ABSTRACT
To compete for the dynamic stream of nutrients flowing into their ecosystem, colonic bacteria must respond rapidly to new resources and then catabolize them efficiently once they are detected. The Bacteroides thetaiotaomicron starch utilization system (Sus) is a model for nutrient acquisition by symbiotic gut bacteria, which harbor thousands of related Sus-like systems. Structural investigation of the four Sus outer membrane proteins (SusD, -E, -F, and -G) revealed that they contain a total of eight starch-binding sites that we demonstrated, using genetic and biochemical approaches, to play distinct roles in starch metabolism in vitro and in vivo in gnotobiotic mice. SusD, whose homologs are abundant in the human microbiome, is critical for the initial sensing of available starch, allowing sus transcriptional activation at much lower concentrations than without this function. In contrast, seven additional binding sites across SusE, -F, and -G are dispensable for sus activation. However, they optimize the rate of growth on starch in a manner dependent on the expression of the bacterial polysaccharide capsule, suggesting that they have evolved to offset the diffusion barrier created by this structure. These findings demonstrate how proteins with similar biochemical behavior can serve orthogonal functions during different stages of cellular adaptation to nutrients. Finally, we demonstrated in gnotobiotic mice fed a starch-rich diet that the Sus binding sites confer a competitive advantage to B. thetaiotaomicron in vivo in a manner that is dependent on other colonizing microbes. This study reveals how numerically dominant families of carbohydrate-binding proteins in the human microbiome fulfill separate and sometimes cooperative roles to optimize gut commensal bacteria for nutrient acquisition.
IMPORTANCE
Our intestinal tract harbors trillions of symbiotic microbes. A critical function contributed by this microbial community is the ability to degrade most of the complex carbohydrates in our diet, which not only change from meal to meal but also cannot be digested by our own bodies. A numerically abundant group of gut bacteria called the Bacteroidetes plays a prominent role in carbohydrate digestion in humans and other animals. Currently, the mechanisms that allow this bacterial group to rapidly respond to available carbohydrates and then digest them efficiently are unclear. Here, we present novel functions for four carbohydrate-binding proteins present in one member of the Bacteroidetes, revealing that these proteins serve unique and separable roles in either initial nutrient sensing or subsequent digestion. Because the protein families investigated are numerous in other gut bacteria colonizing nearly all humans and animals, our findings are fundamentally important to understanding how symbiotic microbes assist human digestion.
INTRODUCTION
A critical symbiotic function of the dense community of bacteria (microbiota) that inhabits the human gut is to break down complex carbohydrates (glycans) that our own digestive enzymes cannot degrade. Short-chain fatty acids and other products from bacterial glycan fermentation are a significant source of nutrition for the host, improving the health of intestinal tissue and directly modulating lymphocyte development (1–3). The collection of carbohydrates available in the colon and the ability of particular bacteria to degrade them shape the membership and abundance of the microbial community (4–6). Since alterations in the microbiota have been linked to a number of health conditions, including inflammatory bowel diseases (7, 8), colon cancer (9, 10), and susceptibility to pathogens (11, 12), the ability to manipulate the composition and physiology of this ecosystem through noninvasive routes like diet or prebiotics is attractive to promote or restore health. For such interventions to be applied, the rules governing diet-microbiota interactions must first be elucidated in mechanistic detail.
Complex carbohydrates may be the most abundant class of nutrients flowing into the colonic ecosystem, but the precise identities and amounts of these molecules change from meal to meal and wane in between feedings. Not surprisingly, competition for glycans in the densely populated colon has driven some gut bacteria to evolve complex systems to sense and scavenge available forms of these nutrients (13). Individual members of the Bacteroidetes, one of the most abundant bacterial taxa in the human gut (14), encode numerous multiprotein complexes with machinery to bind, degrade, and import glycans (15). These complexes are termed Sus-like systems, after the prototypic starch utilization system (Sus) expressed by Bacteroides thetaiotaomicron (16, 17), and the gene clusters encoding them are known as polysaccharide utilization loci (PULs). Sus-like systems have so far been found in all sequenced gut Bacteroidetes species, constituting as much as 18 to 20% of the genome (18, 19). The B. thetaiotaomicron Sus was the first such system described (20) and has become a model for studying glycan acquisition by the many homologous Sus-like systems that degrade a variety of chemically diverse dietary and host-derived glycans (4, 21–23).
A major unresolved aspect of the function of Sus-like systems is how the component proteins function during various stages of the catabolic process. Sus is required for B. thetaiotaomicron to utilize starch, a highly abundant component of the human diet composed solely of α-1,4- and α-1,6-linked glucose that exhibits substantial variability in secondary/tertiary structure based on the relative positions of these linkages. Starch is the only highly polymeric glycan that can be digested by human enzymes. However, several forms of enzyme-resistant starch (RS), as well as highly branched substructures that occur within larger polymers, are inaccessible to these enzymes and likely transit to the distal gut, where microbial colonization is highest (24). As a consequence of this structural variation and partial digestion by host enzymes, the forms of starch that impact the gut microbiota are not well understood. B. thetaiotaomicron Sus is able to target several forms of soluble starch from different plant sources (16), demonstrating that it can accommodate some of the inherent structural diversity. Moreover, when B. thetaiotaomicron is first exposed to high-molecular-weight starch, the transcription of sus is rapidly activated, reaching near-maximum levels in just 5 min (25). This system remains highly responsive to starch at concentrations as low as 0.01 mg/ml, revealing exquisite sensitivity to low substrate levels.
We have previously solved X-ray crystallographic structures of the four peripheral Sus outer membrane proteins (OMPs) (26–28). These studies enigmatically revealed that there are a total of eight distinct starch-binding sites spread across these four OMPs, in addition to a single catalytic site in the enzyme SusG (Fig. 1). SusD, a component with homologs in all Sus-like systems that target polymers other than starch, contains a single binding site, and deletion of the susD gene eliminates B. thetaiotaomicron growth on starch polymers longer than 5 glucose units (26). In addition to its catalytic site, the amylase SusG contains two nonenzymatic starch-binding sites (27). A SusG surface-binding site is contained within the catalytic domain and is distinct from the active site. The second noncatalytic binding site in SusG is contained within a separate carbohydrate-binding module (CBM) domain, with the binding surface oriented ~180° away from the catalytic and surface binding sites. In vitro, the SusG CBM domain decreases enzymatic activity on soluble starch but enhances the degradation of insoluble starch (27). The final two Sus OMPs, SusE and SusF, are multidomain binding proteins, with SusE containing two tandem CBMs and SusF containing three (28). These two proteins are dispensable for growth and, despite a high level of structural similarity between the SusE and SusF CBMs, display distinct oligosaccharide substrate preferences and affinities at individual binding sites (28). The differences in binding site preference, along with variable phenotypes when they are genetically eliminated, suggest that the eight Sus starch-binding sites may play different roles in starch metabolism. On the other hand, the general binding preferences of all sites for unbranched α-1,4 oligosaccharides could suggest that the functions of these eight sites are largely redundant.
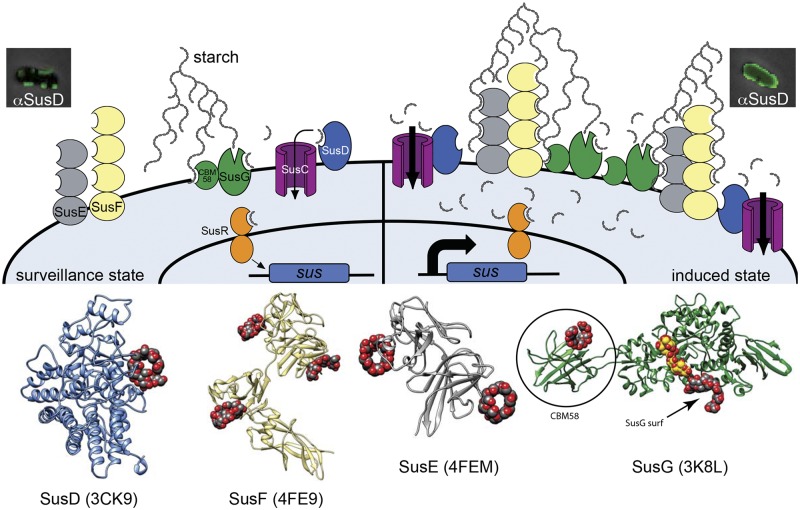
FIG 1 .
Model of the B. thetaiotaomicron starch utilization system (Sus) incorporating findings from this study. Four outer membrane starch-binding lipoproteins, SusD (blue), SusE (gray), SusF (yellow), and SusG (green), contribute to starch binding, along with a TonB-dependent transporter, SusC (purple). Ribbon diagrams of the crystal structures of SusD, -E, -F, and -G are displayed at the bottom to highlight the positions and number of binding sites in each protein (note that an N-terminal domain that is uninvolved in starch binding is missing from the SusE structure). Ligands bound to each of the eight binding sites are shown in red/gray space fill; a ligand in the catalytic site of SusG is shown in red/yellow space fill. The SusG CBM58 and surface (SusG surf) binding sites are labeled. All corresponding binding sites are schematized as open crescents in the cartoon, and the SusG catalytic site is shown as an open “V.” In the uninduced “surveillance state,” SusG cleaves starch to release malto-OS. The SusD binding activity is critical for the binding and import of this malto-OS signal, which is then sensed by SusR, leading to increased expression of the sus locus (all seven functional genes are schematized as a blue box) and, thus, initiating the induced state. During the induced state, the cell surface is flooded with Sus machinery and the SusE, -F, and -G binding sites become cooperatively important for binding starch molecules that have penetrated the B. thetaiotaomicron capsule (not shown in illustration). In this phase, SusD binding activity is no longer essential (shown by lack of ligand occupancy in SusD), but another SusD function is required, based on the results for a strain created using site-directed mutagenesis.
In this study, we systematically explore how these eight different Sus OMP binding sites contribute to B. thetaiotaomicron starch metabolism in living cells, both in culture and in vivo using gnotobiotic mice. We show that the binding site contained in SusD is critical for B. thetaiotaomicron to sense available starch and respond transcriptionally and that this protein has an additional function that is separable from its binding site. In contrast, we show that SusE and SusF, which are dispensable for growth on starch when deleted from the cell surface either alone or together are required in combination with either of the two SusG binding sites in a starch substrate-dependent fashion. Most dramatically, a mutant lacking both SusE and -F and the SusG surface site is completely unable to grow on high-molecular-weight cornstarch, and this and other phenotypes can be compensated by the loss of this symbiont’s surface polysaccharide capsule. Thus, it appears that one role of these carbohydrate-binding proteins is to assist with the capture of external carbohydrates in spite of the diffusion barrier imposed by these species’ ubiquitous production of protective capsules (29). Our results provide an important new layer of mechanistic insight into the function of this family of glycan acquisition systems that is abundantly represented in the human microbiome. By demonstrating separable functions for the apparently redundant Sus OMPs, we provide insight into how gut commensal bacteria have evolved to become more competitive for nutrients in their densely populated habitat.
RESULTS
The SusD binding site is required for growth on large starch molecules.
Homologs of SusD, associated with systems for the degradation of starch and other polysaccharides, can be identified in most if not all sequenced gut Bacteroidetes (18, 30), so we reasoned that these proteins must play a critical role(s). Based on structural and biochemical analysis, SusD contains a single starch binding site and has no enzymatic activity, but the results of deleting it show it to be required for growth on starch molecules longer than 5 glucose units (see Fig. S1 in the supplemental material) (26). To address whether the binding activity of SusD per se is required for starch growth, we constructed a strain carrying a mutant allele of susD where three critical binding residues (tryptophan at position 98 [W98], N101, and W320) (26) were mutated to alanine, abolishing all measurable affinity for starch (Fig. S2A). Identically to the susD deletion (ΔsusD) strain, the binding-deficient SusD (SusD*) mutant was unable to grow on amylopectin (AP) from either maize or potato (Fig. 2A; Fig. S2B), despite being trafficked to the cell surface similarly to native SusD (Fig. S2C). Thus, we conclude that the binding capacity of SusD is directly required for growth on large starch molecules.
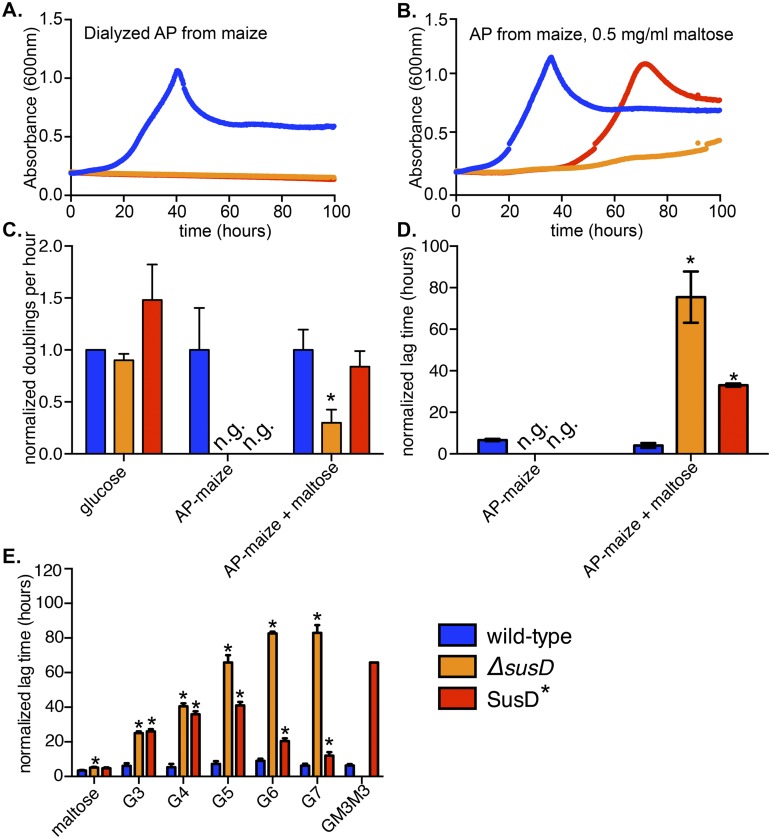
FIG 2 .
Requirement for SusD binding during growth on large starches is overcome with low levels of maltose. (A, B) Representative growth curves of wild-type, ΔsusD, and SusD* B. thetaiotaomicron strains with 5 mg/ml maize amylopectin (AP) (A) or 5 mg/ml maize AP plus 0.5 mg/ml maltose (B) as the sole carbon source. (C) Exponential rates from three replicate growth curves (including those shown in panels A and B) were measured and normalized to the rate of growth on glucose and then to the wild-type growth rate. n.g., no growth observed up to 100 h. (D) Lag times (time for absorbance to reach or surpass 0.35) were measured for three biological replicates and normalized to the corresponding rate of growth on glucose for each replicate. (E) Normalized lag times for B. thetaiotaomicron grown on linear malto-OS ranging from 2 glucose units (maltose) to 7 glucose units (G7) or on GM3M3, a branched glucose heptamer containing two α-1,6 linkages. In all panels, error bars indicate standard errors across three replicates. Statistically significant differences from growth of the wild type (*, P < 0.05) were determined using the one-tailed unpaired Student’s t test. Note that statistics could not be performed for the growth of the SusD* strain on GM3M3, as only one replicate reached sufficient growth density.
Interestingly, we found that SusD* growth could be restored if a small amount of maltose, which alone is too low a concentration to support substantial growth (see Fig. S2B in the supplemental material), was added to high-molecular-weight starch cultures (Fig. 2B). This observation was initially made with commercially available starches, some of which contained minor amounts of contaminating maltooligosaccharides (malto-OS) (Table S1), which we hypothesized could bypass the SusD* defect. Consistent with this idea, only when these starch preparations were dialyzed to remove contaminants did the SusD* mutant exhibit a complete defect (Fig. S2B). We were also surprised to observe that the addition of a small amount of maltose was insufficient to restore growth to the ΔsusD mutant, indicating that the binding site of SusD and the presence of the protein itself (irrespective of binding capacity) are likely to play separable roles (Fig. 2B). This difference in response to maltose is unlikely to be due to effects of the ΔsusD in-frame deletion on up- or downstream sus genes, since we have previously shown that in the ΔsusD mutant, the remaining genes are expressed to wild-type levels in response to maltose (26).
Quantification of two aspects of the growth of B. thetaiotaomicron revealed that the exponential doubling time of the SusD* mutant in maize AP plus maltose was indistinguishable from that of the wild type (Fig. 2C) but that the lag phase of this strain was significantly lengthened (Fig. 2D). Based on this, we hypothesized that SusD binding is important for the efficient import of liberated malto-OS derived from maize AP, which in turn acts as the signal to induce sus transcription. In the absence of normal SusD function, these signals fail to be transported to the periplasm at the concentration at which they are released by bacteria upon early exposure to starch. However, if this signaling blockade is bypassed by the provision of an inducer that does not require SusD function (maltose), Sus is fully activated and the signal transport role of SusD is no longer required to catabolize starch. To further explore whether SusD binding was important for growth on a particular size range of malto-OS, we tested the growth of the two SusD mutants on a panel of malto-OS ranging in length from 2 to 7 glucose units (herein abbreviated as G2 for maltose, G3 for maltotriose, etc.) (Fig. 2E). We observed that, without the entire SusD protein (ΔsusD), the lag time continued to increase with the substrate size (note that we have previously attributed the growth of the ΔsusD mutant on G6 and G7 after ~100 h to spontaneous suppressor mutants [26]; see the legend to Fig. S1 in the supplemental material for additional discussion of this phenomenon). In contrast, the lag time of the SusD* strain on malto-OS increased with the substrate size up to G5 and then began to decrease on longer chains as growth improved. Based on this, we conclude that SusD binding is optimized for the utilization of midrange malto-OS (G3 to G5) and is less important on shorter or longer oligosaccharides. Interestingly, SusD binding also plays a critical role during growth on the double-branched oligosaccharide glucosyl-maltotriosyl-maltotriose (GM3M3), a likely by-product of pullulan degradation, and the observed growth defect does not occur during growth on the isomeric linear maltoheptaose (Fig. S1F and G). Although it remains to be explored in detail, our observations suggest that other components of the Sus machinery are able to compensate for a lack of SusD binding in the presence of longer linear substrates in a fashion that is dependent on the presence of SusD protein but not its binding site.
SusD binding enhances transcriptional sensitivity to starch.
In the experiments described above, we reasoned that increased lag time (always normalized to lag time during growth in glucose to control for culture variations) equates to decreased ability to sense or process available substrates. To directly test this idea and determine if the SusD binding site is important for efficient sensing of malto-OS, we examined the transcriptional responses of wild-type and SusD* strains to concentrations of starch or malto-OS spanning several orders of magnitude. Naive B. thetaiotaomicron cells, grown to mid-exponential phase in glucose (a condition that does not activate sus expression), were washed anaerobically and introduced into medium containing G3, G7, or maize AP in concentrations ranging from 250 ng/ml to 2.5 mg/ml. Samples were collected 30 min after the change of medium, when sus transcription reaches its maximum (25), and the susC message was quantified (Fig. 3).
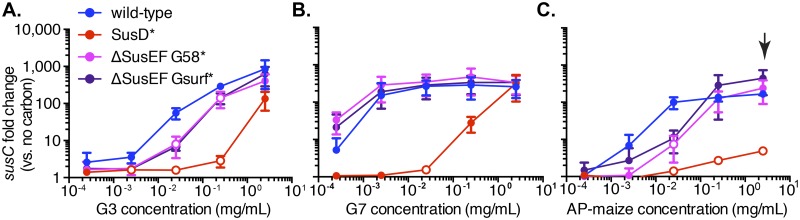
FIG 3 .
SusD binding is critical for sensing starch. Data show the expression of susC transcripts in wild-type and mutant B. thetaiotaomicron cells abruptly exposed to malto-OS or starch for 30 min. (A) Maltotriose (G3); (B) maltoheptaose (G7); (C) maize AP. Fold changes were calculated relative to expression in the glucose-grown, washed cells used to inoculate the starch/malto-OS cultures. Average results and standard deviations of three individual replicates are shown. P values were calculated using the one-tailed unpaired Student’s t test, and statistically significant differences compared to wild-type expression (P < 0.05) are shown by open circles.
Consistent with our hypothesis that SusD enhances sensory responses that promote the transition from nutrient surveillance to active starch degradation, sus expression in bacteria exposed to both oligosaccharides and starch was significantly attenuated in the SusD* mutant compared to its expression in the wild type. At most substrate concentrations tested, sus transcription was substantially lower (sometimes over 100-fold) in the SusD* mutant, highlighting the sensory advantage that SusD provides, especially in low starch/oligosaccharide concentrations (Fig. 3). Even at the highest concentration of 2.5 mg/ml, the SusD* mutant showed lower levels of sus expression on G3, which, although not statistically significant, could explain the increased lag time compared to that of the wild type during growth on this substrate.
In our growth experiments, we observed that the SusD* strain had improved growth on G7 compared to its growth on G3. Indeed, we observed that the SusD* mutant had a more robust transcriptional response to G7 than to G3, although it was still significantly attenuated compared to the wild type (Fig. 3B). The sus expression of the SusD* mutant was ~10-fold higher at 250 µg/ml G7 than at the same concentration of G3 (the latter presenting a 2.3-times-higher molar concentration). Additionally, at the highest concentration, the SusD* mutant exhibited sus levels that were indistinguishable from that in the wild type, which likely explains the lack of defect observed when SusD* is grown on a comparable (5 mg/ml) G7 concentration. These data demonstrate that SusD binding is critical for B. thetaiotaomicron to mount an optimal transcriptional response to available malto-OS and allows the bacterium to sense nutrients at concentrations several orders of magnitude lower than without this function. As expected, given the lack of growth of the SusD* mutant on maize AP and this strain’s defect in sensing small malto-OS, this mutant exhibited a severe defect in sus expression in response to maize AP (Fig. 3C). Since SusD homologs are a defining feature of Bacteroidetes Sus-like systems that target dozens of other glycans (15), this feature may be a fundamental and conserved aspect of this molecular mechanism.
The SusE, -F, and -G binding sites work together to enhance B. thetaiotaomicron’s growth rate in a substrate-dependent manner.
SusD represents just one of the eight starch binding sites contained within the four Sus outer membrane proteins. Across SusE, -F, and -G, there are seven additional noncatalytic binding sites (Fig. 1). To investigate the role of these additional binding sites, we created a series of B. thetaiotaomicron mutants lacking between one and six binding sites in the SusE, -F, and -G proteins. To abolish surface expression of SusE and SusF, we used a strain (ΔSusEF) that contains two mutant alleles in which the cysteine residue at the mature N terminus (after signal peptide cleavage) of each protein was mutated to prevent lipidation and trafficking to the outer membrane (28). To abrogate the binding activity of the SusG CBM58, the SusG58* mutant was created by mutating three critical binding residues (W287, W299, and N330) to alanine, which abolished the binding ability of the CBM (see Fig. S3 in the supplemental material). Similarly, we mutated three critical residues (W460, Y469, and D473) of the SusG surface (SusGsurf) site to alanine to create the SusGsurf* binding-deficient mutant. Of note, we could not directly verify that this mutation eliminated binding at the SusGsurf site because the catalytic site is still present in this same domain and would hydrolyze substrate during a binding experiment. If the catalytic activity was inactivated, the mutated catalytic site, which also contains residues to bind and coordinate malto-OS, would itself become a binding site. However, previous in vitro studies with SusG revealed that this mutation decreases both the binding and the degradation of insoluble cornstarch (27), and as discussed below, we show that the loss of this surface site reduces growth in intact bacteria.
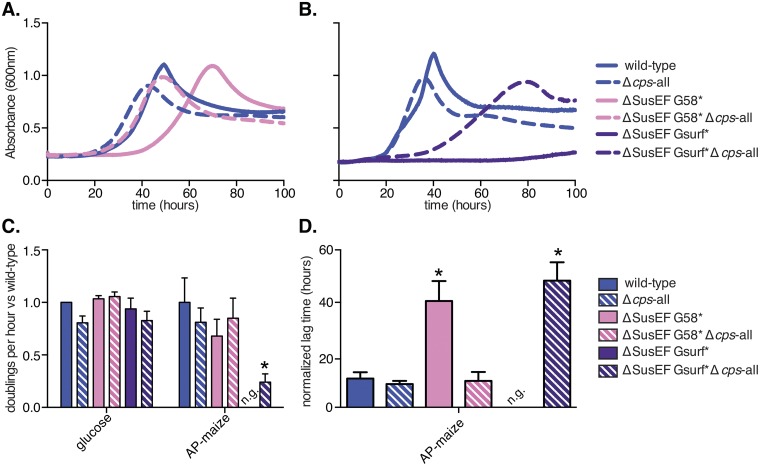
Growth experiments with dialyzed starches as the sole carbon source revealed that, while the ΔSusEF and SusG58* mutants did not demonstrate any growth attenuation compared to the growth of the wild type, the loss of all six Sus CBMs in the ΔSusEF G58* combined mutant resulted in a decreased exponential growth rate (normalized doublings per hour) on both substrates (Fig. 4A). Due to variability between replicates, this was not statistically significant; however, ΔSusEF G58* bacteria did display a significantly increased lag time on maize AP compared to that of the wild type (Fig. 4B).
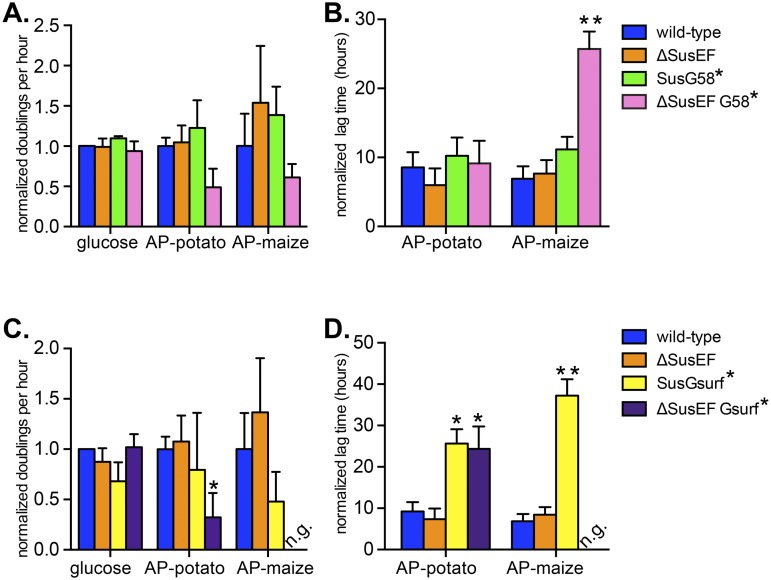
FIG 4 .
SusE, -F, and -G binding sites enhance B. thetaiotaomicron starch growth via overlapping roles. Growth assays were performed with B. thetaiotaomicron wild-type and mutant strains grown on the sole carbon sources noted. Normalized doublings per hour and normalized lag times were calculated as described in the legend to Fig. 2 for strains lacking one or more Sus CBMs (A, B) or strains lacking SusE, SusF, and/or the SusG surface site (C, D). Average results and standard errors across three replicates are shown. Statistically significant differences versus growth of the wild type were calculated using the one-tailed unpaired Student’s t test (*, P < 0.05; **, P < 0.01).
Loss of the SusG surface site alone (SusGsurf*) resulted in a significant growth defect (Fig. 4C and D), with the mutant showing a decrease in the exponential rate on maize AP and a significantly longer lag time on both substrates. Interestingly, the SusGsurf* defect was further exacerbated—resulting in complete loss of growth on maize AP—by the additional combined loss of SusE and SusF (ΔSusEF Gsurf*). This suggests that the SusG surface site is the most critical of the seven SusE, -F, and -G binding sites for growth on high-molecular-weight starch and also that it works cooperatively with the sites contained in SusE and -F. Control staining of whole cells with antisera specific for SusE, -F, and -G suggested that the observed defects were not associated with decreased trafficking of the mutant proteins to the cell surface (see Fig. S4 in the supplemental material; representative growth curves from the experiments described above are displayed in Fig. S5). Finally, the results for separate mutant strains in which either SusE or SusF were removed singly in combination with either of the two different SusG binding sites supported few significant effects compared with the effects of the combined loss of these proteins, suggesting that the partially homologous SusE and -F proteins are sufficient surrogates for one another and need to be lost in tandem along with the SusG binding sites to reveal growth defects (Fig. S5).
Since our results described above suggest that SusD has a critical role(s) independent of its binding function (Fig. 2B), we also tested whether the binding sites in SusE and SusF are solely responsible for their contribution to growth on starch or if restoring the presence of binding-deficient proteins to the cell surface would recover some of the growth defect. To this end, we created binding-deficient alleles of SusE and SusF by using site-directed mutagenesis to target critical binding residues in each of the SusE and SusF CBMs. The resulting alleles encode proteins that show no measurable binding to starch (28) but are expressed on the surface of the cell to levels similar to the expression of susE and -F in the wild type (see Fig. S4 in the supplemental material). The resulting mutant (SusE*F*G58*) displayed a growth profile nearly identical to that of the ΔSusEF G58* strain (Fig. S6), suggesting that, unlike SusD, it is the ability of SusE and SusF to bind maltOS, and not other aspects of their presence on the cell surface, that contribute to their role in growth on starch. This conclusion is further corroborated by the growth of a SusE*F*Gsurf* strain, which was nearly identical to that of the ΔSusEF Gsurf* strain, including the most severe loss-of-growth phenotype on maize AP (Fig. S6).
The combined SusE, -F, and -G binding sites play little role in starch sensing.
Since we determined that the SusD binding site is primarily involved in sensing available malto-OS and is not required for growth on starch when its blockade is bypassed with maltose (Fig. 2B), we sought to determine whether the binding sites in SusE, -F, and -G play a similar role or if they have a mostly separate downstream role. To test this, we performed the same starch, G3, and G7 exposure experiments done previously with the SusD* mutant with the ΔSusEF G58* and ΔSusEF Gsurf* strains. Compared to the SusD* mutant, both strains were substantially more responsive to limited concentrations of malto-OS, and on G3 only, both strains showed significant defects relative to the growth of the wild type, albeit this defect was much less severe than that of the SusD* mutant (Fig. 3A and B). These data suggest that the SusE, -F, and -G binding sites play a far less critical role than does SusD in enhancing the bacterium’s ability to sense and respond to malto-OS.
Because substantial growth defects were observed for the ΔSusEF G58* and ΔSusEF Gsurf* mutants on larger starches, we also tested the transcriptional response of these mutants to dialyzed maize AP. At the highest maize AP concentration, wild-type sus transcription was over 30-fold higher than that of the SusD* mutant, but the sus transcription levels of both of the combined SusE, -F, and -G mutants were indistinguishable from that of the wild type, although both of the combined SusE, -F, and -G mutants displayed significantly lower sus expression at a single intermediate concentration of 250 µg/ml (Fig. 3C). The normal sus expression levels in both of the combined SusE, -F, and -G mutants at the highest starch concentration (Fig. 3C, black arrow) are particularly striking since, at comparable maize AP concentrations of 5 mg/ml (2 times the concentration used here), the ΔSusEF G58* mutant had a significant growth defect and the ΔSusEF Gsurf* mutant was completely unable to grow. Thus, we conclude from these experiments that the SusE, -F, and -G binding sites serve a function(s) that is largely distinct from that of the SusD binding site and that they optimize the growth of B. thetaiotaomicron on starch (e.g., by enhancing growth rate) independent of enhancing the transcriptional response.
Given the multiplicity of binding sites contained in SusE, -F, and -G, we hypothesized that one reason for the observed growth defects could be that strains lacking these sites cannot efficiently sequester oligosaccharides that are released during catalysis and that this may result in diffusion of malto-OS away from the cell surface. To address this question, the various B. thetaiotaomicron strains were grown to mid-exponential phase on either maize AP or maltose (positive control), and cell-free supernatants were collected and immediately denatured by heating to 100°C. Oligosaccharides in the supernatants were labeled with 2-aminobenzamide, and individual malto-OS of 7 glucose units or less were quantified using high-pH anion-exchange chromatography (HPAEC) (see Table S2 in the supplemental material). Only a very low level of malto-OS release, primarily comprised of glucose and maltose, could be detected in the supernatants of wild-type B. thetaiotaomicron cells. Neither the ΔSusEF G58* nor the ΔSusEF Gsurf* strain displayed increased malto-OS release, and in fact, they had slightly decreased malto-OS levels compared to those of the wild type. These data do not support a role for the SusE, -F, and -G binding sites in promoting growth by virtue of their ability to sequester starch breakdown products. Rather, they highlight the exquisite efficiency of this system to scavenge catalyzed starch products, even in the absence of these functions. It is worth noting that SusD is still present in the strains tested and, given the complete loss of growth of the SusD* mutant on maize AP, we could not perform a parallel experiment using the SusD* mutant without adding maltose to the culture. Given our results connecting SusD to enhanced malto-OS sensing, it is plausible that SusD plays the predominant role in sequestering released malto-OS, even during active catalysis.
SusE, -F, and -G binding sites are important for growth on high-molecular-weight starch.
Interestingly, the loss of the SusE, -F, and -G binding sites generally resulted in more severe growth defects on maize AP than on potato AP (Fig. 4). Starch structures can differ significantly between plant sources, so we hypothesized that the SusE, -F, and -G binding sites were required for specific structural aspects of starch that are more prevalent in maize AP, which tends to have a higher degree of α-1,6 branching (31) and a higher molecular weight (32) than potato AP. To investigate which of these structural aspects required the SusE, -F, and -G binding sites, we performed growth assays on two enzyme-treated substrates in which the molecular weight and branching density were varied reciprocally. Waxy cornstarch (WCS), a high-amylopectin starch similar to the maize AP previously used, was treated with β-amylase (BA) and/or branching enzyme (BE). Both of these enzymes modify starch to increase the number of α-1,6 linkages while decreasing the average molecular weight (33) (see Fig. S6C in the supplemental material). The growth rate of the ΔSusEF G58* mutant improved and lag time was significantly decreased as the molecular weight decreased by ~103 but the branch density nearly doubled (Fig. S6D and E). Thus, we conclude that this constellation of binding sites is more important for adapting the cell to higher-molecular-weight substrates than to those with more branches. The ΔSusEF Gsurf* strain had a severe defect on the WCS substrate, as was seen with maize AP; in fact, in only one of three replicates was any growth observed. The growth of this strain improved on both enzyme-treated starch preparations, which is seen most notably as a decrease in lag time. Again, we conclude that since the ΔSusEF Gsurf* strain is more adept at growth on lower-molecular-weight yet more highly branched starches, this collection of binding sites is suited to aiding growth on high-molecular-weight starches.
Requirement of the SusE, -F, and -G binding sites is dependent on polysaccharide capsule.
Among human-associated members of the phylum Bacteroidetes, the ability to produce a polysaccharide capsule is enriched specifically in gut species compared to this ability in oral isolates (29), suggesting that capsule production provides a competitive advantage specifically in the intestinal environment. This capsule layer can be up to several hundred nanometers thick, homogenously covering the cell surface (34), and may represent a significant barrier for large extracellular carbohydrates to penetrate and reach the cell surface Sus machinery. We hypothesized that the multiple SusE, -F, and -G binding sites may have evolved to offset this barrier and increase the overall affinity of the B. thetaiotaomicron cell surface for starch, for example, by holding on to starch chains as they are being degraded. To address this question, we created ΔSusEF G58* and ΔSusEF Gsurf* strains in a B. thetaiotaomicron mutant that does not express a polysaccharide capsule, (ΔSusEF G58* Δcps-all and ΔSusEF Gsurf* Δcps-all strains) to test whether the loss of capsule would reduce the growth defects observed in these strains (25).
Strikingly, we observed that the growth defect associated with loss of the Sus CBMs (ΔSusEF G58* strain) was abolished in the acapsular form of this mutant (Fig. 5A and D). When the ΔSusEF Gsurf* mutation, which led to complete inability to grow on maize AP, was tested in an acapsular background, growth was substantially restored, albeit to less than wild-type levels (Fig. 5B and C). These data suggest that the SusE, -F, and -G binding sites play a measurable role in the presence of polysaccharide surface capsule and may have evolved to counteract the diffusion barrier created by this surface structure. This conclusion is also in agreement with our data suggesting that the SusE, -F, and -G binding sites are particularly important for growth on high-molecular-weight starch, as one would expect that it would be more difficult for starch molecules with a high degree of polymerization to penetrate the capsule layer than it is for smaller substrates. Consistent with the SusD binding site playing a role distinct from that of the SusE, -F, and -G binding sites, the loss of the B. thetaiotaomicron capsule did not restore any growth to the SusD* mutant on maize AP (data not shown).
FIG 5 .
Contributions of the SusE, -F, and -G binding sites are capsule dependent. (A, B) Representative growth curves of B. thetaiotaomicron strains grown with maize AP as the sole carbon source. (C, D) Normalized doublings per hour (C) and normalized lag times (D) were calculated as described in the legend to Fig. 2 for three replicates (including those whose results are shown in panels A and B). Average results and standard errors from three biological replicates are depicted. Statistically significant differences versus the growth of the wild type were calculated using the one-tailed unpaired Student’s t test (*, P < 0.05).
Sus binding sites confer a fitness advantage in vivo on a starch-rich diet.
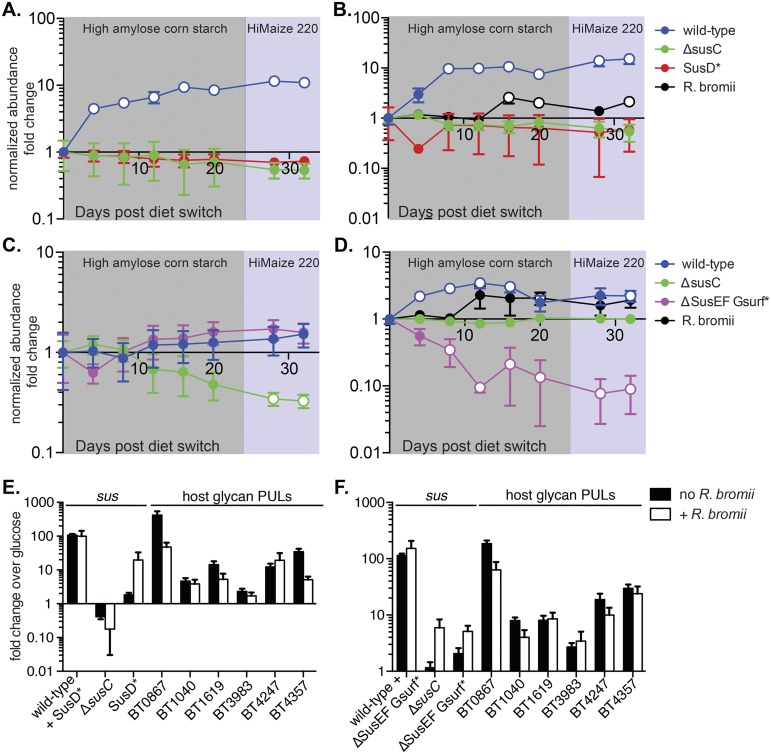
The Sus proteins have evolved in the context of selective pressures encountered in the gut; therefore, we expect that the functions of the Sus binding sites are particularly important in the intestinal environment in the presence of dietary starch. To test this, we performed a competition experiment in gnotobiotic mice. Groups of germfree C57BL/6 mice were inoculated with equal amounts of each of three B. thetaiotaomicron strains: the wild type, the ΔsusC strain (as a control that cannot use starch), and either the SusD* or the ΔSusEF Gsurf* strain, as these two mutants had the most severe growth defects. To ensure that a significant amount of starch escaped host digestion and reached the colon, we used a diet high in resistant starch (RS), which is not easily degraded by mammalian amylases. The results of previous in vitro studies showed that B. thetaiotaomicron cannot degrade RS but that in coculture with Ruminococcus bromii, a species that degrades RS very well, B. thetaiotaomicron’s growth on RS was enhanced (35). Therefore, we colonized half of the groups with R. bromii to investigate whether its addition would increase the amount and types of starch available to B. thetaiotaomicron and perhaps exacerbate any competitive defects of the mutants. After colonization with the appropriate strains was established, the mice were switched to a sequential feeding regimen of two diets that each contained a 50% concentration of one of two different high-amylose RS preparations. DNA was extracted from fecal samples over time, and the relative abundance of each B. thetaiotaomicron strain was determined by quantitative PCR (qPCR) directed at unique genomic tags inserted into each mutant (18).
In the mice colonized with wild-type, ΔsusC, and SusD* strains, the fitness of wild-type B. thetaiotaomicron was enhanced on the RS-rich diets, with the normalized abundance of the bacteria increasing more than 10-fold over the course of the experiment (Fig. 6A and B). The abundance of the SusD* and ΔsusC strains stayed relatively stable throughout the experiment, with modest fold change decreases that were not statistically significant. We did not see any perturbations in strain abundance associated with switching from one type of RS to another. In contrast to the results of the in vitro growth studies (35), the presence of R. bromii did not appear to significantly alter the fitness of B. thetaiotaomicron on RS in vivo (Fig. 6A and B, compare the blue curves). It is important to note that while starch is the only dietary carbohydrate present in this experiment, host mucosal glycans, for which B. thetaiotaomicron encodes many Sus-like degradative systems (18), are a constant nutrient source. On both starch-free and RS diets, it is likely that these host glycans provide a constant alternative for B. thetaiotaomicron, explaining why the ΔsusC and SusD* strains are able to maintain colonization (this is explored in more detail below). Nevertheless, the significant expansion of the wild-type strain suggests that some dietary starch derivatives, presumably from host digestion, are available to B. thetaiotaomicron irrespective of the presence of R. bromii.
FIG 6 .
The Sus binding sites enhance wild-type B. thetaiotaomicron fitness in vivo on a starch-rich diet. Germfree mice fed a starch-free diet were randomly segregated into four treatment groups (n = 5 per group) and colonized with a mixture of wild-type and mutant B. thetaiotaomicron strains. All groups were colonized with wild-type, ΔsusC, and either SusD* (A, B) or ΔSusEF Gsurf* (C, D) strains. Half of the groups (B, D) were also colonized with the keystone starch-degrading species Ruminococcus bromii. Once colonization was established, mice were switched to a diet rich in high-amylose resistant cornstarch for 24 days (shaded in gray) and then switched to a diet rich in Hi-Maize 220 (shaded in purple) for the remainder of the experiment. The relative abundance of each strain was determined by quantifying unique genomic tags using qPCR and was normalized to its abundance on the day of the initial diet switch (day 0). Average results and standards errors across the five mice are shown. Open circles represent significant changes (P < 0.05) in normalized abundance versus the abundance on day 0, calculated using the two-tailed Student’s t test. (E and F) sus transcript levels from cecal contents collected at the end of the experiment were probed using qPCR, and fold changes relative to the levels of B. thetaiotaomicron grown in MM-glucose were calculated. (E) Transcripts from groups whose abundance results are shown in panels A and B. (F) Data from groups whose abundance results are shown in panels C and D. To probe for strain-specific sus expression, primers were designed such that sus transcripts would be amplified from only a subset of the B. thetaiotaomicron strains present (noted on the x axis). sus expression levels were normalized to the relative abundance of the strain from which they were amplified. Transcript levels of PULs targeting host mucosal glycans were probed as well.
Interestingly, in the groups colonized with the ΔSusEF Gsurf* strain, we did observe differences in strain abundance between the B. thetaiotaomicron-only and the R. bromii-colonized group, most notably in the abundance of the ΔSusEF Gsurf* strain itself. In the absence of R. bromii, the ΔSusEF Gsurf* strain behaved very similarly to the wild type, with both strains achieving modest, nonsignificant fold increases in normalized abundance over the course of the experiment (Fig. 6C). However, in the presence of R. bromii, the normalized abundance of the ΔSusEF Gsurf* strain decreased significantly over the course of the experiment, while the abundance of the wild type increased (Fig. 6D).
At the end of the experiment, cecal contents were collected from all mice, total RNA was extracted, and the corresponding cDNA was probed for sus transcript levels, as well as the expression of PULs directed at host mucosal glycans. Because each B. thetaiotaomicron strain differed in the sequence of its sus locus due to the susD, -E, -F, and -G mutations, we designed primers that allowed us to examine strain-specific sus levels. The levels of sus transcription in mice colonized with the SusD* mutant amid wild-type and ΔsusC competitors indicated that most transcripts were produced by wild-type B. thetaiotaomicron, regardless of the presence of R. bromii (Fig. 6E). Notably, the level of sus expression by the SusD* mutant increased ~18-fold in the presence of R. bromii. This result suggests that the presence of this species liberates additional products that are not liberated by host digestion and that activate sus expression in the SusD* mutant. As anticipated, the combined B. thetaiotaomicron community exhibited the expression of several PULs previously associated with the degradation of host glycans (18), confirming that this alternative nutrient pool is being accessed under these conditions. Similar results were observed in mice colonized with the ΔSusEF Gsurf* strain, suggesting that most of the expression of specific sus transcripts also derived from wild-type bacteria under this condition and that host glycans are targeted as alternatives.
Taken together, these data highlight that the degradation of dietary starch by host amylases and/or R. bromii liberates different forms of this nutrient that require distinct Sus binding proteins. SusD likely contributes to the utilization of saccharides, possibly smaller malto-OS, that are released by host digestion. In contrast, the combined presence of SusE, -F, and -Gsurf proteins contributes to the utilization of starch released in the presence of R. bromii, which may correspond to longer pieces of starch, for the metabolization of which these functions are essential in vitro.
DISCUSSION
Microorganisms that thrive in the densely colonized and competitive gut ecosystem undoubtedly have evolved features to enhance their ability to recognize and scavenge nutrients. In this study, we demonstrate that the abundant gut symbiont, B. thetaiotaomicron, has evolved multiple starch-binding proteins that, via unique and sometimes cooperative roles, optimize this organism for starch acquisition. We present a model where the SusD starch-binding site is critical for the initial sensing of starch by enhancing the utilization of medium-length malto-OS, leading to efficient and rapid induction of the sus locus. The seven remaining binding sites spread across SusE, -F, and -G contribute far less to the starch-induced transcriptional response but, instead, optimize the growth rate of B. thetaiotaomicron on starch in a capsule-dependent manner, suggesting that they act to offset the loss of affinity created by this barrier. In our model, the SusE, -F, and -G binding sites are most critical once the Sus machinery is highly expressed and function to keep local concentrations of starch chains high around the B. thetaiotaomicron cell so that catalysis can occur with maximum efficiency (Fig. 1).
We have demonstrated that, despite the absolute requirement for SusD in B. thetaiotaomicron starch utilization, the need for its binding ability per se can be circumvented with small malto-OS, demonstrating a critical function for SusD that is independent from binding. A potential binding-independent role for SusD is to mediate interactions with other Sus proteins. Indeed, previous cross-linking evidence suggests physical interaction between SusC and SusD (36). SusD may promote additional interactions between SusE, -F, and -G (which are needed to efficiently acquire and degrade substrate) and SusC (through which malto-OS are imported). This hypothesis is supported by the presence of a tetratricopeptide repeat domain in SusD (26), a motif associated with protein-protein interactions, although these additional roles for SusD remain to be explored in detail.
The amounts and forms of starch that reach the colonic microbiota are difficult to predict as, unlike other plant polysaccharides, human enzymes in the upper digestive tract degrade a significant portion of starches. However, studies monitoring the digestion of starch as it passes through the digestive tract have reported that approximately 20% of digestible starch and 50% of resistant starch reach the human colon (37, 38). Furthermore, starch-degrading enzymes are among the most common carbohydrate-active enzymes in sequenced representatives from the human microbiome (13), suggesting that it is an important nutrient for the gut microbiota. We found that the SusD binding site increased B. thetaiotaomicron’s ability to sense available starch, by allowing sus expression at starch concentrations several orders of magnitude lower than without SusD binding. The ability to sense and respond rapidly to available nutrients is critical in the gut, where there is intense competition for nutrients, as well as a constantly changing carbohydrate landscape due to meal-to-meal variation. SusD, unlike SusE, -F, or -G, is a conserved component of all Sus-like systems (18). This conservation may be reflective of the fact that the SusD binding site serves a unique function that cannot be compensated by binding sites in the other proteins. We hypothesize that SusD homologs in other systems serve a similar function in enhancing sensitivity to their cognate substrates, but this remains to be tested.
In contrast to the dramatic phenotype associated with loss of SusD binding, eliminating between one and five of the CBMs in SusE, F, and/or G does not significantly affect B. thetaiotaomicron starch growth. However, loss of all six CBMs contained in these three proteins does cause a significant growth defect. Additionally, the defect associated with the loss of the SusG surface-binding site, which is substantial by itself, is exacerbated by the further loss of SusE and SusF. Unlike SusD, there must be overlapping roles for these Sus binding sites, as they each appear to be able to compensate for the loss of others. SusE, -F, and -G are not conserved members of Sus-like systems. However, emerging studies suggest that there is functional conservation of cell surface carbohydrate-binding proteins in other systems studied (4, 21). These divergent carbohydrate-binding proteins may fill roles similar to those of their functional counterparts in Sus. However, it is unlikely that binding proteins in other systems will universally exhibit cooperation or overlap binding sites contained in their accompanying surface enzymes, because in a recent study on xyloglucan degradation, the X-ray crystallographic structure of an essential endo-acting xyloglucanase failed to reveal any additional carbohydrate-binding sites associated with this enzyme (21).
Experiments with an acapsular B. thetaiotaomicron strain suggest that the SusE, -F, and -G binding sites have evolved redundant roles to offset a loss of affinity that is imposed by the production of protective surface polysaccharides. The capsule-dependent role of these binding proteins may be particularly critical for starch acquisition, since it is a large and potentially highly branched plant polysaccharide (on average, 107 to 109 Da in unprocessed cornstarch). However, the forms of other host and dietary polysaccharides that are attacked by living Bacteroides cells in vivo may be similar or greater in complexity, owing to their incorporation in plant cell wall particles or high-molecular-weight secreted mucin glycoproteins. One can imagine that these larger nutrient scaffolds are more difficult to interact with through the capsular polysaccharide mesh. In light of the many emerging studies on Sus-like systems required for the degradation of other polysaccharides by gut and environmental bacteria, it will be interesting in the future to determine whether binding functions akin to those contributed by SusE, -F, and -G play similar or different roles in other systems.
This study provides another layer of mechanistic understanding to a polysaccharide degradation paradigm that has been markedly expanded by studies of bacterial members of the human gut microbiota and for which the B. thetaiotaomicron Sus is the best understood example. We demonstrate that individual binding proteins, with similar biochemical specificities when analyzed in pure form in vitro, play unique roles in the context of a multiprotein complex expressed on the surface of a symbiotic gut bacterium. Investigating these molecular mechanisms in great detail not only contributes to our understanding of the fundamental physiology of our gut microbial symbionts but may also offer clues about how to intervene in their biology and the food webs in which they participate. The latter is the goal of pre- and probiotic approaches that aim to stabilize or alter the function of the gut microbial community and its potential contributions to inflammation and colorectal cancer, various metabolic diseases (including obesity and diabetes), and invasion by outside pathogens.
MATERIALS AND METHODS
Bacterial strains, culture conditions, genetic manipulation, and protein staining in whole cells.
B. thetaiotaomicron ATCC 29148 (VPI-5482) strains were routinely grown in tryptone-yeast extract-glucose (TYG) medium (39), minimal medium (MM) (18), or brain heart infusion (Becton Dickinson) agar that included 10% horse blood (Colorado Serum Co.). Carbon sources were added for a final concentration of 5 mg/ml unless otherwise stated. R. bromii L2-63 was grown in YCFA medium (35) supplemented with 2 mg/ml each of glucose, cellobiose, and soluble starch. Cultures were grown at 37°C in an anaerobic chamber (10% H2, 5% CO2, and 85% N2; Coy Manufacturing, Grass Lake, MI). Mutations were introduced via counterselectable allelic exchange as previously described (26). The primers used in this study are listed in Table S3 in the supplemental material. The presence of proteins on the surfaces of fixed B. thetaiotaomicron cells or in whole-cell lysates was probed with rabbit polyclonal antibodies (Cocalico Biologicals) in nonpermeabilized, formaldehyde-fixed B. thetaiotaomicron cells (immunofluorescence) or whole-cell lysates (Western blots) as previously described (28).
To quantify growth parameters, the increase in culture absorbance (600 nm) in 200-µl cultures was measured every 10 min on an automated plate reading device as described previously (19). To calculate normalized doublings per hour, we used the portion of the graph corresponding to absorbance readings between 0.6 and 0.8 for all data except those shown in Fig. 5; for those data, 0.4 to 0.6 was used to account for the lower maximum absorbance observed in the Δcps-all mutant. Data points were fit to an exponential growth equation, and doublings per hour were normalized to the growth of the wild type on glucose; because some mutants could exhibit defects in growth on glucose, this allowed visualization of the variation for each strain on glucose. The growth of each strain was then normalized to that of the wild type (set at 1.0) for each substrate. Lag time was defined as the time required for the absorbance reading for a particular strain to reach or exceed 0.35 at 600 nm. To account for variations in inoculum size or environment between experiments, the lag time on glucose for each individual experiment was subtracted from the lag time for the substrate of interest; in all cases, cultures had shorter lag times on glucose than on starch or malto-OS.
Confirmation of SusG CBM58 and SusD binding-deficient mutants.
Isothermal titration calorimetry on a NanoITC SV (TA Instruments) was performed to confirm the lack of binding by the SusG CBM58* and SusD* proteins to α-cyclodextrin. The data were plotted and fit to a one-site binding model with NanoAnalyze (TA instruments).
Monitoring transcriptional response to malto-OS and starch.
Mid-exponential-phase B. thetaiotaomicron cultures grown on MM plus glucose were washed and then introduced into prereduced MM containing the appropriate concentration of malto-OS or starch. Cells were collected after 30 min, and qPCR was used to compare the sus transcript levels to those in the reference, B. thetaiotaomicron cells grown in MM glucose then washed with MM no carbon.
Analysis of malto-OS concentrations by HPAEC.
The malto-OS contents in starch stocks and B. thetaiotaomicron culture supernatants were analyzed by 2-aminobenzamide labeling followed by high-pH anion-exchange chromatography (HPAEC) at the UCSD Glycotechnology Core. Quantification was determined by comparison to the retention times of authentic standards for each degree of polymerization.
Gnotobiotic mouse experiments.
All animal experiments, including euthanasia by carbon dioxide asphyxiation, were approved by the University Committee on Use and Care of Animals at the University of Michigan (NIH Office of Laboratory Animal Welfare number A3114-01) and were supervised by a veterinarian. Six-week-old male C57BL/6 germfree mice fed a polysaccharide-free diet (Harlan-Teklad TD.130280) were colonized with tagged B. thetaiotaomicron strains (18) and with R. bromii if appropriate. After colonization was established (day 14), mice were switched to a low-glycemic control diet (TD.120455; Harlan-Teklad) containing 50% (wt/wt) high-amylose resistant cornstarch, and on day 38, mice were switched to a different low-glycemic control diet (TD.08810; Harlan-Teklad) containing 50% (wt/wt) Hi-Maize 220 resistant starch. DNA was extracted from fecal pellets throughout the experiment, and strains were enumerated as previously described (18). The relative abundance of each strain was normalized to its abundance on the day of the diet switch (day 0). Postsacrifice, cecal contents were collected from mice and gene transcript levels were quantified using qPCR.
Additional details of these experimental procedures are provided in Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Includes supplemental materials and methods and references. Download
SusC, -D, and -G mutant growth curves on malto-OS. Representative growth curves of various SusC, SusD, and/or SusG B. thetaiotaomicron mutants on minimal medium with 5 mg/ml maltose (A), maltotriose (B), maltotetraose (C), maltopentaose (D), maltohexaose (E), maltoheptaose (F), or glucosyl-maltotriosyl-maltotriose (GM3M3), a branched glucose heptamer with two α-1,6 linkages (G). As observed previously with a ΔsusD strain (26), after 100 h, we began to see the occasional emergence of presumed suppressor mutants that, despite lacking required Sus components, had regained the ability to grow on longer substrates. We isolated single colonies from growth-positive cultures (SusC, -D, and -G mutants alone or in the combinations listed) after ~100 h and passaged these strains through medium that did not contain starch. When these isolated strains were retested in MM containing maize AP, all exhibited growth without an extended lag time (not shown), revealing a heritable ability to circumvent the mutant defect. For this reason, we only analyzed growth up to 100 h. Download
The SusD* protein is unable to bind starch, stable on the B. thetaiotaomicron surface, and leads to a phenotype distinct from that of wild-type or ΔsusD B. thetaiotaomicron. (A) Isothermal titration calorimetry was performed with α-cyclodextrin to verify that the SusD* binding mutant fails to bind starch. (Top) SusD wild-type protein (positive control, for reference); (bottom) SusD* protein. The data for wild-type SusD were fit to an independent one-site binding model, fixing the number of binding sites (N) to one due to the known stoichiometry of binding. The data for the SusD* mutant were fit to a blank constant (no binding) model. (B) Representative growth curves with 5 mg/ml potato AP (APP) or maize AP (APM) either pre- or postdialysis or 0.5 mg/ml maltose as the sole carbon source. (C) Wild-type and SusD* B. thetaiotaomicron cells were stained (green) using anti-SusD antibodies. Similar levels of surface SusD expression were observed. Western blot assays were also performed using anti-SusD antibodies on whole-cell lysates from wild-type and SusD* cells; again, similar SusD levels were observed. Note that, despite normal trafficking to the cell surface, the SusD* protein runs at a slightly higher apparent molecular weight under these denaturing SDS-PAGE conditions. Despite this altered mobility, sequencing of the entire modified SusD* allele that was integrated back into this strain failed to reveal any point mutations or frameshifts that could explain this phenomenon. Thus, we conclude that the altered mobility is due to an unknown effect of mutating the three binding site residues in this modified SusD protein. Download
The SusG CBM58* mutant is deficient in binding. Isothermal titration calorimetry was performed with α-cyclodextrin to verify that the SusG CBM58* binding mutant fails to bind starch. (Top) Wild-type CBM58; (bottom) CBM58* mutant. The wild-type CBM58 data were fit to an independent one-site binding model, fixing n = 1 due to the known stoichiometry of binding. The data for the CBM58* mutant were fit to a blank constant (no binding) model. Download
Mutated SusE, SusF, and SusG alleles are expressed appropriately in B. thetaiotaomicron cells. (Top) Wild-type, ΔSusEF G58*, ΔSusEF Gsurf*, SusE*F*G58*, and SusE*F*Gsurf* B. thetaiotaomicron cells were grown on maltose to mid-exponential phase, fixed, and stained for SusE, SusF, or SusG using the appropriate antibodies. Bright-field and corresponding fluorescence images are shown side by side. Consistent with mutation of their lipidation sites, the ΔSusE and ΔSusF alleles were not detected on the cell surface, but SusE*, SusF*, SusG58*, and SusGsurf* alleles were detected on the B. thetaiotaomicron cell surface at levels similar to the levels of the wild-type proteins (bottom). Whole-cell lysates from the strains listed above were collected and probed for the expression of SusE, SusF, and SusG using Western blotting (see numbered key below blots). Consistent with mutation of their lipidation sites, only the ΔSusE and ΔSusF proteins are still expressed; their molecular weights are slightly lower than those of the wild-type proteins, consistent with loss of the lipid tail. Although degradation of some of the proteins is observed (most notably the SusE* mutant, whose expression alone does not exhibit a diminished growth phenotype), we believe this can be tolerated, as levels similar to those in the wild-type are observed in surface staining of these proteins. Download
Growth of SusE, -F, and -G mutants on starch. (A to F) Growth curves were performed with B. thetaiotaomicron strains grown on minimal medium with the sole carbon sources noted. We only observed growth up to 100 h so as to exclude the appearance of suppressor mutants as described in the legend to Fig. S1. (G to J) Quantified growth rates and lag times for B. thetaiotaomicron strains grown on the sole carbon sources noted. Normalized doublings per hour and normalized lag times, respectively, for strains lacking SusE and/or SusF and/or the SusG CBM58 (G, H) or strains lacking SusE and/or SusF and/or the SusG surface site (I, J) were calculated as described in the legend to Fig. 2 and Materials and Methods. Average results and standard errors across three replicates are shown. P values (versus the results for the wild type) were calculated using the one-tailed unpaired Student’s t test (*, P < 0.05; **, P < 0.01). Download
The binding ability of SusE and SusF is solely responsible for their contributions to growth of B. thetaiotaomicron on starch, and the SusEFG binding sites enhance growth of B. thetaiotaomicron on high-molecular-weight starch. (A, B) Growth assays were performed with B. thetaiotaomicron strains either not expressing SusE and SusF (ΔSusEF G58* and SusEF Gsurf*) or expressing binding-deficient SusE and SusF mutants (SusE*F*G58* and SusE*F*Gsurf*) on the sole carbon sources noted. Normalized doublings per hour (A) and normalized lag times (B) were calculated as described in the legend to Fig. 2 and Materials and Methods. Average results and standard errors across three replicates are shown. Statistically significant differences versus growth of the wild type were calculated using the one-tailed unpaired Student’s t test (*, P < 0.05; **, P < 0.01). (C) Growth assays were performed with the indicated B. thetaiotaomicron strains on untreated waxy cornstarch (WCS), branching enzyme-treated WCS (BE WCS), or branching enzyme plus β-amylase-treated WCS (BEBA WCS). Percentages of total α-1,6 linkages and average molecular weights are shown for each substrate. (D, E) Normalized doublings per hour (D) and normalized lag times (E) from growth assays were calculated as described in the legend to Fig. 2 and Materials and Methods. Average results and standard errors across three replicates are shown. Statistically significant differences versus growth of the wild type were calculated using the one-tailed unpaired Student’s t test. Download
Maltooligosaccharide concentrations in amylopectin (ng malto-OS per μg total carbohydrate) pre- and postdialysis by high-pH anion-exchange chromatography (HPAEC).
Maltooligosaccharide concentrations in B. thetaiotaomicron supernatants (ng malto-OS per μl).
Oligonucleotides used in this study.
ACKNOWLEDGMENTS
This article is dedicated to Abigail Salyers (1942 to 2013), whose laboratory discovered the B. thetaiotaomicron starch utilization system and pioneered the genetic manipulation and experimental interrogation of many Bacteroides organisms.
We thank Harry J. Flint (University of Aberdeen, Scotland) for kindly providing Ruminococcus bromii L2-63.
This work was supported by National Institutes of Health grants DK084214 and GM099513 and a Global Probiotics Council young investigator grant for probiotics research (all to E.C.M.). E.A.C. was supported by a University of Michigan Genetics Training grant (GM07544) and a Rackham Distinguished predoctoral fellowship.
Footnotes
Citation Cameron EA, Kwiatkowski KJ, Lee B-H, Hamaker BR, Koropatkin NM, Martens EC. 2014. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio 5(5):e01441-14. doi:10.1128/mBio.01441-14.
REFERENCES
- 1. McNeil NI. 1984. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 39:338–342 [DOI] [PubMed] [Google Scholar]
- 2. McIntyre A, Gibson PR, Young GP. 1993. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut 34:386–391. 10.1136/gut.34.3.386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. 10.1016/j.cell.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046. 10.1371/journal.pone.0015046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. 10.1038/ismej.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kostic AD, Xavier RJ, Gevers D. 2014. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146:1489–1499. 10.1053/j.gastro.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. 2010. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8:292–300. 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sears CL, Garrett WS. 2014. Microbes, microbiota, and colon cancer. Cell Host Microbe 15:317–328. 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726–4736. 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114. 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11:497–504. 10.1038/nrmicro3050 [DOI] [PubMed] [Google Scholar]
- 14. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284:24673–24677. 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson KL, Salyers AA. 1989. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 171:3199–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson KL, Salyers AA. 1989. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 171:3192–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. 10.1371/journal.pbio.1001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shipman JA, Berleman JE, Salyers AA. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182:5365–5372. 10.1128/JB.182.19.5365-5372.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. 2014. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506:498–502. 10.1038/nature12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dodd D, Mackie RI, Cann IK. 2011. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol. Microbiol. 79:292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Renzi F, Manfredi P, Mally M, Moes S, Jenö P, Cornelis GR. 2011. The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLoS Pathog. 7:e1002118. 10.1371/journal.ppat.1002118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin AH, Lee BH, Nichols BL, Quezada-Calvillo R, Rose DR, Naim HY, Hamaker BR. 2012. Starch source influences dietary glucose generation at the mucosal alpha-glucosidase level. J. Biol. Chem. 287:36917–36921. 10.1074/jbc.M112.378331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol. Microbiol. 88:876–890. 10.1111/mmi.12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. 10.1016/j.str.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koropatkin NM, Smith TJ. 2010. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure 18:200–215. 10.1016/j.str.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 28. Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. 2012. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J. Biol. Chem. 287:34614–34625. 10.1074/jbc.M112.397380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coyne MJ, Comstock LE. 2008. Niche-specific features of the intestinal Bacteroidales. J. Bacteriol. 190:736–742. 10.1128/JB.01559-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ellrott K, Jaroszewski L, Li W, Wooley JC, Godzik A. 2010. Expansion of the protein repertoire in newly explored environments: human gut microbiome specific protein families. PLoS Comput. Biol. 6:e1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morell MK, Samuel MS, O’Shea MG. 1998. Analysis of starch structure using fluorophore-assisted carbohydrate electrophoresis. Electrophoresis 19:2603–2611. 10.1002/elps.1150191507 [DOI] [PubMed] [Google Scholar]
- 32. Yokoyama W, Renner-Nantz JJ, Shoemaker CF. 1998. Starch molecular mass and size by size-exclusion chromatography in DMSO-LiBr coupled with multiple angle laser light scattering. Cereal Chem. 75:530–535. 10.1094/CCHEM.1998.75.4.530 [DOI] [Google Scholar]
- 33. Lee BH, Yan L, Phillips RJ, Reuhs BL, Jones K, Rose DR, Nichols BL, Quezada-Calvillo R, Yoo SH, Hamaker BR. 2013. Enzyme-synthesized highly branched maltodextrins have slow glucose generation at the mucosal alpha-glucosidase level and are slowly digestible in vivo. PLoS One 8:e59745. 10.1371/journal.pone.0059745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martens EC, Roth R, Heuser JE, Gordon JI. 2009. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J. Biol. Chem. 284:18445–18457. 10.1074/jbc.M109.008094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. 10.1038/ismej.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cho KH, Salyers AA. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 183:7224–7230. 10.1128/JB.183.24.7224-7230.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vonk RJ, Hagedoorn RE, de Graaff R, Elzinga H, Tabak S, Yang YX, Stellaard F. 2000. Digestion of so-called resistant starch sources in the human small intestine. Am. J. Clin. Nutr. 72:432–438 [DOI] [PubMed] [Google Scholar]
- 38. Champ MM, Molis C, Flourié B, Bornet F, Pellier P, Colonna P, Galmiche JP, Rambaud JC. 1998. Small-intestinal digestion of partially resistant cornstarch in healthy subjects. Am. J. Clin Nutr. 68:705–710 [DOI] [PubMed] [Google Scholar]
- 39. Holdeman LV, Cato ED, Moore WEC. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, VA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Includes supplemental materials and methods and references. Download
SusC, -D, and -G mutant growth curves on malto-OS. Representative growth curves of various SusC, SusD, and/or SusG B. thetaiotaomicron mutants on minimal medium with 5 mg/ml maltose (A), maltotriose (B), maltotetraose (C), maltopentaose (D), maltohexaose (E), maltoheptaose (F), or glucosyl-maltotriosyl-maltotriose (GM3M3), a branched glucose heptamer with two α-1,6 linkages (G). As observed previously with a ΔsusD strain (26), after 100 h, we began to see the occasional emergence of presumed suppressor mutants that, despite lacking required Sus components, had regained the ability to grow on longer substrates. We isolated single colonies from growth-positive cultures (SusC, -D, and -G mutants alone or in the combinations listed) after ~100 h and passaged these strains through medium that did not contain starch. When these isolated strains were retested in MM containing maize AP, all exhibited growth without an extended lag time (not shown), revealing a heritable ability to circumvent the mutant defect. For this reason, we only analyzed growth up to 100 h. Download
The SusD* protein is unable to bind starch, stable on the B. thetaiotaomicron surface, and leads to a phenotype distinct from that of wild-type or ΔsusD B. thetaiotaomicron. (A) Isothermal titration calorimetry was performed with α-cyclodextrin to verify that the SusD* binding mutant fails to bind starch. (Top) SusD wild-type protein (positive control, for reference); (bottom) SusD* protein. The data for wild-type SusD were fit to an independent one-site binding model, fixing the number of binding sites (N) to one due to the known stoichiometry of binding. The data for the SusD* mutant were fit to a blank constant (no binding) model. (B) Representative growth curves with 5 mg/ml potato AP (APP) or maize AP (APM) either pre- or postdialysis or 0.5 mg/ml maltose as the sole carbon source. (C) Wild-type and SusD* B. thetaiotaomicron cells were stained (green) using anti-SusD antibodies. Similar levels of surface SusD expression were observed. Western blot assays were also performed using anti-SusD antibodies on whole-cell lysates from wild-type and SusD* cells; again, similar SusD levels were observed. Note that, despite normal trafficking to the cell surface, the SusD* protein runs at a slightly higher apparent molecular weight under these denaturing SDS-PAGE conditions. Despite this altered mobility, sequencing of the entire modified SusD* allele that was integrated back into this strain failed to reveal any point mutations or frameshifts that could explain this phenomenon. Thus, we conclude that the altered mobility is due to an unknown effect of mutating the three binding site residues in this modified SusD protein. Download
The SusG CBM58* mutant is deficient in binding. Isothermal titration calorimetry was performed with α-cyclodextrin to verify that the SusG CBM58* binding mutant fails to bind starch. (Top) Wild-type CBM58; (bottom) CBM58* mutant. The wild-type CBM58 data were fit to an independent one-site binding model, fixing n = 1 due to the known stoichiometry of binding. The data for the CBM58* mutant were fit to a blank constant (no binding) model. Download
Mutated SusE, SusF, and SusG alleles are expressed appropriately in B. thetaiotaomicron cells. (Top) Wild-type, ΔSusEF G58*, ΔSusEF Gsurf*, SusE*F*G58*, and SusE*F*Gsurf* B. thetaiotaomicron cells were grown on maltose to mid-exponential phase, fixed, and stained for SusE, SusF, or SusG using the appropriate antibodies. Bright-field and corresponding fluorescence images are shown side by side. Consistent with mutation of their lipidation sites, the ΔSusE and ΔSusF alleles were not detected on the cell surface, but SusE*, SusF*, SusG58*, and SusGsurf* alleles were detected on the B. thetaiotaomicron cell surface at levels similar to the levels of the wild-type proteins (bottom). Whole-cell lysates from the strains listed above were collected and probed for the expression of SusE, SusF, and SusG using Western blotting (see numbered key below blots). Consistent with mutation of their lipidation sites, only the ΔSusE and ΔSusF proteins are still expressed; their molecular weights are slightly lower than those of the wild-type proteins, consistent with loss of the lipid tail. Although degradation of some of the proteins is observed (most notably the SusE* mutant, whose expression alone does not exhibit a diminished growth phenotype), we believe this can be tolerated, as levels similar to those in the wild-type are observed in surface staining of these proteins. Download
Growth of SusE, -F, and -G mutants on starch. (A to F) Growth curves were performed with B. thetaiotaomicron strains grown on minimal medium with the sole carbon sources noted. We only observed growth up to 100 h so as to exclude the appearance of suppressor mutants as described in the legend to Fig. S1. (G to J) Quantified growth rates and lag times for B. thetaiotaomicron strains grown on the sole carbon sources noted. Normalized doublings per hour and normalized lag times, respectively, for strains lacking SusE and/or SusF and/or the SusG CBM58 (G, H) or strains lacking SusE and/or SusF and/or the SusG surface site (I, J) were calculated as described in the legend to Fig. 2 and Materials and Methods. Average results and standard errors across three replicates are shown. P values (versus the results for the wild type) were calculated using the one-tailed unpaired Student’s t test (*, P < 0.05; **, P < 0.01). Download
The binding ability of SusE and SusF is solely responsible for their contributions to growth of B. thetaiotaomicron on starch, and the SusEFG binding sites enhance growth of B. thetaiotaomicron on high-molecular-weight starch. (A, B) Growth assays were performed with B. thetaiotaomicron strains either not expressing SusE and SusF (ΔSusEF G58* and SusEF Gsurf*) or expressing binding-deficient SusE and SusF mutants (SusE*F*G58* and SusE*F*Gsurf*) on the sole carbon sources noted. Normalized doublings per hour (A) and normalized lag times (B) were calculated as described in the legend to Fig. 2 and Materials and Methods. Average results and standard errors across three replicates are shown. Statistically significant differences versus growth of the wild type were calculated using the one-tailed unpaired Student’s t test (*, P < 0.05; **, P < 0.01). (C) Growth assays were performed with the indicated B. thetaiotaomicron strains on untreated waxy cornstarch (WCS), branching enzyme-treated WCS (BE WCS), or branching enzyme plus β-amylase-treated WCS (BEBA WCS). Percentages of total α-1,6 linkages and average molecular weights are shown for each substrate. (D, E) Normalized doublings per hour (D) and normalized lag times (E) from growth assays were calculated as described in the legend to Fig. 2 and Materials and Methods. Average results and standard errors across three replicates are shown. Statistically significant differences versus growth of the wild type were calculated using the one-tailed unpaired Student’s t test. Download
Maltooligosaccharide concentrations in amylopectin (ng malto-OS per μg total carbohydrate) pre- and postdialysis by high-pH anion-exchange chromatography (HPAEC).
Maltooligosaccharide concentrations in B. thetaiotaomicron supernatants (ng malto-OS per μl).
Oligonucleotides used in this study.