ABSTRACT
Conjugative plasmids have been identified in a wide variety of different bacteria, ranging from proteobacteria to firmicutes, and conjugation is one of the most efficient routes for horizontal gene transfer. The most widespread mechanism of plasmid conjugation relies on different variants of the type IV secretion pathway. Here, we describe the identification of a novel type of conjugative plasmid that seems to be unique for mycobacteria. Interestingly, while this plasmid is efficiently exchanged between different species of slow-growing mycobacteria, including Mycobacterium tuberculosis, it could not be transferred to any of the fast-growing mycobacteria tested. Genetic analysis of the conjugative plasmid showed the presence of a locus containing homologues of three type IV secretion system components and a relaxase. In addition, a new type VII secretion locus was present. Using transposon insertion mutagenesis, we show that in fact both these secretion systems are essential for conjugation, indicating that this plasmid represents a new class of conjugative plasmids requiring two secretion machineries. This plasmid could form a useful new tool to exchange or introduce DNA in slow-growing mycobacteria.
IMPORTANCE
Conjugative plasmids play an important role in horizontal gene transfer between different bacteria and, as such, in their adaptation and evolution. This effect is most obvious in the spread of antibiotic resistance genes. Thus far, conjugation of natural plasmids has been described only rarely for mycobacterial species. In fact, it is generally accepted that M. tuberculosis does not show any recent sign of horizontal gene transfer. In this study, we describe the identification of a new widespread conjugative plasmid that can also be efficiently transferred to M. tuberculosis. This plasmid therefore poses both a threat and an opportunity. The threat is that, through the acquisition of antibiotic resistance markers, this plasmid could start a rapid spread of antibiotic resistance genes between pathogenic mycobacteria. The opportunity is that we could use this plasmid to generate new tools for the efficient introduction of foreign DNA in slow-growing mycobacteria.
INTRODUCTION
Horizontal gene transfer (HGT) is a major driving force of bacterial diversification and speciation. Together with phage transduction and natural transformation, the exchange of DNA by the exchange of plasmids is one of the best known mechanisms for HGT. Transmissible plasmids are classified as being either mobilizable or conjugative (1). Mobilizable plasmids carry the so-called mobility (MOB) region, needed for the actual recognition, binding, and exchange of the DNA. However, they lack genes coding for the coupling protein and/or the mating pair formation complex, needed for contacting the recipient cell and production of a transport channel. The MOB region usually consists of an oriT, i.e., a short cis-acting DNA sequence, and a gene encoding a TraA relaxase. Relaxase recognizes the oriT region, cleaves the strand that will be transferred, and brings it toward the mating pair formation complex. In addition, the relaxase facilitates the final stage of conjugation by ligating the DNA in the recipient cell. The coupling protein and mating pair complex is usually formed by a variant of the type IV secretion (T4S) system, which can be supplied in trans. In contrast, conjugative plasmids contain both the MOB functions and the T4S locus, which allows them to be repeatedly transferred to new strains or even new species.
T4S systems are unique secretion systems, due to their ability to transfer both proteins and nucleic acids (2, 3). Furthermore, they also show an extraordinary host range, with variants of T4S systems found in Gram-negative and Gram-positive bacteria and even Archaea (2, 4). As such, the T4S systems show a remarkable substrate and unique cell envelope variability, which is also reflected in the variable number of components that are present within the system. In fact, although T4S machineries can be composed of 12 different proteins, only homologues of 5 proteins are present in all different systems (2). Probably the best known and most remarkable T4S conjugative system is the VirB/VirD4 system of Agrobacterium tumefaciens, which is required for the transfer of a DNA fragment, coding for oncogenic and metabolic proteins, directly from pathogenic bacteria into plant cells (5). Although T4S-dependent conjugative plasmids have been identified in many different bacteria, they have thus far not been identified in mycobacteria.
Mycobacteria have a highly unusual cell envelope that differs from both Gram-negative and Gram-positive bacteria. The mycobacterial cell envelope contains an outer membrane that is composed mainly of long-chain fatty acids, known as mycolic acids, which are partially linked to the arabinogalactan-peptidoglycan complex and partially present as trehalose-linked dimers (6). Together with a range of other unique lipids, these mycolic acids form a second hydrophobic layer (7) of extremely low fluidity. This outer membrane is a nice example of convergent evolution. To transport proteins across this unusual cell envelope, the mycobacteria use a specialized secretion system known as ESX or type VII secretion (T7S) (8–10). Thus far, five T7S subtypes have been described, ESX-1 to -5. The paradigm ESX system is ESX-1, which is crucial for virulence of Mycobacterium tuberculosis and several other mycobacterial pathogens (10). ESX-1 is responsible for the secretion of a number of crucial virulence factors that enable these pathogens to escape from the phagosome into the cytosol of host macrophages (11, 12). Interestingly, the homologous ESX-1 system of the nonpathogenic fast-growing Mycobacterium smegmatis has a completely different role; it has been implicated in an atypical conjugation mechanism that transfers random genomic fragments to other M. smegmatis strains (13, 14). Also, T7S systems seem to have broad substrate specificity.
Although plasmids are generally absent in M. tuberculosis, they have been identified in several other mycobacterial species (15–18). These plasmids can either be circular or linear. Also, conjugation of plasmids in mycobacteria under artificial laboratory conditions has been described previously, albeit with low efficiencies (19, 20). Recently, a study by Rabello et al. (21) described efficient conjugation of a plasmid between different mycobacterial species. This process was also observed in natural conditions. However, the (complete) sequence of the conjugating plasmid and the mechanism involved were not elucidated.
In this study, we report the presence of a new circular plasmid in Mycobacterium marinum. Genetic analysis showed that this plasmid codes for a putative T7S system, for several genes indicative of a T4S system, and for a relaxase. This plasmid represents the first fully characterized conjugative plasmid in mycobacteria.
RESULTS
Sequence and widespread presence of pRAW-like plasmids.
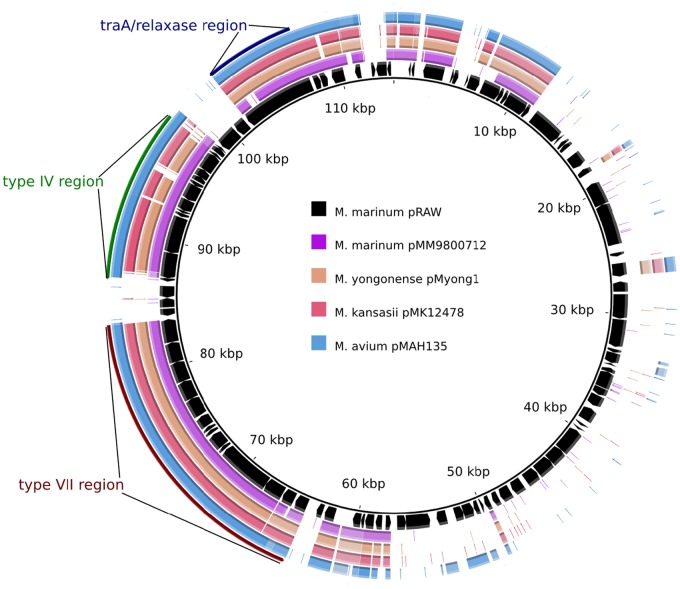
Upon completion of the genome sequence of our model organism, Mycobacterium marinum E11 (E. M. Weerdenburg, A. M. Abdallah, A. Pain, and W. Bitter, submitted for publication), we noticed the presence of a large circular plasmid of 114,229 bp. The plasmid, designated pRAW, has an overall GC content of 64%, which is similar to that of the M. marinum genome. Automated annotation of this plasmid resulted in the prediction of 99 putative coding sequences. pRAW showed no extended sequence homology with other known plasmid sequences of M. marinum or the closely related species Mycobacterium ulcerans. However, highly similar sequences were identified in a number of plasmids from other slow-growing mycobacteria, including Mycobacterium avium subsp. hominis suis, Mycobacterium kansasii, and Mycobacterium yongonense (Table 1, Fig. 1), which indicates that this type of plasmid is widespread within the slow-growing mycobacteria. Closer analysis of these sequences showed that they all contain a core segment of approximately 55 kb (Fig. 1; see also Table S1 in the supplemental material). This conserved region contains a locus coding for a TraA relaxase and a locus with both a virB4 and a virD4 homologue (respectively, mmarE11p_00780 and mmarE11p_00860 in pRAW; see Table S1). This virB4/D4 locus also contained a gene coding for a TcpC homologue (mmarE11p_00810; see Table S1), which is a protein involved in plasmid conjugation in Clostridium perfringens and a structural homologue of VirB8 (22). Finally, this locus contains another small gene coding for a protein with some homology to VirD4 (mmarE11p_00880). We could not identify homologues coding for other T4S components, including the generally conserved components VirB3 and VirB6. The presence of this combination of genes indicates that this plasmid could be conjugative, although some conserved T4S components seem to be absent.
TABLE 1 .
Characteristics of the pRAW-like plasmids analyzed in this study
| Host species | Plasmida | Size (bp) | Reference/source |
|---|---|---|---|
| M. marinum E11 | pRAW | 114,229 | E. M. Weerdenburg et al., submitted |
| M. avium subsp. hominis suis TH135 | pMAH135 | 194,711 | K. Uchiya and K. Ogawa, unpublished data |
| M. kansasii ATCC 12478 | pMK12478 | 144,951 | F. J. Veyrier and M. A. Behr, unpublished data |
| M. yongonense 05-1390 | pMyong1 | 122,976 | 33 |
| M. marinum 9800712 | pMM98 | ~127,300 | This study |
FIG 1 .
pRAW-like plasmids all have a conserved locus for T4S, T7S, and relaxase. Multiple alignment of pRAW with the 4 other mycobacterial plasmids mentioned in Table 1. Coding sequences (CDS) of pRAW are indicated with black arrows, and colored regions indicate sequence identity with other plasmids. Clusters coding for putative T4S and T7S and relaxase are indicated.
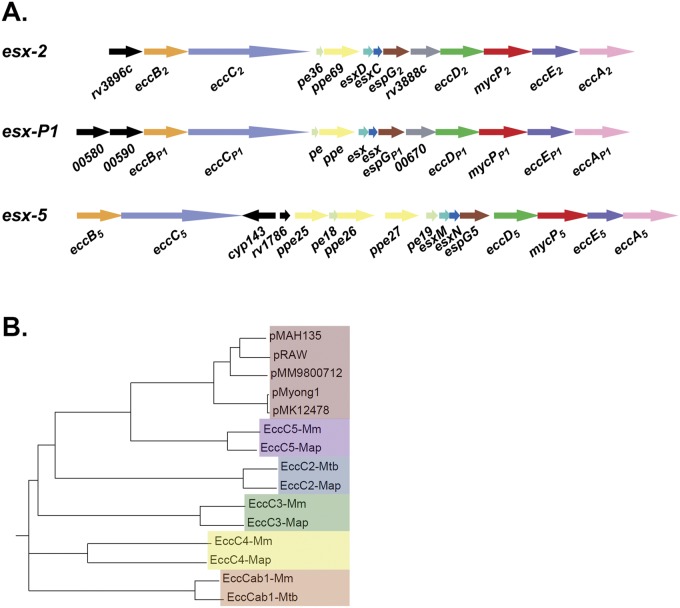
In addition to the genes described above, the conserved plasmid segment also contained a locus coding for a T7S system (Fig. 1 and 2). Mycobacteria contain up to 5 different T7S systems, designated ESX-1 through ESX-5. This new putative ESX system shows the most homology with ESX-5 (Fig. 2; see also Fig. S1 in the supplemental material), although gene order and presence were more like those of ESX-2 (Fig. 2A). The only major difference with known ESX systems is the presence of two new genes (mmarE11p_00580 and mmarE11p_00590) putatively coding for members of the NLP/P60 family of peptidoglycan-associated glycoside hydrolases (Fig. 2A; see also Table S1). These genes are located at the start of the locus and could indicate that this ESX system has a novel function compared to those of the known ESX systems. Because this new system is located on a plasmid and seems to be different from all known ESX systems, we decided to name it ESX-P1.
FIG 2 .
Plasmid-encoded ESX-P1 systems form a homogeneous group that is most homologous to the ESX-5 systems. (A) Gene map showing the esx-P1 locus from pRAW using esx-2 and esx-5 of M. tuberculosis as the reference. Gene colors are similar to those used in reference 9. The gray gene, which is exclusively present in esx-1, esx-2, and esx-P1, codes for a member of the MinD/ParA protein family. Black genes are unique and have no homologue within the esx loci. (B) Phylogenetic tree based on ClustalW2 alignment showing EccC proteins of the different Esx-P1 T7S systems and different EccC proteins from the known mycobacterial T7S systems (ESX-1 through ESX-5). For comparison, we used the EccC proteins from M. marinum (M) and M. avium subsp. paratuberculosis and EccC1 and EccC2 from M. tuberculosis H37Rv.
Together, these data show that there is a group of plasmids in slow-growing mycobacteria that have a highly conserved region coding for a putative relaxase and two different secretion systems putatively involved in plasmid conjugation. To examine the distribution of these types of plasmids in natural isolates of M. marinum and other mycobacteria in more detail, we performed pRAW-specific PCRs for a collection of M. marinum strains. The primers were directed to conserved sequences of the 3 different regions, i.e., the gene encoding relaxase, the virB4-like gene, and two different primer pairs for the esx-P1 locus (see Table S2 in the supplemental material), targeting the conserved eccC or eccA homologues. As expected, we could amplify a DNA fragment of the expected size with all primer pairs for E11, whereas the previously sequenced M strain was negative (Table 2). Out of the 14 other M. marinum strains, 7 tested positive for at least one of these primer pairs. Furthermore, 6 out of the 7 positive strains showed a positive reaction for all PCRs. The PCR-positive M. marinum strains showed a highly diverse origin, with both animal isolates and clinical isolates (Table 2). Also, one other slow-growing mycobacterium (Mycobacterium parascrofulaceum) showed the presence of all these genes. Genome sequencing of one of these strains, M. marinum 9800712, confirmed the presence of a core segment with all three described clusters, which are probably located on a pRAW-like plasmid (Fig. 1; see also Table S1). These data show that (i) pRAW-like plasmids are widely present in M. marinum and several other slow-growing mycobacteria (M. yongonense, M. parascrofulaceum, M. kansasii, and M. avium) and (ii) the presence of these three different gene clusters seem to be strongly linked.
TABLE 2 .
Presence of pRAW-conserved loci in different M. marinum strains and other slow-growing mycobacteria
| Mycobacterium species | Strain | Source | PCR resulta |
|||
|---|---|---|---|---|---|---|
| eccCP1 | eccAP1 | virB4-like | traA/relaxase | |||
| M. marinum | E11 | Fish | + | + | + | + |
| M. marinum | MUSA | Human | − | − | − | − |
| M. marinum | E6 | Fish | − | − | − | − |
| M. marinum | E7 | Fish | − | − | + | − |
| M. marinum | E12 | Fish | + | + | + | + |
| M. marinum | E15 | Fish | + | + | + | + |
| M. marinum | E16 | Fish | + | + | + | + |
| M. marinum | 14641 | Human | − | − | − | − |
| M. marinum | 18347 | Human | − | − | − | − |
| M. marinum | 551962 | Reptile | + | + | + | + |
| M. marinum | 9800607 | Human | − | − | − | − |
| M. marinum | 9800712 | Human | + | + | + | + |
| M. marinum | 9801756 | Human | − | − | − | − |
| M. marinum | 9900036 | Human | − | − | − | − |
| M. marinum | 2000-01053 | Human | − | − | − | − |
| M. marinum | 2001-00796 | Human | + | + | + | + |
| M. chelonae | RIVM1010700496 | − | − | + | − | |
| M. fortuitum | RIVM082273 | − | − | − | − | |
| M. kansasii | 852 | Human | − | − | − | − |
| M. parascrofulaceum | RIVM600526 | + | + | + | + | |
| M. triviale | St5 | − | − | − | − | |
| M. xenopi | RIVM1010700367 | − | − | − | − | |
+, DNA fragment amplified of roughly the expected size; −, no DNA fragment amplified.
pRAW is a conjugative plasmid and is efficiently transferred.
To determine whether pRAW is indeed a conjugative plasmid, we had to introduce a selection marker. Because this plasmid is too large to be manipulated by recombinant DNA methods and homologous recombination is notoriously difficult in mycobacteria, we decided to use another approach. We used a large Himar1 transposon library of M. marinum E11. Hypothetically, these transposons, containing a kanamycin resistance gene, will randomly insert in the genome and in the pRAW plasmid. Subsequently, we performed conjugation experiments with another M. marinum strain as the recipient, i.e., M. marinum M supplemented with a plasmid encoding hygromycin resistance and DsRed. Selection was on plates containing both antibiotics, and colonies were examined for the presence of red fluorescence. The prediction is that colonies growing on these selection plates will belong to strain M and contain a pRAW-like plasmid with a transposon insertion. Because both parent strains have been sequenced (23) (Weerdenburg et al., submitted), we could develop primers specific for genes unique for both strains (see Table S2). The obtained putative transconjugant colonies were analyzed using these species-specific primers, which showed that all positive colonies belonged to the M strain, as expected. Subsequently, we determined the location of the transposon for 15 independent transconjugant colonies by performing ligation-mediated PCR. All colonies had a transposon insertion within the pRAW plasmid, in total at 5 different locations (Table 3). This experiment shows not only that the pRAW plasmid is conjugative but also that chromosomal regions are not (co)transferred with high frequency; otherwise, we would have identified transposon insertions in chromosomal DNA as well. Although this experiment showed transfer of the pRAW plasmid, still some essential transfer functions could have been provided by the donor genome. Therefore, we performed a second conjugation experiment using one of the newly created strains, i.e., M. marinum M strain with pRAW containing a transposon insertion at bp 47,881 (colony 4 in Table 3), as the donor and M. marinum M with a streptomycin resistance marker integrated on the genome as the recipient. Since we now had a version of the pRAW plasmid with a selectable marker, we could also determine conjugation efficiency in the same experiment. This experiment showed that the transposon-containing pRAW plasmid could indeed be conjugated to another M strain, indicating that this plasmid contains all information required for conjugation (Fig. 3). Furthermore, conjugation efficiency, after filter-based conjugation and overnight incubation, was on average 6.3 (±6.0) × 10−2 (n = 5), which means that 1 out of 16 recipient cells had received the pRAW plasmid, indicating highly efficient transfer.
TABLE 3 .
Transposon insertion sites in pRAW of M. marinum M recipient colonies
| Colony | Transposon insertion site (bp) | Gene affected |
|---|---|---|
| MUSA 1 | 47881-47882 | mmarE11p_00410 |
| MUSA 2 | 46290-46291 | mmarE11p_00400 |
| MUSA 3 | 8752-8753 | mmarE11p_00080 |
| MUSA 4 | 58700-58701 | Intergenic |
| MUSA 5 | 58664-58665 | Intergenic |
| Type VII 1 | 68840-68841 | eccCP1 (mmarE11p_00610) |
| Type IV 1 | 90304-90305 | mmarE11p_00780 (virB4-like) |
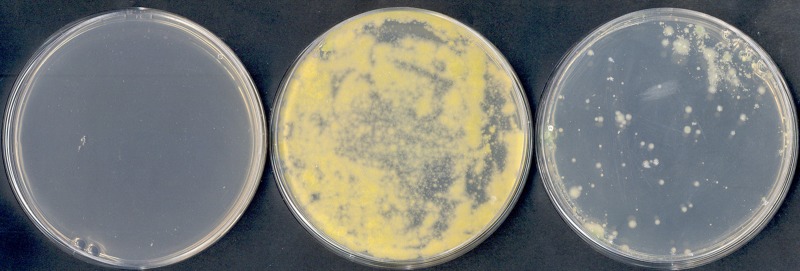
FIG 3 .
Conjugation of pRAW between two different M. marinum M strains, i.e., a pRAW containing donor and empty acceptor strain. The plate on the left shows a control selection plate with only recipient cells that have not been in contact with donor cells; the other two plates show recipient cells that have been incubated for 48 h with two different donor strains carrying pRAW with a transposon insertion at bp 58700 (middle plate) or bp 46290 (right plate). Donor cells do not grow on these selection plates.
Because we used a pRAW variant with a transposon insertion that was selected after a conjugation procedure, we could have selected for mutants with increased conjugation efficiency. Therefore, we determined conjugation efficiency of an isolate with a different transposon insertion (i.e., colony number 2 in Table 3). Although this isolate conjugated efficiently to a new M. marinum strain, the conjugation frequency was lower, i.e., 1 in 250 (n = 3) (Fig. 3). The transposon in the high-efficiency conjugation mutant is inserted at bp 58700, which is an intergenic region just in front of hypothetical gene p00530. Possibly, this gene plays a role in the regulation of conjugation. In conclusion, the conjugation frequency of transposon-containing pRAW variants is high.
pRAW has a restricted host range.
Genome sequencing had shown that pRAW-like plasmids are present in different slow-growing mycobacteria (Table 1, Fig. 1). Furthermore, we also identified close homologues of the conserved plasmid domain in three unfinished genome sequencing of slow-growing mycobacteria, including Mycobacterium parascrofulaceum ATCC BAA614 and two M. avium strains (11-0986 and 10-5581). These data show that these types of plasmids are widespread in slow-growing mycobacteria. To examine the host range of the pRAW conjugative plasmid, we performed conjugation experiments with a number of fast-growing mycobacteria and some slow-growing species (Table 4). Interestingly, successful conjugation could be established for the slow-growing species M. tuberculosis and Mycobacterium bovis BCG. In contrast, we obtained only occasional spontaneous resistant colonies and no transconjugants for any of the six fast-growing mycobacteria tested and for the slow-growing species Mycobacterium triviale. Apparently, pRAW seems to be limited to (some) slow-growing mycobacteria. To further examine whether the pRAW plasmid is indeed self-sufficient for conjugation, we next tested whether the newly created M. tuberculosis strain containing pRAW could function as a donor. In a new conjugation experiment, using three independent M. tuberculosis colonies containing pRAW as the donor, efficient transfer to an empty M. marinum M acceptor strain could be observed.
TABLE 4 .
Mycobacterial species examined as recipient for pRAW
| Mycobacterium species | Strain | Slow/fast growing | Conjugationa |
|---|---|---|---|
| M. smegmatis | MC2 155 | Fast | − |
| M. fortuitum | RIVM082273 | Fast | − |
| M. phlei | RIVM1010601174 | Fast | − |
| M. chelonae | RIVM1010700496 | Fast | − |
| M. holsaticum | RIVM1010601271 | Fast | − |
| M. gilvum | NCTC10942 | Fast | − |
| M. triviale | st5 | Slow | − |
| M. tuberculosis | MC2 6020 | Slow | + |
| M. bovis | BCG Pasteur | Slow | + |
+, multiple colonies on selection plates that belong to the acceptor strain and do contain the pRAW plasmid (as checked by PCR); −, no colonies on the selection plates or only colonies that do not contain the pRAW plasmid (false positives).
Both the virB4 homologue and eccCP1 are essential for plasmid conjugation.
The conserved region of this new type of plasmid contains both a locus for a T7S system (esx-P1) and a variant T4S system. Since both these systems have been linked to conjugation previously (2, 13), we wondered which of these systems was involved in pRAW conjugation. To study this, we identified mutants in the eccCP1 gene (mmarE11p_00610) and the virB4 homologue (mmarE11p_00780), which are usually coding for essential components of the T7S and T4S systems, respectively. We selected these mutants by screening batchwise our random transposon library using one transposon-specific primer and one primer located near our gene of interest (see Materials and Methods). Once a batch tested positive, single colonies from this batch were tested. Using this approach, we identified transposon insertions in both genes (Table 3). Subsequent testing of these mutants in conjugation experiments showed that both genes were in fact crucial for conjugation, i.e., no transconjugants could be identified. To confirm these results, we complemented these mutations by introducing a plasmid with a functional copy of the disrupted genes. As expected, complementation of these mutations restored conjugation of the pRAW plasmid. These data show that both the variant T4S system and T7S system ESX-P1 are essential for conjugation, indicating that a unique conjugation mechanism is probably involved.
DISCUSSION
The unique outer membrane of mycobacteria is not evolutionary related to the outer membrane of Gram-negative bacteria. As a consequence, outer membrane proteins and cell envelope transport systems are completely different between these classes of bacteria. This difference is exemplified by the only mycobacterial outer membrane porin with known structure, MspA (24). Although this protein forms a beta-barrel, similar to Gram-negative outer membrane porins, the actual structure and sequence is completely different. Therefore, at first sight, it seems surprising that a variant T4S system is present in mycobacteria to produce a mating pair complex. There are two important aspects that could help to explain this ambiguity. The first aspect is that T4S systems have an extraordinary variability, both in the nature of the secreted substrate and in the adaptation to different cell envelope architectures; T4S systems are involved in plasmid conjugation in a wide variety of species ranging from Gram-negative bacteria to Archaea (2, 3). Another special feature showing the variability of T4S systems is that, although these systems usually operate as a one-step mechanism, some variants secrete substrates from the periplasm (25). The second aspect that could indicate why T4S could function in mycobacteria is that we, thus far, have identified only inner membrane or cytoplasmic components of the T4S system, i.e., homologues of the ATPase VirB4 and VirD4 and a structural homologue of the inner membrane protein VirB8, whereas the proteins that form the periplasmic/outer membrane complex (VirB7, VirB9, and VirB10) and even some conserved components of the inner membrane (VirB3, VirB6) are missing. We hypothesize two different possibilities. The first is that the outer membrane components have yet to be identified; they might, for instance, be encoded by the other genes of the variant T4S locus on the conjugative plasmids. This locus contains more than 10 genes, and, apart from the three genes mentioned, no clear functions can be assigned to these genes based on homology searches, but some of them are coding for proteins with putative transmembrane domains. In this scenario, the ESX-P1 system or its substrates could function as an adhesion structure, required for the initiation of cell-cell contact, while the variant T4S system forms the mating pair complex. The other possibility is that the variant T4S system forms a mating pair complex together with the ESX-P1 complex. If this second hypothesis is true, one would expect structural adaptations to the ESX-P1 machinery to accommodate interaction with the T4S system. Apart from the presence of two new members of the NLP/P60 protein family, we see no obvious differences between ESX-P1 and other T7S systems. Interestingly, one of the missing components that is usually conserved in the T4S system is VirB1 (26), which is in fact also an NLP/P60 protein. Therefore, perhaps these NLP/P60 proteins encoded by the ESX-P1 cluster form the bridge between the two secretion systems.
Because both the variant T4S and ESX-P1 are required for conjugation, the total domain required for this function, including the relaxase-encoding gene locus, accounts for more than 40 kb. This means that in fact most of the 55-kb conserved region of the 5 sequenced pRAW-like plasmids is linked to conjugation (Fig. 1; see also Table S1 in the supplemental material). The other conserved genes are coding mainly for hypothetical (plasmid) proteins. In addition to this large conserved region, each plasmid also contains unique loci. The functions of the nonconserved genes of plasmid pRAW-like plasmids seem to be highly variable, indicating that these plasmids evolve relatively quickly. Thus far, we have been unable to link any putative physiological function to the specific region of pRAW, although we noticed a large number of genes with a predicted regulatory function. We also tested the virulence of M. marinum M with and without the pRAW plasmid in zebrafish embryos and could not find any clear differences (results not shown).
Apart from the observation that two secretion systems are both essential for conjugation, the most surprising finding is that the presence and transfer of pRAW-like plasmids seem to be limited to slow-growing mycobacteria. The genus Mycobacterium can be roughly divided in two clades based on the appearance or not of visible colonies within 7 days of incubation (27). Genomic analysis showed that this growth difference also marks the difference between two genetic clades, although the actual physiological difference has not been identified yet. Our results with the pRAW plasmid could indicate that the cell envelopes of fast- and slow-growing mycobacteria have a fundamental difference, leading to host variation. The only exception seems to be the slow-growing Mycobacterium triviale, which was not able to receive the plasmid. Interestingly, M. triviale is one of the earliest branching slow-growing mycobacteria, and its classification shows some problematic characteristics (28). However, perhaps the host variation of mycobacterial conjugative plasmids is more variable; genome BLAST analysis did show weaker homologues of the pRAW T4S system in the genome sequences of the fast-growing Mycobacterium chubuense NBB4 and Mycobacterium rhodesiae JS60. Therefore, different version of these plasmids could perhaps be able to cross the boundary between fast- and slow-growing mycobacteria.
Based on our genetic analysis of different M. marinum strains and the identification of similar pRAW-like plasmids in other species (Tables 1 and 2), we conclude that the presence of these conjugative plasmids is widespread in slow-growing mycobacteria. The notable exception is M. tuberculosis, which is, with thousands of strains sequenced (29, 30), the most studied mycobacterial species. We have demonstrated in this study that M. tuberculosis can be efficiently targeted by this plasmid. Furthermore, it has also been demonstrated previously that conjugal transfer of plasmids between slow-growing mycobacteria, i.e., M. avium and M. kansasii, can occur in mixed infections in a patient (21). Therefore, perhaps the most probable explanation is that these conjugative plasmids are sometimes transferred to, but not stably maintained in, M. tuberculosis. Of course, this situation could change if one of these conjugative plasmids would acquire antibiotic resistance markers, enabling M. tuberculosis to survive antibiotic treatments. Therefore, the spread and evolution of pRAW-like plasmids are a potential future threat that has to be studied and monitored in detail. Another reason to study this conjugative plasmid is that it could provide a highly efficient way to introduce genetic material in these difficult bacteria, using more genetically amenable mycobacteria such as M. marinum as an intermediate, and could further extend the genetic toolbox of tuberculosis researchers.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. marinum wild-type strain MUSA (23) and E11 (31) were routinely grown in Middlebrook 7H9 liquid medium or Middlebrook 7H10 agar supplemented with 10% Middlebrook albumin-dextrose-catalase (ADC) or oleic acid-albumin-dextrose-catalase (OADC), respectively (BD Biosciences), and 0.05% Tween 80. The recipient MUSA strains contain either a pSMT3 plasmid with hygromycin resistance marker and DsRed (31) or a streptomycin resistance cassette and a lacZ reporter gene integrated on its genome using integration vector pSM128 (32). When required, antibiotics were added at the following concentration: kanamycin, 25 mg/ml; hygromycin, 50 mg/ml; and streptomycin, 35 mg/ml. To determine the distribution of pRAW-like plasmids, we used a large collection of animal and human isolates of M. marinum. Most of these strains have been described previously (31), and the human isolates were derived from the RIVM National Mycobacteria Reference Laboratory. For the conjugation experiments, we used the mycobacterium species mentioned in Table 2, which were obtained from the same reference laboratory. Escherichia coli strain DH5α was used for all cloning procedures and cultured at 37°C in LB liquid broth or solid LB agar.
Conjugation.
For mating experiments, the different strains were first freshly grown to an optical density at 600 nm (OD600) between 1.2 and 1.5. Cells of a 1-ml culture were isolated by centrifugation and resuspended in 50 µl 7H9 broth without Tween 80. The suspensions of both donor and recipient cells were pooled and pipetted on a membrane filter (0.45 µm, type HA; Merck Millipore) that were placed on top of a 7H10 plate. The plates where then incubated at 30°C for different time periods (24 h, 48 h, and 72 h). After incubation, the filter, together with the dried pellet, was gently placed into a sterile 15-ml falcon tube. A total of 2 ml 7H9 broth supplemented with 0.05% Tween 80 was added to the filter, and the cell pellet was resuspended by vortexing. Different dilutions of this suspension were plated on 7H10 plates supplemented with the appropriate antibiotics. For the determination of the conjugation efficiency, plates were used to select for all recipient cells. Based on the genome sequences of MUSA and E11, we generated two specific PCR primer pairs for strain-specific regions (primers in Table 4).
Complementation of mutants.
To complement the mutants, three different constructs were made. The eccCP1 gene, the virB4-like gene, or the eccBCP1 operon were amplified by PCR with Phusion DNA polymerase (Thermo Scientific) using M. marinum E11 isolated DNA as the template and the primers indicated in Table S2 in the supplemental material. The primers also contained restriction sites for AflII, HindIII, or PacI, as indicated. After amplification by PCR, DNA fragments of the correct size were extracted from gel, digested with the appropriate enzymes, and used for cloning in digested pSMT3-enhanced green fluorescent protein (eGFP). Resulting clones were analyzed by colony PCR and sequencing. Correct plasmids were transformed to M. marinum E11 containing pRAW with the transposon insertion in the eccCP1 and virB4-like genes. Subsequently, conjugation experiments were performed as described above.
Genome sequencing.
One microgram of DNA from M. marinum strain 9800712 was used to prepare libraries for next-generation sequencing (NGS) according to the manufacturer’s instruction using the Illumina genomic DNA sample preparation kit (Illumina, Inc., San Diego, CA). The DNA libraries were sequenced on Illumina HiSeq2000 technology (Illumina, Inc., San Diego, CA) according to the manufacturer’s specifications.
Illumina assembly and annotation process.
For the assembly of the M. marinum 9800712 genome and the plasmid, an Illumina paired-end library was used. First, the short reads were assembled with Velvet. We further manually improved the plasmid assembly by using the pRAW plasmid of the M. marinum E11 strain sequence of the orthologous genes and some directed PCR amplification experiments to connect different contigs. For the pMM98 plasmid function annotation, we used PROKKA (http://bioinformatics.net.au/prokka-manual.html) for ab initio annotation. A Bespoke PERL script merged both annotations, choosing the RATT transferred gene models if in a region the PROKKA gene model overlapped the RATT model. The pRAW plasmid has 98 predicted protein-coding genes.
Nucleotide sequence accession number.
The plasmid sequence of pRAW was deposited in the GenBank database (accession number HG917973).
SUPPLEMENTAL MATERIAL
Comparison of the pRAW esx-P1 cluster with the M. marinum esx-5 cluster using the Artemis comparison tool (ACT). Conserved regions are shown in red. Download
Homologues and nonhomologues of pRAW genes in the different pRAW-like plasmids.
Primers used for various purposes in this study.
ACKNOWLEDGMENTS
We thank Shoaib Amini for data analysis and Edith N. G. Houben, Joen Luirink, and Ben J. Appelmelk for valuable discussions.
Footnotes
Citation Ummels R, Abdallah AM, Kuiper V, Aâjoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5(5):e01744-14. doi:10.1128/mBio.01744-14.
REFERENCES
- 1. Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74:434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 164:620–639. 10.1016/j.resmic.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christie PJ. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694:219–234. 10.1016/j.bbamcr.2004.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. 2013. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70:289–302. 10.1016/j.plasmid.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lacroix B, Tzfira T, Vainstein A, Citovsky V. 2006. A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet. 22:29–37. 10.1016/j.tig.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 6. Kaur D, Guerin ME, Skovierová H, Brennan PJ, Jackson M. 2009. Chapter 2: biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv. Appl. Microbiol. 69:23–78. 10.1016/S0065-2164(09)69002-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963–3967. 10.1073/pnas.0709530105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883–891. 10.1038/nrmicro1773 [DOI] [PubMed] [Google Scholar]
- 9. Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5:e1000507. 10.1371/journal.ppat.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simeone R, Bottai D, Brosch R. 2009. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 12:4–10. 10.1016/j.mib.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 11. Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8:e1002507. 10.1371/journal.ppat.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell. Microbiol. 14:1287–1298. 10.1111/j.1462-5822.2012.01799.x [DOI] [PubMed] [Google Scholar]
- 13. Coros A, Callahan B, Battaglioli E, Derbyshire KM. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 69:794–808. 10.1111/j.1365-2958.2008.06299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. U. S. A. 101:12598–12603. 10.1073/pnas.0404892101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bachrach G, Colston MJ, Bercovier H, Bar-Nir D, Anderson C, Papavinasasundaram KG. 2000. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology 146:297–303 [DOI] [PubMed] [Google Scholar]
- 16. Kirby C, Waring A, Griffin TJ, Falkinham JO, III, Grindley ND, Derbyshire KM. 2002. Cryptic plasmids of Mycobacterium avium: Tn552 to the rescue. Mol. Microbiol. 43:173–186. 10.1046/j.1365-2958.2002.02729.x [DOI] [PubMed] [Google Scholar]
- 17. Labidi A, Mardis E, Roe BA, Wallace RJ., Jr. 1992. Cloning and DNA sequence of the Mycobacterium fortuitum var. fortuitum plasmid pAL5000. Plasmid 27:130–140. 10.1016/0147-619X(92)90013-Z [DOI] [PubMed] [Google Scholar]
- 18. Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PD, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. U. S. A. 101:1345–1349. 10.1073/pnas.0305877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gormley EP, Davies J. 1991. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J. Bacteriol. 173:6705–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leão SC, Matsumoto CK, Carneiro A, Ramos RT, Nogueira CL, Lima JD, Jr, Lima KV, Lopes ML, Schneider H, Azevedo VA, da Costa da Silva A. 2013. The detection and sequencing of a broad-host-range conjugative IncP-1β plasmid in an epidemic strain of Mycobacterium abscessus subsp. bolletii. PLoS One 8:e60746. 10.1371/journal.pone.0060746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabello MC, Matsumoto CK, Almeida LG, Menendez MC, Oliveira RS, Silva RM, Garcia MJ, Leão SC. 2012. First description of natural and experimental conjugation between mycobacteria mediated by a linear plasmid. PLoS One 7:e29884. 10.1371/journal.pone.0029884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, Lyras D, Whisstock JC, Rood JI. 2012. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol. Microbiol. 83:275–288. 10.1111/j.1365-2958.2011.07930.x [DOI] [PubMed] [Google Scholar]
- 23. Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18:729–741. 10.1101/gr.075069.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faller M, Niederweis M, Schulz GE. 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189–1192. 10.1126/science.1094114 [DOI] [PubMed] [Google Scholar]
- 25. Pantoja M, Chen L, Chen Y, Nester EW. 2002. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol. Microbiol. 45:1325–1335. 10.1046/j.1365-2958.2002.03098.x [DOI] [PubMed] [Google Scholar]
- 26. Höppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151:3469–3482. 10.1099/mic.0.28326-0 [DOI] [PubMed] [Google Scholar]
- 27. Rastogi N, Legrand E, Sola C. 2001. The mycobacteria: an introduction to nomenclature and pathogenesis. Rev. Sci. Tech. 20:21–54 [DOI] [PubMed] [Google Scholar]
- 28. Tortoli E. 2012. Phylogeny of the genus Mycobacterium: many doubts, few certainties. Infect. Genet. Evol. 12:827–831. 10.1016/j.meegid.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, Liu Z, Li C, Galwey N, Deng J, Zhou Y, Zhu Y, Gao Y, Wang T, Wang S, Huang Y, Wang M, Zhong Q, Zhou L, Chen T, Zhou J, Yang R, Zhu G, Hang H, Zhang J, Li F, Wan K, Wang J, Zhang XE, Bi L. 2013. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45:1255–1260. 10.1038/ng.2735 [DOI] [PubMed] [Google Scholar]
- 30. Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45:1176–1182. 10.1038/ng.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van der Sar AM, Abdallah AM, Sparrius M, Reinders E, Vandenbroucke-Grauls CM, Bitter W. 2004. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect. Immun. 72:6306–6312. 10.1128/IAI.72.11.6306-6312.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dussurget O, Timm J, Gomez M, Gold B, Yu S, Sabol SZ, Holmes RK, Jacobs WR, Jr, Smith I. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim BJ, Kim BR, Lee SY, Seok SH, Kook YH, Kim BJ. 2013. Whole-genome sequence of a novel species, Mycobacterium yongonense DSM 45126T. Genome Announc. 1:e00604-13. 10.1128/genomeA.00604-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the pRAW esx-P1 cluster with the M. marinum esx-5 cluster using the Artemis comparison tool (ACT). Conserved regions are shown in red. Download
Homologues and nonhomologues of pRAW genes in the different pRAW-like plasmids.
Primers used for various purposes in this study.