ABSTRACT
Clostridium difficile is an important human-pathogenic bacterium causing antibiotic-associated nosocomial infections worldwide. Mobile genetic elements and bacteriophages have helped shape C. difficile genome evolution. In many bacteria, phage infection may be controlled by a form of bacterial immunity called the clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) system. This uses acquired short nucleotide sequences (spacers) to target homologous sequences (protospacers) in phage genomes. C. difficile carries multiple CRISPR arrays, and in this paper we examine the relationships between the host- and phage-carried elements of the system. We detected multiple matches between spacers and regions in 31 C. difficile phage and prophage genomes. A subset of the spacers was located in prophage-carried CRISPR arrays. The CRISPR spacer profiles generated suggest that related phages would have similar host ranges. Furthermore, we show that C. difficile strains of the same ribotype could either have similar or divergent CRISPR contents. Both synonymous and nonsynonymous mutations in the protospacer sequences were identified, as well as differences in the protospacer adjacent motif (PAM), which could explain how phages escape this system. This paper illustrates how the distribution and diversity of CRISPR spacers in C. difficile, and its prophages, could modulate phage predation for this pathogen and impact upon its evolution and pathogenicity.
IMPORTANCE
Clostridium difficile is a significant bacterial human pathogen which undergoes continual genome evolution, resulting in the emergence of new virulent strains. Phages are major facilitators of genome evolution in other bacterial species, and we use sequence analysis-based approaches in order to examine whether the CRISPR/Cas system could control these interactions across divergent C. difficile strains. The presence of spacer sequences in prophages that are homologous to phage genomes raises an extra level of complexity in this predator-prey microbial system. Our results demonstrate that the impact of phage infection in this system is widespread and that the CRISPR/Cas system is likely to be an important aspect of the evolutionary dynamics in C. difficile.
INTRODUCTION
The bacterium Clostridium difficile is a major nosocomial pathogen (1), which can also be carried asymptomatically (2), is present in environmental and zoonotic reservoirs (3), and can transmit between livestock and humans (4). The evolution of C. difficile is shaped by the acquisition and loss of mobile elements in its genome (5). A critical component of the mobilome is bacteriophages, which can mediate horizontal gene transfer (HGT) and impact upon the evolution of their hosts (6). Prophages are common within C. difficile genomes, and several temperate phages have been described which can also infect specific strains by following a lytic cycle (7). Either temperate or lytic phage infection could promote the HGT of novel genetic material, as exemplified by a recent demonstration of phage-mediated transduction in C. difficile (8). Typically, the reported lytic host ranges of C. difficile phages are narrow, and phages can differentially infect strains of the same ribotype (see the review by Hargreaves and Clokie [9]). The contribution that phages make to C. difficile evolution will clearly depend on the breadth of the hosts that they are able to infect.
The specificity of phage-host interactions is dependent on mechanisms used by bacteria to resist phage infection. One is the clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) system, which consists of arrays of short (24- to 47-bp) direct repeats (DRs) separated by variable spacer regions (26 to 72 bp) and Cas proteins. These recognize and degrade foreign DNA that is homologous to a spacer (10–15). Spacers are heritable, and can also be acquired through the incorporation of foreign DNA sequences. This means the CRISPR/Cas system is considered to be a form of adaptive immunity, and the spacer content of CRISPR arrays is a record of past infections (13). Examining spacers can provide insights into phage-host dynamics that have occurred within bacterial populations (12, 16, 17). One “cost” of an effective CRISPR/Cas system to a bacteria is that HGT is suppressed, which in turn reduces the gain of new, potentially useful genes introduced by phages or plasmids (18). The impact of the CRISPR/Cas system in limiting interactions with phages and plasmids differs by species and has been previously described (19–21); however, relatively few species have been studied in detail, and very little has been published for C. difficile.
All C. difficile phages sequenced to date carry integrases, which suggests that they can access the temperate life cycle and are therefore not truly lytic (22–27). Although prophage infection is prevalent in C. difficile (28), relatively few free or inducible prophages have been found that can propagate in a lytic manner on tested strains, despite large-scale screens for such phages (24, 27, 29). The mechanisms controlling these phage-host dynamics in C. difficile are unknown. Interestingly, an unusual situation exists in C. difficile in which CRISPR arrays are also found in C. difficile prophages (5). Transcription and processing of CRISPR RNA (crRNA) from prophage-carried arrays have been detected in C. difficile strain CD630 (30). If these prophage-carried arrays are fully functional CRISPR elements, they represent a mechanism for prophage elements to influence secondary phage infection and, subsequently, the extent of HGT. An example of phages impacting the CRISPR dynamics can be seen in the ICR1 phage that infects Vibrio cholerae. This phage has been shown to carry a functioning CRISPR/Cas system that targets a phage inhibitory chromosomal island, thus permitting infection of the host (31). The relationship between prophages and the CRISPR/Cas system is therefore not unidirectional.
In order to examine the potential significance of the CRISPR/Cas system in C. difficile, we used bioinformatic approaches to identify how the CRISPR/Cas system in diverse C. difficile strains relates to 31 phage and prophage genomes. To do this, we first examined phage and prophage genomic diversity to determine their relatedness. Second, we examined these 31 genomes for CRISPR arrays and established their DR and spacer diversity. We then searched the same phage genomes against a database of C. difficile spacers (including those from arrays in prophages). The locations of protospacers were identified, and the targets of the CRISPR/Cas system were examined in regard to gene function and conservation. Finally, putative protospacer adjacent motifs (PAMs) were identified using protospacer sequences from this analysis. Taken together, our analyses suggest that the CRISPR/Cas system is an important determinant of phage infection in C. difficile.
RESULTS
The CRISPR/Cas system in C. difficile strain CD630.
In order to characterize the CRISPR/Cas system within C. difficile strains, we examined the seven strains present in the CRISPRdb database of CRISPRfinder (32, 33). These carry multiple and diverse arrays, with the total number of spacers per strain ranging from 43 to 153, as determined using the CRISPRcompar tool (34). The C. difficile strain CD630 carries two sets of cas-like genes. One set belongs to type I-B (Tneap subtype) according to the typing system of Makarova et al. (35) and is composed of cas2 (CD2975), cas1 (CD2976), cas4 (CD2977), cas3 (CD2978), cas5 (CD2979), cas7 (or cst2) (CD2980), cas8b (or cst1) (CD2981), and cas6 (CD2982), as assigned by the Genome Properties Report from the JCVI Comprehensive Microbial Resource. These genes are near to the largest of the CD630 CRISPR arrays, NC_009089_17, which contains 19 spacers. The second set of predicted cas genes is distinct from the described types and has genes homologous to cas3 (CD2451), cas5 (CD2452), and cas6 (CD2455) but lacks a homolog of cas1, which is a nearly universal component of the cas system. The DR sequences from strain CD630 arrays are predicted to be folded, and a comparison of the DR sequences to those in Kunin et al. (36) shows they cluster closest to groups 4 (folded bacterial) and 11 (unfolded bacterial).
Whole-phage-genome analysis supports subgrouping according to particle morphology.
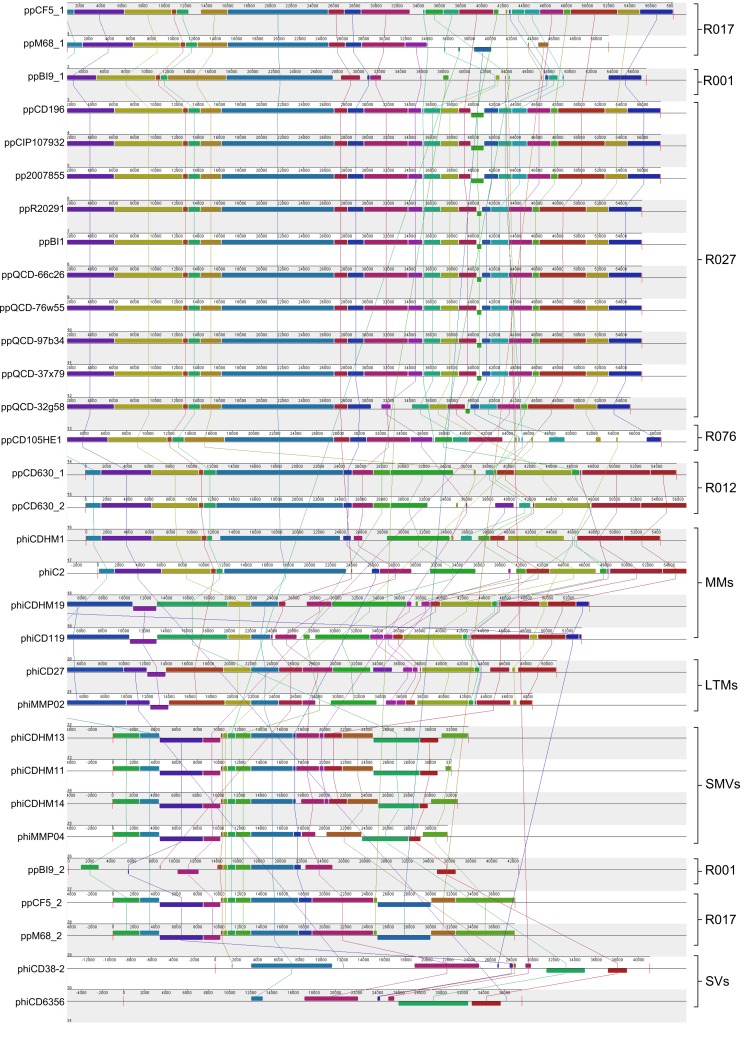
Several studies have produced sequence data for C. difficile phages and prophages within C. difficile genomes which were included in our analysis. In order to determine the genomic diversity across C. difficile phages, we compared the genomes of 12 phages and 19 prophages (see Table S1 in the supplemental material). Genomes were aligned in MAUVE, which identifies locally colinear blocks (LCBs), which are shared genomic regions free from homologous recombination (Fig. 1). The presence or absence of LCBs can be used to determine patterns of evolutionary relatedness, and several conserved orthologous LCBs were identified in multiple genomes. The phages can be separated into distinct lineages and have been arranged according to genomic similarity, genome size, and particle morphology, if known (37, 38). The majority of prophages examined in this study are similar to ϕC2, and representatives are present in all 15 of the C. difficile strains examined. Four of these strains carry two predicted prophages. Strain CD630 (R012) carries two ϕC2-like prophages, and strain BI9 (R001) carries a ϕC2-like prophage and one phage that appears similar to the small myovirus type. Lastly, strain M68 (R017) and strain CF5 (R017) carry ϕC2-like prophages that are closely related to one another. There are several instances where LCBs are shared across divergent lineages, and two were identified in all the examined sequences.
FIG 1 .
Whole-genome alignment of 31 phages and prophages infecting C. difficile displays genomic lineages and homologous recombination occurring between phage subtypes. Alignments were performed in MAUVE using the progressiveMAUVE algorithm (36). Colored blocks indicate regions free from homologous recombination, and lines to colored blocks show homologous regions in other genomes. The phages have been grouped according to morphology if known; medium myoviruses (MMs), long-tailed myoviruses (LTMs), small myoviruses (SMVs) and siphoviruses (SVs), and prophages, by the ribotype of their lysogen and genomic similarity to the phage genomes.
Prophages are a source of diverse CRISPR spacers in the C. difficile cell.
Because the identification of CRISPR arrays on prophages in C. difficile is unusual, we tested how widespread and diverse they are in the 19 prophage and 12 phage genomes using CRISPRfinder (32). All of the ϕC2-like prophages carry multiple predicted CRISPR arrays (n = 2 to 4), with the number of spacers varying between 7 and 24 (Fig. 2). The arrays are located in the structural region of the phage genomes between the xkdN gene (whose product is a predicted structural protein) and the tape measure protein (TMP) gene. In one, ppBI9_1, an additional single predicted array is located in the DNA replication region. Interestingly, no CRISPR arrays were identified in the ϕC2-like phage genomes which had been isolated following lytic propagation. However, we detected a PCR product of the expected size of the CRISPR arrays using primers to specifically amplify the CRISPR arrays from released free phage particles (see Fig. S1 in the supplemental material). This was established by performing transmission electron microscopy (TEM) to visualize phage particles and PCR on DNase-treated phage lysates which were negative for a bacterial 16S rRNA PCR product.
FIG 2 .
Locations of CRISPR arrays on prophage genomes and spacer content. (A) The CRISPR arrays (boxes) and adjacent genes (arrows showing orientation) were mapped to each prophage genome (scale in bp). Gene annotations were assigned following identification of protein domains in Pfam and by performing blastp searches against the NCBI nr/nt database. The consensus DR sequences of the arrays are color coded. Spacer content of the arrays revealed that ppCD196 represents all the R027 prophages, as they have identical CRISPR contents. ppCD630_1 also represents the CRISPR content of ppCD630_2, as both are identical. (B) Alignment of the consensus DR from each CRISPR array identified with single-nucleotide polymorphisms (SNPs; red boxes). Color coding reflects conserved groups. (C) The spacer content for each CRISPR array is shown with the corresponding DR with use of color to show identical sequences. White indicates spacers which are unique.
Six distinct prophage-carried CRISPR arrays can be resolved based on their consensus DR sequence, spacer content, and adjacent coding DNA sequences (CDSs). The strain CD630 prophages have CRISPR regions that are identical to one another, as do the strain R027 prophages; they are represented in Fig. 2 by ppCD630_1 and ppCD196, respectively. Across the prophages, there is at least one array with a conserved DR sequence, and all the DRs belong to the same family (Fig. 2), which is also present in C. difficile chromosomal arrays. Most spacers are unique, but eight are present in more than one array type and in different locations within the arrays which carry them. No cas genes were identified in the prophage genomes, and it is likely that they are processed by the bacterial Cas proteins. There are several CDSs adjacent to the arrays. They encode predicted proteins with putative regulatory or DNA binding roles, for example, ORF6N (39), Bro-N (40), and ribosomal_L12 (41). These proteins may be involved in transcription of the prophage-carried CRISPR arrays.
C. difficile CRISPR spacer homology supports various host-phage interactions.
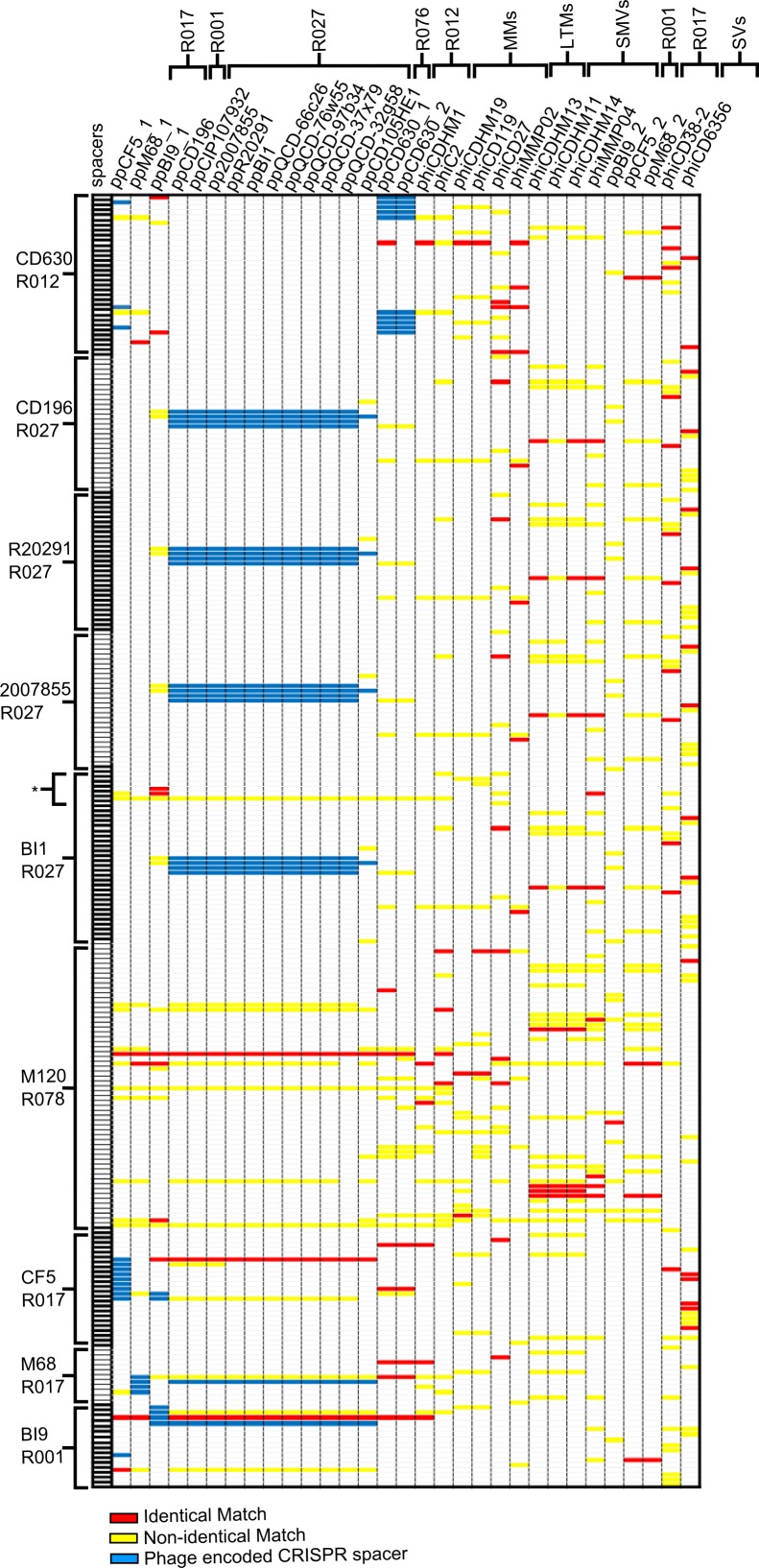
To determine if the CRISPR/Cas system of C. difficile could target known phages, the 31 genomes were searched against the CRISPRdb database, containing spacers (33). In total, 758 matches between spacers and phage sequences were identified, of which 162 were identical (Fig. 3). This large number is despite there being only nine C. difficile strains in the CRISPRdb database (as of September 2013), four of which are the same ribotype and therefore likely to represent a small proportion of the total C. difficile CRISPR spacer diversity. The spacers we identified represent the minority of total spacers, with 17 to 38% in each strain. All phages and prophages have a spacer which matches to them in at least one C. difficile strain, and similarly every strain has at least one spacer with an identical match to a phage sequence. The number of spacers with matches for each phage or prophage ranges from 12 to 53 (with between 1 and 16 having identical matches) and in each strain from 38 to 300 (with identical matches ranging between 4 and 55 for each). The CRISPR profiles generated are the same for the R027 strains, with the exception of strain BI1. This strain has additional spacers in CRISPR arrays on a large extrachromosomal piece of DNA ~300 kbp in size (GenBank accession no. NC_017177), and the seven spacers account for a further 30 matches (three identical) in this data set.
FIG 3 .
CRISPR profiles of 31 phages and 9 strains indicating homologous matches between spacers and genome sequences. All significantly scoring matches (E value of >0.005) between spacers and phage sequences are shown following searches using CRISPRfinder (see Table S5 for sequences). Spacers (in rows) are in numerical order for each C. difficile strain and include only those which had significant matches to the searched phage sequences (not all spacers in the strains). Of the nine strains, four belong to R027 and two to R017. Asterisk indicates spacers carried on strain BI1 extrachromosomal DNA. Phage and prophages (columns) are arranged according to their order in the MAUVE alignment. Solid red boxes represent identical matches between spacer and genome sequence, yellow boxes represent nonidentical matches, and blue boxes are prophage CRISPR spacers.
Multiple strains carry spacers which match to the same phage, e.g., 8/9 strains have spacers that identically match to sequences in the genomes of ϕCD27. In contrast, spacers of one strain match to ϕC2, which suggests less widespread predicted immunity to this phage. From a bacterial perspective, individual strains have multiple spacers which match to several phages (e.g., those in strain M120 match to all but two phages), whereas other strains have fewer spacers with matches (e.g., those in strain M68 match to only four phages). If the CRISPR/Cas system can impart immunity, our data suggest that this mechanism would result in some phages being able to infect in a generalist manner, but others would be more specialized.
Despite the occurrence of identical matches between spacers and sequences, the CRISPR/Cas system can be evaded by divergence in the PAM (42). Using the identical spacer matches detected in the analysis, the corresponding protospacer sequences in the phage and prophage genomes were located. The upstream and downstream nucleotide (nt) sequences were compared, and a putative PAM sequence was identified, CCN, but no motif was identified in the downstream sequences (see Fig. S2 in the supplemental material). Multiple PAMs have been reported in other type I-B systems (43), and our alignment suggests that this may also be the case in C. difficile, as a motif of CCA was present in 46% and a motif of CCT was present in 23% of these upstream regions.
The complexity of the C. difficile CRISPR/Cas system is highlighted by the surprising identification of spacers with matches to their own prophage sequences. An example of this is in strain CD630, which has one chromosomally carried spacer that matches to the genomic sequence of one of its prophages, ppCD630_1. Avoidance of chromosomal self-targeting has been described in other bacterial species by mutation of the PAM sequence (44). The protospacer sequence in CD630 is identical to that of the spacer, and it may avoid recognition by the CRISPR/Cas system, as it has a divergent PAM sequence, CTA.
CRISPR arrays carried by C. difficile prophages may provide widespread phage resistance.
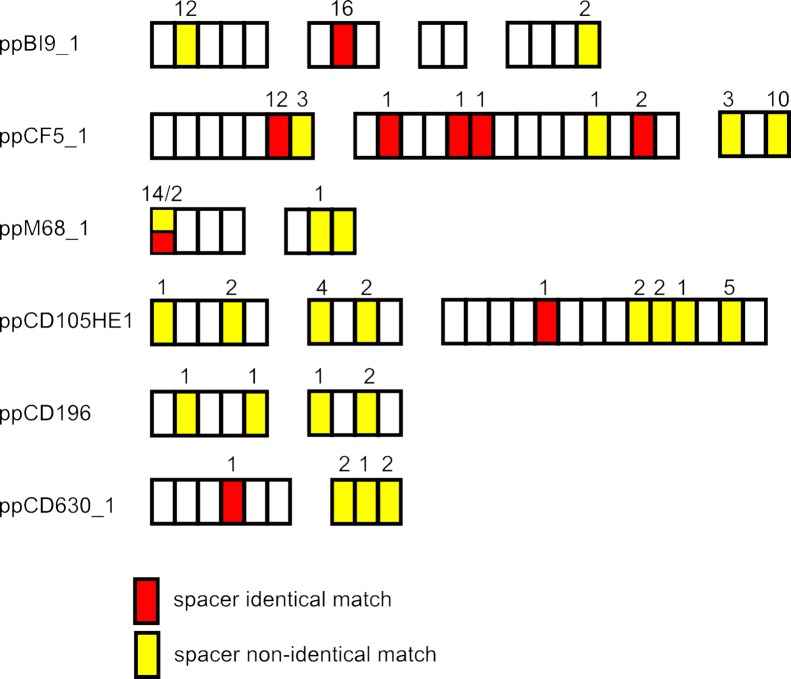
The nine C. difficile strains in the CRISPRdb database all harbor at least one prophage with CRISPR arrays, as shown in Fig. 2. As part of our analysis, these 10 prophages were included in the searches against the CRISPRdb database in order to identify protospacers in their genomes (Fig. 2). We detected spacers in these prophage arrays which match to other phage sequences used in the analysis. For example, ppCF5_1 has four spacers, located across its arrays, which match to 16 phages (Fig. 4). For the 31 spacers with matches, 11 of the corresponding protospacer sequences are located in the TMP gene, but identical matches were also identified to protospacer sequences located in a predicted endonuclease, hypothetical proteins, and tail sheath proteins. Several more spacers were found to match in a nonidentical manner to all phage sequences except the small myoviruses, ΦMMP02 and ppCF5_2, illustrating the potential immunity conferred by these prophage-carried spacers across phage lineages.
FIG 4 .
Prophage-carried spacers match to C. difficile phage and prophage genomes. The spacers from the multiple CRISPR arrays identified in the legends to Fig. 2 and 3 are shown with respective matches to the 31 phage and prophage genomes. They include identical and nonidentical matches, as well as those matching to multiple sequences and those matching to single sequences. The distributions of spacers with matches to the searched genomes are throughout the arrays.
The number of shared protospacer sequences is higher in related phages than in less-related phages.
In order to determine if the level of genetic similarity of phages could predict host ranges based on CRISPR/Cas immunity, we identified shared protospacer sequences between phage sequences. If host immunity is controlled by the CRISPR/Cas system, then shared protospacers would predict similar host ranges. We plotted the number of shared protospacers against a whole-genome blastn score in pairwise comparisons (see Fig. S3 and Tables S2 and S3 in the supplemental material). The two values positively correlate, R = 0.7467, supporting this suggestion. However, differences in host ranges between related phages can be explained by unshared spacer matches as well as sequence differences in protospacers or PAM. The resulting nucleotide differences in the phage sequences include both synonymous and nonsynonymous mutations. An example is in ppCF5_1 and ppM68_1. The endolysin genes in each prophage share a similarity of 98.40% at the nucleotide level, and each gene contains two protospacer sequences. In each case, these differ by one nucleotide; in one, this difference results in an identical match between the spacer and protospacer in ppCF5_1 and a nonidentical match in ppM68_1. Whether this confers evasion against the immune system is unknown, but when the nucleotide sequence is translated, a nonsynonymous change occurs. The amino acid sequence of the endolysin differs by 6 residues between the two prophages, 2 of which result from the protospacer sequences, and we suggest that CRISPR evasion likely impacts on the conserved endolysin gene.
Protospacer containing CDSs by function tested relative to size and frequency.
In order to investigate whether specific genes were more frequently observed to contain protospacers than others in our analysis, the location of each protospacer was identified. Protospacers are present in genes with predicted functions that encompass all essential processes associated with a temperate phage life cycle (see Fig. S4 in the supplemental material). This includes genes encoding structural proteins, involved in the control of lysogeny, and involved in the lytic life cycle. No protospacers were identified in genes whose predicted products are potential lysogenic conversion factors, such as AbiF encoded in some of the phage genomes (e.g., phiC2p37 in ϕC2), the agrDBC-like cassette in phiCDHM1, or the VirE protein (e.g., CD_1450 in ppCD196), but protospacers were found in genes encoding hypothetical proteins located in the predicted lysogeny conversion modules of some phage genomes, downstream of the endolysin genes and on the negative-sense orientation. Protospacers are also present in many hypothetical proteins whose functions are unknown, as well as some which are located outside predicted CDSs.
In this analysis, we also see that the number of times a specific gene is targeted varies, as does the number of phages or prophages with the same protospacers (see Fig. S4). Examples where we can see this relative bias are within the lysis and structural genes. To examine this further, we investigated the TMP and the endolysin gene, both of which are present in all the examined genomes. We identified 23 unique spacers across all the strains which are homologous to sequences in TMP genes; the majority of spacers match only to a single phage’s copy of the gene. In contrast, four spacers in two C. difficile strains, M120 and BI1, match to phage and prophage endolysin genes, with 23, 20, 5, and 1 matches across the panel examined. The different number of spacers identified for each gene may result from gene length and/or PAM content. To test this, we found 192 instances of either CCA or CCT across the 2,343-nt TMP gene and 62 in the 813-nt endolysin gene of phiCDHM1, with frequencies of 8.19% and 7.62%, respectively, but the relative proportions of spacers in this data set are 11.98% and 6.45%, respectively. Phylogenetic analysis of these genes and the mapping of spacer matches illustrate the conservation for each and the distribution of matches with this phage set (see Fig. S5 in the supplemental material). The overall mean distances for each gene when aligned are 0.116 and 0.776 for the endolysin and TMP genes, respectively, which suggests that sequence conservation is likely to account for bias observed in our data set.
The positions of the spacers were mapped to the translated endolysin sequences. Two are positioned in the C-terminal region of the protein and two within the amidase 3 protein domain (PF01520) in the N-terminal region. The crystal structure of this has been solved for CD27L, the endolysin of ϕCD27, so it is possible to determine how the position of one spacer corresponds to amino acid residues 56 to 67. These are located in a loop extension and alpha-2 helix of the protein (45) and could explain how the function of the endolysin is retained following sequence mutation. The other spacer is homologous to one phage, and the sequence it matches to overlaps the predicted start of the CDS. This sequence is divergent to that in the other phage endolysin sequences (data not shown).
The holin gene is another highly conserved phage gene which is targeted by two spacers in strain M120. One of these matches to the holin gene of ΦMMP02, whereas the second matches to a sequence in the holin genes of seven phages. Previously, the sequence similarity between the tcdE gene and phage-encoded holin gene led to the suggestion that there is a phage origin of the toxin carrying PaLoc in C. difficile (23). To determine if the CRISPR system differentiates between these two genes, we searched the tcdE gene from strain CD630 (CD630_06610) against the CRISPRdb. The first of these spacers has a nonidentical match to tcdE, reflecting the sequence similarity of these genes and the theory that this gene has a xenologous origin.
DISCUSSION
Although phage CRISPR/Cas system dynamics have been explored in other systems, little analysis has been published examining the possible role of the CRISPR/Cas system in C. difficile. To test the hypothesis that the C. difficile CRISPR/Cas system could contribute to phage infection dynamics in this species, we examined the potential for C. difficile spacers to target known C. difficile phage genomes.
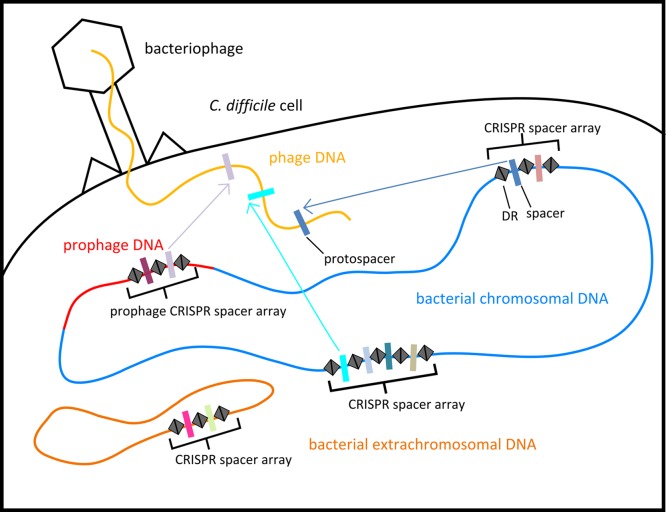
The CRISPR/Cas system of C. difficile is shown to be diverse between strains, and in this respect it is similar to the functional systems that have been reported and studied for other species (30, 31). Our results show that multiple C. difficile spacers were identified that are homologous to known phage and prophage sequences (Fig. 5). They include spacers located on prophages, the extrachromosomal DNA of strain BI1, and the chromosomes of all examined strains. The spacer content of CRISPR arrays can provide insights into recent and predominant phage predation; Díez-Villaseñor et al. (46) found that the most recently incorporated spacers in Escherichia coli had more matches to known “extant” phage genomes. In contrast, we found spacers that match to the known phages positioned throughout the bacterial or prophage CRISPR arrays. This may mean spacer acquisition does not occur primarily at the leader region or that the C. difficile strains examined have been more recently challenged by unknown phages or plasmids. This is pertinent when considering the potential role of phages in driving the evolution of epidemic strains.
FIG 5 .
Locations of CRISPR arrays in the C. difficile cell. Multiple CRISPR arrays are in C. difficile genomes, on the bacterial chromosome, and on extrachromosomal DNA and prophages.
Spacer content has been used to estimate the abundance and diversity of phage populations for specific species: examples include the M120-like phages infecting Streptococcus (47) and the diversity of phages infecting Microcystis aeruginosa (16). In our data set for C. difficile, we identified multiple spacer matches to single phages, with multiple shared and distinct spacers between C. difficile strains. These observations are consistent with a scenario resulting from a model of coevolutionary dynamics of bacteria and phage populations using evolving CRISPR defense, where multiple hosts are present in a coalition and have immunity conferred by different spacers against similar viruses (48). In the model, these coalitions are dominant but fall when a newly divergent phage emerges to which no strains have immunity. When the CRISPR spacers of natural bacterial populations have been examined, genotypes were detected which had multiple specific phage immunities (16). Similarly, in our analysis, the specific patterns of matches between host spacers and phage sequences suggest there are groupings of susceptible hosts to groups of phages. However, while CRISPR analysis showed that the genetic relatedness of phages and shared protospacers were positively correlated, there were also unique protospacers between these related phages. Also, the locations of several protospacers suggest that there may be bias in this data set, as highly conserved genes are targeted by multiple spacers which could confer wide immunity. Importantly, the CRISPR system is unlikely to be the only phage resistance mechanism in C. difficile. The use of the CRISPR system to predict phage interactions is currently limited by unknown factors, such as the rates of escape mutants and of spacer acquisition. Published host range data available for C. difficile phages suggest that phage-host interactions do not depend solely on the CRISPR/Cas system (e.g., differences in absorption [49]). A host range analysis which included ϕCD38-2, ΦMMP04, and ΦMMP02, also used in the CRISPR analysis here, showed that ϕCD38-2 could infect strains CD196 and R20291 (50), but these strains have a spacer which identically matches to the sequence in its genome with an intact CCA motif. Although infective, the infection is reported on the lowest scoring, indicating that there may be predominantly lysogenic infection occurring. These findings suggest that this phage may have a mechanism to evade the CRISPR/Cas system. Notably, the CRISPR/Cas system has recently been found to be evaded by Pseudomonas phages that carry anti-CRISPR proteins (51).
Similarly, a chromosomal spacer in strain CD630 is identical to the sequence of one of its prophages, ppCD630_1. The control of prophage insertion and excision in Pseudomonas aeruginosa has been previously suggested to occur, as this species also has spacers which match to temperate rather than virulent phage genomes (52, 53). However, the targeting of an established prophage has been shown in E. coli mutants to be highly lethal (54). In our example of a chromosomal spacer matching an established prophage, it appears likely to avoid recognition via a mutation in its PAM, which is known to avoid chromosomal self-targeting (44). Other sequence differences in specific regions of the protospacer, such as the seed regions identified in E. coli, can also interfere with the CRISPR/Cas pathway (55). Mutations in the protospacers located outside the seed region do not inhibit CRISPR spacer recognition, and if this is the case also in C. difficile, it would alter the predicted interactions and warrant future research.
This analysis has also shown that several related prophages carry CRISPR arrays which are diverse in composition. Their presence may be the result of chromosomal scattering of the CRISPR arrays, and subsequent loss and gain of spacers occurred. A partial transposase gene is located upstream of the CRISPR array regions, which supports the theory that these have been transferred via one or more HGT events. Homologs of genes adjacent to the CRISPR arrays are present in some of the other C. difficile phage genomes, for example, phiC2p19 in ϕC2. Although in prophage genomes, the CRISPR arrays may be transferred, as the CD630 prophages can also access the lytic pathway (23), and we show that the prophage of C. difficile CD105HE1 was released spontaneously and retains the CRISPR arrays in its genome. This is consistent with the report from a metagenomic study of the human virome that found that CRISPR arrays are present in DNA sequence data from free viral particles, suggesting that there is the potential for exchange of arrays via HGT in the human gut microbial population (56). In this study and a subsequent study, the researchers report the detection of virus-carried spacers targeting viral sequences, specifically, from a temperate phage infecting Ruminococcus bromii (57).
Whether the prophage CRISPR arrays function to confer immunity has not been established, but in strain CD630, both prophage arrays are transcribed, and the processing of the pre-crRNA was detected in arrays 15 and 16 (on ppCD630_2) (30). Our analysis found that the spacer content could be highly divergent between prophages, suggesting that continued spacer acquisition and loss have occurred. Typically, spacers are acquired from the leader region, but HGT and other mechanisms of incorporation occur in some CRISPR/Cas systems (31, 58–60). While CRISPR array evolution is thought to be primarily rapid, the spacer contents of the R027 prophages are identical despite originating from isolates obtained over decadal timescales (61), which agrees with a slower reported rate of CRISPR change in E. coli (19, 21). Mechanisms for the loss of CRISPR spacers are not fully understood but have been observed following a pathogenic host shift in Mycoplasma gallisepticum (62). Similarly, CRISPR arrays in prophages from an environmental isolate (CD105HE1) (this study) and an isolate from asymptomatic human (CF5) (63) carry more spacers than those in prophages from clinical strains, which have on average 38% fewer spacers. The numbers of CRISPR spacers in surviving populations of E. coli have been found to increase during phage infection but decrease during instances where lateral gene transfer was beneficial in an in vitro test using antibiotic selection and a plasmid carrying the resistance gene (64). The different numbers of arrays and spacers between prophages, and between strains of C. difficile, may have arisen following different selection pressures in the natural and clinical environments.
Our findings suggest that specific prophages in C. difficile could confer immunity to invading phages via the spacers in their CRISPR arrays. This is important to consider, as phage infection in this species has been found to influence bacterial physiology, such as toxin production (26, 65–68), and is also being explored as novel therapeutics (68). Prophage carriage in the ribotypes examined show how they could influence phage susceptibility, as the R027 strains carry similar prophages with identical CRISPR spacers; but in contrast, two strains belonging to R017 carry prophages with distinct CRISPR contents. Differential carriage of prophages could explain differences in phage susceptibility in specific ribotype groups and presents a highly novel facet of the CRISPR immune system in phage-phage wars. We suggest that an advantage to retaining prophages in the highly lysogenized C. difficile may be due to the fact that some are a source of spacers. Further work will evaluate the activity of this CRISPR/Cas system.
MATERIALS AND METHODS
Whole-genome alignments to assess phage and prophage diversity.
The genomes of 31 phages and prophages were used in the analysis, as they represent morphologically diverse types (including medium myoviruses, long-tailed myoviruses, small myoviruses and siphoviruses, and prophages in epidemic and nonepidemic, recent and historical strains) (see Table S1 in the supplemental material). Prophage sequences were predicted using PHAST (69). CDSs were predicted using FGENESV (Softberry Inc., United States) and annotated based on results of searches against the online Pfam database (70) and the NCBI nt/nr database using blastp accessed at http://www.ncbi.nlm.nih.gov/Blast.cgi. Genome sequences were visualized using Artemis Genome Browser (71), and prophage sequences were rearranged to start with the terminase genes. Whole-genome alignment was performed using MAUVE v2.3.1 (37) by using the progressiveMAUVE algorithm (38).
C. difficile CRISPR array analysis and protospacer identification in phage and prophage genomes.
The same set of phages and prophages were searched for CRISPR arrays using CRISPRfinder (32) and against the CRISPRdb database (33). DR and gene alignments were performed using Clustal Omega (72). A relative measure of phage genetic relatedness was calculated from the blastn results generated from pairwise comparison of genomes using Double Act v2 accessed at http://www.hpa-bioinfotools.org.uk/pise/double_act.html#. The score value was defined as the total number of bases aligned in sequences that had ≥80% identity and were ≥20 nt in length. Shared protospacer analysis used unique identical and nonidentical matches. Data analysis and correlation tests were performed in Microsoft Office Excel and MiniTab. Protospacer adjacent motifs (PAMs) were identified from comparison of 10-nt upstream and downstream sequences for 52 unique protospacers which had a perfect match to spacers using WebLogo (73, 74). Cas genes in strain CD630 were identified using the Genome Properties search tool (accessed 31 January 2014 at http://cmr.jcvi.org/tigr-scripts/CMR/shared/MakeFrontPages.cgi?page=genome_property). RNA structure was predicted using Vienna RNAalifold (accessed at http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi). DR sequence alignment using MUSCLE and unweighted pair group method with arithmetic mean (UPGMA) analysis was performed in MEGA v5.2 (75). Matches to the spacers in arrays in ppCD105HE1 were identified using CRISPRTarget (76). Overall mean composite distances were calculated for the nucleotide sequences of the tail tape measure (TMP) and endolysin genes (excluding partial CDSs) in MEGA v6.06 (77). Sequences were first aligned using MUSCLE, and the distance estimation was performed using the Poisson model with uniform rates and pairwise deletions. Maximum likelihood analysis was performed at the amino acid level using the Jones-Taylor-Thornton (JTT) model with gamma distribution and 500 interactions for bootstrapping.
Release of ppCD105HE1 particles and PCR-based detection of CRISPR arrays.
Release of phage particles was assessed using TEM from culture lysates using methods described previously (28). Primers targeting each of the three CRISPR arrays were designed using Primer3 v0.4.0 (78) (see Table S2 in the supplemental material for the oligonucleotide sequences). Lysates of strain CD105HE1 grown overnight in brain heart infusion broth (BHI; Oxoid, United Kingdom) were centrifuged at 3,398 × g for 10 min, and the supernatant was filtered through a 0.22-µm-pore-size filter (Millipore, United Kingdom). The lysate was then treated with TurboDNase (Life Technologies, United Kingdom) according to the manufacturer’s guidelines. Contamination with bacterial DNA was detected using primers targeting the 16S rRNA gene as described previously (79). Uncontaminated lysate was then used as the template in PCRs with each of the CRISPR array primer sets. Reactions were performed in 25 µl, containing template DNA, 0.6 mM forward and reverse primers, 0.25 mM deoxynucleoside triphosphates (dNTPs), 3 mM MgCl2, 1× PCR buffer, and 0.5 U of BioTaq polymerase (Bioline, United Kingdom) under conditions of 95°C for 5 min, 30 cycles of 95°C for 45 s, 48°C for 45 s, and 72°C for 60 s, with a final extension step of 5 min at 72°C. Products were separated using 110 V for 30 min on a 1% agarose Tris-acetate-EDTA (TAE) gel stained using ethidium bromide. Product size estimates were based on a 1-kbp GeneRuler ladder (Thermo Scientific, United Kingdom).
SUPPLEMENTAL MATERIAL
PCR-based detection of CRISPR arrays retained following excision of ppCD105HE1 from the bacterial chromosome. DNA from free viral particles of ppCD105HE1 following spontaneous release was assayed for the presence of the CRISPR arrays identified in the prophage genome using three sets of primers flanking each array in a PCR. Lanes 2 to 4 are no-template DNA control, gDNA of CD105HE1 and ppCD105HE1 DNA with primers targeting array 1, with both CD105HE1 and ppCD105HE1 producing the expected ~499-bp product; lanes 5 to 7, the same order of templates for array 2, also with CD105HE1 and ppCD105HE1 samples showing the same expected 399-bp product; and lanes 8 to 10, showing CD105HE1 and ppCD105HE1 samples with the expected ~1-kbp product for array 3. Download
Conserved motif prediction upstream and downstream of protospacers. Sequences 10 nt in length either upstream (A) or downstream (B) of identical protospacer matches were aligned. In the upstream alignment, two cytosines were identified as highly conserved across 51 upstream regions, and putative PAM sequences of CCA and CCT were predicted. No highly conserved motif was identified in the downstream regions. Only unique, identical matches were included in the WebLogo analysis in an effort to eliminate sequence bias. Download
Pairwise analyses of phages show that the number of shared protospacers positively correlates to the relatedness of phages. A plot is shown of the number of protospacers shared by pairs of phages (see Table S3 in the supplemental material) against a relative value of their relatedness calculated from results of ACT comparison between the whole phage genomes (see Table S4). All combinations of phages were analyzed in a pairwise manner. The number of shared spacers was calculated from matches to the CD630, BI1, M120, and CF5 spacers, as these represented all the unique hits available to avoid bias by including the R20291, CD196, and 2007855 strains, which all have the same spacer hits to phages as BI1, with the exception of those on BI1’s extrachromosomal DNA. Download
Relative proportions of CDSs with protospacers in phage and prophage genomes. Three pie charts show the relative proportions of CDSs in the genome of phiCDHM1 (top) and proportion of CDSs containing protospacer sequence (unique hits, bottom left; total hits, bottom right). The CDSs have assigned groups according to putative gene product functions: clockwise, packaging (including predicted portal and terminase genes), structural (including tape measure protein and tail sheath genes), lysis/attachment (including endolysin and tail fiber genes), lysogeny conversion, lysogeny control (including integrase and repressor genes), replication (including DNA methylase and DNase/helicase genes), unknown (which includes hypothetical genes and non-CDSs and/or significant hit was detected). The relative proportion changes between analyzing unique spacers and the total number of matches, reflecting a difference in results when examining diversity (unique hits) versus frequency (total hits). The analysis included identical and nonidentical matches, and the proportions are relative to the size of the gene of all matches for each analysis. To avoid bias, genomes which had a >95% similarity score using blastn were excluded from the analysis, and one genome was used in each case. Download
ML phylogenetic analysis of the TMP and endolysin genes with spacer matches. Phylogenetic trees for the TMP (A) and endolysin (B) are shown with spacer matches indicated by circles, and where there are shared matches by the same spacer, circles are connected. Download
List of phages and prophages used, with accession numbers and strain details.
CRISPR array flanking oligonucleotide sequences.
Pairwise shared protospacer analysis.
Pairwise whole-genome BLASTN score analysis.
C. difficile spacer sequence with homologous protospacer sequence.
ACKNOWLEDGMENTS
This work was supported by an MRC Centenary Fellowship awarded to K.R.H. and an MRC New Investigator award (G0700855) awarded to M.R.J.C. C.O.F. is supported by a CONACyT (Mexico) graduate fellowship.
We thank Julie Pratt for her useful comments on the manuscript. We also thank Joshua Weitz for his useful discussion, comments, and expert advice when we were writing the manuscript.
Footnotes
Citation Hargreaves KR, Flores CO, Lawley TD, Clokie MRJ. 2014. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. mBio 5(5):e01045-13. doi:10.1128/mBio.01045-13.
REFERENCES
- 1. Bouza E. 2012. Consequences of Clostridium difficile infection: understanding the healthcare burden. Clin. Microbiol. Infect. 18:5–12. 10.1111/j.1469-0691.2012.03862.x [DOI] [PubMed] [Google Scholar]
- 2. Hall IC, O’Toole E. 1935. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. Am. J. Dis. Child. 49:390–402. 10.1001/archpedi.1935.01970020105010 [DOI] [Google Scholar]
- 3. al Saif N, Brazier JS. 1996. The distribution of Clostridium difficile in the environment of South Wales. J. Med. Microbiol. 45:133–137. 10.1099/00222615-45-2-133 [DOI] [PubMed] [Google Scholar]
- 4. He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 45:109–113. 10.1038/ng.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786. 10.1038/ng1830 [DOI] [PubMed] [Google Scholar]
- 6. Siefert JL. 2009. Defining the mobilome. Methods Mol. Biol. 532:13–27. 10.1007/978-1-60327-853-9_2 [DOI] [PubMed] [Google Scholar]
- 7. Goh S, Riley TV, Chang BJ. 2005. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 71:1079–1083. 10.1128/AEM.71.2.1079-1083.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goh SH, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. 2013. Phage ϕC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 4(6):e00840-13. 10.1128/mBio.00840-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hargreaves KR, Clokie MRJ. 2014. Clostridium difficile phages: still difficult? Front. Microbiol. 5:184. 10.3389/fmicb.2014.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575. 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- 11. Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- 12. Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 13. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 14. Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186. 10.1038/nrmicro1793 [DOI] [PubMed] [Google Scholar]
- 15. Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62–70. 10.1016/j.ijfoodmicro.2008.05.030 [DOI] [PubMed] [Google Scholar]
- 16. Kuno S, Yoshida T, Kaneko T, Sako Y. 2012. Intricate interactions between the bloom-forming cyanobacterium Microcystis aeruginosa and foreign genetic elements, revealed by diversified clustered regularly interspaced short palindromic repeat (CRISPR) signatures. Appl. Environ. Microbiol. 78:5353–5360. 10.1128/AEM.00626-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stern A, Mick E, Tirosh I, Sagy O, Sorek R. 2012. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 22:1985–1994. 10.1101/gr.138297.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barrangou R, Horvath P, Doyle M, Klaenhammer T. 2012. CRISPR. New horizons in phage resistance and strain identification. Annu. Rev. Foods Sci. Technol 3:143–162. 10.1146/annurev-food-022811-101134 [DOI] [PubMed] [Google Scholar]
- 19. Touchon M, Rocha EP. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. 10.1371/journal.pone.0011126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. 2011. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One 6:e19543. 10.1371/journal.pone.0019543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Touchon M, Charpentier S, Clermont O, Rocha EP, Denamur E, Branger C. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J. Bacteriol. 193:2460–2467. 10.1128/JB.01307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Govind R, Fralick JA, Rolfe RD. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage Phi CD119. J. Bacteriol. 188:2568–2577. 10.1128/JB.188.7.2568-2577.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goh S, Ong P, Song K, Riley T, Chang B. 2007. The complete genome sequence of Clostridium difficile phage phi C2 and comparisons to phi CD119 and inducible prophages of CD630. Microbiology 153:676–685. 10.1099/mic.0.2006/002436-0 [DOI] [PubMed] [Google Scholar]
- 24. Horgan M, O’Sullivan O, Coffey A, Fitzgerald GF, van Sinderen D, McAuliffe O, Ross RP. 2010. Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family. Gene 462:34–43. 10.1016/j.gene.2010.04.010 [DOI] [PubMed] [Google Scholar]
- 25. Mayer MJ, Narbad A, Gasson MJ. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190:6734–6740. 10.1128/JB.00686-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sekulovic O, Meessen-Pinard M, Fortier LC. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 193:2726–2734. 10.1128/JB.00787-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meessen-Pinard M, Sekulovic O, Fortier LC. 2012. Evidence of in vivo prophage induction during Clostridium difficile infection. Appl. Environ. Microbiol. 78:7662–7670. 10.1128/AEM.02275-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hargreaves KR, Colvin HV, Patel KV, Clokie JJ, Clokie MR. 2013. Genetically diverse Clostridium difficile strains harboring abundant prophages in an estuarine environment. Appl. Environ. Microbiol. 79:6236–6243. 10.1128/AEM.01849-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shan J, Patel KV, Hickenbotham PT, Nale JY, Hargreaves KR, Clokie MR. 2012. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Appl. Environ. Microbiol. 78:6027–6034. 10.1128/AEM.01311-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 9:e1003493. 10.1371/journal.pgen.1003493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. 10.1038/nature11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a Web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. 10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grissa I, Vergnaud G, Pourcel C. 2008. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 36:W145–W148. 10.1093/nar/gkn228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kunin V, Sorek R, Hugenholtz P. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61. 10.1186/gb-2007-8-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iyer LM, Koonin EV, Aravind L. 2002. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 3:R0012. 10.1186/gb-2002-3-3-research0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zemskov EA, Kang W, Maeda S. 2000. Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J. Virol. 74:6784–6789. 10.1128/JVI.74.15.6784-6789.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leijonmarck M, Liljas A. 1987. Structure of the C-terminal domain of the ribosomal protein-L7/L12 from Escherichia coli at 1.7 A. J. Mol. Biol. 195:555–580. 10.1016/0022-2836(87)90183-5 [DOI] [PubMed] [Google Scholar]
- 42. Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJJ. 2013. Type I-E CRISPR-Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 9:e1003742. 10.1371/journal.pgen.1003742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shah SA, Erdmann S, Mojica FJ, Garrett RA. 2013. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 10:891–899. 10.4161/rna.23764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9:e1003454. 10.1371/journal.pgen.1003454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. 2011. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J. Bacteriol. 193:5477–5486. 10.1128/JB.00439-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156:1351–1361. 10.1099/mic.0.036046-0 [DOI] [PubMed] [Google Scholar]
- 47. van der Ploeg JR. 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966–1976. 10.1099/mic.0.027508-0 [DOI] [PubMed] [Google Scholar]
- 48. Childs LM, Held NL, Young MJ, Whitaker RJ, Weitz JS. 2012. Multiscale model of CRISPR-induced coevolutionary dynamics: diversification at the interface of Lamarck and Darwin. Evolution 66:2015–2029. 10.1111/j.1558-5646.2012.01595.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramesh V, Fralick JA, Rolfe RD. 1999. Prevention of Clostridium difficile-induced ileocecitis with bacteriophage. Anaerobe 5:69–78. 10.1006/anae.1999.0192 [DOI] [Google Scholar]
- 50. Sekulovic O, Garneau JR, Néron A, Fortier L-C. 2014. Characterization of temperate phages infecting Clostridium difficile Isolates from human and animal origin. Appl. Environ. Microbiol. 80:2555–2563. 10.1128/AEM.00237-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Essoh C, Blouin Y, Loukou G, Cablanmian A, Lathro S, Kutter E, Thien HV, Vergnaud G, Pourcel C. 2013. The susceptibility of Pseudomonas aeruginosa strains from cystic fibrosis patients to bacteriophages. PLoS One 8:e60575. 10.1371/journal.pone.0060575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 194:5728–5738. 10.1128/JB.01184-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Edgar R, Qimron U. 2010. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J. Bacteriol. 192:6291–6294. 10.1128/JB.00644-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U. S. A. 108:10098–10103. 10.1073/pnas.1104144108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616–1625. 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U. S. A. 110:12450–12455. 10.1073/pnas.1300833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85:1044–1056. 10.1111/j.1365-2958.2012.08171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. 2009. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72:259–272. 10.1111/j.1365-2958.2009.06641.x [DOI] [PubMed] [Google Scholar]
- 60. Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412. 10.1128/JB.01415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. 10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Delaney N, Balenger S, Bonneaud C, Marx C, Hill G, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. 2012. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 8:e1002511. 10.1371/journal.pgen.1002511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297–7305. 10.1128/JB.00664-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 9:e1003844. 10.1371/journal.pgen.1003844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Goh S, Chang BJ, Riley TV. 2005. Effect of phage infection on toxin production by Clostridium difficile. J. Med. Microbiol. 54:129–135. 10.1099/jmm.0.45821-0 [DOI] [PubMed] [Google Scholar]
- 66. Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. 2009. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J. Virol. 83:12037–12045. 10.1128/JVI.01256-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Meader E, Mayer MJ, Gasson MJ, Steverding D, Carding SR, Narbad A. 2010. Bacteriophage treatment significantly reduces viable Clostridium difficile and prevents toxin production in an in vitro model system. Anaerobe 16:549–554. 10.1016/j.anaerobe.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 68. Meader E, Mayer MJ, Steverding D, Carding SR, Narbad A. 2013. Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe 22:25–30. 10.1016/j.anaerobe.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 69. Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39:W347–W352. 10.1093/nar/gkq1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 72. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100. 10.1093/nar/18.20.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Biswas A, Gagnon JN, Brouns SJJ, Fineran PC, Brown CM. 2013. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 10:817–827. 10.4161/rna.24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 79. Hargreaves KR, Colvin HV, Patel KV, Clokie JJ, Clokie MR. 2013. Genetically diverse Clostridium difficile strains harbouring abundant prophages in an estuarine environment. Appl. Environ. Microbiol. 79:6236–6243. 10.1128/AEM.01849-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR-based detection of CRISPR arrays retained following excision of ppCD105HE1 from the bacterial chromosome. DNA from free viral particles of ppCD105HE1 following spontaneous release was assayed for the presence of the CRISPR arrays identified in the prophage genome using three sets of primers flanking each array in a PCR. Lanes 2 to 4 are no-template DNA control, gDNA of CD105HE1 and ppCD105HE1 DNA with primers targeting array 1, with both CD105HE1 and ppCD105HE1 producing the expected ~499-bp product; lanes 5 to 7, the same order of templates for array 2, also with CD105HE1 and ppCD105HE1 samples showing the same expected 399-bp product; and lanes 8 to 10, showing CD105HE1 and ppCD105HE1 samples with the expected ~1-kbp product for array 3. Download
Conserved motif prediction upstream and downstream of protospacers. Sequences 10 nt in length either upstream (A) or downstream (B) of identical protospacer matches were aligned. In the upstream alignment, two cytosines were identified as highly conserved across 51 upstream regions, and putative PAM sequences of CCA and CCT were predicted. No highly conserved motif was identified in the downstream regions. Only unique, identical matches were included in the WebLogo analysis in an effort to eliminate sequence bias. Download
Pairwise analyses of phages show that the number of shared protospacers positively correlates to the relatedness of phages. A plot is shown of the number of protospacers shared by pairs of phages (see Table S3 in the supplemental material) against a relative value of their relatedness calculated from results of ACT comparison between the whole phage genomes (see Table S4). All combinations of phages were analyzed in a pairwise manner. The number of shared spacers was calculated from matches to the CD630, BI1, M120, and CF5 spacers, as these represented all the unique hits available to avoid bias by including the R20291, CD196, and 2007855 strains, which all have the same spacer hits to phages as BI1, with the exception of those on BI1’s extrachromosomal DNA. Download
Relative proportions of CDSs with protospacers in phage and prophage genomes. Three pie charts show the relative proportions of CDSs in the genome of phiCDHM1 (top) and proportion of CDSs containing protospacer sequence (unique hits, bottom left; total hits, bottom right). The CDSs have assigned groups according to putative gene product functions: clockwise, packaging (including predicted portal and terminase genes), structural (including tape measure protein and tail sheath genes), lysis/attachment (including endolysin and tail fiber genes), lysogeny conversion, lysogeny control (including integrase and repressor genes), replication (including DNA methylase and DNase/helicase genes), unknown (which includes hypothetical genes and non-CDSs and/or significant hit was detected). The relative proportion changes between analyzing unique spacers and the total number of matches, reflecting a difference in results when examining diversity (unique hits) versus frequency (total hits). The analysis included identical and nonidentical matches, and the proportions are relative to the size of the gene of all matches for each analysis. To avoid bias, genomes which had a >95% similarity score using blastn were excluded from the analysis, and one genome was used in each case. Download
ML phylogenetic analysis of the TMP and endolysin genes with spacer matches. Phylogenetic trees for the TMP (A) and endolysin (B) are shown with spacer matches indicated by circles, and where there are shared matches by the same spacer, circles are connected. Download
List of phages and prophages used, with accession numbers and strain details.
CRISPR array flanking oligonucleotide sequences.
Pairwise shared protospacer analysis.
Pairwise whole-genome BLASTN score analysis.
C. difficile spacer sequence with homologous protospacer sequence.