ABSTRACT
Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) was recognized in Europe and worldwide in the late 1990s. Within a decade, several genetically and geographically distinct CA-MRSA lineages carrying the small SCCmec type IV and V genetic elements and the Panton-Valentine leukocidin (PVL) emerged around the world. In Europe, the predominant CA-MRSA strain belongs to clonal complex 80 (CC80) and is resistant to kanamycin/amikacin and fusidic acid. CC80 was first reported in 1993 but was relatively rare until the late 1990s. It has since been identified throughout North Africa, the Middle East, and Europe, with recent sporadic reports in sub-Saharan Africa. While strongly associated with skin and soft tissue infections, it is rarely found among asymptomatic carriers. Methicillin-sensitive S. aureus (MSSA) CC80 strains are extremely rare except in sub-Saharan Africa. In the current study, we applied whole-genome sequencing to a global collection of both MSSA and MRSA CC80 isolates. Phylogenetic analyses strongly suggest that the European epidemic CA-MRSA lineage is derived from a PVL-positive MSSA ancestor from sub-Saharan Africa. Moreover, the tree topology suggests a single acquisition of both the SCCmec element and a plasmid encoding the fusidic acid resistance determinant. Four canonical SNPs distinguish the derived CA-MRSA lineage and include a nonsynonymous mutation in accessory gene regulator C (agrC). These changes were associated with a star-like expansion into Europe, the Middle East, and North Africa in the early 1990s, including multiple cases of cross-continent imports likely driven by human migrations.
IMPORTANCE
With increasing levels of CA-MRSA reported from most parts of the Western world, there is a great interest in understanding the origin and factors associated with the emergence of these epidemic lineages. To trace the origin, evolution, and dissemination pattern of the European CA-MRSA clone (CC80), we sequenced a global collection of strains of the S. aureus CC80 lineage. Our study determined that a single descendant of a PVL-positive methicillin-sensitive ancestor circulating in sub-Saharan Africa rose to become the dominant CA-MRSA clone in Europe, the Middle East, and North Africa. In the transition from a methicillin-susceptible lineage to a successful CA-MRSA clone, it simultaneously became resistant to fusidic acid, a widely used antibiotic for skin and soft tissue infections, thus demonstrating the importance of antibiotic selection in the success of this clone. This finding furthermore highlights the significance of horizontal gene acquisitions and underscores the combined importance of these factors for the success of CA-MRSA.
INTRODUCTION
A remarkable change was observed in the epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in the late 1990s with the emergence of new MRSA lineages causing infections in the community in otherwise healthy individuals with no reported contact with the health care system (1–4). Within a very few years, community-acquired MRSA (CA-MRSA) strains were observed worldwide, involving a number of different geographically distinct lineages, including the Southwest Pacific clone (sequence type 30 [ST30], associated with the staphylococcal cassette chromosome mec element [SCCmec] type IV [ST30-IV]) in East Asia and Oceania (5–8), USA400 (ST1-IV) and USA300 (ST8-IV) in the United States (9, 10), and the European CA-MRSA clone (ST80-IV) in Europe, North Africa, and the Middle East (5, 11–14).
The European CA-MRSA clone belonging to clonal complex 80 (CC80) was recognized first in Denmark in 1997 as EDK-97 (15) and then soon after in Finland, Greece, and France (16–18), and it subsequently spread to become the predominant CA-MRSA lineage throughout Europe (19–22). Retrospectively, the first known isolate has been traced back to 1993 in Denmark (23, 24). Epidemiological data suggested that the clone may have had its origin outside Europe, as many of the European CA-MRSA cases in Scandinavia were associated with import by individuals with relations to the Middle East and Africa (24, 25). Concurrently, multiple reports showed that the European CA-MRSA clone was prevalent in North Africa and the Middle East (13, 14, 26–28).
The typical molecular features of the European CA-MRSA clone include the presence of a type IVc SCCmec, a φSa2 prophage carrying the lukS and lukF genes, encoding the Panton-Valentine leukocidin (PVL) (referred to as lukS/F-PV genes), and an agr type III quorum-sensing system. Resistance to fusidic acid (fusB), tetracycline [tet(K)], and kanamycin/amikacin (aadK and aphA) are all strongly associated with the European CA-MRSA clone. These antimicrobials are widely used topical agents for skin infections (29, 30). It is notable that CC80 isolates have historically been found to be predominantly methicillin resistant, and a recent European multicenter study showed that they account for >30% of all CA-MRSA isolates (31). In contrast, CC80 isolates are rarely found in cases of S. aureus bacteremia or among healthy carriers (28, 31–37).
PVL-positive CC80 MSSA isolates have recently been reported from France and from countries in sub-Saharan Africa, including Gabon, Nigeria, São Tomé e Príncipe, Togo, and Uganda (38–43). These reports could indicate a geographically larger dissemination of CC80 with a distinct cluster of methicillin- and fusidic acid-susceptible CC80 isolates. This raises questions about the origin of the European CA-MRSA lineage, its dissemination, and the selective forces driving its epidemic expansion.
The origin, epidemiology, and dispersal of CA-MRSA have been primarily based on localized epidemiological studies or national surveillance monitoring. Whole-genome sequencing (WGS) has greatly expanded the understanding of bacterial evolution and has been used to investigate health care-associated clones such as ST22 and ST239 and the livestock-associated ST398 lineage (44–46) and to trace and investigate the diversification of USA300 in the community (47). In the current study, we applied WGS to an extensive and geographically diverse collection of MSSA and MRSA CC80 isolates collected over almost two decades to reconstruct the origin and evolution of the European CA-MRSA lineage.
RESULTS
Phylogenetic relationship of the CC80 complex.
We determined the genome sequences of 97 CC80 S. aureus isolates (74 MRSA and 23 MSSA strains) covering the period of 1993 to 2010 obtained from 21 countries in Africa, Europe, the Middle East, and Malaysia (see Table S1 and Fig. S1 in the supplemental material). The genomic DNA was sequenced to an average depth of >90-fold (33- to 240-fold) coverage using the ST80 11819-97 genome as a reference.
Phylogenetic analysis determined S. aureus CC1 to be the outgroup most closely related to CC80 when the CC80 sequence was compared to published genome sequences representing CC1, CC5, CC8, CC30, CC45, CC59, CC93 and CC398 (data now shown); therefore, S. aureus MW2 (CC1) was subsequently used for rooting. Once mobile genetic elements were excluded, a total of 3,493 single nucleotide polymorphisms (SNPs) identified within the conserved core genome were used to reconstruct the phylogenetic structure of the CC80 lineage. Neighbor nets were inferred in order to detect putative recombination signatures that would result in networks rather than trees. No major splits were found. However, the pairwise homoplasy index (PHI) test predicted significant evidence for recombination events (P < 0.001). Based on visual inspection, contiguous SNPs consisting of three or more SNPs, mirroring homologous recombination, were detected. A total of 32 independent possible recombination events involving 298 SNPs, ranging from 3 to 24 consecutive SNPs, were identified and removed (see Table S2 in the supplemental material). A rerun of the PHI test on the pruned data set detected no traces of recombination (P = 0.142).
After this preliminary stage, the phylogenetic content of the SNP data set was evaluated with likelihood mapping analyses. With the majority of all quartet points localized in the central region (74.1%), the result pointed to a rather mild phylogenetic signal with a large degree of star-like evolution in the CC80 lineage (48). Thus, the phylogeny seemed to be resolved only in certain parts of the tree (see Fig. S2 in the supplemental material).
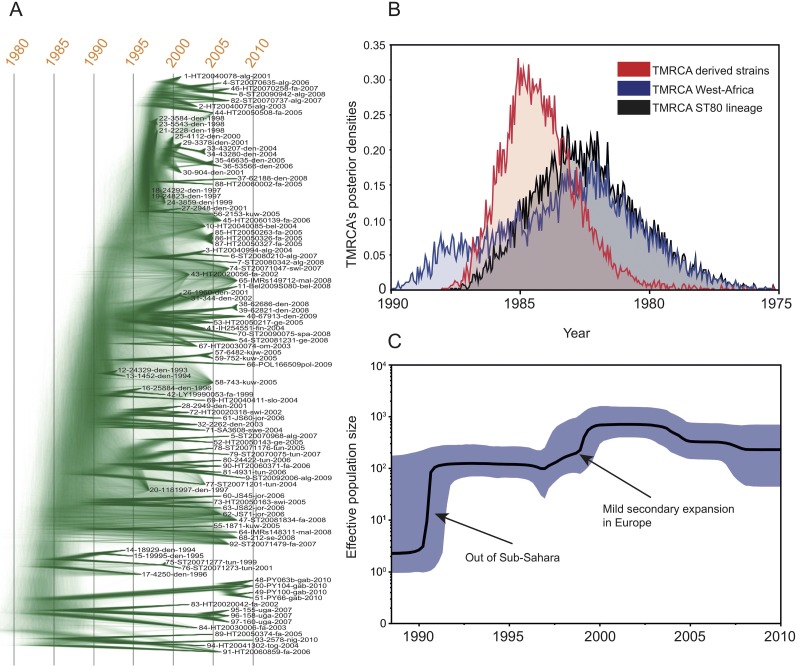
The rooted maximum-likelihood phylogeny identified two distinct clades: a basal clade of 13 MSSA isolates mostly from sub-Saharan Africa and a highly polytomic derived clade with 84 isolates that contained the European CA-MRSA isolates from Europa, North Africa, and the Middle East (Fig. 1). Four French isolates clustered in the sub-Saharan basal clade. For two of these, 89-HT20050374-fra-2005 and 91-HT20060859-fra-2006, epidemiological information could be obtained, and in both cases they were linked to sub-Saharan Africa either by military service or by ethnicity.
FIG 1 .
Rooted maximum-likelihood phylogeny of 97 CC80 isolates based on 3,493 SNPs, including 739 parsimony-informative SNPs. Strains are labeled with isolate number, isolate ID, country of origin, and year of sampling. Countries of origin: alg, Algeria; fa, France; swi, Switzerland; bel, Belgium; jo, Jordan: den, Denmark; swe, Sweden; mal, Malaysia; kuw, Kuwait; spa, Spain; ge, Germany; gr, Greece; fin, Finland; om, Romania; se, Serbia; slo, Slovenia; pol, Poland; tun, Tunesia; tog; Toga; nig, Nigeria; uga, Uganda; gab, Gabon. Columns 1 to 3 show the presence of the methicillin resistance determinant mecA, the fusidic acid resistance determinant fusB, and the tetracycline resistance determinant tet(K). The presence and absence of genetic elements are indicated by black and white, respectively. Gray bars in the SCCmec column indicate remnants of SCC in MSSA isolates, and hatched bars in the tet(K) column denote the pT181 and pT45 plasmids.
A total of four canonical SNPs distinguished the basal sub-Saharan African clade from the derived clade with one nonsynonymous SNP (reference genome position 2130486) positioned in the agr receptor gene agrC, one intergenic SNP (position 2480591), and two synonymous SNPs (positions 225883 and 431761). The SNP in agrC resulted in an amino acid change (L184I) in transmembrane region 6 (TM6) (49, 50) within the sensor domain of the AgrC receptor, to which the agr autoinducing peptide (AIP) binds. Seven spa types were observed. spa type t044 dominated the derived clade (73/84), whereas spa types t934 and t5941 were found exclusively in isolates (12/13) of the basal clade. The remaining isolate in the basal clade carried spa type t376, similar to two isolates in the derived clade.
Dating analysis and demographic expansion.
In order to investigate if the S. aureus CC80 lineage corresponded to a measurably evolving population (51), we plotted the genetic distance from the common ancestor against sampling time (see Fig. S3 in the supplemental material). A significant correlation was observed, indicating that CC80 is a measurably evolving population. Using this basic approach, the time to the most recent common ancestor (TMRCA) of the CC80 lineage was dated to 1988, with a mean nucleotide substitution rate of 1.39 × 10−6 substitution per site per year.
In a second step, we applied the more sophisticated Bayesian coalescent method using both relaxed and strict molecular clock models to infer the phylogeny and the rate of evolution of the CC80 complex and its sublineages. The BEAST package is based on “time” trees, i.e., oriented toward time-measured phylogenies that differ from those obtained via maximum-likelihood approaches. However, the phylogeny obtained using the Bayesian method (Fig. 2A) did not differ in topology from the maximum-likelihood tree (Fig. 1). Interestingly, a DensiTree representation of the Bayesian analysis strongly highlights, in both a graphical and qualitative manner, the low topological support of the most recent divergent clade, with fuzzy connections, whereas the deepest nodes of the tree display much stronger support.
FIG 2 .
Bayesian analyses of the CC80 complex. (A) DensiTree representation of the Bayesian coalescent trees using a strict clock model based on 3,493 SNPs. Tips of the trees are constrained by year of isolation; the time scale is shown at the top. (B) Posterior estimates of the TMRCA for the derived and sub-Saharan African strains under the strict clock model. (C) Effective population size through time (Bayesian skyline) of the S. aureus CC80 lineage. The shaded area represents the 95% confidence intervals, and the arrows point to potential socioeconomic events that might have impacted the demography of the MRSA population.
The mean nucleotide substitution rate within CC80 using the Bayesian coalescent method was 1.29 × 10−6 substitutions per site per year (95% highest posterior densities [HPDs], 1.10 × 10−6 to 1.51 × 10−6), which varied marginally depending on the choice of tree prior (see Table S3 in the supplemental material). Given the rate of molecular evolution, we were able to extrapolate the TMRCA for the full lineage and for the derived clade. According to the best model under the strict clock (constant population size; see the Bayes factor analysis in Table S3 in the supplemental material), the TMRCA of the 97 clones was estimated to 1982 (95% HPDs, 1986 to 1978). The posterior estimates of the TMRCA for the derived strains confirmed that the expansion from sub-Saharan Africa started two to three years later (Fig. 2B). Importantly, the TMRCA of the sub-Saharan lineages overlapped the TMRCA of the full lineage, confirming the ancestral state of the sub-Saharan African samples. Multiple independent runs using the relaxed clock model (lognormal uncorrelated) were executed; however, those runs were poorly mixing and are not presented here. This situation might reflect the departure from a strict clock model suggested in Fig. S3 in the supplemental material.
The Bayesian skyline plot (Fig. 2C) indicated a sharp increase of the effective CC80 population size starting in the early 1990s. At this early stage, the population size increased by two orders of magnitude, followed by a secondary mild expansion (in year 2000) and a more recent stepwise slow decrease. Though Bayesian skylines should not be overinterpreted, it is worth noting that both the demographic expansions and the timings are in good agreement with the European CA-MRSA epidemiology reported in the literature in Europe, North Africa, and the Middle East.
SCCmec acquisition and diversity.
Seven distinct SCCmec IV elements were found among the 74 MRSA isolates (SCCmec IVc1 to IVc7) (see Fig. S4 in the supplemental material), with the predominant type, IVc1, having a length of 37 kb. Five of these subtypes (IVc1 to IVc5) were found in 71 of the 74 isolates and had ~29 kb of conserved content. The remaining two subclasses (IVc6 and IVc7) had major deletions in the SCCmec region spanning the J1 and J2 regions (see Fig. S4 in the supplemental material), but SNPs in the 5′ end of SCCmec clustered these with SCCmec types IVc1 to IVc5 (data not shown). A comparison of types IVc1 to IVc5 with six non-CC80 SCCmec type IV variants (IVa, IVb, IVc [n = 2], IVe, and IVg) identified 244 SNPs, and a phylogenetic analysis showed that the CC80 isolates clustered distinctly together near the two SCCmec type IVc variants and were distant from the other four variants (see Fig. S5 in the supplemental material). Only four SNPs were identified to differentiate subtypes IVc1 to IVc5 among the 71 isolates. Two MSSA isolates from the derived clade had SCCmec remnants. Comparison of their remnant regions with SCCmec IVc1 to IVc5 revealed a high degree of similarity within the conserved regions (data not shown).
All isolates except SCCmec IVc7 and Remnant2 shared an identical ~4,200-bp region integrated into orfX (see Fig. S4 in the supplemental material); however, no other MSSA isolates in the collection contained SCCmec remnants. These results strongly suggest that the IVc SCCmec element was acquired only once and subsequently underwent multiple unrelated deletions.
Fusidic acid resistance determinants.
In 69 of the 71 fusidic acid-resistant strains, all from the derived clade, the resistance was due to fusB located on the plasmid p11819-97 (52). Analysis of the p11819-97 diversity in comparison with that of the two plasmids (pWBG753 and SAP103A) that had the highest degree of homology to p11819-97 identified 309 SNPs (see Fig. S5C in the supplemental material) and showed the p11819-97 plasmid to be highly clonal. A total of 28 SNPs were observed in the p11819-97 sequences. Similar to the SCCmec element, this strongly suggests that the p11819-97 plasmid was acquired once in the CC80 background. The p11819-97 plasmid was closed by insertion of ~800 bp encoding a TnpA transposase and found to contain only two resistance determinants (fusB and blaZ). One fusidic acid-susceptible isolate (88-HT20060002-fa-2005) carried p11819-97 with a complete fusB gene with no mutations in the gene or promoter region. Subsequent microarray analysis and susceptibility testing confirmed the genotypic carriage of fusB associated with a sensitive phenotype. Three isolates had mutations associated with fusidic acid resistance in the fusA gene (see Table S1 in the supplemental material).
In total, 24 different resistance profiles were detected (see Table S1 in the supplemental material). Major differences were found between the basal MSSAs and the MSSAs and MRSAs within the derived clade (Table 1). Regarding the MRSAs in the derived clade, 88% were resistant to fusidic acid and 77% to tetracycline. The most common phenotype, found in 44 of the 74 MRSA isolates, displayed resistance to fusidic acid, kanamycin, and tetracycline (FKT). Resistance to erythromycin and clindamycin was observed in 18% and 15% of the isolates, respectively. Single isolates were resistant to either mupirocin or rifampin.
TABLE 1 .
Phenotypic resistance of CC80 isolatesa
| Clade and organism (n) | No. (%) of isolates |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive | Resistant to: |
|||||||||
| Cef | Ery | Clin | Fus | Tet | Kan | Mup | Nor | Rif | ||
| Basal | ||||||||||
| MSSA (13) | 6 (46) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (54) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Derived | ||||||||||
| MSSA (10) | 0 (0) | 0 (0) | 2 (20) | 1 (10) | 6 (60) | 5 (50) | 1 (10) | 0 (0) | 0 (0) | 1 (10) |
| MRSA (74) | 0 (0) | 74 (100) | 15 (20) | 14 (19) | 65 (88) | 57 (77) | 68 (92) | 1 (1) | 4 (5) | 0 (0) |
| Total (97) | 6 (6) | 74 (76) | 17 (18) | 15 (15) | 71 (73) | 69 (71) | 69 (71) | 1 (1) | 4 (4) | 1 (1) |
Antimicrobial-resistant CC80 isolates from the basal and derived clades in relation to mecA status. Cef, cefoxitin; Ery, erythromycin; Clin, clindamycin; Fus, fusidic acid; Tet, tetracycline; Kan, kanamycin; Mup, mupirocin; Nor, norfloxacin; Rif, rifampin.
Resistance determinants.
The tet(K) gene was found in all 69 tetracycline-resistant isolates and in one susceptible isolate (88-HT20060002-fa-2005). No mutations in the gene or promoter region of this sensitive isolate were found, and concurrent microarray analysis and susceptibility testing further confirmed these observations. The tet(K) gene was found in three different plasmids: the 4.4-kb plasmid pT181 (53) and two novel plasmids, pT45 and pT49 (4.5 kb and 4.9 kb). pT181 was exclusively associated with the basal clade, whereas pT49 was exclusive to the derived clade. pT45 was found in only two isolates from Tunisia (derived clade) (Fig. 1). The pT49 and pT45 plasmids appeared to be derivatives of pT181 and could be explained by insertion of a transposase in pT181 (pT49) and subsequent deletion of a plasmid recombination gene (mobE), leading to pT45.
SCCmec subtypes IVc1 to IVc3 and Remnant1 (n = 68) (see Fig. S4 in the supplemental material) all contained the aadK [ant(6)-Ia] and aphA [aph(3′)-III] genes, which are responsible for aminoglycoside resistance (they were kanamycin/amikacin resistant but susceptible to tobramycin and gentamicin). These genes were absent in the MRSA isolate 7-ST20080342-alg-2008, which carried the aac(6')-aph(2'') gene, associated with resistance against all aminoglycosides.
Panton-Valentine leukocidin.
Ninety-six of the isolates carried the ΦSa2 prophage integrated in the same site of the chromosome. In 95 of these isolates, ΦSa2 harbored lukS/F-PV, and one had a deletion of lukS/F-PV. In addition, isolate 47-ST20081834-fa-2008 carried remnant parts of a ΦSa2 prophage (see Table S1 in the supplemental material). Comparison of the 96 CC80 ΦSa2 prophages to those from other CA-MRSA lineages (CC1, CC8, CC30, CC59, and CC93) showed that the CC80 ΦSa2 prophages are distinct and highly clonal (see Fig. S5C in the supplemental material). In contrast to both the SCCmec and the fusidic acid resistance plasmid (p11819-97), the ΦSa2 prophage seems to be an ancestral trait of the CC80 lineage.
Accessory genome.
The core and accessory genome of the CC80 isolates comprised 2,151 and 1,575 gene families, respectively. Within the accessory genome, a total of 669 gene families were found only in single isolates. Of the 906 remaining gene families, 27 were associated with the derived clade, all of which were related to the SCCmec region (see Fig. S6 in the supplemental material). No gene families were found to be associated with loss in the derived clade.
DISCUSSION
Combining phylogenomics, molecular clock analyses, and epidemiological data provided a compelling depiction of the emergence of CC80 from its proposed origins in sub-Saharan Africa in the early 1980s to its status as the dominant CA-MRSA clone in Europe today. Bayesian and maximum-likelihood phylogenetic analyses of this measurably evolving population (Fig. 1 and 2A; also, see Fig. S3 in the supplemental material) showed a distinct basal clade primarily consisting of sub-Saharan MSSA isolates and a derived clade dominated by MRSA isolates from Europe, North Africa, and the Middle East.
The molecular clock analyses as well as the phylogenetic structure of the Bayesian tree are supported by known human migration patterns. The coalescence analyses indicate that the TMRCA of the CC80 lineage dated to the early 1980s, with epidemiological data suggesting a geographically confined dissemination in western sub-Saharan Africa. Traditionally, movements of pastoralists, such as the Fula or Fulani tribe, have shaped these movements across the region (54), and this is consistent with dissemination of the CC80 lineage. Though the majority of African migration is intracontinental, many of the countries on the Guinea coast tend to have a higher rate of international mobility, with France being the most common destination country (55), which may explain the presence of the methicillin-sensitive isolates in France. Recently, an intensification of migratory movement from the Upper Guinea coast, along the coast, or through the Sahara (Sahel), on the part of migrants pushing north into Europe due to economic crisis and political conflicts has also been observed (55–57). The TMRCA of the derived European CA-MRSA clade dated to the mid-1980s, and this is consistent with increased migration from West Africa to Europe. It also matches increased transnational movement from Europe to West Africa through networks established over the years by past migration (56, 57) as well as by a thriving tourism industry in several parts of this region (58). Such migration-associated dissemination has also been described for other human pathogens, such as Helicobacter pylori, Mycobacterium tuberculosis, Mycobacterium leprae, and Salmonella enterica serovar Typhi (59–62).
Furthermore, the star-like phylogeny of the derived clade suggests a rapid expansion in the early 1990s, reflecting the dissemination of CC80 around the Mediterranean and the rest of Europe after the introduction of the progenitor strain. Despite the general lack of phylogenetic structure, a small group of older Danish and Tunisian isolates formed a distinct subcluster within the CA-MRSA clade, representing an unsuccessful subpopulation not commonly found today.
The first reported case of infection with the European CA-MRSA was a 1993 community-associated skin and soft tissue infection in Denmark (23, 24). While observed in Denmark through the 1990s, it was first described outside Denmark after 1997 in multiple countries around Europe (16–21). Our analysis of the genotypic data shows a highly unresolved population structure in the derived CA-MRSA clade. This indicates a rapid clonal expansion that could easily have been overlooked in countries with no systematic surveillance or collection of community MRSA. Surveillance data from Denmark, Norway, and the Netherlands indicate an increased prevalence of the CA-MRSA clone after 2001 (19, 21, 24, 63). Together, these data are in agreement with the skyline plot (Fig. 2C), which indicates that the European CA-MRSA spread rapidly across Europe, North Africa, and the Middle East prior to 1993 and had a smaller secondary increase in the effective population size around the turn of the century.
Our data suggest that the acquisition of highly specific canonical SNPs as well as resistance to methicillin, kanamycin/amikacin, and fusidic acid coincided with the emergence of the CA-MRSA clone and that these traits could be essential factors driving its expansion. Analysis of the accessory genome showed that these features were absent in the basal clade of the phylogenetic tree but prevalent in the derived clade. Interestingly, various mutations in the agrC gene have been documented to be beneficial for S. aureus to overcome reduced fitness often caused by larger mobile genetic elements or mutations causing reduced fitness (64, 65). A recent study of the S. aureus identified an SNP (G55R) in agrC among most contemporary CC30 isolates distinguishing them from historic CC30 isolates of phage type 80/81 or the southwest Pacific CA-MRSA clone. In most cases, these contemporary isolates had reduced levels of RNAIII expression; such variants were associated with reduced lethality in a murine bacteremia model (66), and no studies have specifically addressed the agrC gene of agr type III present in the CC80 lineage (67). The nonsynonymous replacement in AgrC (L184I) is located in the last transmembrane helix (TM6) of the sensor domain, which has been shown to be of critical importance in signaling the binding of the AIP agonist to the histidine kinase domain (68). The nonsynonymous replacement, resulting in a change from a leucine to an isoleucine, occurs within the same class of amino acids (aliphatic) with the addition of a CH3 side chain. The impact of this subtle change may play a role in increasing the fitness of the derived CC80 isolates, but this remains to be tested. No other resistance markers or virulence factors besides SCCmec IVc, fusB, and the variation of agrC were associated with the derived clade. The genes encoding exfoliative toxin D and epidermal cell differentiation inhibitor (etd and edin, respectively) have previously been linked to the virulence of European CA-MRSA clone; however, we found a pathogenicity island containing these two genes conserved among all isolates in the collection, including the basal isolates.
Our analyses strongly suggest that the SCCmec and the p11819-97 plasmid were acquired only once as part of the geographic expansion of CC80 (see Fig. S5 in the supplemental material). In contrast, the ΦSa2 prophage carrying the PVL-encoding genes was found to be an ancestral component of this lineage. All but one of the CC80 isolates described here carried a ΦSa2 prophage, with 99% (95/96) of these encoding PVL (see Table S1 in the supplemental material). When analyzed independently, the ΦSa2 phylogeny was consistent with that of the core genome of CC80, indicating that this prophage was introduced to the lineage before its transition out of sub-Saharan Africa (see Fig. S5C in the supplemental material). This is corroborated by the high rate of PVL-positive MSSA isolates in sub-Saharan Africa. Thus, Africa may serve as a reservoir for PVL-positive lineages. Indeed, our study reveals several unrelated imports of basal PVL-positive MSSA CC80 isolates into France. Similarly, a sub-Saharan African origin of the PVL-positive ST152 MRSA cluster in Central Europe has been hypothesized (69, 70). The ΦSa2 phylogeny also revealed that CC80-associated ΦSa2 represents a unique subgroup of this important prophage.
Among the included CC80 isolates, the SCCmec type IVc element is exclusively associated with the derived clade, with no MRSA isolates related to sub-Saharan Africa. In-depth analysis of the SCC element revealed seven SCCmec IVc subtypes and two different SCC remnants, with the majority of these being single insertion/deletion variants of the predominant SCCmec IVc1 subtype (see Fig. S4 in the supplemental material). In contrast to data from other S. aureus clonal lineages, such as CC5, CC8, CC22, and CC398 (46, 71–73), a single acquisition of the SCCmec element in the CC80 lineage was observed (see Fig. S5A in the supplemental material). An ~4-kb insertion into the att SCC integration site in orfX was observed in the vast majority (95/97) of the CC80 isolates. The type IV SCCmec elements associated with successful CA-MRSA lineages have been described as relatively short (<30 kb), with an almost absent J2 region (74). These shorter SCCmec elements have been proposed to impose a lower fitness disadvantage in community lineages than the larger elements associated with hospital lineages (75, 76). However, with a 37-kb type IVc1 cassette being the most prevalent in this study, CC80 seems to carry an unusually large community-associated SCCmec variant harboring a 17-kb J2 region. This region includes two resistance genes, aadK and aphA, conferring resistance against kanamycin and amikacin (see Fig. S4 in the supplemental material). Kanamycin and amikacin have primarily been used in hospitals, which may coincide with the reported presence of CC80 in hospitals in Africa, the Middle East, and Southern Europe (13, 14, 16, 77).
The fusidic acid resistance determinant fusB was the sole resistance determinant on the 23-kb p11819-97 plasmid that has been acquired once by the CC80 lineage prior to the expansion out of sub-Saharan Africa (see Fig. 1). fusB-mediated resistance to fusidic acid, a phenotypic characteristic of the European CA-MRSA lineage (30), has previously been identified in S. aureus and is encoded on two plasmids, pUB101 and pUB102 (GenBank accession numbers NC_005127 and DQ269019, respectively). Neither of these was present in our collection. Fusidic acid has been a widely used topical agent against S. aureus-related skin and soft tissue infections in Europe, the Middle East, and North Africa since the early 1980s (Jim Frater [LEO Pharma A/S], personal communication). These three regions also define the geographical dissemination of the fusidic acid-resistant European CA-MRSA clone, and they coincide historically with our time estimate of the clonal expansion of the fusidic acid-sensitive ancestral clone into these areas around 1990.
The antimicrobial resistance profiles observed in the derived clade of both MSSA and MRSA isolates exhibited striking differences compared to those in the basal MSSA clade (Table 1; also, see Table S1 in the supplemental material). The predominant resistance profile, beta-lactams, fusidic acid, kanamycin/amikacin, and tetracycline, was present in 57% (48/84) of the isolates in the derived clade, with sporadic additional resistances to mupirocin, norfloxacin, and rifampin. The ancestral lineage was mostly susceptible to all antimicrobials tested, including beta-lactams and fusidic acid, but carried the tetracycline resistance determinant tet(K) on a small plasmid, pT181. In the derived clade, the tet(K) gene was present on two small plasmids related to pT181 (Fig. 1) but not present on the same plasmid as the compared fusidic acid determinant, as previously reported (78). Thus, isolates in the derived clade generally carry two resistance plasmids. Interestingly, a recent study showed that positive epistasis between coexisting large and small plasmids (79) may be supportive for the observed long-term plasmid persistence. Tetracycline is commonly used in Africa in both humans and animals (80, 81). In North Africa and the Middle East, S. aureus strains are commonly resistant to tetracycline (39, 77, 82). In Europe, tetracycline has also been used in the treatment of various S. aureus indications and long-term treatment of acne as well as in livestock. Contrary to population studies on S. aureus CC8 and CC22 that showed a time-dependent increase in resistance over time (72, 73), the predominant resistance profile of isolates in the derived clade of CC80 has been highly stable over time (data not shown).
The phylogeny of the S. aureus CC80 lineage is in many ways unique compared to other investigated S. aureus lineages. This study presents evidence of a parallel spread of the European CA-MRSA lineage to most areas of present-day distribution soon after the successful dominant clone emerged. The lack of phylogeographic signal is likely the result of extensive human travel activities within Europe and through immigration from primarily North Africa and the Middle East into Europe (24, 25, 83). Put together, both phenomena shaped the dispersal pattern of the CA-MRSA clone.
We found no evidence of specific virulence factors associated with either of the two major clades of CC80 (see Fig. S6 in the supplemental material) but did observe notable acquisitions of genes responsible for resistance to methicillin, fusidic acid, and aminoglycosides and a noteworthy change in the element carrying the tetracycline resistance determinant tet(K). Combined, our results indicate that the European CA-MRSA clone developed within a short time span and only spurred the dispersal of a single successful multidrug-resistant descendant. This is in contrast to the evolution of the ST22-A and USA300 clones (72, 73), where the clonal expansions have been linked to an increase in antibiotic resistance traits over time.
An important limitation of this study and others of its kind is the inevitable bias that comes with selecting isolates. In order to minimize this inherent limitation, we established a large strain collection covering 21 countries, spanning a period of almost 20 years, and enriching for the rare MSSA isolates. Proportionally, a large number of Danish and French isolates were included in the study; however, this would not inherently influence the results. The other major limitation was our inability to determine the true geographic origin of isolates, because in most cases we were unable to inquire about the recent travel/immigration history of the human subjects from whom samples were collected. This is highlighted by the French isolates belonging to the ancestral clade for which direct connections with West Africa (military activities, African origin) have been determined.
In conclusion, the analyses that we conducted provide a proposed depiction of the natural history of one of the most successful CA-MRSA clones described to date, the so-called European CA-MRSA lineage. Phylogeographic reconstruction and molecular clock analyses presented here are supported by known human migration and clinical antimicrobial use patterns that may have driven the epidemic spread of this lineage. Finally, while this study shows the importance of horizontal gene flow to the adaptation of S. aureus to new host environments, it also underscores the importance of clonal expansion in MRSA epidemiology when a genetic combination of adapted and successful traits has been obtained. Future studies may elucidate the functional impact of the agrC codon change and its importance in making CC80 a highly successful clone.
MATERIALS AND METHODS
Bacterial isolates.
A total of 97 S. aureus CC80 isolates sampled in Europe (Belgium, Denmark, Finland, France, Germany, Greece, Poland, Romania, Serbia, Slovenia, Spain, Sweden, and Switzerland), North Africa (Algeria and Tunisia), sub-Saharan Africa (Gabon, Nigeria, Togo, and Uganda), the Middle East (Kuwait and Jordan), and Asia (Malaysia) between 1993 and 2010 were included in the study (Table 1; also, see Table S1 and Fig. S1 in the supplemental material). In total, 23 MSSA isolates and 74 MRSA isolates were included. All isolates were spa typed as previously described (84). Danish isolates constituted a large proportion of the MRSA isolates, as the Danish MRSA collection contains a consecutive complete national collection since 1988, including the oldest reported CC80 isolates.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility was determined using disc diffusion according to EUCAST methodology (http://www.eucast.org) for 11 antimicrobials: clindamycin, cefoxitin, erythromycin, fusidic acid, kanamycin, linezolid, mupirocin, norfloxacin, rifampin, sulfamethoxazole-trimethoprim, and tetracycline.
Genome sequencing.
DNA sequencing was performed as previously described (46). Briefly, for each isolate, 1 to 5 µg DNA was sheared using a SonicMAN (Matrical BioScience, WA) sonicator to a size range of 200 to 1,000 bp. After ligation of the adaptors, 500- to 600-bp fragments were isolated and purified using gel electrophoresis. The pooled paired-end libraries were sequenced on an Illumina IIx genome analyzer (Illumina, Inc., CA) to a read length of 101 bp, except for isolate 93-2578-nig-2010, for which read lengths of 251 bp were obtained using a MiSeq instrument (Illumina Inc.).
DNA microarray.
A single isolate, 88-HT20060002-fa-2005, was characterized using the Alere S. aureus genotyping kit (Alere Technologies, Jena, Germany) according to the manufacturer’s instructions.
SNP calling.
SNPs were detected by aligning Illumina reads from individual isolates against the chromosome of S. aureus 11819-97 (GenBank accession number NC_017351) (52), a complete reference genome of a Danish clinical MRSA isolate from the European CA-MRSA lineage, using SHRiMP 2.0 (85). SNPs in the conserved core genome of the strain collection were identified using Nesoni v 0.52 (http://www.bioinformatics.net.au). Custom Perl script was used to retain only those SNPs that were supported by at least 90% of base calls in all the analyzed strains. Indications of horizontal gene transfer events were defined as calling of ≥3 unique SNPs in succession in a single isolate. For SNP identification in specific elements (see “Genetic elements” below), Illumina WGS data sets were aligned against the 11819-97 reference genome using the short-read alignment component of the Burrows-Wheeler aligner (86), and external genomes were aligned using MUMmer (87). Each alignment was analyzed for SNPs using SolSNP (http://sourceforge.net/projects/solsnp/) and excluded all SNPs that did not meet a minimum coverage of 10 or if the variant was present in <90% of the base calls for that position. SNPs identified in duplicated regions on the reference genome were removed.
Recombination detection.
Most of the analyses developed in our analytical framework (phylogenetics and Bayesian inference) are based on the assumption that S. aureus evolution is mostly clonal and that recombination can be neglected. Therefore, in a preliminary step, we tested for the presence of mosaic genomes with the algorithm SplitsTree v4.13.1 (88). Putative recombination signatures were inferred using Neighbor-Net (89), and each data set was analyzed for the presence of recombinant sequences using the PHI test in SplitsTree with an alpha value of 0.001.
Likelihood mapping.
The phylogenetic signal of the data set was investigated with the likelihood mapping method implemented in TREE-PUZZLE v 5.2 (90) by analyzing 10,000 random quartets. This method proceeds by evaluating, using maximum likelihood, groups of four randomly chosen sequences (quartets). The three possible unrooted tree topologies for each quartet are weighted, and the posterior weights are then plotted using triangular coordinates, such that each corner represents a fully resolved tree topology. The resulting distribution of the points therefore shows whether the data are suitable for a phylogenetic reconstruction.
Phylogenetic inferences.
Genome-wide phylogenetic relationships were reconstructed using the maximum-likelihood approach implemented in PhyML v 3.412 (91). The robustness of the maximum-likelihood tree topology was assessed with bootstrapping analyses of 1,000 pseudoreplicated datasets. A generalized time-reversible (GTR) substitution model (92) with gamma-distributed rate heterogeneity was selected based on Akaike’s information criterion using jModelTest2 (93). Phylogenetic analysis determined CC1 to be the outgroup most closely related to CC80 when the CC80 sequence was compared to published genome sequences representing CC1, CC5, CC8, CC30, CC45, CC59, CC93, and CC398 (GenBank accession numbers BA000033, BA000018, CP000255, BX571856, CP006044, NC_016928, NC_017338, and AM990992, respectively) as previously described (46). Therefore, the phylogenies were rooted with the community-associated strain MW2 (GenBank accession number BA000033). Phylogenetic relationships for genetic elements were reconstructed using the maximum-likelihood approach implemented in CLCbio’s Genomics Workbench v 6.5 (CLCbio, Aarhus, Denmark) based on Akaike’s information criterion.
Coalescence-based analyses.
During analysis, evolutionary rates and tree topologies were analyzed using the GTR and Hasegawa-Kishino-Yano (HKY) (94) substitution models with gamma-distributed among-site rate variation with four rate categories (Γ4). The molecular clock was calibrated under either a strict molecular clock, which assumes the same evolutionary rates for all branches in the tree, or a relaxed clock, which allows different rates. Constant-sized, logistic, exponentially growing coalescent models were used. We also used the Bayesian skyline plot model (95), based on a general, nonparametric prior that enforces no particular demographic history. We used a piecewise linear skyline model with 10 groups and then compared the marginal likelihood for each model using Bayes factors estimated in Tracer v 1.5 (http://tree.bio.ed.ac.uk/software/tracer/). For each analysis, two independent runs of 100 million steps were performed, and the chain was sampled every 10,000th generation. Examination of the Markov chain Monte Carlo (MCMC) samples with Tracer indicated convergence and adequate mixing of the Markov chains with effective sample sizes (ESS) for each parameter in the hundreds or thousands. The first 10% of each chain was discarded as burn-in. The MCMC samples were summarized using the maximum clade credibility topology found with TreeAnnotator v 1.7.5 (96), with branch lengths reported in years (median number of of branches that were present in at least 50% of the sampled trees). The Bayesian skyline plot was reconstructed using the posterior tree sample and Tracer.
Evaluation of competing models.
Models were compared by calculating the Bayes factor (BF), which is the ratio of the marginal likelihoods of the two models being compared. Approximate marginal likelihoods for each coalescent model were calculated via importance sampling (1,000 bootstraps) using the harmonic mean of the sampled likelihoods. The ratio of the marginal likelihoods between any two models is the BF. Evidence against the null model (i.e., the one with lower marginal likelihood) is indicated by the formulas 2×ln(BF) > 3 (positive) and 2×ln(BF) > 10 (strong) (97). The calculations were performed with BEAST v 1.7.5 and Tracer.
Genetic elements.
Analyses of the genetic diversity of the SCCmec cassette, the PVL-encoding prophage ΦSa2, and the fusB plasmid were investigated by SNP diversity in conserved regions among the 97 isolates and selected references as described below. Closing of seven of the ten distinct SCCmec elements inserted in the attB site, the plasmid p11819-97, and two novel plasmids, pT45 and pT49 tet(K), was performed by de novo assembly with subsequent gap closure in silico using Genomics Workbench (CLCbio).
For analysis of the SCCmec diversity, six SCCmec type IV variants were initially compared for overall similarity of content using Mauve (98) prior to SNP calling; type IVa, isolate JCSC1968; type IVb, isolate JCSC1978; type IVc, isolate MR108; type IVc, isolate 2314, type IVe, isolate AR43/3330.1; type IVg, isolate M03-68, with GenBank accession numbers AB063172, AB063173, AB096217, AY271717, AJ810121, and DQ106887, respectively.
Plasmid p11819-97 (GenBank accession number NC_017350) resembled two other available S. aureus plasmids from Australia, the complete plasmid pWBG753 (GenBank accession number GQ900395), from an ST8 isolate, and the draft plasmid SAP103A (GenBank accession number GQ900497) (99), based on BLASTN analysis against GenBank. Both were included in the SNP analysis in order to determine the diversity of p11819-97 within the CC80 collection.
ΦSa2 diversity analysis within the CC80 strains was performed using seven lukS/F-PV-containing ΦSa2 prophages from five distinct community-associated S. aureus genetic lineages as part of the analysis: a CC1 isolate (MW2), two CC8 isolates (FPR3757 and TCH1516), two CC30 isolates (55_2053 and TCH60), a CC59 isolate (M013), and a CC93 isolate (JKD6159), with GenBank accession numbers NC_003923, CP000255, CP000730, CP002388, CP002110, NC_016928, and CP002114, respectively.
The presence or absence of mecA, fusB, lukS/F-PV, tet(K), tet(L), tet(M), and ileS2 was examined using local BLASTN against the generated contigs from the genome data of the 97 CC80 isolates (Genomics Workbench; CLCbio). The query sequences used were mecA, fusB, lukS/F-PV (GenBank accession number NC_017351), tet(K) (GenBank accession number NC_017331), tet(L) (GenBank accession number FN390947), tet(M) (GenBank accession number AM990992), and ileS2 (GenBank accession number EU366902).
Accessory genome.
Protein sequences were extracted from the genome sequences using Prodigal (100). All protein sequences were searched against the Pfam-A database (101, 102) using HMMER3 (103). The protein sequences were binned by the absence or presence of Pfam protein domains. Within each bin, all sequences were compared pairwise using BLAST 2.2.26 (104) without low-complexity filtering and using an E-value cutoff of 0.05. The sequences within each bin were subsequently clustered based on the BLAST matches using the Markov cluster (MCL) algorithm (105, 106) with a set I value of 5. Stable clusters were considered gene families. Identified gene families were divided into three groups: core genes (gene families present in all isolates), singleton genes (gene families present in only one isolate), and accessory genes (all remaining gene families). Acquisition of genes related to the derived clade was examined by identifying gene families present in ≤2 (15%) of the basal isolates and absent in ≤13 (15%) of the derived isolates. Gene families absent in ≤2 basal isolates and present in ≤13 of the derived isolates were used to identify gene loss associated with the derived clade.
Sequence data accession numbers.
The accession numbers for the Illumina sequences generated from the 97 S. aureus CC80 isolates described in this study are available in the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under the primary identification number PRJEB6777. Sequences can also be located in the ENA using the following study title: “Origin and Evolution of the European Community-Acquired Methicillin-Resistant Staphylococcus aureus.” The sequence of the SCCmec type IVc1 element has been deposited in GenBank under accession number KM281804; the sequences of plasmids pT45 and pT49 have been deposited in GenBank under accession numbers KM281802 and KM281803.
SUPPLEMENTAL MATERIAL
Geographic distribution of the countries from which CC80 strains have been identified and included. Download
Likelihood mapping analysis for the full S. aureus CC80 SNP data set (n = 97). Download
Linear regression plot of the root-to-tip distances of the CC80 phylogenetic tree against the year of sampling of the isolates. Download
Integrated genetic elements in orfX. Download
Unrooted maximum-likelihood phylogeny of the SNP variation in the SCCmec, p11819-97, and ΦSa2 elements. Download
Heat map of the presence of selected gene families associated with gene uptake in the derived CC80 clade. Download
S. aureus CC80 strain collection. Detailed information regarding the country of origin, phenotypic resistance profiles, and presence or absence of selected genetic markers. Antimicrobial resistance profiles include abbreviations for kanamycin (K), cefoxitin (M), clindamycin (C), erythromycin (E), norfloxacin (N), tetracycline (T), fusidic acid (F), and mupirocin (Mup). SCCmec type indicates determined subtypes of the integrated content into the attB site (see Fig. S4 in the supplemental material).
Homologous recombinations in CC80.
Date estimate according to coalescent priors.
ACKNOWLEDGMENTS
Stine-Frese Madsen, Alexandra Medina, Mille Weismann Poulsen, Lone Ryste Kildevang Hansen, Elvira Chapka, and Julie Hindsberg Nielsen (Statens Serum Institut), Tania Contente-Cuemo and Katerina Soldanova (Translational Genomics Research Institute), and Michele Bes (CIRI, French National Reference Center for Staphylococci) are thanked for excellent technical assistance.
This work was supported by Pfizer (Grant Europe ASPIRE 2010 Research Awards) and the Foundation pour la Recherche Médicale (grant number ING20111223510).
We declare that we have no conflicts of interest.
Footnotes
Citation Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simões PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirković I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade J, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. mBio 5(5):e01044-14. doi:10.1128/mBio.01044-14.
REFERENCES
- 1. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chatterjee SS, Otto M. 2013. Improved understanding of factors driving methicillin-resistant Staphylococcus aureus epidemic waves. Clin. Epidemiol. 5:205–217. 10.2147/CLEP.S3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598. 10.1001/jama.279.8.593 [DOI] [PubMed] [Google Scholar]
- 4. Hunt C, Dionne M, Delorme M, Murdock D, Erdrich D, Wolsey D, Groom A, Cheek J, Jacobson J, Cunningham B, Shireley L, Belani K, Kurachek S. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123–1125. 10.1001/jama.282.12.1123 [DOI] [PubMed] [Google Scholar]
- 5. Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978–984. 10.3201/eid0908.030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coombs GW, Nimmo GR, Bell JM, Huygens F, O’Brien FG, Malkowski MJ, Pearson JC, Stephens AJ, Giffard PM, Australian Group for Antimicrobial Resistance 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735–4743. 10.1128/JCM.42.10.4735-4743.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho PL, Tse CW, Mak GC, Chow KH, Ng TK. 2004. Community-acquired methicillin-resistant Staphylococcus aureus arrives in Hong Kong. J. Antimicrob. Chemother. 54:845–846. 10.1093/jac/dkh426 [DOI] [PubMed] [Google Scholar]
- 8. Hsu LY, Koh YL, Chlebicka NL, Tan TY, Krishnan P, Lin RT, Tee N, Barkham T, Koh TH. 2006. Establishment of ST30 as the predominant clonal type among community-associated methicillin-resistant Staphylococcus aureus isolates in Singapore. J. Clin. Microbiol. 44:1090–1093. 10.1128/JCM.44.3.1090-1093.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tenover FC, Goering RV. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64:441–446. 10.1093/jac/dkp241 [DOI] [PubMed] [Google Scholar]
- 10. Roberts JC. 2013. Community-associated methicillin-resistant epidemic clone USA100; more than a nosocomial pathogen. Springerplus 2:133. 10.1186/2193-1801-2-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsen AR, Stegger M, Böcher S, Sørum M, Monnet DL, Skov RL. 2009. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus [sic] aureus infections in Denmark, 1999 to 2006. J. Clin. Microbiol. 47:73–78. 10.1128/JCM.01557-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Witte W, Strommenger B, Cuny C, Heuck D, Nuebel U. 2007. Methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leucocidin gene in Germany in 2005 and 2006. J. Antimicrob. Chemother. 60:1258–1263. 10.1093/jac/dkm384 [DOI] [PubMed] [Google Scholar]
- 13. Tokajian ST, Khalil PA, Jabbour D, Rizk M, Farah MJ, Hashwa FA, Araj GF. 2010. Molecular characterization of Staphylococcus aureus in Lebanon. Epidemiol. Infect. 138:707–712. 10.1017/S0950268810000440 [DOI] [PubMed] [Google Scholar]
- 14. Antri K, Rouzic N, Dauwalder O, Boubekri I, Bes M, Lina G, Vandenesch F, Tazir M, Ramdani-Bouguessa N, Etienne J. 2011. High prevalence of methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clin. Microbiol. Infect. 17:526–532. 10.1111/j.1469-0691.2010.03273.x [DOI] [PubMed] [Google Scholar]
- 15.Skov R, Elsberg CS, Møller JK, Garhn-Hansen B, Schønheyder HC, Westh H.Abstract 221, 9th International Symposium for Staphylococci and Staphylococcal. 2000. Infections, Kolding, Denmark.
- 16. Aires de Sousa M, Bartzavali C, Spiliopoulou I, Sanches IS, Crisostomo MI, de Lencastre H. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027–2032. 10.1128/JCM.41.5.2027-2032.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dufour P, Gillet Y, Bes M, Lina G, Vandenesch F, Floret D, Etienne J, Richet H. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819–824. 10.1086/342576 [DOI] [PubMed] [Google Scholar]
- 18. Salmenlinna S, Lyytikäinen O, Vuopio-Varkila J. 2002. Community-acquired methicillin-resistant Staphylococcus aureus, Finland. Emerg. Infect Dis. 8:602–607. 10.3201/eid0806.010313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanssen AM, Fossum A, Mikalsen J, Halvorsen DS, Bukholm G, Sollid JU. 2005. Dissemination of community-acquired methicillin-resistant Staphylococcus aureus clones in northern Norway: sequence types 8 and 80 predominate. J. Clin. Microbiol. 43:2118–2124. 10.1128/JCM.43.5.2118-2124.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Witte W, Braulke C, Cuny C, Strommenger B, Werner G, Heuck D, Jappe U, Wendt C, Linde HJ, Harmsen D. 2005. Emergence of methicillin-resistant Staphylococcus aureus with Panton-Valentine leukocidin genes in central Europe. Eur. J. Clin. Microbiol. Infect. Dis. 24:1–5. 10.1007/s10096-004-1262-x [DOI] [PubMed] [Google Scholar]
- 21. Stam-Bolink EM, Mithoe D, Baas WH, Arends JP, Möller AV. 2007. Spread of a methicillin-resistant Staphylococcus aureus ST80 strain in the community of the northern Netherlands. Eur. J. Clin. Microbiol. Infect. Dis. 26:723–727. 10.1007/s10096-007-0352-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brauner J, Hallin M, Deplano A, De Mendonça R, Nonhoff C, De Ryck R, Roisin S, Struelens MJ, Denis O. 2013. Community-acquired methicillin-resistant Staphylococcus aureus clones circulating in Belgium from 2005 to 2009: changing epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 32:613–620. 10.1007/s10096-012-1784-6 [DOI] [PubMed] [Google Scholar]
- 23. Faria NA, Oliveira DC, Westh H, Monnet DL, Larsen AR, Skov R, de Lencastre H. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 43:1836–1842. 10.1128/JCM.43.4.1836-1842.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larsen AR, Böcher S, Stegger M, Goering R, Pallesen LV, Skov R. 2008. Epidemiology of European Community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J. Clin. Microbiol. 46:62–68. 10.1128/JCM.01381-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larsson AK, Gustafsson E, Johansson PJ, Odenholt I, Petersson AC, Melander E. 2014. Epidemiology of MRSA in southern Sweden: strong relation to foreign country of origin, health care abroad and foreign travel. Eur. J. Clin. Microbiol. Infect. Dis. 33:61–68. 10.1007/s10096-013-1929-2 [DOI] [PubMed] [Google Scholar]
- 26. Khalil W, Hashwa F, Shihabi A, Tokajian S. 2012. Methicillin-resistant Staphylococcus aureus ST80-IV clone in children from Jordan. Diagn. Microbiol. Infect. Dis. 73:228–230. 10.1016/j.diagmicrobio.2012.03.012 [DOI] [PubMed] [Google Scholar]
- 27. Udo EE, O’Brien FG, Al-Sweih N, Noronha B, Matthew B, Grubb WB. 2008. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J. Clin. Microbiol. 46:3514–3516. 10.1128/JCM.00966-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ben Nejma M, Mastouri M, Bel Hadj Jrad B, Nour M. 2013. Characterization of ST80 Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn. Microbiol. Infect. Dis. 77:20–24. 10.1016/j.diagmicrobio.2008.02.010 [DOI] [PubMed] [Google Scholar]
- 29. Dobie D, Gray J. 2004. Fusidic acid resistance in Staphylococcus aureus. Arch. Dis. Child. 89:74–77. 10.1136/adc.2003.019695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilkinson JD. 1998. Fusidic acid in dermatology. Br. J Dermatol. 139(Suppl 53):37–40. 10.1046/j.1365-2133.1998.1390s3037.x [DOI] [PubMed] [Google Scholar]
- 31. Rolo J, Miragaia M, Turlej-Rogacka A, Empel J, Bouchami O, Faria NA, Tavares A, Hryniewicz W, Fluit AC, de Lencastre H, CONCORD Working Group 2012. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS One 7:e34768. 10.1371/journal.pone.0034768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ben Slama K, Gharsa H, Klibi N, Jouini A, Lozano C, Gomez-Sanz E, Zarazaga M, Boudabous A, Torres C. 2011. Nasal carriage of Staphylococcus aureus in healthy humans with different levels of contact with animals in Tunisia: genetic lineages, methicillin resistance, and virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 30:499–508. 10.1007/s10096-010-1109-6 [DOI] [PubMed] [Google Scholar]
- 33. Andersen PS, Pedersen JK, Fode P, Skov RL, Fowler VG, Jr, Stegger M, Christensen K. 2012. Influence of host genetics and environment on nasal carriage of Staphylococcus aureus in Danish middle-aged and elderly twins. J. Infect. Dis. 206:1178–1184. 10.1093/infdis/jis491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katopodis GD, Grivea IN, Tsantsaridou AJ, Pournaras S, Petinaki E, Syrogiannopoulos GA. 2010. Fusidic acid and clindamycin resistance in community-associated, methicillin-resistant Staphylococcus aureus infections in children of central Greece. BMC Infect. Dis. 10:351. 10.1186/1471-2334-10-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Staphylococcus Laboratory, Statens Serum Institut. 2011. Annual report on Staphylococcus aureus bacteraemia cases in Denmark, 2011. Statens Serum Institut, Copenhagen, Denmark: http://bit.ly/H7DgZL [Google Scholar]
- 36. Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, European, Staphylococcal Reference Laboratory Working Group 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. 10.1371/journal.pmed.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sangvik M, Olsen RS, Olsen K, Simonsen GS, Furberg AS, Sollid JU. 2011. Age- and gender-associated Staphylococcus aureus spa types found among nasal carriers in a general population: the Tromso Staph and Skin Study. J. Clin. Microbiol. 49:4213–4218. 10.1128/JCM.05290-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Conceição T, Santos Silva I, de Lencastre H, Aires-de-Sousa M. 2014. Staphylococcus aureus nasal carriage among patients and health care workers in Sao Tome and Principe. Microb. Drug Resist. 20:57–66. 10.1089/mdr.2013.0136 [DOI] [PubMed] [Google Scholar]
- 39. Shittu AO, Okon K, Adesida S, Oyedara O, Witte W, Strommenger B, Layer F, Nübel U. 2011. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 11:92. 10.1186/1471-2180-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rasigade JP, Laurent F, Lina G, Meugnier H, Bes M, Vandenesch F, Etienne J, Tristan A. 2010. Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981-2007. J. Infect. Dis. 201:1589–1597. 10.1086/652008 [DOI] [PubMed] [Google Scholar]
- 41. Schaumburg F, Kock R, Friedrich AW, Soulanoudjingar S, Ngoa UA, von Eiff C, Issifou S, Kremsner PG, Herrmann M, Peters G, Becker K. 2011. Population structure of Staphylococcus aureus from remote African Babongo Pygmies. PLoS Negl. Trop Dis. 5:e1150. 10.1371/journal.pntd.0001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schaumburg F, Mugisha L, Peck B, Becker K, Gillespie TR, Peters G, Leendertz FH. 2012. Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 74:1071–1075. 10.1002/ajp.22067 [DOI] [PubMed] [Google Scholar]
- 43. Schaumburg F, Alabi AS, Köck R, Mellmann A, Kremsner PG, Boesch C, Becker K, Leendertz FH, Peters G. 2012. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ. Microbiol. Rep 4:141–146. 10.1111/j.1758-2229.2011.00316.x [DOI] [PubMed] [Google Scholar]
- 44. Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 13:130–136. 10.1016/S1473-3099(12)70268-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uhlemann AC, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MT, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc. Natl. Acad. Sci. U. S. A. 111:6738–6743. 10.1073/pnas.1401006111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Strimmer K, von Haeseler A. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. U. S. A. 94:6815–6819. 10.1073/pnas.94.13.6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen LC, Tsou LT, Chen FJ. 2009. Ligand-receptor recognition for activation of quorum sensing in Staphylococcus aureus. J. Microbiol. 47:572–581. 10.1007/s12275-009-0004-2 [DOI] [PubMed] [Google Scholar]
- 50. Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. 2008. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J. Mol. Biol. 381:300–309. 10.1016/j.jmb.2008.06.018 [DOI] [PubMed] [Google Scholar]
- 51. Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo A. 2003. Measurably evolving populations. Trends Ecol. Evol. 19:481–488. 10.1016/S0169-5347(03)00216-7 [DOI] [Google Scholar]
- 52. Stegger M, Price LB, Larsen AR, Gillece JD, Waters AE, Skov R, Andersen PS. 2012. Genome sequence of Staphylococcus aureus strain 11819-97, an ST80-IV European Community-acquired methicillin-resistant isolate. J. Bacteriol. 194:1625–1626. 10.1128/JB.06653-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khan SA, Novick RP. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus Process. Plasmid 10:251–259 [DOI] [PubMed] [Google Scholar]
- 54. Stenning DJ. 1957. Transhumance, migratory drift, migration; patterns of pastoral Fulani nomadism. J. R. Anthropol. Inst. 87:57–73 [Google Scholar]
- 55. Shimeles A. 2010. Migration patterns, trends and policy issues in Africa. Working paper no. 119. Working paper series. African Development Bank Group, Tunis, Tunisia [Google Scholar]
- 56. Vigh H. 2009. Conflictual motion and political inertia: on rebellions and revolutions in Bissau and beyon. Afr. Stud. Rev. 52:143–164. 10.1353/arw.0.0171 [DOI] [Google Scholar]
- 57. Vigh H. 2009. Wayward migration: on imagined futures and technological voids. Ethnos 74:91–109. 10.1080/00141840902751220 [DOI] [Google Scholar]
- 58. Fair D. 1997. West African tourist trends. Afr. Insight 27:146–152 [Google Scholar]
- 59. Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915–918. 10.1038/nature05562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Monot M, Honoré N, Garnier T, Araoz R, Coppée JY, Lacroix C, Sow S, Spencer JS, Truman RW, Williams DL, Gelber R, Virmond M, Flageul B, Cho SN, Ji B, Paniz-Mondolfi A, Convit J, Young S, Fine PE, Rasolofo V, Brennan PJ, Cole ST. 2005. On the origin of leprosy. Science 308:1040–1042. 10.1126/science/1109759 [DOI] [PubMed] [Google Scholar]
- 61. Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, Achtman M. 2006. Evolutionary history of Salmonella typhi. Science 314:1301–1304. 10.1126/science.1134933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fossum AE, Bukholm G. 2006. Increased incidence of methicillin-resistant Staphylococcus aureus ST80, novel ST125 and SCCmecIV in the south-eastern part of Norway during a 12-year period. Clin. Microbiol. Infect. 12:627–633. 10.1111/j.1469-0691.2006.01467.x [DOI] [PubMed] [Google Scholar]
- 64. Schwan WR, Langhorne MH, Ritchie HD, Stover CK. 2003. Loss of hemolysin expression in Staphylococcus aureus agr mutants correlates with selective survival during mixed infections in murine abscesses and wounds. FEMS Immunol. Med. Microbiol. 38:23–28. 10.1016/S0928-8244(03)00098-1 [DOI] [PubMed] [Google Scholar]
- 65. Zorzet A, Andersen JM, Nilsson AI, Møller NF, Andersson DI. 2012. Compensatory mutations in agrC partly restore fitness in vitro to peptide deformylase inhibitor-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 67:1835–1842. 10.1093/jac/dks168 [DOI] [PubMed] [Google Scholar]
- 66. DeLeo FR, Kennedy AD, Chen L, Bubeck Wardenburg J, Kobayashi SD, Mathema B, Braughton KR, Whitney AR, Villaruz AE, Martens CA, Porcella SF, McGavin MJ, Otto M, Musser JM, Kreiswirth BN. 2011. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 108:18091–18096. 10.1073/pnas.1111084108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564. 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- 68. Geisinger E, Muir TW, Novick RP. 2009. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc. Natl. Acad. Sci. U. S. A. 106:1216–1221. 10.1073/pnas.0807760106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pérez-Roth E, Alcoba-Florez J, Lopez-Aguilar C, Gutiérrez-González I, Rivero-Pérez B, Méndez-Alvarez S. 2010. Familial furunculosis associated with community-acquired leukocidin-positive methicillin-susceptible Staphylococcus aureus ST152. J. Clin. Microbiol. 48:329–332. 10.1128/JCM.00622-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ruimy R, Maiga A, Armand-Lefevre L, Maiga I, Diallo A, Koumaré AK, Ouattara K, Soumaré S, Gaillard K, Lucet JC, Andremont A, Feil EJ. 2008. The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J. Bacteriol. 190:3962–3968. 10.1128/JB.01947-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 105:14130–14135. 10.1073/pnas.0804178105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlickova H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramirez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nubel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genet. Res. 23:653–664. 10.1101/gr.147710.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Strommenger B, Bartels MD, Kurt K, Layer F, Rohde SM, Boye K, Westh H, Witte W, De Lencastre H, Nübel U. 2014. Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. J. Antimicrob. Chemother. 69:616–622. 10.1093/jac/dkt413 [DOI] [PubMed] [Google Scholar]
- 74. International Working Group on the Classification of Staphylococcal Cassette Chromosome E 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ender M, McCallum N, Adhikari R, Berger-Bächi B. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2295–2297. 10.1128/AAC.48.6.2295-2297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bächi B, Cook GM. 2007. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob. Agents Chemother. 51:1497–1499. 10.1128/AAC.01239-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Udo EE, Sarkhoo E. 2010. The dissemination of ST80-SCCmec-IV community-associated methicillin resistant Staphylococcus aureus clone in Kuwait hospitals. Ann. Clin Microbiol. Antimicrob. 9:31. 10.1186/1476-0711-9-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Monecke S, Slickers P, Hotzel H, Richter-Huhn G, Pohle M, Weber S, Witte W, Ehricht R. 2006. Microarray-based characterisation of a Panton-Valentine leukocidin-positive community-acquired strain of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12:718–728. 10.1111/j.1469-0691.2006.01420.x [DOI] [PubMed] [Google Scholar]
- 79. San Millan A, Heilbron K, Maclean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J. 8:601–612. 10.1038/ismej.2013.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Laxminarayan R, Bhutta Z, Duse AG, Jenkins P, O’Brien T, Okeke IN, Pablo-Mendez A, Klugman KP. 2006. Disease control priorities in developing countries, p 55 In Jamison DT, Breman JG, Measham AR. (ed), Drug resistance, 2nd ed. World Bank, Washington, DC [Google Scholar]
- 81. Darwish WS, Eldaly EA, El-Abbasy MT, Ikenaka Y, Nakayama S, Ishizuka M. 2013. Antibiotic residues in food: the African scenario. Jpn. J. Vet. Res. 61(Suppl):S13–S22 [PubMed] [Google Scholar]
- 82. Vlieghe E, Phoba MF, Tamfun JJ, Jacobs J. 2009. Antibiotic resistance among bacterial pathogens in Central Africa: a review of the published literature between 1955 and 2008. Int. J. Antimicrob. Agents 34:295–303. 10.1016/j.ijantimicag.2009.04.015 [DOI] [PubMed] [Google Scholar]
- 83. Zanger P, Nurjadi D, Schleucher R, Scherbaum H, Wolz C, Kremsner PG, Schulte B. 2012. Import and spread of Panton-Valentine leukocidin-positive Staphylococcus aureus through nasal carriage and skin infections in travelers returning from the tropics and subtropics. Clin. Infect. Dis. 54:483–492. 10.1093/cid/cir822 [DOI] [PubMed] [Google Scholar]
- 84. Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Skov R, Larsen AR. 2012. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect 18:395–400. 10.1111/j.1469-0691.2011.03715.x [DOI] [PubMed] [Google Scholar]
- 85. David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 27:1011–1012. 10.1093/bioinformatics/btr046 [DOI] [PubMed] [Google Scholar]
- 86. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-6-p12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 89. Bryant D, Moulton V. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255–265. 10.1093/molbev/msh018 [DOI] [PubMed] [Google Scholar]
- 90. Schmidt HA, Strimmer K, Vingron M, von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504. 10.1093/bioinformatics/18.3.502 [DOI] [PubMed] [Google Scholar]
- 91. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 92. Rodríguez F, Oliver JL, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485–501. 10.1016/S0022-5193(05)80104-3 [DOI] [PubMed] [Google Scholar]
- 93. Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174. 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 95. Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192. 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- 96. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kass RE, Raftery AE. 1995. Bayes factors. J. Am. Stat. Assoc. 90:773–795 [Google Scholar]
- 98. Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O’Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 1:581–591. 10.1534/g3.111.000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sonnhammer EL, Eddy SR, Durbin R. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420. [DOI] [PubMed] [Google Scholar]
- 103. Eddy SR. 2011. Accelerated profile HMM Searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol. Direct 7:12. 10.1186/1745-6150-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. 10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dongen SV. 2008. Graph clustering via a discrete uncoupling Process. SIAM J. Matrix Anal. Appl. 30:121–141. 10.1137/040608635 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic distribution of the countries from which CC80 strains have been identified and included. Download
Likelihood mapping analysis for the full S. aureus CC80 SNP data set (n = 97). Download
Linear regression plot of the root-to-tip distances of the CC80 phylogenetic tree against the year of sampling of the isolates. Download
Integrated genetic elements in orfX. Download
Unrooted maximum-likelihood phylogeny of the SNP variation in the SCCmec, p11819-97, and ΦSa2 elements. Download
Heat map of the presence of selected gene families associated with gene uptake in the derived CC80 clade. Download
S. aureus CC80 strain collection. Detailed information regarding the country of origin, phenotypic resistance profiles, and presence or absence of selected genetic markers. Antimicrobial resistance profiles include abbreviations for kanamycin (K), cefoxitin (M), clindamycin (C), erythromycin (E), norfloxacin (N), tetracycline (T), fusidic acid (F), and mupirocin (Mup). SCCmec type indicates determined subtypes of the integrated content into the attB site (see Fig. S4 in the supplemental material).
Homologous recombinations in CC80.
Date estimate according to coalescent priors.