ABSTRACT
Large-scale rearrangements may be important in evolution because they can alter chromosome organization and gene expression in ways not possible through point mutations. In a long-term evolution experiment, twelve Escherichia coli populations have been propagated in a glucose-limited environment for over 25 years. We used whole-genome mapping (optical mapping) combined with genome sequencing and PCR analysis to identify the large-scale chromosomal rearrangements in clones from each population after 40,000 generations. A total of 110 rearrangement events were detected, including 82 deletions, 19 inversions, and 9 duplications, with lineages having between 5 and 20 events. In three populations, successive rearrangements impacted particular regions. In five populations, rearrangements affected over a third of the chromosome. Most rearrangements involved recombination between insertion sequence (IS) elements, illustrating their importance in mediating genome plasticity. Two lines of evidence suggest that at least some of these rearrangements conferred higher fitness. First, parallel changes were observed across the independent populations, with ~65% of the rearrangements affecting the same loci in at least two populations. For example, the ribose-utilization operon and the manB-cpsG region were deleted in 12 and 10 populations, respectively, suggesting positive selection, and this inference was previously confirmed for the former case. Second, optical maps from clones sampled over time from one population showed that most rearrangements occurred early in the experiment, when fitness was increasing most rapidly. However, some rearrangements likely occur at high frequency and may have simply hitchhiked to fixation. In any case, large-scale rearrangements clearly influenced genomic evolution in these populations.
IMPORTANCE
Bacterial chromosomes are dynamic structures shaped by long histories of evolution. Among genomic changes, large-scale DNA rearrangements can have important effects on the presence, order, and expression of genes. Whole-genome sequencing that relies on short DNA reads cannot identify all large-scale rearrangements. Therefore, deciphering changes in the overall organization of genomes requires alternative methods, such as optical mapping. We analyzed the longest-running microbial evolution experiment (more than 25 years of evolution in the laboratory) by optical mapping, genome sequencing, and PCR analyses. We found multiple large genome rearrangements in all 12 independently evolving populations. In most cases, it is unclear whether these changes were beneficial themselves or, alternatively, hitchhiked to fixation with other beneficial mutations. In any case, many genome rearrangements accumulated over decades of evolution, providing these populations with genetic plasticity reminiscent of that observed in some pathogenic bacteria.
INTRODUCTION
Large-scale chromosomal rearrangements have played important roles in long-term organismal evolution (1, 2), including in processes of speciation (3, 4) and genome reduction (5). On shorter time scales, even single rearrangement events such as duplications, amplifications, inversions, deletions, and translocations can have profound effects on organismal phenotypes, typically by altering gene regulation or disrupting genes. In bacteria, some genome rearrangements have led to traits important for virulence (6), and rearrangements are sometimes even developmentally regulated (7).
Large-scale rearrangements have been identified in diverse bacteria—Gram negative, Gram positive, pathogenic, and nonpathogenic—including Escherichia coli (8), Salmonella enterica var. Typhi, Yersinia pestis, Helicobacter pylori, Mycobacterium leprae (9, 10), Pseudomonas stutzeri (11), Pseudomonas aeruginosa (12), Francisella tularensis (13), Neisseria gonorrhoeae (14), Lactococcus lactis (15), and Staphylococcus aureus (16). Besides their effects on chromosome structure, bacterial DNA rearrangements may cause phenotypic changes by the incorporation of foreign DNA into host genomes through horizontal gene transfer (17, 18), by changes in gene expression (19), or by genome reduction through the loss of nonessential genes (20). The major mechanisms producing chromosomal rearrangements are recombinational exchanges between homologous sequences that include ribosomal operons (21) as well as mobile genetic elements such as transposons (16), insertion sequence (IS) elements (15), and prophages (8). Comparisons of related genomes often reveal numerous DNA inversions, among other rearrangements (9, 22, 23).
Some constraints may influence the occurrence of DNA rearrangements. First, selection may preserve symmetry in the size of the two replichores of a circular chromosome between the origin and terminus of replication (9, 24). This selection may help explain the strong conservation of gene order (synteny) between E. coli and Salmonella, although large inversions have been observed under laboratory conditions where such selection may be relaxed. Second, the structural organization of the E. coli chromosome can affect rearrangements (25). Specifically, the genome is organized into distinct macrodomains (26), and rearrangements affecting the replication origin or terminus domain and inversions between the left and right macrodomains have been shown to be detrimental owing to their effects on replication-fork progression (25).
Large-scale rearrangements occur spontaneously at measurable frequencies, although the rates at which they occur are uncertain. In an older study, duplications were reported to arise at frequencies of between 10−2 and 10−5 per cell division, depending on their chromosomal location (21). A more recent whole-genome sequencing study (27) of a population of S. enterica var. Typhimurium that was propagated in a chemostat found duplications, inversions, and small deletions in ≥20% of the cells after only 50 generations. There are several possible explanations for these differences. First, it is generally difficult to disentangle underlying mutation rates from the effects of selection that may cause some mutants to replicate faster or slower than nonmutant cells. Second, the earlier study involved plating to isolate clonal genotypes, which may induce the loss of unstable rearrangements, including duplications, thereby underestimating their true frequency (28). Third, earlier studies were based on observable phenotypes that were produced after the rearrangements occurred and might have missed many other events (29–32).
Chromosomal rearrangements have been discovered in many evolution experiments in which bacterial populations were propagated under various laboratory conditions. Deletions, duplications, and large-scale inversions have been detected in E. coli populations propagated in batch (33–35) and chemostat (28, 36) cultures as well as under stressful conditions (37). Rearrangements have also been found to occur in P. aeruginosa populations evolving in cystic fibrosis patients (38). In some cases, specific rearrangements have been shown to confer increased fitness or phenotypic innovations in these evolution studies (28, 34, 35, 37). To date, however, no study has attempted to provide an exhaustive analysis of the multiple, large-scale chromosomal rearrangements that have arisen during a long evolution experiment.
Rearrangements have usually been investigated using methods such as pulsed-field gel electrophoresis, microarray-based hybridization experiments, and whole-genome sequencing. Despite the name, even whole-genome sequencing typically relies on sequencing short DNA fragment libraries and therefore cannot detect certain large-scale rearrangements, including inversions and other events involving long sequence repeats. To complement sequencing data, whole-genome mapping (referred to here as “optical mapping”) techniques have been developed that produce a high-resolution, physically ordered restriction map of bacterial genomes (39–41). We produced optical chromosomal maps for clones isolated from a long-term evolution experiment (LTEE) with E. coli. In this ongoing experiment, 12 populations have been independently propagated from a common ancestor in the same glucose-limited minimal medium for more than 25 years and 50,000 cell generations. These evolving populations have adapted to the experimental environment and have increased in competitive fitness relative to the ancestor by more than 70%, on average (42).
Some chromosomal rearrangements have been previously detected in the LTEE populations by using other techniques. However, with the exception of deletions involving the ribose operon found in all 12 populations (34), few rearrangements have been analyzed in detail. The ribose deletions were shown to occur at a high rate as well as to confer a slight fitness benefit under the LTEE conditions. In addition, other rearrangements have been detected in three populations (designated Ara+1, Ara–1, and Ara–3). In the Ara+1 population, an analysis of the distribution of IS elements by Southern blotting revealed an inversion of about one-third of the chromosome by recombination between two copies of IS150 (33). A substantial increase in the IS150 copy number also occurred in this population (43). In Ara–1, analysis of the distribution of IS elements and whole-genome sequencing found four large deletions (ranging from ~8 to ~23 kbp) and an inversion of one-third of the chromosome that is different from the inversion in Ara+1 (33, 44). The fitness consequences of these rearrangements are unknown. In Ara–3, numerous deletions, duplications, and amplifications were detected in evolved clones by using genome sequencing data, including a specific tandem duplication and further amplification events involved in the production of a novel Cit-positive (Cit+) phenotype (35).
In an effort to obtain a more complete picture of the number and types of rearrangements that were substituted over time in all 12 LTEE populations, we combined optical mapping, genome sequencing, and targeted PCR and Sanger sequencing to analyze a total of 19 clones, including a single clone sampled at 40,000 generations from each population as well as additional clones sampled at each of 7 other time points from population Ara–1. The resolution of optical maps cannot reliably detect rearrangements smaller than ~5 kbp (including, for example, new insertions of IS elements) unless they alter restriction sites. Nevertheless, we found that all 12 populations experienced large-scale chromosomal rearrangements and that most of these involved IS elements or other repeated sequences. Moreover, we saw many cases of parallel evolution across the populations in the genes that were affected by these rearrangements. Also, three populations had undergone complex rearrangements that involved successive inversion events.
RESULTS
Chromosomal rearrangements in the twelve populations of the long-term evolution experiment.
We combined optical mapping and genome sequence analyses to identify the precise location and borders of all large-scale chromosomal rearrangements that occurred in one clone sampled at 40,000 generations from each of the 12 E. coli populations of the LTEE (Fig. 1; see also Table S1 and Text S1 in the supplemental material). Combining these approaches allowed us to resolve rearrangements between large repeated elements, which is difficult or impossible with genome sequencing data alone, and to map the borders of the rearrangements with single-nucleotide resolution, which is impossible with optical mapping data alone. We also verified the rearrangement borders for two evolved clones from populations Ara+1 and Ara+2, in which we detected more complex rearrangements, using PCR and Sanger sequencing. Primer pairs that were adjacent to repeat sequences, including IS elements and rRNA-encoding genes, were designed for these assays. The results agreed with our predictions in all cases, giving us confidence that our inferences concerning events in other clones are also accurate. Note, however, that optical mapping cannot detect most IS insertion events because most rearrangements smaller than ~5 kbp are too small to resolve unless they affect restriction sites.
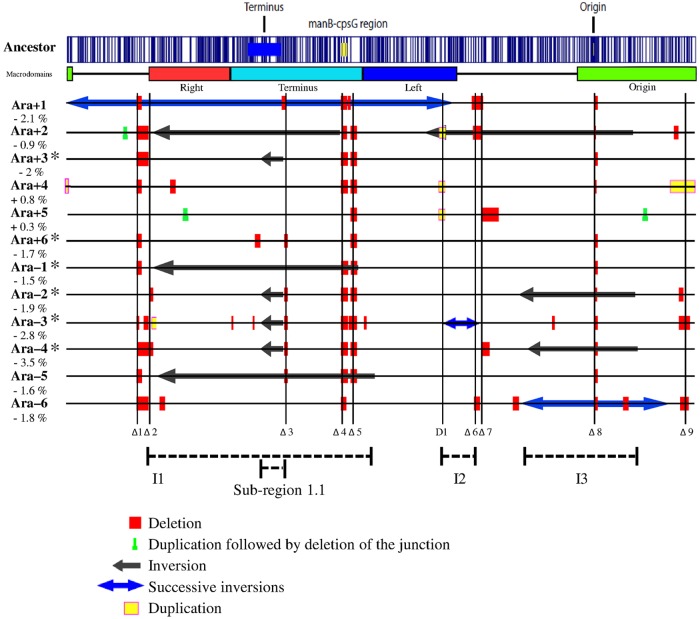
FIG 1 .
Large-scale chromosomal rearrangements in evolved clones sampled after 40,000 generations from each of the 12 populations of the long-term evolution experiment. Each clone is indicated by the name of the population from which it was sampled. Asterisks mark clones that evolved point mutation rates higher than those seen with the ancestor. The percentage value shown below each clone designation indicates the change in its genome size relative to the ancestor. The optical map of the ancestral strain, computed from its genome sequence (70), is shown on the top, with the vertical blue lines showing the locations of the NcoI restriction sites used for this procedure. The locations of the replication origin and terminus are shown on the ancestral map, together with the manB-cpsG region that was affected by deletions in 10 evolved clones. The chromosomal macrodomains (26) are indicated below the ancestral map. All large rearrangements are shown, relative to the ancestral genome for easier comparison, using the color key below the figure. New IS element insertions cannot be detected by optical mapping because they generally produce rearrangements too small to be resolved by this method. Vertical lines labeled Δ1 to Δ9 indicate regions affected repeatedly (in two or more populations) by deletions; the D1 vertical line indicates a region affected repeatedly by duplication events. Three chromosomal intervals, shown as I1 to I3, and a subregion (1.1) within I1 were affected repeatedly by inversions. We describe the boundaries of all chromosomal rearrangements according to the ancestral map shown here. As a consequence, three inversions (inversion 1 in Ara+1 and inversions 1 and 2 in Ara–3) have sizes larger than one-half of the chromosome (see Table S1 in the supplemental material). These inversions could have been described alternatively as inversions of the other part of the chromosome, with their sizes then being smaller than one-half of the chromosome. For example, we describe inversion 1 as being ~2.8 Mbp, whereas its size would be ~1.8 Mbp according to the alternative description. We use the coordinates according to the ancestral map for internal consistency, and this choice does not affect any conclusions.
We identified a total of 110 rearrangement events in the 12 40,000-generation clones, including 82 deletions, 19 inversions, and 9 duplications (Fig. 1; see also Table S1 and Text S1 in the supplemental material). Among the inversions, nine were involved in successive series of events that occurred over time in three populations (see the next section). Among the duplications, three apparently involved successive events in which a typical tandem duplication was followed by deletion of the junction between the duplicated copies, thereby resulting in an imperfect duplication (see the next section).
Large deletions were the most frequent type of rearrangement, and they were found in all 12 populations, ranging in size up to ~55 kbp. Prophage remnants were often affected by these deletions; 30 (36.6%) of the 82 large deletions resulted in the loss of prophage DNA, although these regions cover only ~4% of the ancestral genome. This overrepresentation of prophage DNA is highly significant (binomial test, P = 3 × 10−21). The 19 inversions were found in nine populations, ranging in size from ~164 kbp to ~1.8 Mbp (see Fig. 1 legend for explanation of inversion sizes). In seven cases (one each in populations Ara+1, Ara–1, and Ara–5 and two each in Ara+2 and Ara–3), more than a quarter of the chromosome was included within the inversions (Fig. 1). Successive inversions were inferred in some cases, and they were confirmed by examining multiple clones from different generations (see the next section). Nine duplications, ranging in size from ~3 kbp to ~180 kbp, were found in clones from four populations, including three with further deletions of the copy junctions (see the next section).
Most rearrangements occurred by recombination between repeated sequences, including 76 between homologous IS copies, 7 between the manB and cpsG genes (which share 96% sequence identity), and 5 between rRNA-encoding operons (see Table S1 and Fig. S1 in the supplemental material). The other 22 rearrangements occurred by unknown mechanisms not involving any repeated sequences, and 11 of these changes resulted in the loss of prophage remnants. Thus, IS elements were the main drivers of large-scale chromosomal rearrangements in these populations. In fact, IS-mediated events accounted for at least half of the rearrangements in the 40,000-generation clones from every population, including all of them in Ara+1 (see Fig. S1).
Complex rearrangements.
In several cases, we observed genomic rearrangements that appear to have involved multiple successive events. A total of nine inversion events could have generated the complex rearrangements seen in the 40,000-generation clones from populations Ara+1, Ara–3, and Ara–6. A total of three duplications in the clones from populations Ara+2 and Ara+5 were imperfect, with the junctures between the duplicate copies apparently having been deleted following the duplication events (Fig. 1). To evaluate these hypotheses more thoroughly, we analyzed the genome sequences of the corresponding regions in these clones as well as other clones sampled from earlier generations, with a particular focus on the rearrangements observed in population Ara+1.
Optical mapping suggested that the complex rearrangements in population Ara+1 resulted from four successive inversions (Fig. 2). By using PCR experiments, we analyzed additional clones sampled earlier in this population (Table 1; see also Tables S2 and S3 and Text S1 in the supplemental material). The resulting data support the scenario shown in Fig. 2 that outlines the chronology of the inversions, which we number 1 through 4 in the order of their inferred appearance. In a similar vein, the optical maps of the clones sampled at 40,000 generations from populations Ara–3 and Ara–6 imply two and three successive inversions, respectively (see Fig. S2).
FIG 2 .
Successive inversions in population Ara+1. (A) Optical map of the genome of the evolved clone sampled at 40,000 generations from population Ara+1 compared to the ancestor. Dark blue lines indicate NcoI restriction sites. White boxes show discrepant regions. The variously colored arrows indicate homologous regions of the two genomes with their corresponding locations in the two chromosomes. Red boxes indicate deletions. Black lines connecting the two genomes show alignment. (B) Chronology of the four inversions that occurred over evolutionary time in population Ara+1. Time points (2K, 20K, and 30K for 2,000, 20,000, and 30,000 generations, respectively) indicate the earliest detected occurrence of each inversion. “IND” indicates an inversion that was not detected in any of the evolved clones that were analyzed but which represents one of the two possible intermediate steps leading to the genome observed at 40,000 generations. The variously colored arrows are as described for panel A. Black lines indicate the inversions, with the names and locations of primers used during the PCR experiments also shown.
TABLE 1 .
Evolutionary dynamics of four successive inversions in population Ara+1
| Generation | Clone | Inversiona |
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 0 | REL607 | ||||
| 2,000 | REL1158A | ||||
| REL1158B | |||||
| REL1158C | + | ||||
| 15,000 | REL7183A | + | |||
| REL7183B | + | ||||
| REL7183C | |||||
| 20,000 | REL9282A | + | + | ||
| REL9282B | + | + | |||
| REL9282C | + | + | |||
| 25,000 | REL10241 | + | + | ||
| 30,000 | REL10450 | + | + | + | + |
| REL10451 | + | + | |||
| REL10452 | + | + | |||
| 35,000 | REL10796 | + | Alt | ||
| REL10797 | + | Alt | |||
| REL10798 | + | Alt | |||
| 40,000 | REL11008 | + | + | + | + |
| REL11009 | + | Alt | |||
| REL11010 | + | Alt | |||
Plus signs indicate the presence of a given inversion; “Alt” indicates the presence of an alternative rearrangement instead of inversion 2 (see text).
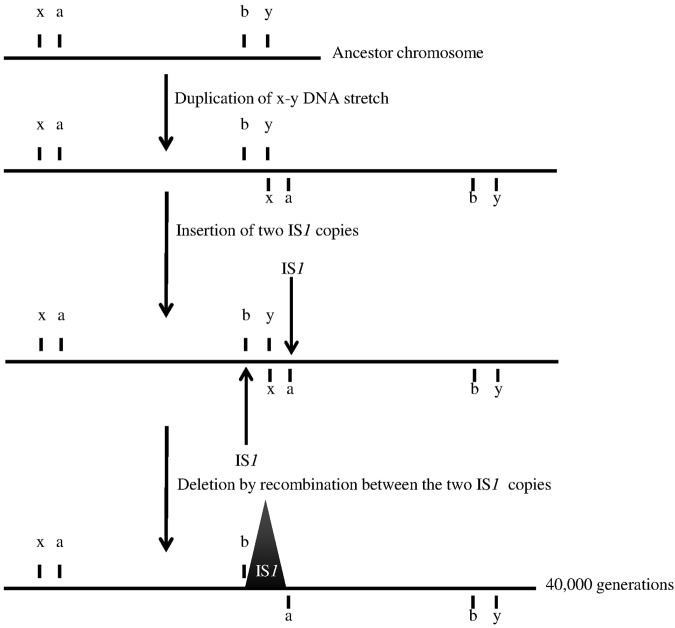
Optical maps suggested the presence of three imperfect duplications in the 40,000-generation samples, one in the clone from population Ara+2 and two in the clone from Ara+5 (Fig. 1), with deletions of the conjoined regions between the duplicated copies. The sequence of that region in the Ara+2 clone showed the absence of the junction sequence between the copies and the presence instead of an IS1 element (see Table S1 in the supplemental material), suggesting the scenario depicted in Fig. 3. Two copies of IS1 likely inserted, in the same orientation, at the end and start of the first and second tandem copies of the duplication, respectively. A subsequent recombination event between these IS1 elements resulted in the deletion of the intervening region, generating the junction seen with the imperfect duplication (Fig. 3; see also Table S1). The two imperfect duplications in population Ara+5 seem to have occurred by the same mechanism, with deletions of the junctions again being associated with a new IS1 element (see Table S1).
FIG 3 .
Hypothetical mechanism for the imperfect duplication seen in the evolved clone from population Ara+2. The horizontal line represents a section of the genome. The letters x and y show the borders of the duplication; a and b are the future insertion sites of two IS1 elements. The two IS1 insertions are indicated by arrows. The IS1-mediated deletion event is indicated by the large cone, with only one IS1 copy remaining in the 40,000-generation clone.
Effects of rearrangements on genome size and structure.
We analyzed both the optical maps and the genome sequences to estimate the genome size of each evolved clone sampled at 40,000 generations. Genome size was reduced in 10 of the 12 clones by amounts ranging from 0.9% to 3.5% of the ancestral genome size (Fig. 1). However, the clones from populations Ara+4 and Ara+5 showed slight increases in genome size of 0.8% and 0.3%, respectively, that resulted from duplications, including one encompassing ~4% of the genome (~180 kbp) in Ara+4. The overall tendency toward reduced genome size reflected the fact that large deletions were much more common than large duplications.
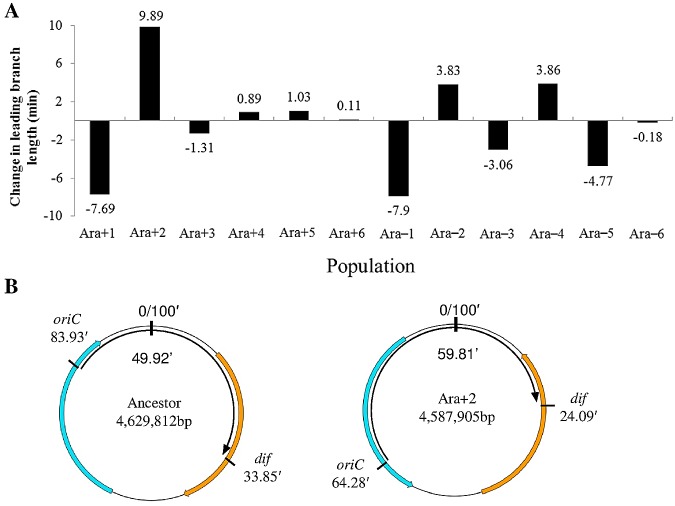
Two types of structural constraints have been hypothesized to influence genome structure. One hypothesizes a requirement for symmetry between the origin and terminus of replication in a circular chromosome (9, 24), and the other is based on the organization of the chromosome into distinct macrodomains (25). Imbalances of less than ~10% in the lengths of the two replichores have been reported to have little or no effect on E. coli growth (25). In contrast, inversions that disrupt the replication terminus macrodomain such that the replication forks meet far from the ancestral replichore junction have negative effects on growth, as do inversions between the replication origin and right macrodomains. The inversion events that we identified in the LTEE populations affected the symmetry of the evolved genomes to various extents (Fig. 4A). Five populations (Ara+3, Ara+4, Ara+5, Ara+6, and Ara–6) showed little change; four populations (Ara–2, Ara–3, Ara–4, and Ara–5) showed moderate changes ranging from ~3 to ~5 min of the leading replication branch; and three populations (Ara+1, Ara+2, and Ara–1) showed larger changes of ~8 to ~10 min. In Ara+2, one inversion affected both the replication terminus and right macrodomains, while another affected the replication origin and left macrodomains (Fig. 4B). Inversions in Ara–1 and Ara–5 also affected the replication terminus and right macrodomains, while inversions in Ara–2 and Ara–4 affected the replication origin and the adjacent nonstructured domains. The four successive inversions in Ara+1 had the most dramatic effects on macrodomain organization, spanning the right, replication terminus, and left macrodomains. All of the LTEE populations became much more fit than their common ancestor based on competition assays (42), and thus none of these inversions had any highly deleterious effects; however, we do not know whether the inversions were beneficial mutations or, alternatively, were selectively neutral or even weakly deleterious mutations that hitchhiked with other beneficial mutations.
FIG 4 .
Changes in genome symmetry. (A) Changes in genome symmetry between the oriC and dif loci along the leading replication branch for the evolved clones sampled after 40,000 generations from each of the 12 populations are shown in minutes. (B) Circular maps of the chromosome show the near-perfect symmetry of the ancestor (left) and the imbalance in the evolved clone from population Ara+2 (right). The curved black arrow inside the circle corresponds to the leading strand, and the colored arrows show the two large inversions. The positions of oriC and dif are shown in minutes outside the circle; the length of the leading strand and the genome size (bp) are shown inside the circles.
Parallel rearrangements across populations.
The optical maps revealed a high level of parallel evolution—that is, similar large-scale rearrangements—across the 12 populations (Fig. 1). We define parallel rearrangements as those involving chromosomal regions that were affected by the same type of rearrangement event (i.e., deletion, inversion, or duplication) in at least two populations. Based on this criterion, nine distinct chromosomal regions were repeatedly affected by deletions (numbered Δ1 to Δ9 from left to right in Fig. 1), three by inversions (called intervals I1 to I3 in Fig. 1), and one by duplications (numbered D1 in Fig. 1).
Chromosomal region Δ8 was deleted in all 12 populations (Fig. 1; see also Tables S1 and S4 in the supplemental material), causing the loss of part or all of the rbs operon, which encodes proteins required for growth on ribose. These deletions have been described previously and have been shown to contribute a small but consistent fitness benefit in the glucose-limited environment of the LTEE (34). Region Δ4, which encompasses the DNA between the manB and cpsG genes, was deleted in the clones from 10 populations (Fig. 1; see also Tables S1 and S4 and Text S1). All of these deletions occurred by recombination between repeated elements. Overall, a common set of 12 genes associated with O antigen biosynthesis was lost in the 10 populations affected by these deletions, and a set of six additional genes associated with colanic acid biosynthesis was eliminated in 8 of the populations.
Four of the other repeatedly deleted chromosomal regions (Δ1, Δ3, Δ5, and Δ7) contained prophage remnants (Fig. 1; see also Tables S1 and S4 in the supplemental material). The DLP12-like locus (region Δ1) and the prophage 2 locus (region Δ5) were each lost in 10 populations, the Qin-like locus (region Δ3) was deleted in 6 populations, and the CP-44-like locus (region Δ7) was lost in 3 populations. Eleven populations lost either two or three of these prophage regions. Most of these deleted genes have unknown functions; it is quite possible that they were not expressed in or useful to the ancestral strain. In population Ara–4, the deletion that included the DLP12-like prophage was larger and overlapped a deletion found in Ara–2 (region Δ2 in Fig. 1; see also Table S4). Region Δ2 was also affected in population Ara+1.
Region Δ2 contained several genes that encode proteins involved with the production and regulation of enterobactin, an iron-scavenging siderophore; it was deleted in populations Ara+1, Ara–2, and Ara–4. Region Δ6, deleted in populations Ara+1, Ara+2, and Ara–6, spans 17 genes, including 10 having unknown functions; the other 7 genes are annotated as phage proteins, which suggests that region Δ6 also corresponds to a phage remnant, although it has not been annotated as such. Region Δ9 was deleted in populations Ara–3 and Ara–6, resulting in the loss of the hsdSM genes that encode a type 1 restriction-modification complex.
For inversions, we view as representing parallel changes the chromosomal intervals containing genes that were inverted in at least two populations, regardless of the overall length of the inversions. Three intervals fulfilled this rule (Fig. 1; see also Tables S1 and S4 in the supplemental material): I1 in populations Ara+1, Ara+2, Ara–1, and Ara–5; I2 in Ara+2 and Ara–3; and I3 in Ara+2, Ara–2, Ara–4, and Ara–6. Within the I1 interval, a subregion denoted 1.1 was affected by smaller inversions in populations Ara+3, Ara–2, Ara–3, and Ara–4; this subregion was thus inverted in 8 of the 12 populations. All of these inversions contained many genes and were mediated by recombination between repeated elements (see Text S1 in the supplemental material).
A single chromosomal region, D1, underwent parallel duplications in three populations, Ara+2, Ara+4, and Ara+5 (Fig. 1, see also Tables S1 and S4 in the supplemental material). These three duplications ranged in size from ~11 to ~60 kbp, but they shared ~11 kbp and 14 genes (see Tables S1 and S4). The rpoS gene that encodes an alternative sigma factor is present in all three duplications, as are pcm and surE, both essential for survival in stationary phase, and the cysDNC operon involved in sulfur metabolism.
Temporal dynamics of rearrangements in population Ara–1.
We analyzed the optical maps and genome sequences of eight clones sampled from population Ara–1 at 2,000, 5,000, 10,000, 15,000, 20,000, 30,000, 40,000, and 50,000 generations. We detected 6 deletion events ranging from ~7 to ~23 kbp in size (Table 2), three of which went to fixation, including one that arose before 2,000 generations and two that occurred between generations 5,000 and 10,000. The ~1.5-Mbp inversion that we detected in the 40,000-generation clone (Fig. 1, see also Table S1 in the supplemental material) occurred between generations 5,000 and 10,000 and was then fixed in the population. Two translocation events were present in the 5,000-generation clone but were not seen in any of the later samples. Of the five rearrangements detected at 50,000 generations, four were already present at 10,000 generations (Table 2). Assuming a uniform rate of 5/50,000, one is unlikely to observe 4 or more mutations by generation 10,000 (one-tailed Poisson test; P = 0.019), suggesting heterogeneity in the evolution of these large-scale rearrangements over time. This heterogeneity might reflect a change in the underlying mutational processes that generate these rearrangements. Alternatively, the rate of fitness improvement and the corresponding rate at which beneficial mutations went to fixation were much higher early in the experiment than later on (42, 44), and this difference may explain the greater number of rearrangements fixed in the early generations.
TABLE 2 .
Rearrangements detected in clones sampled over time from population Ara–1
| Mutation | Start positiona | End positiona | Size (bp) | Mechanismb | Observed at generation(s): |
|---|---|---|---|---|---|
| Deletion | 547,701 | 555,877 | 8176 | RRE–IS1 | 10K, 15K, 20K, 30K, 40K, 50K |
| Deletion | 1,599,000 | 1,628,000 | 8600 | Unknown | 50K |
| Deletion | 2,031,703 | 2,054,996 | 23,293 | RRE–manB-cpsG | 10K, 15K, 20K, 30K, 40K, 50K |
| Deletion | 2,100,286 | 2,122,432 | 22,146 | Unknown | 40K |
| Deletion | 2,129,369 | 2,137,411 | 8042 | RRE–IS1 | 40K |
| Deletion | 3,894,996 | 3,901,921 | 6927 | RRE–IS150 | 2K, 5K, 10K, 15K, 20K, 30K, 40K, 50K |
| Inversion | 634,745 | 2,128,599 | 1,493,854 | RRE–IS1 | 10K, 15K, 20K, 30K, 40K, 50K |
| Translocation | 430,000 | 590,715 | 160,715 | RRE–IS186 | 5K |
| Translocation | 1,721,000 | 1,849,000 | 128,000 | Unknown | 5K |
All positions are shown according to the genomic coordinates of the ancestral strain (70).
RRE, recombination between repeated elements, with the identity of the repeated element indicated after the hyphen.
DISCUSSION
We combined optical mapping and genome sequencing to identify chromosomal rearrangements that occurred in each of 12 populations during 40,000 generations of experimental evolution. We detected a total of 110 rearrangements, of which 75% were deletions, 17% were inversions, and 8% were duplications. However, the resolution of the optical mapping did not allow the detection of many small deletions and insertions, including new IS element insertion events. Some of the complex rearrangements were shown to involve a succession of events, including multiple inversions as well as duplications followed by deletions overlapping the junction of the two copies. Most (~70%) rearrangements occurred by recombination between IS elements, and many chromosomal regions were repeatedly affected by similar rearrangements in two or more populations. In most populations, the overall chromosomal organization was maintained without a large imbalance of the symmetry between the origin and terminus of replication or any major disruption of the chromosomal macrodomains. However, three populations evolved rather substantial asymmetry between the replication origin and terminus.
The dynamics of the rearrangements over time were examined in one population, and they showed that most of the rearrangements were substituted in the early generations of the experiment, when the rate of fitness increase was highest, suggesting that these rearrangements may have contributed to the genetic adaptation of these populations. However, it is also possible that many of these rearrangements are nonadaptive events that occur at high rates (relative to point mutations) and then spread through a population by hitchhiking with beneficial mutations. Chromosomal rearrangements have been detected at high frequencies when bacterial cells are exposed to stresses such as starvation (45). In the LTEE, the bacteria deplete the limiting nutrients each day and enter stationary phase, but in general they do not experience any discernible mortality (46), and the nutrients are replenished when the bacteria are transferred into fresh medium. Therefore, the fact that many chromosomal rearrangements have occurred during the LTEE suggests that prolonged starvation is not necessary for substantial genome restructuring to occur. Of course, it is also important to emphasize that the large rearrangements detected in this study, which averaged ~9 per population, occurred over many years.
The 12 experimental populations have been evolving in and adapting to the same environment for tens of thousands of generations. Therefore, one might expect them to lose unused functions and evolve smaller genomes, as observed for bacteria adapting to stable environments, such as endosymbionts adapting to their hosts (47, 48). As predicted, deletions were indeed the predominant rearrangements detected in the LTEE. We also observed a high level of parallelism, with nine chromosomal regions deleted in at least two populations (Fig. 1; see also Table S4 in the supplemental material). These parallel deletions removed genes from the rbs operon, genes involved in O antigen and colanic acid biosynthesis, and prophage-related genes. In a previous study (34), we demonstrated that rbs deletions occurred at a very high frequency owing to the presence of an IS150 element adjacent to the operon and that they conferred a small but significant fitness increase. More generally, the deleted genes have functions that are not used under the conditions prevailing during the LTEE. These deletions might have conferred higher fitness by eliminating unnecessary and costly gene expression (49), or they might have been effectively neutral if the affected genes were already not expressed. The involvement of IS elements in producing many of these deletions probably reflects increased local mutagenesis caused by homologous recombination between two identical elements, similar to the process demonstrated for the rbs operon (34). Deletions have also been reported in evolution experiments with other bacteria and environments (27, 38, 50).
In the LTEE, the size of the E. coli genome declined after 40,000 generations in 10 of the 12 populations, with the reductions ranging from 0.9% to 3.5% relative to the ancestor; two populations showed slight increases in genome size of 0.3% and 0.8%. Over all 12 populations, we recorded 70 reductive events with sizes of 1 kbp or larger, with an average of ~17 kbp and a median of ~11 kbp. These values are far below the reductions inferred for pathogenic bacteria, including Mycobacterium leprae, Yersinia pestis, and Mycoplasma ulcerans (51); of course, the time periods over which these pathogens evolved their reduced genomes were much longer. On a time scale more commensurate with the LTEE, a study of P. aeruginosa adapting to the lungs of human patients with cystic fibrosis found that up to 8% of the ancestral genome was lost over the course of 35 years (38); in that study, the average and median deletion sizes were 44.5 kbp and 26.6 kbp, respectively, for 27 deletion events with sizes of at least 1 kbp. Most deletions in the P. aeruginosa study occurred through illegitimate and homologous recombination events, but IS elements were not involved. In contrast with that dramatic reduction, an analysis of 11 natural isolates of E. coli O157:H7 found only very limited genome reductions of up to 3.7 kbp, or ~0.1%, and slight increases in chromosome size were detected in several of those isolates (39). Note, however, that these estimates exclude the effect of new insertions of IS elements, which are not detected by optical mapping.
Three evolutionary factors have often been suggested as drivers of reduced genome sizes in pathogenic and endosymbiotic bacteria: severe population bottlenecks, the absence of horizontal gene transfer, and the elimination of selection for various functions owing to the availability of nutrients and other services provided by the host (5, 47, 48). Bottlenecks are not severe in the LTEE, with more than 106 cells transferred each day to fresh medium. There is no horizontal transfer in the LTEE, as plasmids and functional phages are absent and E. coli does not undergo natural transformation. The nutritional environment of the LTEE consists of a minimal medium with glucose and ammonium, providing carbon and nitrogen, respectively. Owing to this simple environment, certain functions cannot be lost, including, for example, the production of amino acids. However, some functions are dispensable, including those involved with using alternative resources (e.g., the loss of the ability to grow on ribose) and those necessary for thriving in natural environments (e.g., loss of genes involved with O antigen and colanic acid biosynthesis). Thus, the simple flask environment—like a host organism—provides environmental constancy and protection that allow certain functions to be discarded. In doing so, the cells may save energy, thereby providing a competitive advantage; even without that benefit, any unused functions will tend to decay or be deleted by ongoing mutations (20, 52, 53). Another factor that contributed to genome reductions in the LTEE is homologous recombination, especially that mediated by IS elements. The majority of rearrangements detected in this study involved IS elements, and these elements often flank noncore genes that were acquired by horizontal gene transfer in the distant past, i.e., prior to E. coli B being brought into the laboratory (54). These horizontally acquired genes would thus be both dispensable and prone to deletion.
Besides deletions, we detected two other types of large-scale rearrangements, namely, duplications and inversions. As for deletions, some of these duplications and inversions affected the same chromosomal regions in multiple populations. Parallel evolution is often interpreted as indirect evidence of positive selection (34, 44, 55, 56), and from that perspective these mutations might therefore be beneficial in the context of the LTEE. However, while this interpretation has been frequently confirmed for point mutations by constructing and competing isogenic strains, it is more difficult to support this inference for large rearrangements because they affect many genes. Instead, the parallel evolution of these large rearrangements might reflect their high frequency of occurrence, especially since most of these rearrangements involved recombination events between repeated sequences, including IS elements and rRNA operons.
Duplications were rare, with only nine events detected among the 12 populations after 40,000 generations. The paucity of duplications compared to deletions probably reflects the intrinsic instability of duplications, which readily collapse back to a single copy when cells are propagated under conditions that do not favor having multiple copies of the relevant genes (28). The nine duplications we detected range in size from ~3 to ~180 kbp. Owing to the number and diversity of the genes found in the duplicated regions, it is difficult to know whether and how the duplications affected the fitness of the bacteria. However, one region was duplicated in three populations, and it spans an ~11-kbp region containing 14 genes, including rpoS, which encodes the alternative sigma factor involved in the transition into stationary phase (57). Previous work showed that the LTEE populations underwent changes in the regulatory networks involved in the transitions between exponential and stationary phases, which they experience on a daily basis (58, 59). The parallel duplications might affect the expression of rpoS and confer a competitive advantage during these transitions, although this possibility remains untested.
We detected a total of 19 inversions, including 7 that affected more than a quarter of the chromosome, and several populations underwent multiple successive inversions. Three chromosomal regions were inverted in multiple populations, whereas only one region (spanning genome positions 4,453,625 to 146,102) was not affected by any of the inversions. Owing to the large number of genes in these inversions, it is difficult to predict their effects, if any, on the fitness or other phenotypes of the evolved cells. Chromosomal inversions have been found in natural isolates of many bacterial species, including E. coli (39), Staphylococcus aureus (16, 40), Enterococcus faecium (60), Francisella tularensis (13), and Bacillus anthracis (61). Some of these inversions have been related to phenotypic changes, including colony morphology (16) and virulence (61), but in most cases their effects are unknown. Experiments have shown that some inversions adversely affect cell growth because they substantially reduce the symmetry between the origin and terminus of replication (24) or because they disrupt the overall organization of the chromosome into macrodomains, including especially the structure of the terminus (25). The inversions and other rearrangements observed in the evolved genomes of the LTEE had variable effects on the symmetry between the origin and terminus of replication: five populations showed almost no change in symmetry, four had increased asymmetry leading to imbalances of the replication arms of a few percent, and three evolved imbalances of ~8% to ~10% (following the numerical scheme for calculating imbalance used in reference 25). None of the evolved clones we studied, however, had imbalances as great as the 15% imbalances previously shown to impair cell growth (25). In any case, the variations in chromosomal organization produced by the rearrangements we detected are clearly well tolerated under the conditions of the LTEE.
The strong conservation of gene order between E. coli and Salmonella chromosomes, which have diverged for over 100 million years (62), led to the suggestion that selection constrains gene order (29). In that respect, the extent of inversions in the LTEE over just 2 decades is surprising. One possible explanation for this discrepancy is that horizontal gene transfer—in particular, its importance for adaptation to changing environments—generates the constraint. In the LTEE, there is no gene transfer and the experimental environment does not change, relieving the constraint and thus allowing gene order to vary more freely. It is also possible that selection constraints that are important over very long time spans and in very large populations are less important at the smaller scale of the LTEE. Also, the LTEE environment is obviously very different from natural conditions; an alternative hypothesis, therefore, is that these inversions increase fitness in the LTEE environment, perhaps by changing the distribution of genes on the leading and lagging strands. During replication, the DNA and RNA polymerase complexes move along the same DNA molecule, and their physical interactions depend on gene orientation (63). Collisions between DNA polymerase and RNA polymerase transcribing genes from the lagging strand occur with a higher probability, causing the replication machinery to stall and potentially also generating truncated transcripts. In contrast, when genes are located on and transcribed from the leading strand, such collisions merely slow down the replication complex and the transcript is released after completion. Thus, essential (as well as highly expressed) genes tend to be located on the leading strand (64). We analyzed essential genes (65) present in the ancestral genome to see whether their proportion on the leading strand changed as a result of inversions in the 40,000-generation clones (see Text S1 in the supplemental material). The results do not support the hypothesis that the inversions improved fitness by reducing collisions between the DNA and RNA polymerase complexes.
About 70% of the large-scale rearrangements we detected in the evolved clones occurred by homologous recombination between IS elements, and that proportion does not include new insertions of IS elements (because optical mapping cannot resolve such events). These elements have previously been shown to contribute to evolution in the LTEE in three ways. First, IS elements have generated some beneficial mutations, including the deletions of the rbs operon (34). Second, some new IS insertions occurred in genes that were mutated in many or all of the LTEE populations (33, 56), and that genetic parallelism strongly suggests that these insertions were also beneficial. Third, population Ara+1 has undergone a striking increase in the IS150 copy number (43), including some insertions inferred to be beneficial based on the previous criterion. Here, we have further shown that IS elements, by providing a substrate for homologous recombination, played the predominant role in large-scale rearrangements that restructured the genomes during this long-term experiment. IS elements have also been shown to contribute to genomic plasticity in other studies, including both evolution experiments in the laboratory (36, 37, 66) and analyses of natural isolates (13, 67, 68).
In summary, we used optical mapping to find large-scale chromosomal rearrangements that occurred during a long-term evolution experiment with E. coli. The rearrangements thus discovered had substantial effects on the size and structure of the chromosome, demonstrating the impressive plasticity of bacterial genomes. Several lines of evidence, including parallel changes observed in independently evolving populations, suggest that at least some of the rearrangements conferred higher fitness in the experimental environment. However, we cannot exclude the alternative explanation that these rearrangements occurred at high rates and then hitchhiked to fixation. Consistent with the latter possibility, IS elements mediated most of the large-scale rearrangements by providing a substrate for recombination. While new sequencing technologies make it increasingly easy to find point mutations and other small changes in the genomes of experimentally evolving populations, our results demonstrate the value of also analyzing large-scale chromosomal rearrangements in these studies.
MATERIALS AND METHODS
Bacterial strains.
All strains came from the E. coli long-term evolution experiment (42, 69). Twelve populations, named Ara+1 to Ara+6 and Ara–1 to Ara–6, were founded from the same ancestral strains, REL606 and REL607 (a spontaneous Ara+ mutant of REL606). The populations have been propagated by daily transfers in Davis minimal medium containing 25 µg/ml glucose (DM25) as a limiting carbon source (69). Samples from each population have been taken at 500-generation intervals and stored at −80° C. For optical mapping, we used one clone isolated from each population at 40,000 generations, as well as one clone from population Ara–1 at each of 2,000, 5,000, 10,000, 15,000, 20,000, and 50,000 generations. Additional clones were sampled at several time points from Ara+1 to investigate specific rearrangements. All strains used in this study are listed in Table S2 in the supplemental material.
Optical mapping.
The optical-mapping procedure was performed by OpGen (Gaithersburg, MD), as described elsewhere (40). Clones were revived from stocks kept at −80°C in 15% glycerol by overnight growth in LB medium. Genomic DNA was extracted using an OpGen sample preparation kit (OpGen, Inc., Gaithersburg, MD) and an Agencourt Genfind v2 kit (Beckman Coulter, Miami, FL). Single DNA molecules were captured on an Argus surface within a MapCard, digested with the NcoI restriction enzyme, and stained with JOJO-1 on an Argus MapCard Processor. They were analyzed by automated fluorescence microscopy using an Argus Optical Mapper. This software records the size and order of restriction fragments for each DNA molecule. Collections of single-molecule restriction maps for each genome were assembled according to overlapping fragment patterns to produce a whole-genome optical-map assembly. The consensus optical-map assemblies for each evolved clone were then compared to the predicted restriction map of the ancestral strain’s genome (70) to identify large-scale rearrangements using the MapSolver software. Rearrangements smaller than ~5 kbp are too small to resolve. Thus, IS insertion events were not detected in this study.
Characterization of rearrangement borders.
The precise locations of the rearrangement borders were identified (see Table S1 in the supplemental material) by analyzing the genome sequences of the evolved clones. The genomes of population Ara–1 clones from generations 2,000 to 40,000 were previously sequenced (44, 71), as were those of the 40,000-generation clones from populations Ara–3, Ara–5, Ara–6, Ara+1, Ara+2, Ara+4, and Ara+5 (72). The additional genomes analyzed in this study were the 40,000-generation clones from populations Ara–2, Ara–4, Ara+3, and Ara+6, and they were sequenced on the Illumina Genome Analyzer platform at the Centre National de Séquençage, Genoscope (Évry, France), with one lane of single-end 36-bp reads per genome. Sequence reads were compared to the genome of the REL606 ancestral strain (70), using both breseq, a computational pipeline for analyzing resequenced bacterial genomes (35, 44, 71), and a customized pipeline (http://www.genoscope.cns.fr/agc/microscope/expdata/sniperRes.php [73]). The four new genome sequences have been deposited in the National Center for Biotechnology Information Sequence Read Archive (see below). The borders of the rearrangements detected by optical mapping were further checked by PCR experiments for two populations, Ara+1 and Ara+2 (see Table S1). PCR was performed using 1× reaction buffer, 3 mM MgCl2, 0.2 mM deoxyribonucleotide triphosphate (dNTP), 0.2 mM (each) primer, 50 ng of genomic DNA, and 1.25 unit of Taq DNA polymerase (Invitrogen, Carlsbad, CA) in a 25-µl reaction volume. Reaction mixtures were heated at 95°C for 2 min and then subjected to 32 cycles of 30 s at 94°C, 90 s at 55°C, and 3 min at 72°C before a final step of 10 min at 72°C. Table S3 lists the primers used to amplify the borders of specific rearrangements. The PCR products were separated by agarose (0.8%) gel electrophoresis in 1× Tris-acetate-EDTA (TAE) buffer, purified using a Qiagen gel purification kit, and sequenced (GATC-Biotech, Germany) with the same primers used for PCR assays.
Sequence analysis.
Sequences of PCR products containing the borders of rearrangements were analyzed with BioEdit (version 7.0.9.0), and the resulting FASTA files were analyzed using CLC Sequence Viewer software (v 7.0.2; CLC Bio). All sequences were compared to the ancestral genome sequence (70) and checked to confirm the rearrangements deduced from the optical maps. A Python script was written to construct FASTA files containing the reconstructed genome sequences of the evolved clones; we have deposited the script at the Dryad Digital Repository (doi:10.5061/dryad.283pp). Point mutations and deletions detected in the evolved clones were automatically inserted; other rearrangements were added by hand in CLC Sequence Viewer. The leading-strand branch length was calculated from the origin of replication oriC (74) to the middle of the terminus region defined by the dif locus (75). For the evolved clones, the branch-length measurements were based on the rearranged genomes using the new locations of these two loci.
Nucleotide sequence accession number.
The four new genome sequences have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP045228).
SUPPLEMENTAL MATERIAL
Supplemental Results with respect to chromosomal rearrangements in the 12 populations of the LTEE, complex rearrangements, and parallel rearrangements; Supplemental Discussion on distribution of essential genes on the leading strand. Download
Number of rearrangements in clones sampled from each of the 12 populations at 40,000 generations. Colors indicate the different types of rearrangements according to the genetic mechanism of production (see text). Download
Successive inversions leading to genomes observed in populations Ara–3 and Ara–6 at 40,000 generations. Optical maps and hypothetical chronologies of the two and three successive inversions found in populations Ara–3 (A) and Ara–6 (B), respectively, compared to the ancestor genome are shown. Symbols are identical to those described in the Fig. 2 legend. Download
Rearrangements detected at 40,000 generations in clones sampled from each of the 12 populations of the LTEE.
Clones from the LTEE used in this study.
Primers used in this study.
Parallel rearrangements in the LTEE.
ACKNOWLEDGMENTS
This work was supported by European Union program FP7-ICT-2013-10 project EvoEvo grant 610427 to D.S., the Université Grenoble Alpes (D.S.), Centre National de la Recherche Scientifique (D.S.), U.S. National Institutes of Health grant R00-GM087550 to J.E.B., U.S. National Science Foundation grant DEB-1019989 to R.E.L., and U.S. National Science Foundation support (DBI-0939454) for the BEACON Center for the Study of Evolution in Action. C.R. was supported by a fellowship from the French Ministry of Education and Research. D.E.D. was supported by a traineeship from the Cancer Prevention Research Institute of Texas.
Footnotes
Citation Raeside C, Gaffé J, Deatherage DE, Tenaillon O, Briska AM, Ptashkin RN, Cruveiller S, Médigue C, Lenski RE, Barrick JE, Schneider D. 2014. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. mBio 5(5):e01377-14. doi:10.1128/mBio.01377-14.
REFERENCES
- 1. Ohno S. 1970. Evolution by gene duplication. Springer Verlag, New York, NY. [Google Scholar]
- 2. Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. 10.1038/nature06967 [DOI] [PubMed] [Google Scholar]
- 3. Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. 2003. Engineering evolution to study speciation in yeasts. Nature 422:68–72. 10.1038/nature01418 [DOI] [PubMed] [Google Scholar]
- 4. Navarro A, Barton NH. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57:447–459 http://dx.doi.org.gate1.inist.fr/10.1554/0014-3820(2003)057[0447:APIGIP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 5. Sloan DB, Moran NA. 2013. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol. Evol. 5:783–793. 10.1093/gbe/evt044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morrow CA, Lee IR, Chow EWL, Ormerod KL, Goldinger A, Byrnes EJ, III, Nielsen K, Heitman J, Schirra HJ, Fraser JA. 2012. A unique chromosomal rearrangement in the Cryptococcus neoformans var. grubii type strain enhances key phenotypes associated with virulence. mBio 3:e00310-11. 10.1128/mBio.00310-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abe K, Yoshinari A, Aoyagi T, Hirota Y, Iwamoto K, Sato T. 2013. Regulated DNA rearrangement during sporulation in Bacillus weihenstephanensis KBAB4. Mol. Microbiol. 90:415–427. 10.1111/mmi.12375 [DOI] [PubMed] [Google Scholar]
- 8. Iguchi A, Iyoda S, Terajima J, Watanabe H, Osawa R. 2006. Spontaneous recombination between homologous prophage regions causes large-scale inversions within the Escherichia coli O157:H7 chromosome. Gene 372:199–207. 10.1016/j.gene.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 9. Eisen JA, Heidelberg JF, White O, Salzberg SL. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:RESEARCH0011. 10.1186/gb-2000-1-6-research0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darling AE, Miklós I, Ragan MA. 2008. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 4:e1000128. 10.1371/journal.pgen.1000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ginard M, Lalucat J, Tümmler B, Römling U. 1997. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int. J. Syst. Bacteriol. 47:132–143. 10.1099/00207713-47-1-132 [DOI] [PubMed] [Google Scholar]
- 12. Kresse AU, Dinesh SD, Larbig K, Römling U. 2003. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 47:145–158. 10.1046/j.1365-2958.2003.03261.x [DOI] [PubMed] [Google Scholar]
- 13. Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, Gallagher LA, Manoil C, Ernst RK, Drees B, Buckley D, Haugen E, Bovee D, Zhou Y, Chang J, Levy R, Lim R, Gillett W, Guenthener D, Kang A, Shaffer SA, Taylor G, Chen J, Gallis B, D’Argenio DA, Forsman M, Olson MV, Goodlett DR, Kaul R, Miller SI, Brittnacher MJ. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. 10.1186/gb-2007-8-6-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spencer-Smith R, Varkey EM, Fielder MD, Snyder LA. 2012. Sequence features contributing to chromosomal rearrangements in Neisseria gonorrhoeae. PLoS One 7:e46023. 10.1371/journal.pone.0046023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daveran-Mingot ML, Campo N, Ritzenthaler P, Le Bourgeois P. 1998. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J. Bacteriol. 180:4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui L, Neoh HM, Iwamoto A, Hiramatsu K. 2012. Coordinated phenotype switching with large-scale chromosome flip-flop inversion observed in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:1647–1656. 10.1073/pnas.1204307109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skippington E, Ragan MA. 2011. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 35:707–735. 10.1111/j.1574-6976.2010.00261.x [DOI] [PubMed] [Google Scholar]
- 18. Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Médigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M. 2010. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. 10.1371/journal.pone.0010433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR. 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. U. S. A. 103:17319–17324. 10.1073/pnas.0608309103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee MC, Marx CJ. 2012. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 8:e1002651. 10.1371/journal.pgen.1002651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson P, Roth J. 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl. Acad. Sci. U. S. A. 78:3113–3117. 10.1073/pnas.78.5.3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tillier ER, Collins RA. 2000. Genome rearrangement by replication-directed translocation. Nat. Genet. 26:195–197. 10.1038/79918 [DOI] [PubMed] [Google Scholar]
- 23. Zivanovic Y, Lopez P, Philippe H, Forterre P. 2002. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 30:1902–1910. 10.1093/nar/30.9.1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roth JR, Benson N, Galitski T, Haack K, Lawrence JG, Miesel L. 1996. Rearrangements of the bacterial chromosome: formation and applications, p 2256–2276 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 25. Esnault E, Valens M, Espéli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 3:e226. 10.1371/journal.pgen.0030226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boccard F, Esnault E, Valens M. 2005. Spatial arrangement and macrodomain organization of bacterial chromosomes. Mol. Microbiol. 57:9–16. 10.1111/j.1365-2958.2005.04651.x [DOI] [PubMed] [Google Scholar]
- 27. Sun S, Ke R, Hughes D, Nilsson M, Andersson DI. 2012. Genome-wide detection of spontaneous chromosomal rearrangements in bacteria. PLoS One 7:e42639. 10.1371/journal.pone.0042639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maharjan RP, Gaffé J, Plucain J, Schliep M, Wang L, Feng L, Tenaillon O, Ferenci T, Schneider D. 2013. A case of adaptation through a mutation in a tandem duplication during experimental evolution in Escherichia coli. BMC Genomics 14:441–452. 10.1186/1471-2164-14-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Segall A, Mahan MJ, Roth JR. 1988. Rearrangement of the bacterial chromosome: forbidden inversions. Science 241:1314–1318. 10.1126/science.3045970 [DOI] [PubMed] [Google Scholar]
- 30. Zieg J, Kushner SR. 1977. Analysis of genetic recombination between two partially deleted lactose operons of Escherichia coli K-12. J. Bacteriol. 131:123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bierne H, Seigneur M, Ehrlich SD, Michel B. 1997. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol. 26:557–567 [DOI] [PubMed] [Google Scholar]
- 32. Albertini AM, Hofer M, Calos MP, Miller JH. 1982. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell 29:319–328. 10.1016/0092-8674(82)90148-9 [DOI] [PubMed] [Google Scholar]
- 33. Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence mediated mutations and rearrangements. Genetics 156:477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cooper VS, Schneider D, Blot M, Lenski RE. 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183:2834–2841. 10.1128/JB.183.9.2834-2841.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blount ZD, Barrick JE, Davidson CJ, Lenski RE. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489:513–518. 10.1038/nature11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaffé J, McKenzie C, Maharjan RP, Coursange E, Ferenci T, Schneider D. 2011. Insertion sequence-driven evolution of Escherichia coli in chemostats. J. Mol. Evol. 72:398–412. 10.1007/s00239-011-9439-2 [DOI] [PubMed] [Google Scholar]
- 37. Riehle MM, Bennett AF, Long AD. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 98:525–530. 10.1073/pnas.98.2.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rau MH, Marvig RL, Ehrlich GD, Molin S, Jelsbak L. 2012. Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ. Microbiol. 14:2200–2211. 10.1111/j.1462-2920.2012.02795.x [DOI] [PubMed] [Google Scholar]
- 39. Kotewicz ML, Jackson SA, LeClerc JE, Cebula TA. 2007. Optical maps distinguish individual strains of Escherichia coli O157 H7. Microbiology 153:1720–1733. 10.1099/mic.0.2006/004507-0 [DOI] [PubMed] [Google Scholar]
- 40. Shukla SK, Kislow J, Briska A, Henkhaus J, Dykes C. 2009. Optical mapping reveals a large genetic inversion between two methicillin-resistant Staphylococcus aureus strains. J. Bacteriol. 191:5717–5723. 10.1128/JB.00325-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turner PC, Yomano LP, Jarboe LR, York SW, Baggett CL, Moritz BE, Zentz EB, Shanmugam KT, Ingram LO. 2012. Optical mapping and sequencing of the Escherichia coli KO11 genome reveal extensive chromosomal rearrangements, and multiple tandem copies of the Zymomonas mobilis pdc and adhB genes. J. Ind. Microbiol. Biotechnol. 39:629–639. 10.1007/s10295-011-1052-2 [DOI] [PubMed] [Google Scholar]
- 42. Wiser MJ, Ribeck N, Lenski RE. 2013. Long-term dynamics of adaptation in asexual populations. Science 342:1364–1367. 10.1126/science.1243357 [DOI] [PubMed] [Google Scholar]
- 43. Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. U. S. A. 96:3807–3812. 10.1073/pnas.96.7.3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247. 10.1038/nature08480 [DOI] [PubMed] [Google Scholar]
- 45. Lin D, Gibson IB, Moore JM, Thornton PC, Leal SM, Hastings PJ. 2011. Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells. PLoS Genet. 7:e1002223. 10.1371/journal.pgen.1002223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vasi F, Travisano M, Lenski RE. 1994. Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. Am. Nat. 144:432–456. 10.1086/285685 [DOI] [Google Scholar]
- 47. Ochman H, Moran NA. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096–1099. 10.1126/science.1058543 [DOI] [PubMed] [Google Scholar]
- 48. Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, Tichit M, Porter JL, Ryan J, Johnson PD, Davies JK, Jenkin GA, Small PL, Jones LM, Tekaia F, Laval F, Daffé M, Parkhill J, Cole ST. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192–200. 10.1101/gr.5942807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dekel E, Alon U. 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436:588–592. 10.1038/nature03842 [DOI] [PubMed] [Google Scholar]
- 50. Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JC, Andersson DI. 2005. Bacterial genome size reduction by experimental evolution. Proc. Natl. Acad. Sci. U. S. A. 102:12112–12116. 10.1073/pnas.0503654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O’Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32–40. 10.1038/ng1227 [DOI] [PubMed] [Google Scholar]
- 52. Cooper VS, Lenski RE. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407:736–739. 10.1038/35037572 [DOI] [PubMed] [Google Scholar]
- 53. Leiby N, Marx CJ. 2014. Metabolic erosion primarily through mutation accumulation, and not tradeoffs, drives limited evolution of substrate specificity in Escherichia coli. PLoS Biol. 12:e1001789. 10.1371/journal.pbio.1001789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Studier FW, Daegelen P, Lenski RE, Maslov S, Kim JF. 2009. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21(DE3) and comparison of the E. coli B and K-12 genomes. J. Mol. Biol. 394:653–680. 10.1016/j.jmb.2009.09.021 [DOI] [PubMed] [Google Scholar]
- 55. Crozat E, Philippe N, Lenski RE, Geiselmann J, Schneider D. 2005. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics 169:523–532. 10.1534/genetics.104.035717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:9107–9112. 10.1073/pnas.0602917103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hengge-Aronis R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165–168. 10.1016/0092-8674(93)90655-A [DOI] [PubMed] [Google Scholar]
- 58. Philippe N, Crozat E, Lenski RE, Schneider D. 2007. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays 29:846–860. 10.1002/bies.20629 [DOI] [PubMed] [Google Scholar]
- 59. Hindré T, Knibbe C, Beslon G, Schneider D. 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol. 10:352–365. 10.1038/nrmicro2750 [DOI] [PubMed] [Google Scholar]
- 60. Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, Howden BP, Stinear TP. 2012. Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194:2334–2341. 10.1128/JB.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Okinaka RT, Price EP, Wolken SR, Gruendike JM, Chung WK, Pearson T, Xie G, Munk C, Hill KK, Challacombe J, Ivins BE, Schupp JM, Beckstrom-Sternberg SM, Friedlander A, Keim P. 2011. An attenuated strain of Bacillus anthracis (CDC 684) has a large chromosomal inversion and altered growth kinetics. BMC Genomics 12:477. 10.1186/1471-2164-12-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ochman H, Elwyn S, Moran NA. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. U. S. A. 96:12638–12643. 10.1073/pnas.96.22.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rocha EP. 2004. Order and disorder in bacterial genomes. Curr. Opin. Microbiol. 7:519–527. 10.1016/j.mib.2004.08.006 [DOI] [PubMed] [Google Scholar]
- 64. Rocha EP, Danchin A. 2003. Essentiality, not expressiveness, drives gene-strand bias in Bacteria. Nat. Genet. 34:377–378. 10.1038/ng1209 [DOI] [PubMed] [Google Scholar]
- 65. Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D’Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabási AL, Oltvai ZN, Osterman AL. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673–5684. 10.1128/JB.185.19.5673-5684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zinser ER, Schneider D, Blot M, Kolter R. 2003. Bacterial evolution through the selective loss of beneficial genes. Trade-offs in expression involving two loci. Genetics 164:1271–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M, Charette SJ. 2012. An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS One 7:e33725. 10.1371/journal.pone.0033725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mijnendonckx K, Provoost A, Monsieurs P, Leys N, Mergeay M, Mahillon J, Van Houdt R. 2011. Insertion sequence elements in Cupriavidus metallidurans CH34: distribution and role in adaptation. Plasmid 65:193–203. 10.1016/j.plasmid.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 69. Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315–1341. 10.1086/285289 [DOI] [Google Scholar]
- 70. Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Ségurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21 (DE3). J. Mol. Biol. 394:644–652. 10.1016/j.jmb.2009.09.052 [DOI] [PubMed] [Google Scholar]
- 71. Wielgoss S, Barrick JE, Tenaillon O, Wiser MJ, Dittmar WJ, Cruveiller S, Chane-Woon-Ming B, Médigue C, Lenski RE, Schneider D. 2013. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U. S. A. 110:222–227. 10.1073/pnas.1219574110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wielgoss S, Barrick JE, Tenaillon O, Cruveiller S, Chane-Woon-Ming B, Médigue C, Lenski RE, Schneider D. 2011. Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3 (Bethesda) 1:183–186. 10.1534/g3.111.000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Thil Smith AA, Weiman M, Médigue C. 2013. Microscope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41:D636–D647. 10.1093/nar/gks1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kaguni JM. 2011. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15:606–613. 10.1016/j.cbpa.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ip SC, Bregu M, Barre FX, Sherratt DJ. 2003. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 22:6399–6407. 10.1093/emboj/cdg589 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Results with respect to chromosomal rearrangements in the 12 populations of the LTEE, complex rearrangements, and parallel rearrangements; Supplemental Discussion on distribution of essential genes on the leading strand. Download
Number of rearrangements in clones sampled from each of the 12 populations at 40,000 generations. Colors indicate the different types of rearrangements according to the genetic mechanism of production (see text). Download
Successive inversions leading to genomes observed in populations Ara–3 and Ara–6 at 40,000 generations. Optical maps and hypothetical chronologies of the two and three successive inversions found in populations Ara–3 (A) and Ara–6 (B), respectively, compared to the ancestor genome are shown. Symbols are identical to those described in the Fig. 2 legend. Download
Rearrangements detected at 40,000 generations in clones sampled from each of the 12 populations of the LTEE.
Clones from the LTEE used in this study.
Primers used in this study.
Parallel rearrangements in the LTEE.