredity 78, 507–514. ( 10.1038/hdy.1997.79) [DOI] [Google Scholar]
- 50.Ketola T, Mikonranta L, Zhang J, Saarinen K, Örmälä AM, Friman VP, Mappes J, Laakso J. 2013. Fluctuating temperature leads to evolution of thermal generalism and preadaptation to novel environments. Evolution 67, 2936–2944. ( 10.1111/evo.12148) [DOI] [PubMed] [Google Scholar]
- 51.Crill WD, Wichman HA, Bull JJ. 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kassen R. 2002. The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 15, 173–190. ( 10.1046/j.1420-9101.2002.00377.x) [DOI] [Google Scholar]
- 53.Bradshaw AD. 1965. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 13, 115–155. ( 10.1016/S0065-2660(08)60048-6) [DOI] [Google Scholar]
- 54.Pigliucci M. 2005. Evolution of phenotypic plasticity: where are we going now? Trends Ecol. Evol. 20, 481–486. ( 10.1016/j.tree.2005.06.001) [DOI] [PubMed] [Google Scholar]
- 55.Franks SJ, Hoffmann AA. 2012. Genetics of climate change adaptation. Annu. Rev. Genet. 46, 185–208. ( 10.1146/annurev-genet-110711-155511) [DOI] [PubMed] [Google Scholar]
- 56.Bergmann N, Winters G, Rauch G, Eizaguirre C, Gu J, Nelle P, Fricke B, Reusch TBH. 2010. Population-specificity of heat stress gene induction in northern and southern eelgrass Zostera marina populations under simulated global warming. Mol. Ecol. 19, 2870–2883. ( 10.1111/j.1365-294X.2010.04731.x) [DOI] [PubMed] [Google Scholar]
- 57.Fu F-X, Warner ME, Zhang Y, Feng Y, Hutchins DA. 2007. Effects of increased temperature and CO2 on photosynthesis, growth, and elemental ratios in marine Synechococcus and Prochlorococcus (Cyanobacteria). J. Phycol. 43, 485–496. ( 10.1111/j.1529-8817.2007.00355.x) [DOI] [Google Scholar]
- 58.Hutchins DA, Mulholland MR, Fu F. 2009. Nutrient cycles and marine microbes in a CO2-enriched ocean. Oceanography 22, 128–145. ( 10.5670/oceanog.2009.103) [DOI] [Google Scholar]
- 59.Collins S, Sültemeyer D, Bell G. 2006. Rewinding the tape: selection of algae adapted to high CO2 at current and pleistocene levels of CO2. Evolution 60, 1392–1401. ( 10.1111/j.0014-3820.2006.tb01218.x) [DOI] [PubMed] [Google Scholar]
- 60.Watve M, Parab S, Jogdand P, Keni S. 2006. Aging may be a conditional strategic choice and not an inevitable outcome for bacteria. Proc. Natl Acad. Sci. USA 103, 14 831–14 835. ( 10.1073/pnas.0606499103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341. ( 10.1086/285289) [DOI] [Google Scholar]
- 62.Chao L. 2010. A model for damage load and its implications for the evolution of bacterial aging. PLoS Genet. 6, e1001076 ( 10.1371/journal.pgen.1001076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laney SR, Olson RJ, Sosik HM. 2012. Diatoms favor their younger daughters. Limnol. Oceanogr. 57, 1572–1578. ( 10.4319/lo.2012.57.5.1572) [DOI] [Google Scholar]
- 64.de Paula WBM, Agip ANA, Missirlis F, Ashworth R, Vizcay-Barrena G, Lucas CH, Allen JF. 2013. Female and male gamete mitochondria are distinct and complementary in transcription, structure, and genome function. Genome Biol. Evol. 5, 1969–1977. ( 10.1093/gbe/evt147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collins S, Lawrie E, Brickley M. In preparation. Evolutionary changes in mitochondrial function in response to high CO2 in a green alga.
- 66.Egleston ES, Sabine CL, Morel FMM. 2010. Revelle revisited: buffer factors that quantify the response of ocean chemistry to changes in DIC and alkalinity. Glob. Biogeochem. Cycles 24, GB1002 ( 10.1029/2008GB003407) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available at https://datadryad.org/handle/10255/3/submit?workspaceID=93761.