ABSTRACT
N-Acetylglucosamine (GlcNAc), the main component of chitin and a major constituent of bacterial peptidoglycan, is present only in trace amounts in plants, in contrast to the huge amount of various sugars that compose the polysaccharides of the plant cell wall. Thus, GlcNAc has not previously been considered a substrate exploited by phytopathogenic bacteria during plant infection. Xanthomonas campestris pv. campestris, the causal agent of black rot disease of Brassica plants, expresses a carbohydrate utilization system devoted to GlcNAc exploitation. In addition to genes involved in GlcNAc catabolism, this system codes for four TonB-dependent outer membrane transporters (TBDTs) and eight glycoside hydrolases. Expression of all these genes is under the control of GlcNAc. In vitro experiments showed that X. campestris pv. campestris exploits chitooligosaccharides, and there is indirect evidence that during the early stationary phase, X. campestris pv. campestris recycles bacterium-derived peptidoglycan/muropeptides. Results obtained also suggest that during plant infection and during growth in cabbage xylem sap, X. campestris pv. campestris encounters and metabolizes plant-derived GlcNAc-containing molecules. Specific TBDTs seem to be preferentially involved in the consumption of all these plant-, fungus- and bacterium-derived GlcNAc-containing molecules. This is the first evidence of GlcNAc consumption during infection by a phytopathogenic bacterium. Interestingly, N-glycans from plant N-glycosylated proteins are proposed to be substrates for glycoside hydrolases belonging to the X. campestris pv. campestris GlcNAc exploitation system. This observation extends the range of sources of GlcNAc metabolized by phytopathogenic bacteria during their life cycle.
IMPORTANCE
Despite the central role of N-acetylglucosamine (GlcNAc) in nature, there is no evidence that phytopathogenic bacteria metabolize this compound during plant infection. Results obtained here suggest that Xanthomonas campestris pv. campestris, the causal agent of black rot disease on Brassica, encounters and metabolizes GlcNAc in planta and in vitro. Active and specific outer membrane transporters belonging to the TonB-dependent transporters family are proposed to import GlcNAc-containing complex molecules from the host, from the bacterium, and/or from the environment, and bacterial glycoside hydrolases induced by GlcNAc participate in their degradation. Our results extend the range of sources of GlcNAc metabolized by this phytopathogenic bacterium during its life cycle to include chitooligosaccharides that could originate from fungi or insects present in the plant environment, muropeptides leached during peptidoglycan recycling and bacterial lysis, and N-glycans from plant N-glycosylated proteins present in the plant cell wall as well as in xylem sap.
INTRODUCTION
During their life cycle, bacteria must adapt to several environments. To cope with environments containing few nutrients, Gram-negative bacteria have selected efficient transport systems with TonB-dependent transporters (TBDTs) for nutrient uptake through the outer membrane. Originally identified for its role in high-affinity transport of cobalamin and iron-siderophore complexes (1), the TonB machinery, consisting of three inner membrane energy-coupling TonB-ExbB-ExbD proteins and TBDT proteins, is now known for its role in uptake of a large variety of substrates, such as carbohydrates and organic acids (2–10). In marine environments, TBDTs are the dominant membrane proteins identified (11) and are associated with adaptation of bacteria populations to the phytoplankton blooms (12). In human gut symbiont Bacteroides species, TBDTs interact with carbohydrate-binding proteins and therefore participate in the binding of substrates to the outer membrane (13). In Xanthomonas campestris pv. campestris, the causal agent of black rot disease in Brassica plants (14), SuxA TBDT, which is involved in sucrose uptake, is an important pathogenicity determinant (4).
Surveys of TBDT genes present in sequenced bacterial genomes have shown that around 70% contain 1 to 15 TBDT genes, whereas around 15% possess more than 30 TBDT genes (4, 15). Bacteria displaying TBDT overrepresentation all have the ability to exploit complex carbohydrates (4). Among these, Bacteroides thetaiotaomicron adapts the expression of TBDT and degradative enzyme genes to forage either polysaccharides from the diet or host mucus glycans (16). Interestingly, genes coding for TBDTs involved in carbohydrate or polysaccharide utilization are often linked to genes required for sensing, binding, degradation, and transport through the bacterial envelope. These genetic loci have been termed carbohydrate utilization loci containing TBDT (CUT) in X. campestris pv. campestris (4) and polysaccharide utilization loci (PUL) in Bacteroides species (17).
In X. campestris pv. campestris, we previously identified and characterized a CUT system devoted to the exploitation of N-acetylglucosamine (GlcNAc) (8). This system comprises genes coding for (i) a cytoplasmic pathway with NagA, NagB-II, NagK-IIA, and NagK-IIB enzymes, which are involved in GlcNAc catabolism; (ii) the transcriptional repressors NagQ and NagR; (iii) the GlcNAc inner membrane transporter NagP; and (iv) four TBDTs (NixA, -B, -C, and -D [N-acetylglucosamine induced in Xanthomonas]) (8) (Fig. 1A). Expression of all genes belonging to this CUT system is induced in the presence of GlcNAc through NagQ and NagR inactivation (8). Although the expression of Nix TBDTs is under the control of GlcNAc, single mutants are not altered in GlcNAc uptake, indicating that none of these transporters is a major GlcNAc transporter (8).
FIG 1 .
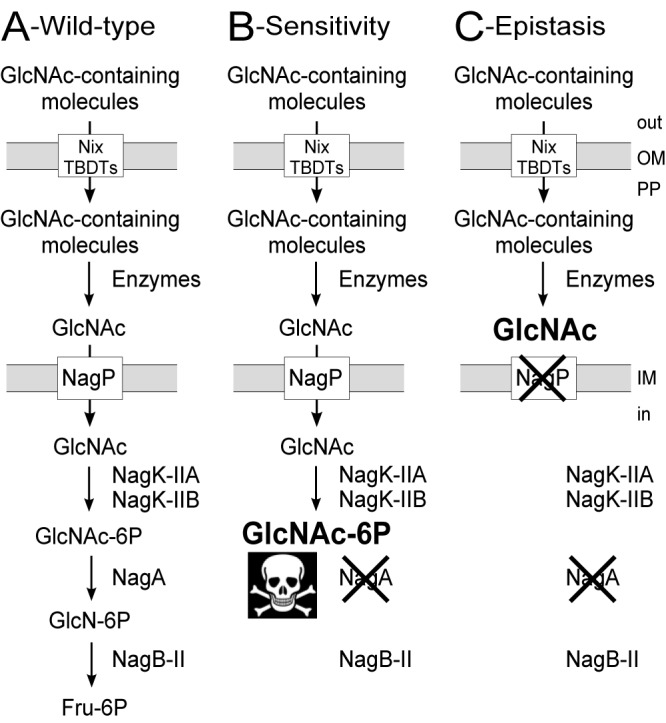
Schematic representation of the proposed N-acetylglucosamine (GlcNAc) utilization pathway in X. campestris pv. campestris. (A) Wild-type strain. (B) ΔnagA mutant. The nagA phenotype is illustrated, i.e., sensitivity to GlcNAc due to accumulation of GlcNAc-6P, which is toxic for X. campestris pv. campestris. (C) ΔnagA ΔnagP double mutant. This mutant displays epistasis—loss of sensitivity of the ΔnagA mutant due to a secondary mutation in a gene coding for a protein located upstream in the GlcNAc utilization pathway. There is no accumulation of GlcNAc-6P. In this example, the secondary mutation (ΔnagP) leads to the nontoxic accumulation of GlcNAc in the periplasm (PP). GlcN, glucosamine; Fru, fructose; OM, outer membrane; IM, inner membrane; TBDT, TonB-dependent transporter; Nix, N-acetylglucosamine induced in Xanthomonas.
X. campestris pv. campestris strains with mutations in the GlcNAc deacetylase gene nagA display impairment in growth in the presence of GlcNAc (8). This GlcNAc sensitivity phenotype (or nagA phenotype), previously observed in Escherichia coli and defined as an inhibition of growth by GlcNAc even in the presence of other carbon sources, is probably due to GlcNAc-6P accumulation (Fig. 1B) (18, 19). Deletion of either the inner membrane transporter gene (nagP) or the two GlcNAc kinase genes (nagK-IIA and nagK-IIB) is epistatic to this sensitivity in vitro (8). Indeed, these genes are located upstream of nagA in the GlcNAc utilization pathway and lead to the nontoxic accumulation of GlcNAc in the periplasm or inside the bacterial cell (see Fig. 1C for epistasis by deletion of nagP). Therefore, in this context, epistasis on nagA mutants can be defined as the loss of GlcNAc sensitivity due to the blocking of the GlcNAc utilization pathway upstream of NagA, leading to the absence of entry of GlcNAc into the cell or to the accumulation of a nontoxic compound (no accumulation of GlcNAc-6P).
GlcNAc has important structural roles as the monomer unit of chitin which composes the exoskeleton of crustaceans and insects and the cell wall of fungi. It is also a key component of bacterial cell wall peptidoglycan and of the extracellular matrix of animal cells. In addition to these structural roles, GlcNAc is important for protein function and location in both mammals and plants as a constituent of N- and O-glycans of glycosylated proteins and is also involved in cell signaling (20). Despite its central role, to our knowledge, GlcNAc was never considered as a putative substrate that could be exploited by phytopathogenic bacteria during infection of host plants. Given the presence of the CUT system in X. campestris pv. campestris, we investigated whether this pathogen does indeed take advantage of GlcNAc during infection. By using the GlcNAc-sensitive ΔnagA mutant and performing epistasis experiments, we indirectly showed that Nix TBDTs are involved in specific uptake of GlcNAc-containing molecules derived from the plant, from the bacterium, and from other organisms present in the plant environment, such as insects or fungi. We also identified a cluster of genes coding for glycoside hydrolases that are predicted to be involved in N-glycan degradation, suggesting that the origin of plant GlcNAc could be N-glycans from plant N-glycoproteins.
RESULTS
A GlcNAc-sensitive ΔnagA mutant is altered in pathogenicity.
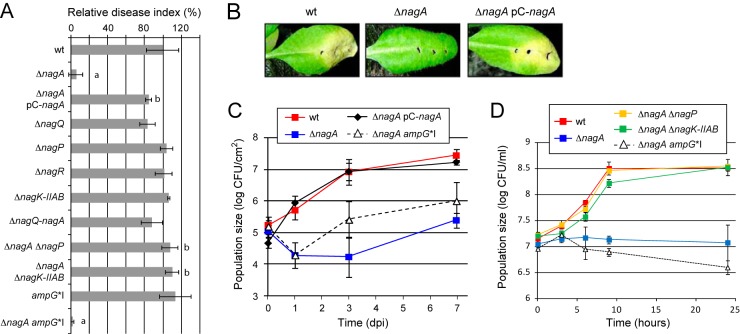
Deletion mutants of genes belonging to the GlcNAc CUT system were tested for pathogenicity by wound inoculation on the susceptible plant Arabidopsis thaliana ecotype Sf-2 (Fig. 2A and B; also, see Table S1 in the supplemental material). Symptoms were analyzed by scoring disease index (from 0 [no symptoms] to 4 [leaf death]), and results were given as the percentage of the value for the reference strain at 8 days postinoculation (dpi). Surprisingly, the nagA deletion mutant (ΔnagA strain) was affected in disease development with around 5 to 10% of symptom intensity retained compared to the wild-type (wt) strain. Complementation of nagA deletion by expression of nagA in trans on a plasmid (pC-nagA) restored symptom development (Fig. 2A and B).
FIG 2 .
The X. campestris pv. campestris ΔnagA mutant is altered in pathogenicity on Arabidopsis thaliana and during growth in xylem sap from cabbage. X. campestris pv. campestris strains (OD600 = 0.05) were inoculated onto A. thaliana ecotype Sf-2 leaves by piercing. (A) Disease index scored at 8 dpi (days postinoculation) and presented as a percentage of the value for the wild-type (wt) strain. Standard deviations were obtained from at least 16 inoculated leaves from four plants. Values significantly different (P < 0.0001) from those for the wt strain (a) and the ΔnagA mutant (b) are indicated. (B) Black rot symptoms observed on leaves at 8 dpi. (C) IGC in the inoculated area. Bacteria were extracted from inoculated leaves at different dpi and counted by plating serial dilutions. Results are given as CFU per cm2 of leaves. Standard deviations were obtained from at least 3 independent samples. (D) Growth of X. campestris pv. campestris strains in xylem sap from Brassica oleracea cv. Bartolo F1 cabbages (OD600 = 0.01 at time zero). At different time points, CFU were counted by plating serial dilutions. Standard deviations were obtained from at least 3 independent experiments.
Deletion of nagQ, nagP, nagR, or nagK-IIAB did not significantly affect disease development, suggesting that on its own, the GlcNAc utilization pathway is not required for symptom development. The ΔnagQ-nagA mutant, which contains deletions of nagQ, nagP, nagR, nagB-II, and nagA, was also not altered in pathogenicity (Fig. 2A), suggesting that the pathogenicity phenotype observed for nagA mutants is intrinsic to nagA and probably linked to GlcNAc sensitivity. This was confirmed by deletion of nagP or nagK-IIAB in the ΔnagA background. Indeed, these deletions restored disease development on A. thaliana (Fig. 2A; also, see Table S1 in the supplemental material). Therefore, the inner membrane transporter and the two GlcNAc kinases, which are located upstream of NagA in the GlcNAc utilization pathway (Fig. 1), are epistatic to the phenotype observed for the nagA mutant in planta. This suggests that the absence of symptoms observed for the ΔnagA strain reflects sensitivity to GlcNAc and/or GlcNAc-containing molecules encountered in planta during the infection process. In regard to substrate specificities of both NagP and GlcNAc kinases (NagK-IIA and NagK-IIB) (8), sensitivity observed for the ΔnagA strain is probably not due to another compound that could be present in planta.
Population size of the ΔnagA strain in planta decreased after inoculation compared to that of the wt strain (Fig. 2C). After 7 days, the population size obtained for the ΔnagA mutant was around 150 times lower than that of wt strain. Growth was restored when nagA was supplemented in trans with the plasmid pC-nagA.
Growth of the ΔnagA mutant is altered in xylem sap.
X. campestris pv. campestris is a bacterium that enters and colonizes xylem vessels through leaf wounds or hydathodes (21). A recent analysis of X. campestris pv. campestris adaptation to xylem sap (XS) from Brassica oleracea cv. Bartolo F1 cabbages demonstrate that XS is a good tool to study in vitro early infection steps (22). We therefore checked whether GlcNAc encountered by X. campestris pv. campestris during infection could be found in XS by analyzing the GlcNAc sensitivity phenotype of the ΔnagA mutant grown in XS (Fig. 2D; also, see Table S2 in the supplemental material). Compared to the wt strain, growth of the ΔnagA mutant was significantly impaired after transfer into XS. Deletion of either nagP or nagK-IIAB in the ΔnagA background restored growth to levels equivalent or close to those reached by the wt strain. This epistatic effect suggests that the phenotype observed for nagA mutants in XS is probably due to GlcNAc sensitivity.
Interestingly, GC-MS analysis of XS compounds did not allow the identification of free GlcNAc (22), suggesting that if GlcNAc is present in XS, the free GlcNAc concentration is below 1 µM. Sensitivity to GlcNAc of the ΔnagA strain is observed for concentrations above 5 µM (8). Therefore, sensitivity observed during growth in XS is probably due not to free GlcNAc but rather to GlcNAc-containing complex molecules.
GlcNAc is generated during stationary growth phase.
To investigate whether GlcNAc sensitivity observed in planta and in XS was due at least in part to GlcNAc that originates from the bacterium, we analyzed the survival of the ΔnagA mutant grown in minimal medium for 48 h (Fig. 3). To compare X. campestris pv. campestris growth in XS with growth in minimal medium, we developed MMC minimal medium (0.5% Casamino Acids), which is derived from MME minimal medium (0.15% Casamino Acids). In MMC medium, log phase and entry into stationary phase are comparable to what is observed during growth in XS (data not shown). Therefore, MMC minimal medium was used to study epistasis during the different growth phases.
FIG 3 .
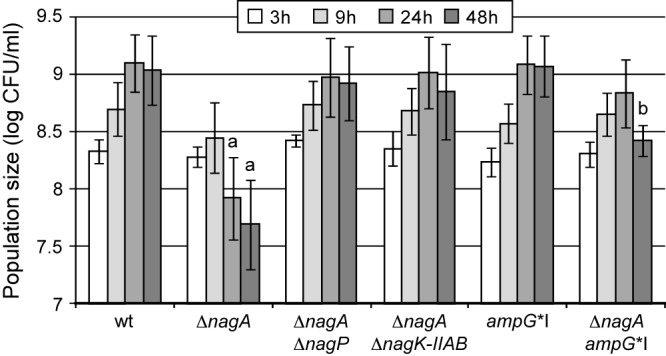
Growth of the X. campestris pv. campestris ΔnagA mutant is altered at the entry into stationary phase. X. campestris pv. campestris strains (OD600 = 0.1 at time zero) were grown in MMC minimal medium for 48 h at 30°C. At different time points, bacterial populations were measured by plating serial dilutions, and results are given as log CFU/ml. Standard deviations were obtained from at least three independent experiments. Values significantly different (P < 0.0001) from those for the wt strain (a) and both the wt and ΔnagA strains (b) are indicated.
Between 3 and 9 h of growth in MMC minimal medium (log phase), ΔnagA and wt strains showed similar growth. This result suggests that during these first 9 h of growth, the ΔnagA strain does not display GlcNAc sensitivity in this medium. Therefore, sensitivity observed from 6 h in XS is probably due to GlcNAc-containing molecules that are specific to the plant. In contrast, from 24 to 48 h, when growth of the wt strain slows down (stationary phase), the ΔnagA mutant displayed a significant decrease in bacterial population. Mutation of either nagP or nagK-IIAB in ΔnagA background restored growth suggesting a link between the phenotype observed for the ΔnagA mutant during growth in vitro and GlcNAc. The MMC minimal medium used in these experiments contains no GlcNAc (see Materials and Methods). Therefore, we can speculate that GlcNAc, which could be responsible for the decrease in the bacterial population observed for the ΔnagA strain from 24 h of growth in MMC minimal medium, may originate from the bacterium.
In X. campestris pv. campestris, GlcNAc is a major component of the peptidoglycan (PG) (23), and in E. coli and probably in other Gram-negative bacteria, almost 50% of the PG is broken down and recycled at each generation (24, 25). In E. coli, PG degradation products, i.e., anhydromuropeptides, are transported across the inner membrane into the cytoplasm via AmpG permease (25, 26). A protein showing homology to AmpG is encoded in X. campestris pv. campestris by XCC3845 (34% identity and 52% similarity at the amino acid level with the E. coli AmpG permease). In contrast, X. campestris pv. campestris does not encode homologs of other transporters of PG recycling products in E. coli (MurP, MppA, and Opp [27]). We therefore analyzed the role of X. campestris pv. campestris AmpG in the uptake of bacterium-derived GlcNAc-containing molecules by performing epistasis experiments on the ΔnagA mutant during growth in MMC minimal medium. Interestingly, bacterial growth of the X. campestris pv. campestris ΔnagA ampG*I double mutant in MMC minimal medium was comparable to the growth observed for the wt strain up to 24 h (Fig. 3), suggesting that from 9 to 24 h (entry into stationary phase), ampG is epistatic to nagA in MMC minimal medium. Therefore, at least part of the GlcNAc sensitivity observed in vitro in minimal medium is due to GlcNAc-containing molecules transported by AmpG and thus probably generated during PG recycling. At 48 h, ampG was partially epistatic to nagA, suggesting that at this time point, bacterium-derived GlcNAc is probably at least in part transported through another inner membrane transporter.
PG recycling cannot completely explain the in planta phenotype of nagA mutants.
As described above, AmpG is proposed to be involved in PG recycling in X. campestris pv. campestris. To check whether PG recycling could be responsible for the release of GlcNAc in planta or during growth in XS, we analyzed the epistasis of the AmpG permease to the nagA phenotype under both sets of conditions. Surprisingly, disease index and growth in XS were similar for ΔnagA ampG*I and ΔnagA mutants, suggesting that AmpG was not epistatic in planta and during growth in XS (Fig. 2A and D, respectively). However, bacterial population densities observed in planta suggest that AmpG was at least partially epistatic (Fig. 2C). These results indirectly suggest that although part of the GlcNAc sensitivity observed in planta could be attributed to bacterium-derived GlcNAc, i.e., peptidoglycan recycling, it cannot completely explain the phenotypes of the ΔnagA mutant observed in planta and in XS.
The nagA phenotype observed in planta probably reflects an addition of sensitivity to GlcNAc molecules that originate from different compounds. Mutation of ampG is presumed to limit entry of only bacterium-derived GlcNAc-containing molecules from PG, and therefore, in planta or during growth in XS, GlcNAc-containing molecules from other origins could still induce sensitivity. Conversely, during growth in minimal medium, sensitivity is due only to bacterium-derived GlcNAc-containing molecules, and mutation of ampG would abolish entry of the main source of GlcNAc-containing molecules. This could explain epistasis observed with ampG in vitro, whereas almost no epistasis was observed in planta or during growth in XS.
Nix TBDTs are epistatic to the nagA phenotype in planta and during growth in XS.
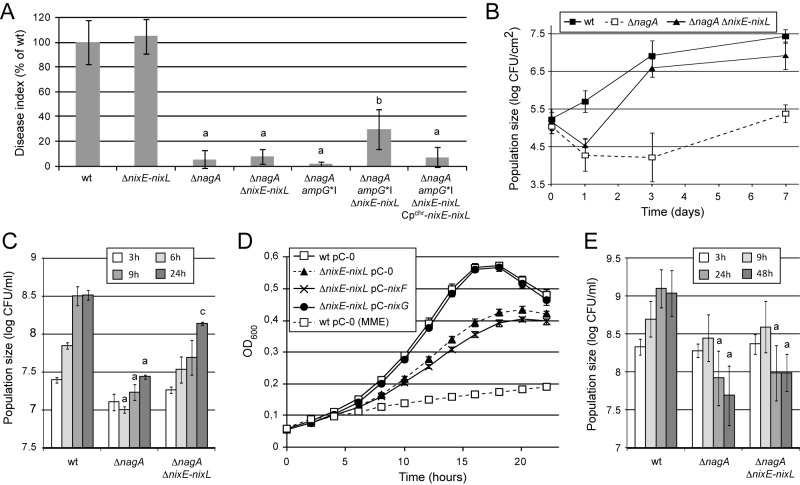
Individual and multiple deletions of the four nix TBDT genes did not affect pathogenicity of the strain (see Table S1A in the supplemental material). However, the quadruple deletion mutant in the ΔnagA background led to an epistasis in planta, as symptoms were partially restored (Fig. 4A) and bacterial growth was nearly restored in Arabidopsis (Fig. 4B). Similarly, an important epistasis was observed during growth in XS from cabbages (Fig. 4C). Interestingly, [14C]GlcNAc transport was not affected in the ΔnixABCD mutant compared to the wt strain (Table 1), and nix TBDTs were not epistatic to the nagA sensitivity phenotype when the strain was grown in minimal medium containing free GlcNAc (see Table S3 in the supplemental material). These results suggest that Nix TBDTs are involved in the uptake not of free GlcNAc but rather of GlcNAc-containing complex molecules.
FIG 4 .
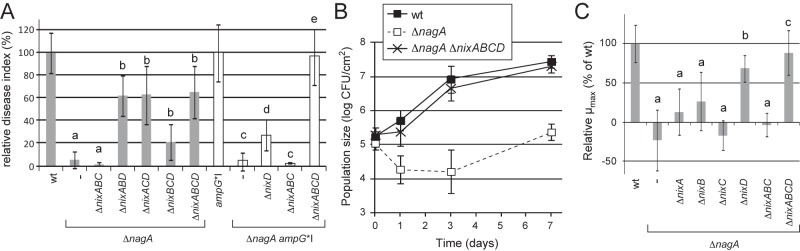
Nix TonB-dependent transporters of X. campestris pv. campestris are epistatic to the nagA phenotype during plant infection and growth in cabbage xylem sap. (A and B) X. campestris pv. campestris strains (OD600 = 0.05) were inoculated onto leaves of Arabidopsis thaliana Sf-2 ecotype by piercing. (A) Disease index scored at 8 days postinoculation (dpi), presented as percentages of the value for the wt strain (gray) or the ampG*I strain (white). Standard deviations were obtained from at least 16 leaves from 4 plants. Values significantly different (P value <0.0001) from those for the wt strain (a; sensitivity), both the wt and ΔnagA strains (b; partial epistasis), the ampG*I strain (c; sensitivity), the ΔnagA ampG*I and ampG*I strains (d; partial epistasis), and the ΔnagA ampG*I strain (e; total epistasis) are indicated. (B) IGC in the inoculated area. Bacteria were extracted from infected leaves at different dpi and counted by plating serial dilutions. Standard deviations were obtained from at least 3 independent samples. (C) Epistasis on the ΔnagA mutant during growth in xylem sap from Brassica oleracea cv. Bartolo F1 cabbages (OD600 = 0.01 at time zero). At different time points, CFU were counted by plating serial dilutions. Maximal specific growth rates (µmax) were calculated between 3 and 9 h of growth from CFU counts. Standard deviations were obtained from at least three independent experiments. Values significantly different (P < 0.0001) from those for the wt strain (a; sensitivity), both the wt and ΔnagA strains (b; partial epistasis), and the ΔnagA strain (c; total epistasis) are indicated.
TABLE 1 .
Rates of 14C-labeled-N-acetylglucosamine transport in X. campestris pv. campestris mutants compared to the wild-type (wt) straina
| Strain genotype | Gene ID | Transport (% of wt)b |
|---|---|---|
| wt | 100.0 ± 10.1 | |
| ΔnixA | XCC3408 | 84.6 ± 9.1 |
| ΔnixB | XCC0531 | 91.4 ± 2.9 |
| ΔnixC | XCC2944 | 91.6 ± 4.7 |
| ΔnixD | XCC2887 | 94.0 ± 5.5 |
| ΔnixABCD | 90.4 ± 5.6 |
Transport rates were measured 60 min after addition of 14C-labeled N-acetylglucosamine.
Values are means and standard deviations, which were calculated from six independent experiments.
Single deletions of nix TBDT genes were not epistatic to the nagA phenotype (see Table S1B in the supplemental material). Interestingly, among double and triple deletion mutants, only deletions containing nixD (i.e., ΔnixAD, ΔnixBD, ΔnixCD, ΔnixABD, ΔnixACD, and ΔnixBCD mutants) resulted in partial epistasis on the nagA phenotype. Conversely, no epistasis was observed with the ΔnixAB, ΔnixAC, ΔnixBC, and ΔnixABC mutants (Fig. 4A; also, see Table S1B in the supplemental material). Therefore, NixD seems to play a major role in the uptake of GlcNAc-containing molecules in planta. Similarly, in the ΔnagA ampG*I background, partial epistasis was observed when nixD was deleted (Fig. 4A), and almost total epistasis was observed when both nixA and nixD were deleted (see Table S1D in the supplemental material), suggesting that NixA also participates in the uptake of GlcNAc-containing molecules in planta.
In XS, partial epistasis on the ΔnagA mutant was observed when nixD was deleted (Fig. 4C), and epistasis was complete when either nixA, nixB, or nixC was deleted in the ΔnixD mutant (ΔnixAD, -BD, and -CD in the ΔnagA background) (see Table S2 in the supplemental material) suggesting that in XS, in addition to NixD, NixA, -B, and -C also contribute to the uptake of GlcNAc-containing molecules.
NixC is epistatic to the nagA phenotype upon entry into stationary phase.
As described above, there is indirect evidence that Nix TBDTs are involved in the uptake of GlcNAc-containing molecules during infection. These molecules could at least in part originate from bacterial PG recycling. Therefore, we analyzed the role of each Nix TBDT in the uptake of GlcNAc-containing molecules during entry into stationary phase by performing epistasis experiments during growth in MMC minimal medium. Interestingly, upon entry into stationary phase, epistasis on the ΔnagA strain was complete when nixC was deleted (Fig. 5A), whereas no epistasis was observed when nixA, nixB, and nixD were deleted individually (data not shown) or simultaneously (Fig. 5A). This suggests that NixC plays a major role in the uptake of GlcNAc-containing molecules derived from the bacterium.
FIG 5 .
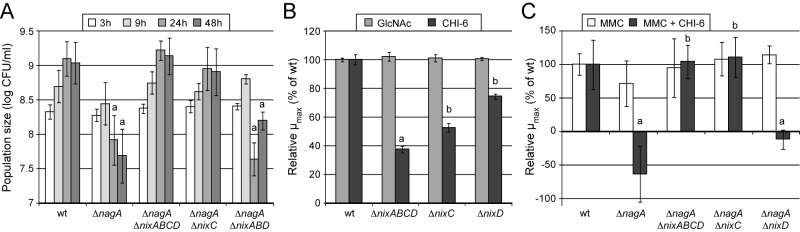
The X. campestris pv. campestris TonB-dependent transporter NixC is epistatic to the nagA phenotype during growth in MMC minimal medium and with NixD is involved in chitooligosaccharide uptake. (A) Epistasis on the ΔnagA mutant during growth in MMC minimal medium. Bacterial populations were measured by plating serial dilutions, and results are given as log CFU/ml. Standard deviations were obtained from at least three independent experiments. Values significantly different (P < 0.0001) from that of the wt strain (a; sensitivity) are indicated. (B) Growth of X. campestris pv. campestris strains in MME minimal medium containing N-acetylglucosamine (GlcNAc) or chitohexaose (CHI-6) at 0.1% (wt/vol). Results are given as maximal specific growth rates (µmax) relative to the wild-type (wt) strain. Specific µmax were calculated during the log phase of the wt strain in GlcNAc-containing medium (between 7 and 12 h) based on OD600 values. Standard deviations were obtained from at least three independent experiments. Values significantly different (P < 0.0001) from that of the wt strain (a) and both the wt and ΔnixABCD strains (b) are indicated. (C) Epistasis on the ΔnagA mutant during growth in MMC minimal medium or MMC containing chitohexaose at 100 µM (MMC + CHI-6). Results are given as specific µmax relative to the wt strain. Specific µmax were calculated between 3 and 9 h of growth from CFU counts. Standard deviations were obtained from at least three independent experiments. Values significantly different (P < 0.0001) from that of the wt strain (a; sensitivity) and the ΔnagA strain (b; total epistasis) are indicated.
NixC and NixD are involved in the uptake of chitooligosaccharides.
In the environment, chitin constitutes a natural source of GlcNAc for chitinolytic bacteria (28, 29). X. campestris pv. campestris is not chitinolytic (8), but it is able to use chitooligosaccharides (CHI) from a degree of polymerization of 2 (CHI-2; chitobiose) (8) to one of 6 (CHI-6; chitohexaose) (see Table S4A in the supplemental material). NagP and NagKII-AB proteins are involved in chitooligosaccharide utilization, as strains with mutations in the corresponding genes had altered growth on these substrates (see Table S4B in the supplemental material). Growth of the ΔnixABCD strain on chitooligosaccharides was impaired (Fig. 5B; also, see Table S4), suggesting that Nix TBDTs are involved in the uptake of chitooligosaccharides. Deletion of nixA or nixB did not affect growth with any of the tested chitooligosaccharides (see Table S4A), and reintroduction of either nixA or nixB into the genome failed to complement the ΔnixABCD strain (see Table S4B). We observed a slight reduction of growth for the ΔnixD mutant on all chitooligosaccharides tested (Fig. 5B; also, see Table S4A), and growth was complemented when nixD was reintroduced in the ΔnixABCD mutant (see Table S4B). Interestingly, the ΔnixC mutant exhibited a significant growth defect on chitohexaose (CHI-6) (Fig. 5B) but not other chitooligosaccharides (see Table S4A). However, growth of the ΔnixABCD strain in the presence of the tested chitooligosaccharides was restored when nixC was reintroduced in the genome (see Table S4B, data for the ΔnixABCD Cpchr-nixC strain). This suggests that the absence of phenotype for ΔnixC on CHI-2 to CHI-5 is due to functional redundancy with nixD. These results were confirmed by epistasis experiments using the ΔnagA mutant during growth in the presence of chitooligosaccharides. Indeed, among single-deletion mutants, only nixC was epistatic to nagA on chitohexaose (Fig. 5C; also, see Table S2). Therefore, both NixC and NixD transport chitooligosaccharides ranging from CHI-2 to CHI-6, but NixC seems to be the major transporter of large molecules (CHI-6).
Altogether, these results are consistent with the idea that NixC is the primary transporter of bacterium-derived GlcNAc-containing molecules and large chitooligosaccharides. NixD also contributes to chitooligosaccharide uptake, whereas neither NixA nor NixB is involved in bacterium-derived GlcNAc-containing molecules and chitooligosaccharide transport. In contrast, during the infection process in planta and during growth in XS, GlcNAc-containing molecules are mainly transported via NixD with a contribution of NixA. NixB and NixC also participate in the uptake of GlcNAc-containing molecules during growth in XS. This specific uptake profile observed for Nix TBDTs suggests that at least part of the GlcNAc-containing molecules encountered in planta and in XS are different from bacterium-derived and chitooligosaccharide molecules and are probably plant-specific GlcNAc-containing molecules.
Plant-specific GlcNAc-containing molecules could be N-glycans from plant N-glycoproteins.
We identified genes coding for several glycoside hydrolases (XCC2888 to XCC2895) (Fig. 6A) downstream of and in the same orientation as nixD (XCC2887). These genes belong to the GlcNAc and NagR regulons, as determined by their expression profile. Indeed, the expression of XCC2888 to XCC2895 and of XCC2888 to XCC2891 was induced in the presence of GlcNAc and in a ΔnagR background, respectively (Table 2). Consequently, these genes were renamed nixE to nixL, in accordance with the nomenclature of nix TBDT genes (8). Furthermore, the expression profiles of these genes in nixD, nixE, nixG, and nixI pVO155 polar insertion mutants (see Fig. S1 in the supplemental material) suggest that nixD to nixL form an operon with a putative promoter localized upstream of nixG. Indeed, a residual expression of genes nixG to nixL was detected in the nixD*I and nixE*I mutants, which was lost in the nixG*I and nixI*I mutants (see Fig. S1).
FIG 6 .
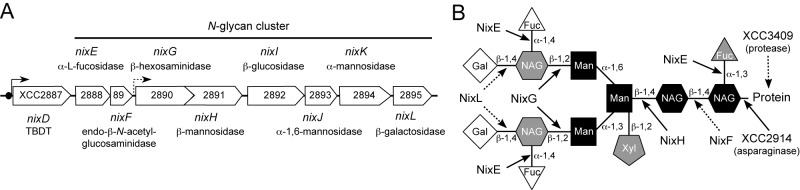
N-Glycans from plant N-glycosylated proteins can be substrates for Nix glycoside hydrolases encoded by the X. campestris pv. campestris N-glycan cluster. (A) Genetic organization of genes belonging to the N-glycan cluster. Black arrows indicate transcription start sites. The dashed black arrow represents a putative transcription start identified in this study. TBDT, TonB-dependent transporter. (B) Proposed cleavage sites by Nix enzymes encoded by the N-glycan cluster on a plant complex-type N-glycan with Lewisa epitopes. Residues conserved in all N-glycans are in black, residues found in all complex-type N-glycans are in gray, and residues specific to complex-type N-glycans with Lewisa epitopes are in white. Cleavage sites proposed in the KEGG pathway database (http://www.genome.jp/kegg-bin/show_pathway?org_name=xcc&mapno=00511&mapscale=&show_description=show) are depicted by solid arrows, whereas cleavage sites proposed in the present study are depicted by dotted arrows. Fuc, fucose; Gal, galactose; Man, mannose; NAG, N-acetylglucosamine; Xyl, xylose.
TABLE 2 .
Relative expression ratios of X. campestris pv. campestris nixE to nixL and aspG genesa
| Gene ID | Gene name | Expression ratio (mean ± SD) |
|
|---|---|---|---|
| wt with GlcNAc/wt in MME | ΔnagR in MME/wt in MME | ||
| XCC2888 | nixE | 77 ± 24b | 31.6 ± 9.75b |
| XCC2889 | nixF | 50.3 ± 19.8b | 23.8 ± 2.5b |
| XCC2890 | nixG | 4.36 ± 1.02b | 2.8 ± 1.3b |
| XCC2891 | nixH | 3.04 ± 0.4b | 2.1 ± 0.2b |
| XCC2892 | nixI | 6.64 ± 0.88b | 2.57 ± 0.99 |
| XCC2893 | nixJ | 2.72 ± 1.4 | 0.95 ± 0.26 |
| XCC2894 | nixK | 3.17 ± 0.7b | 1.6 ± 0.5 |
| XCC2895 | nixL | 3.64 ± 0.55b | 1.76 ± 0.7 |
| XCC2914 | aspG | 1 ± 0.01 | 0.9 ± 0.3 |
From real-time quantitative reverse transcriptase PCR performed in at least three independent experiments. Calculation of relative expression includes normalization against the 16S rRNA endogenous control gene. MME, minimal medium; GlcNAc, N-acetylglucosamine; wt: wild type.
The levels of expression under the conditions compared were significantly different (P < 0.05).
The putative function of NixE to NixL glycoside hydrolases is indicated in Fig. 6A. The KEGG pathway database proposes that the putative α-l-fucosidase (NixE), β-hexosaminidase (NixG), β-mannosidase (NixH), and asparaginase (AspG/XCC2914, which does not belong to the GlcNAc regulon, as determined by qRT-PCR) (Table 2) cleave glycosidic bonds found in N-glycans of N-glycoproteins (Fig. 6B). Furthermore, nixF encodes an enzyme with a putative chitinase and/or endo-β-N-acetylglucosaminidase activity. X. campestris pv. campestris strain ATCC 33913 is not able to use chitin as a carbon source (8), but this enzyme could be involved in the endohydrolysis of the N,N′-diacetylchitobiosyl units found in all N-glycans (Fig. 6B). nixK and nixJ encode putative α-mannosidases potentially involved in the cleavage of α-glycosidic bonds between two mannose residues. nixL codes for a β-galactosidase that could cleave terminal galactose residues found in some complex N-glycans (Fig. 6B). Finally, nixI encodes a protein belonging to the GH-3 family, which includes β-glucosidases. Its role in N-glycan degradation could not be determined. All these proteins possess a signal peptide, suggesting that they are all translocated through the inner membrane to the periplasm and/or to the extracellular environment. NixL is probably not secreted, as no β-galactosidase activity was detected in the culture supernatant of X. campestris pv. campestris strain Xc17 (30).
To distinguish between the locus comprising nag genes (GlcNAc cluster [8]) involved in GlcNAc catabolism and the locus of nix enzyme genes potentially involved in N-glycan degradation, we named the locus from nixE to nixL the N-glycan cluster (Fig. 6A).
Nix enzymes seem to participate in the release of GlcNAc in planta.
Similar to what is observed for the GlcNAc cluster, deletion of the entire N-glycan cluster did not affect the capacity of the strain to develop symptoms on A. thaliana (Fig. 7A), meaning that these enzymes are not important for disease symptoms under the tested conditions. To test whether the N-glycan cluster could participate in the consumption of GlcNAc-containing molecules in planta, we performed epistasis experiments on strains with the nagA phenotype. During plant infection, no epistasis on disease development was observed for the ΔnagA ΔnixE-nixL mutant (Fig. 7A). However, despite the absence of visible symptoms at 7 dpi (days postinoculation), growth was almost restored (Fig. 7B), suggesting that partial epistasis occurs. The 0.5-log difference in bacterial population between wt and ΔnagA ΔnixE-nixL strains at 7 dpi (Fig. 7B) could explain the difference in the appearance of symptoms (Fig. 7A). Deletion of this cluster in a ΔnagA ampG*I background (ΔnagA ampG*I ΔnixE-nixL strain) partially restored disease symptoms (Fig. 7A), and insertion of the entire N-glycan cluster elsewhere in the chromosome (ΔnagA ampG*I ΔnixE-nixL Cpchr-nixE-nixL complemented strain) restored the sensitivity (Fig. 7A). It is worth noting that the ΔnagA ampG*I ΔnixE-nixL strain was still sensitive to free GlcNAc in vitro, as its growth was comparable to the growth observed for the ΔnagA ampG*I mutant in medium containing GlcNAc (see Table S3 in the supplemental material). Furthermore, growth of the ΔnixE-nixL mutant on GlcNAc was comparable to that of the wt strain, suggesting that none of the enzymes encoded by this cluster play a major role in GlcNAc utilization (see Table S4). Deletion of this cluster partially restored growth of the ΔnagA mutant in XS at 24 h (Fig. 7C), and epistasis in XS was complete in the ΔnagA ampG*I background (see Table S2).
FIG 7 .
Nix glycoside hydrolases of X. campestris pv. campestris are partially epistatic to the nagA phenotype during plant infection and growth in cabbage xylem sap. (A and B) X. campestris pv. campestris strains (OD600 = 0.05) were inoculated by piercing leaves of Arabidopsis thaliana ecotype Sf-2. (A) Disease index scored at 8 days postinoculation. Results are given as percentages of the value for the wild-type (wt) strain. Standard deviations were obtained from at least 16 inoculated leaves from four plants. (B) IGC for the inoculated area. Results are given as CFU per cm2 of leaves. Standard deviations were obtained from at least 3 independent samples. (C) Growth of X. campestris pv. campestris strains in xylem sap from Brassica oleracea cv. Bartolo F1 cabbage (OD600 = 0.01 at time zero). (D) Growth of X. campestris pv. campestris strains in MME minimal medium containing chitotetraose (0.1% [wt/vol]). Standard deviations were obtained from at least three independent experiments. (E) Growth of X. campestris pv. campestris strains in MMC minimal medium (OD600 = 0.1 at time zero). (C and E) At different time points, CFU were counted by plating serial dilutions. Standard deviations were obtained from at least three independent experiments. Values significantly different (P < 0.0001) from that for the wt strain (a), both the wt and ΔnagA ampG*I strains (b), and the ΔnagA strain (c) are indicated.
The N-glycan cluster participates in chitooligosaccharide utilization, as growth of the ΔnixE-nixL mutant was altered on these substrates (Fig. 7D; also, see Table S4 in the supplemental material). However, growth observed was intermediate, suggesting that Nix enzymes only partially contribute to chitooligosaccharide utilization. This could explain the absence of epistasis of nixE-nixL on the nagA phenotype in the presence of chitooligosaccharides (see Tables S2 and S3). Two enzymes encoded by the N-glycan cluster could cleave β-N-acetylglucosamine links (i.e., NixF and NixG), but only NixG was able to complement bacterial growth of the ΔnixE-nixL mutant on chitotetraose (Fig. 7D) or chitobiose (see Table S4A).
Finally, no significant epistasis of nixE-nixL on the nagA phenotype was observed during bacterial growth in minimal medium (Fig. 7E).
The absence of epistasis at the early stationary phase and during growth in the presence of chitooligosaccharides while epistasis was observed both in planta and in XS indirectly suggests that enzymes belonging to the N-glycan cluster (NixE to NixL) are involved in the release of plant-specific GlcNAc during the infection process and during growth in XS.
DISCUSSION
X. campestris pv. campestris, the causal agent of black rot disease on Brassica, produces an extensive repertoire of glycoside hydrolases, lyases, and esterases (31) that target linkages present in plant cell wall polysaccharides such as cellulose, mannan, xylan, galacturonan, and pectin. Some of these enzymes contribute to pathogenicity by providing nutrients through degradation of plant cell wall polysaccharides and by facilitating bacterial spread in plant tissues (32). We previously identified in X. campestris pv. campestris a CUT system devoted to N-acetylglucosamine (GlcNAc) utilization (8). In addition to genes coding for two repressors, an inner membrane transporter, and enzymes involved in GlcNAc catabolism (lower pathway, in red in Fig. 8), we determined that this CUT system also codes for 4 TBDTs (NixA, -B, -C, and -D) and 8 glycoside hydrolases (NixE to NixL).
FIG 8 .
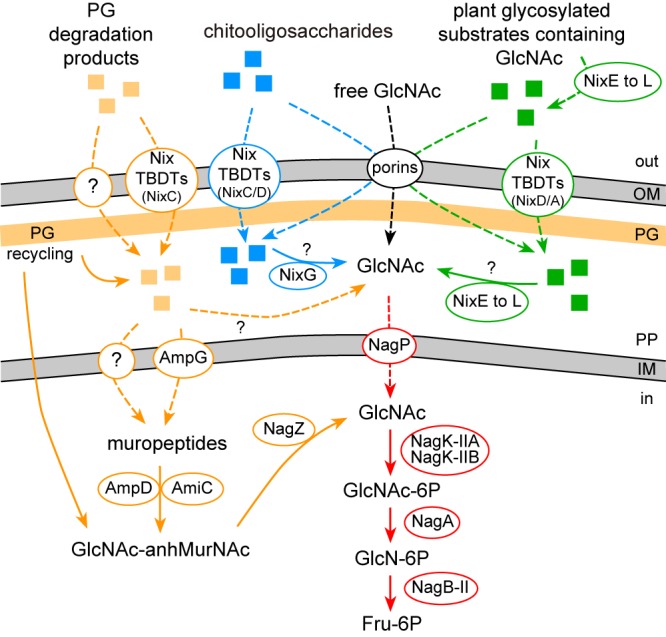
Schematic representation of utilization pathways for N-acetylglucosamine (GlcNAc) and GlcNAc-containing molecules in X. campestris pv. campestris. GlcNAc-containing molecules can originate from bacteria (peptidoglycan degradation products [orange]), from insects or fungi (chitooligosaccharides derived from chitin [blue]), or from plants (plant-glycosylated substrates [green]). The GlcNAc utilization pathway (uptake through NagP and catabolism) is depicted in red. Enzymatic reactions are designated by solid arrows, whereas dashed arrows indicate transport. The Nix TBDT(s) proposed to play a major role in the uptake of specific GlcNAc-containing molecules is indicated. OM, outer membrane; PG, peptidoglycan; PP, periplasm; IM, inner membrane; TBDT, TonB-dependent transporter; GlcN, glucosamine; Fru, fructose; GlcNAc-anhMurNAc, N-acetylglucosaminyl-1,6-anhydro-N-acetylmuramyl.
GlcNAc-sensitive nagA mutants: a tool to study the GlcNAc utilization pathway in X. campestris pv. campestris.
As observed for the xylan CUT system (9), the ability of X. campestris pv. campestris to metabolize GlcNAc is not required for pathogenicity under the conditions tested. However, mutants in the nagA gene coding for the GlcNAc deacetylase are severely impaired in pathogenicity and show a significant decrease in bacterial population due to GlcNAc sensitivity: the presence of GlcNAc or GlcNAc-containing molecules inhibits growth despite the presence of other carbon sources (Fig. 1B). The nagA phenotype was used in this study as a tool to analyze in X. campestris pv. campestris the GlcNAc utilization pathway upstream of the inner membrane transporter NagP, focusing on the role of Nix TBDT transporters and Nix glycoside hydrolases. GlcNAc has a central role in nature as a component of bacterial and fungal cell walls (peptidoglycan and chitin, respectively), insects and crustacean exoskeletons, and the extracellular matrix of animal cells. However, this molecule is not part of plant cell wall polysaccharide matrix and was never considered as a putative substrate that could be utilized by phytopathogens during infection of host plants. Results obtained in this study suggest that X. campestris pv. campestris encounters and metabolizes GlcNAc during plant infection and growth in XS. Nix TBDTs are proposed to be involved in the uptake of complex molecules containing GlcNAc derived from the plant, bacteria, fungi, and/or insects, whereas Nix enzymes mainly contribute to the generation of GlcNAc-containing molecules derived from the plant (Fig. 8).
Besides its usefulness as a tool to study the GlcNAc utilization pathway, the nagA phenotype observed in pathogenicity tests could be used like in vivo expression technology (IVET) (33) as a more generalized tool to study expression pattern of a large range of promoters in planta. Indeed, it is possible to clone X. campestris pv. campestris promoters upstream of the nagA gene in an expression plasmid. These plasmids could be introduced in a ΔnagA strain of X. campestris pv. campestris, and the resulting strains could be used to perform pathogenicity tests. Only bacteria expressing nagA from the plasmid (carrying an active promoter) in planta would be able to produce symptoms on plants.
Indirect evidence that GlcNAc is exploited during peptidoglycan catabolism.
When cultured in minimal medium, growth of the X. campestris pv. campestris ATCC 33913 nagA deletion mutant was severely impaired at the early stationary phase (Fig. 3). We attribute this phenotype to peptidoglycan recycling. Indeed, the AmpG inner membrane permease, involved in the uptake of muropeptides (peptidoglycan degradation products) in E. coli (25, 26), is epistatic to the nagA phenotype until 24 h of growth. Intriguingly, the GlcNAc inner membrane transporter NagP is also epistatic to the nagA phenotype during the early stationary phase. This observation suggests either that NagP could transport muropeptides or that the release of GlcNAc from muropeptides during PG recycling takes place at least part in the periplasm. Based on our results, the outer membrane transporter NixC suppresses the sensitivity of the ΔnagA mutant during early stationary phase. Thus, this TBDT could be involved in the uptake of PG degradation products. To our knowledge, this is the first evidence of a transporter involved in the uptake of PG degradation products through the outer membrane.
X. campestris pv. campestris exploits chitooligosaccharides.
X. campestris pv. campestris is an epiphytic bacterium that can live on leaf surfaces or as a saprophyte in necrotic tissues or in the ground (14). Thus, it could be in close proximity to and in competition with a large range of other microorganisms, like other bacteria or fungi, or even with insects. Therefore, although chitin is not a polymer produced by plants, it can be present in plant environments. X. campestris pv. campestris does not encode a chitinase, but chitin can be degraded into chitooligosaccharides by plant chitinases or by chitinases from chitinolytic microorganisms present in the plant environment. This work has shown that chitooligosaccharides are substrates of the X. campestris pv. campestris GlcNAc CUT system, with the TBDTs NixC and NixD being involved in the uptake of oligomers ranging from 2 to 6 GlcNAc residues, NixC being the major transporter for large oligomers (CHI-6). In Caulobacter crescentus, the TBDT encoded by CC0446 is involved in the passive uptake of GlcNAc and in the active uptake of chitooligosaccharides (6). NixA, which seems not to be involved in the uptake of chitooligosaccharides, has the highest sequence identity with CC0446 (40.7%). However, phylogenetic analysis suggests that NixC (37.8% identity) is the ortholog of CC0446 (see Fig. S2 in the supplemental material), which is in agreement with their role in uptake of chitooligosaccharides. The β-hexosaminidase NixG belonging to the X. campestris pv. campestris GlcNAc CUT system participates in the catabolism of chitooligosaccharides. Interestingly, the NixG ortholog in the Xanthomonadaceae member Stenotrophomonas maltophilia (Hex; 45.8% identity) was recently shown to be an exo-β-N-acetylhexosaminidase releasing GlcNAc from chitooligosaccharides up to hexamers (34). Growth of the ΔnixE-nixL strain on chitooligosaccharides was not abolished, suggesting that X. campestris pv. campestris encodes other β-N-acetylglucosaminidases that could degrade these substrates. In the carbohydrate-active enzyme database (CAZy; http://www.cazy.org), enzymes able to cleave GlcNAc residues can be found in classes EC 3.2.1.14 (chitinases; glycoside hydrolase families GH18, GH19, GH23, and GH48), EC 3.2.1.52 (β-N-acetylhexosaminidases; GH3, GH18, GH20, GH84, and GH116), and EC 3.2.1.96 (mannosyl-glycoprotein endo-β-N-acetylglucosaminidase; GH18, GH20, GH73, and GH85). Eighteen X. campestris pv. campestris proteins belong to these GH families, 3 of which (XCC2889, XCC2890, and XCC2892) are encoded by the N-glycan cluster. Further work is required to identify which X. campestris pv. campestris protein(s), not encoded by the N-glycan cluster, could participate in chitooligosaccharides utilization.
N-Glycans of N-glycoproteins: the plant source of GlcNAc?
Based on the predicted enzymatic activities of the glycosides hydrolases NixE (XCC2888) to NixL (XCC2895), we propose that plant-derived GlcNAc-containing complex molecules metabolized by X. campestris pv. campestris during plant infection could be N-glycans from N-glycosylated proteins. Although the characterization of these enzymes is still under way, this cluster was named the N-glycan cluster. Epistasis experiments on the nagA phenotype in planta confirm the involvement of this cluster in the generation of GlcNAc during plant infection. Interestingly, epistasis experiments indirectly suggest that nixD, which is located in the same operon as the N-glycan cluster, codes for the major transporter of GlcNAc-containing molecules in planta, although NixA, -B, and -C also contribute to the uptake of these molecules during growth in XS. A proteomic study performed on A. thaliana stems revealed that most N-glycosylated proteins are predicted to have signal peptides, allowing secretion to the cell wall or plasma membrane (35). Furthermore, most of the proteins identified in the XS of Brassica oleracea cv. Bartolo F1 were secreted (36). Therefore, N-glycosylated proteins could be accessible to X. campestris pv. campestris during the infection process as constituents of the plant cell wall but also directly in XS. Compared to the huge amount of cell wall polysaccharides, N-glycans represent only a minor proportion of sugar polymers in plants (37). To exploit these molecules, bacteria must be able to recognize and transport them with high specificity and high affinity, which is in agreement with the involvement of active transporters such as TBDTs. Indeed, CUT systems, including degradative enzymes and active outer membrane transporters belonging to the TBDT family, were proposed to be devoted to the scavenging of scarce nutrients (4).
Among bacteria associated with plants, the N-glycan cluster is specific to Xanthomonas and Xylella.
A genome survey of bacteria associated with plants (pathogenic, symbiotic, and plant-associated bacteria) showed that the presence of the N-glycan cluster is a common feature of all Xanthomonas strains, whether these bacteria are vascular (X. campestris pv. campestris) or mesophyll (X. campestris pv. vesicatoria) pathogens (see Table S5 in the supplemental material). The cluster is also present in Xanthomonas albilineans and in Xylella, a xylem-restricted member of the Xanthomonadaceae closely related to Xanthomonas (see Table S5). Both X. albilineans and Xylella displayed an important genome reduction (38). This shows the importance of this cluster for Xanthomonas and Xylella strains during their natural life cycle. Despite its high conservation among Xanthomonas and Xylella strains, the N-glycan cluster is absent in other phytopathogenic bacteria belonging to different genera (the α-proteobacterium Agrobacterium, the β-proteobacterium Ralstonia solanacearum, and the γ-proteobacteria Dickeya dadantii, Erwinia amylovora, and Pseudomonas syringae, for example), in nitrogen-fixing bacteria (such as Sinorhizobium meliloti and Burkholderia phenoliruptrix), and in plant growth-promoting or plant-associated bacteria (such as Azospirillum caulinodans and Pseudomonas fluorescens) (see Table S5). This observation raises the question of the role of these proteins during plant infection.
Deglycosylation: only for feeding?
In the human pathogens Streptococcus pneumoniae and Streptococcus oralis, monosaccharides released during the sequential degradation of complex-type N-glycans from human glycoconjugates are used as a source of fermentable carbon (39, 40). Although the oligosaccharide moiety could be the nutrient source, N-glycan-degradative enzymes could also contribute to the accessibility of the peptide part of the glycoprotein to bacterial proteases, resulting in an optimized nutrient supply. Indeed, glycoproteins become more sensitive to proteases after deglycosylation of N-linked oligosaccharides (41). A role for the X. campestris pv. campestris GlcNAc utilization pathway might be to improve the access to amino acids as nutrients. Another possibility is that degradation of glycoprotein might lead to a weakening of the plant cell wall, leading to a better accessibility of plant cell wall polymers to degradative enzymes. Finally, this cluster could have an impact in signalization by activating or inactivating the function of proteins from the host. Indeed, in eukaryotes, glycosylation plays a crucial role in the proper folding of the protein and in intracellular trafficking related to cell surface expression (42). Glycosylations also regulate many physiological and pathological events, including cell growth (43), cell migration and cancer progression (44, 45), differentiation (46), plant cell wall synthesis (47, 48), and host-pathogen interactions (49), including PAMP (pathogen-associated molecular pattern) recognition by plant PRRs (pattern recognition receptors) (50). Further investigations are now being undertaken to decipher the role of the N-glycan cluster in X. campestris pv. campestris during the epiphytic life and the infection process and to characterize Nix enzymes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The X. campestris pv. campestris strains and plasmids used in this study are listed in Table S6 in the supplemental material.
X. campestris pv. campestris cells were grown on MOKA rich medium (4), on MME minimal medium (51), or on MMC minimal medium [Casamino Acids, 0.5 g/liter; K2HPO4, 10.5 g/liter; KH2PO4, 4.5 g/liter; MgSO4, 0.12 g/liter; (NH4)2SO4, 1 g/liter] at 30°C. Antibiotics were used at 50 µg/ml for rifampin and kanamycin and 5 µg/ml for tetracycline. Pimaricin (30 µg/ml) was added to plates used for internal growth curves.
E. coli strains were grown on Luria-Bertani medium at 37°C. Antibiotics were used at 50 µg/ml for ampicillin and kanamycin, 10 µg/ml for tetracycline, and 40 µg/ml for spectinomycin.
Construction of X. campestris pv. campestris insertion and deletion mutants.
Insertion mutants were constructed using the suicide plasmid pVO155 (52) with ~300-bp PCR fragments internal to each open reading frame (see Table S7 in the supplemental material for primer pairs and Table S6 for insertion sites). Deletion mutants were constructed using the sacB system (53). We obtained amplicons of ~700 bp located upstream and downstream of the region to be deleted by PCR using specific primers (see Table S7) and cloned them into the multiple cloning site (MCS) of pK18mobsacB plasmid. The sequences of all cloned PCR amplicons were verified by sequencing.
Plasmids obtained were introduced into X. campestris pv. campestris by triparental conjugation as described by Turner et al. (54). Insertions were verified by PCR using ProR (a pVO155-specific primer) and gene-specific seq1 primers (see Table S7 in the supplemental material). Deletions were verified by PCR using the left primer specific to the upstream fragment and the right primer specific to the downstream fragment used to amplify upstream and downstream amplicons, respectively (see Table S7).
Complementation plasmid constructs.
Genes corresponding to nagA, nixF, and nixG were PCR amplified using specific primers (see Table S7). PCR amplicons were cloned into the MCS of complementation plasmids pCZ917 (8) or pCZ1016 (9) (see Table S6) and verified by sequencing. Plasmids obtained were introduced into X. campestris pv. campestris by triparental conjugation.
Complementations by chromosomal insertions.
Genes corresponding to nix TBDTs and to the nixE-to-nixL cluster were PCR amplified using specific primers (see Table S7 in the supplemental material). PCR amplicons were cloned into the MCS of the pCZ1034 plasmid (9) under the control of the Ptac promoter and the T7 terminator. Plasmids obtained were introduced into X. campestris pv. campestris by triparental conjugation, and deletion of the XCC0127-to-XCC0128 region combined with the insertion of the desired gene or region was achieved using the sacB protocol (53). Insertion of each gene was verified by PCR using the left primer specific to the upstream fragment and the right primer specific to the downstream fragment used to amplify upstream and downstream amplicons, respectively (see Table S7).
qRT-PCR analysis.
Quantitative RT-PCR (qRT-PCR) was performed as previously described (8) (see Table S7 in the supplemental material for primers used). Briefly, cells were grown in MME minimal medium supplemented or not with GlcNAc at a final concentration of 10 mM and harvested by centrifugation when the optical density at 600 nm (OD600) was between 0.4 and 0.6. RNAs were extracted using the RNeasy minikit (Qiagen) and treated with RNase-free DNase I (Sigma) before being reverse transcribed by Superscript II (Invitrogen) using random hexamers (Biolabs). Quantitative-PCR amplification was performed on a Light Cycler (Roche). Experiments were carried out in three independent biological assays. As a control for real-time PCR, we used the 16S rRNA as previously described (4, 55) (see Table S7 in the supplemental material).
Calculation of maximal growth rate and population estimation, in CFU/ml.
In vitro growth curves were generated using the FLUOstar Omega apparatus (BMG Labtech, Offenburg, Germany) with four independent replicates. Growth rates were measured using 96-well flat-bottom microtiter plates containing 200 µl of MME minimal medium containing the desired substrate inoculated at an OD600 of 0.1 from 4 independent washed overnight precultures in MOKA rich medium. The microplates were shaken continuously at 700 rpm using the linear shaking mode. Specific maximal growth rate was calculated as described in reference 9.
To follow the in vitro bacterial populations, we determined the CFU per ml during bacterial growth. Cells from three independent washed overnight precultures in MOKA rich medium were inoculated at an OD600 of 0.1 in 2 ml MMC minimal medium containing the desired substrate in glass tubes at 30°C with shaking at 200 rpm. Growth experiments in XS were performed in a total volume of 350 µl in glass tubes. To limit dilution of XS, 17.5 µl of bacteria (OD600 = 0.2 in MMC) were added to 332.5 µl of XS, leading to an OD of 0.01 at time zero. CFU were quantified at various time points, by performing serial dilutions in sterile water before spotting 5 µl of these dilutions 3 times on rifampin-containing MOKA plates. After 48 h of growth at 30°C, colonies were enumerated in spots containing 1 to 30 colonies. Bacterial populations were calculated, and results are given in log CFU/ml.
Collection of cabbage xylem sap through root pressure.
Xylem sap was collected from Brassica oleracea cv. Bartolo F1 cabbages as previously described (56). Briefly, XS was collected from 6- to 8-week-old plants after stem decapitation above the first true leaf pair. After the section had been rinsed thoroughly with distilled water, root pressure exudates (herein called XS) were collected throughout the day by manual pipetting. The two first harvests were discarded to avoid contamination by the remaining cell wall debris and phloem compounds. Fractions were frozen immediately after collection and stored at −20°C. For bacterial cultivation, XS was sterilized by filtration using 0.22 µM polytetrafluoroethylene (PTFE) filters.
Pathogenicity assays.
Pathogenicity assays were conducted on Arabidopsis thaliana ecotype Sf-2 by wound inoculation of the central vein as previously described (4, 57) using a bacterial inoculum with an OD600 of 0.05. Similarly, Brassica oleracea cv. Bartolo F1 cabbage leaves of 6-week-old plants were inoculated by piercing the central vein three times using a bacterial inoculum with an OD600 of 0.2. Disease development on A. thaliana leaves was scored at different days after inoculation using a disease index score (0, no symptoms; 1, faint chlorosis; 2, extended chlorosis; 3, necrosis; 4, leaf death). Results are given as percentages of the value for the reference strain.
The stability of pVO155 insertions during pathogenicity tests (8 days without selection pressure) was verified by analyzing the kanamycin resistance of 100 individual bacterial clones purified from leaf tissues 8 days postinoculation.
In planta internal growth curves (IGC).
To determine in planta bacterial population densities during infection, we obtained IGC. Twenty plants were inoculated, and symptoms were scored as described above. Each strain was tested in at least two independent experiments. For each sample, 3 inoculated leaf discs from 3 different plants were sampled using a cork borer (diameter, 0.65 cm; surface area, 0.33 cm2) at days 0, 1, 3, and 7 after inoculation and ground with beads in 500 µl of sterile water. The homogenates were serially diluted in sterile water, and 5-µl drops were spotted three times for each dilution (10−1 to 10−6) on MOKA plates supplemented with rifampin and pimaricin. Plates were incubated at 30°C for 48 h, and colonies were enumerated in spots containing 1 to 30 colonies. Bacterial densities in leaves were calculated, and values are reported in log CFU/cm2.
[14C]GlcNAc transport experiments.
Transport experiments with radiolabeled GlcNAc (specific activity, 2.04 GBq/mmol; PerkinElmer) were performed as described previously (4, 8).
Bioinformatic analyses.
The putative function of Nix glycoside hydrolases was examined using the carbohydrate-active enzyme database (CAZy; http://www.cazy.org/) and the KEGG pathway database (http://www.genome.jp/kegg/pathway.html).
Proteins belonging to the GlcNAc CUT system were sought in Xanthomonas and Xylella proteomes and in representative plant nitrogen-fixing, plant-associated, and plant-pathogenic bacterial proteomes using a BlastP search. Only bacteria with completely sequenced and assembled genomes (available genomes on 4 April 2014) were analyzed, and only proteins with at least 50% coverage and 30% identity or similarity were considered.
Phylogenetic analysis was performed using the Phylogeny.fr web server (http://www.phylogeny.fr/) in default mode (58).
SUPPLEMENTAL MATERIAL
X. campestris pv. campestris nixD to nixL genes form an operon with a putative additional promoter upstream of nixG. (A) Schematic representation of the nixE-nixL cluster with arrowheads indicating pVO155 insertion sites. Light gray, insertion in nixD (nixD*I mutant); white, insertion in nixE (nixE*I mutant); dark gray, insertion in nixG (nixG*I mutant); black, insertion in nixI (nixI*I mutant). NagR boxes are represented by black circles. Black arrows indicate transcription start sites. (B to E) Expression of nix genes relative to the wt strain in nixD*I (B), nixE*I (C), nixG*I (D), and nixI*I (E) insertion mutants. Expressions were analyzed by qRT-PCR with at least three independent experiments on RNA extracted from strains grown in MME minimal medium supplemented with 10 mM N-acetylglucosamine. Calculation of relative expression includes normalization against the endogenous control gene 16S RNA. Download
Neighbor-joining phylogenetic tree of X. campestris pv. campestris NixA, NixB, NixC, and NixD and Caulobacter crescentus CC0446 TonB-dependent transporters. This phylogenetic analysis was performed by using the Phylogeny.fr web server (http://www.phylogeny.fr/). The significance of each node was assessed by approximate-likelihood-ratio tests. Download
Relative black rot symptom intensity, obtained for different strains of X. campestris pv. campestris on Arabidopsis thaliana ecotype Sf-2. Disease index was scored at 8 dpi and is presented as a percentage of the value for the wt strain, except in panel D, where disease index was given as percentage of the value for the ampG*I mutant. (A) Disease index of single mutants and complemented strains. (B) Epistasis of the ΔnagA mutant. (C and D) Epistasis of the ΔnagA ampG*I mutant.
Epistasis during growth of X. campestris pv. campestris in MMC minimal medium containing chitohexaose (CHI-6) or in xylem sap (XS) of Brassica oleracea cv. Bartolo F1 cabbages. Results are given as maximal specific growth rates (µmax [h−1]) relative to the wild-type (wt) strain. Specific μmax were calculated from CFU counts during the log phase (between 3 and 9 h of growth). Standard deviations (SD) were obtained from at least 3 independent experiments. In red are values significantly different from those for the wt strain (sensitivity), in green are values significantly different from those for the ΔnagA strain (total epistasis), and in brown are values significantly different from those for both the wt and ΔnagA strains (partial epistasis), all with a P value of <0.0001. nd, not determined.
Epistasis during growth of X. campestris pv. campestris strains in MMC minimal medium containing N-acetylglucosamine (GlcNAc) or chitotriose (CHI-3) at 100 µM. Results are given as maximal specific growth rates (µmax [h−1]) relative to the wild-type (wt) strain. Specific µmax were calculated from OD600 values during the log phase of the wt strain in MMC minimal medium containing GlcNAc (between 7 and 12 h of growth). Standard deviations (SD) were obtained from at least 3 independent experiments. In red are values significantly different from those for the wt strain (sensitivity), in green are values significantly different from those for the ΔnagA mutant (total epistasis), and in brown are values significantly different from those for both the wt and ΔnagA strains (partial epistasis), all with a P value of <0.0001.
Growth of X. campestris pv. campestris strains MME minimal medium containing chitooligosaccharides. Results are given as maximal specific growth rates (µmax in h−1) relative to the wild-type (wt) strain. Specific µmax were calculated from OD600 values during the log phase of the wt in medium containing N-acetylglucosamine (GlcNAc) (between 7 and 12 h [A] or between 5 and 10 h [B] depending on the experiments). Substrates were added at a final concentration of 0.1% (wt/vol). Standard deviations (SD) were obtained from at least 3 independent experiments. Values underlined in gray indicate strains affected in growth compared to the wt strain, with a P value of <0.0001. CHI-2, chitobiose; CHI-3, chitotriose; CHI-4, chitotetraose; CHI-5, chitopentaose; CHI-6, chitohexaose; CHI-n, chitin; nd, not determined.
Conservation of genes belonging the X. campestris pv. campestris strain ATCC 33913 N-acetylglucosamine (GlcNAc) CUT system in sequenced and assembled genomes of Xanthomonas and Xylella strains and representative plant nitrogen-fixing, plant-associated, and plant-pathogenic bacteria.
List of X. campestris pv. campestris strains and plasmids used or generated in this study.
List of primers used in this study. Sequences corresponding to restriction sites are underlined and sequences nonhomologous to X. campestris pv. campestris are in italics.
ACKNOWLEDGMENTS
We thank Nicolas Hollebecq for technical assistance. We are grateful to Laurent Noël for valuable discussion and critical reading of the manuscript and Deborah Hinton and Mireille Ansaldi for critical reading of the manuscript. A.B. was funded by the French Ministry of Research and Technology.
We gratefully acknowledge financial support from the Département Santé des Plantes et Environnement-Institut National de la Recherche Agronomique and from the French Agence Nationale de la Recherche (grant ANR-08-BLAN-0193-01). This work was supported by the French Laboratory of Excellence project TULIP (ANR-10-LABX-41, ANR-11-IDEX-0002-02).
Footnotes
Citation Boulanger A, Zischek C, Lautier M, Jamet S, Rival P, Carrère S, Arlat M, Lauber E. 2014. The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. mBio 5(5):e01527-14. doi:10.1128/mBio.01527-14.
REFERENCES
- 1. Postle K, Kadner RJ. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869–882. 10.1046/j.1365-2958.2003.03629.x [DOI] [PubMed] [Google Scholar]
- 2. Reeves AR, D’Elia JN, Frias J, Salyers AA. 1996. A bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187:8300–8311. 10.1128/JB.187.24.8300-8311.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. 10.1371/journal.pone.0000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He J, Ochiai A, Fukuda Y, Hashimoto W, Murata K. 2008. A putative lipoprotein of Sphingomonas sp. strain A1 binds alginate rather than a lipid moiety. FEMS Microbiol. Lett. 288:221–226. 10.1111/j.1574-6968.2008.01354.x [DOI] [PubMed] [Google Scholar]
- 6. Eisenbeis S, Lohmiller S, Valdebenito M, Leicht S, Braun V. 2008. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J. Bacteriol. 190:5230–5238. 10.1128/JB.00194-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roy S, Douglas CW, Stafford GP. 2010. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J. Bacteriol. 192:2285–2293. 10.1128/JB.00079-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boulanger A, Déjean G, Lautier M, Glories M, Zischek C, Arlat M, Lauber E. 2010. Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J. Bacteriol. 192:1487–1497. 10.1128/JB.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Déjean G, Blanvillain-Baufumé S, Boulanger A, Darrasse A, Dugé de Bernonville T, Girard AL, Carrére S, Jamet S, Zischek C, Lautier M, Solé M, Büttner D, Jacques MA, Lauber E, Arlat M. 2013. The xylan utilization system of the plant pathogen Xanthomonas campestris pv campestris controls epiphytic life and reveals common features with oligotrophic bacteria and animal gut symbionts. New Phytol. 198:899–915. 10.1111/nph.12187 [DOI] [PubMed] [Google Scholar]
- 10. Kawai F, Kitajima S, Oda K, Higasa T, Charoenpanich J, Hu X, Mamoto R. 2013. Polyvinyl alcohol and polyethylene glycol form polymer bodies in the periplasm of Sphingomonads that are able to assimilate them. Arch. Microbiol. 195:131–140. 10.1007/s00203-012-0859-1 [DOI] [PubMed] [Google Scholar]
- 11. Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G. 2010. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 4:673–685. 10.1038/ismej.2010.4 [DOI] [PubMed] [Google Scholar]
- 12. Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glöckner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- 13. Tancula E, Feldhaus MJ, Bedzyk LA, Salyers AA. 1992. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J. Bacteriol. 174:5609–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vicente JG, Holub EB. 2013. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to Brassica crops. Mol. Plant Pathol. 14:2–18. 10.1111/j.1364-3703.2012.00833.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang K, Jiao N, Liu K, Zhang Y, Li S. 2012. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS One 7:e41204. 10.1371/journal.pone.0041204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- 17. Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284:24673–24677. 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bernheim NJ, Dobrogosz WJ. 1970. Amino sugar sensitivity in Escherichia coli mutants unable to grow on N-acetylglucosamine. J. Bacteriol. 101:384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White RJ. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Konopka JB. 2012. N-Acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo) 2012:489208. 10.6064/2012/489208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hugouvieux V, Barber CE, Daniels MJ. 1998. Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: a system for studying early infection events in bacterial pathogenesis. Mol. Plant Microbe Interact. 11:537–543 [DOI] [PubMed] [Google Scholar]
- 22. Dugé de Bernonville T, Noël LD, SanCristobal M, Danoun S, Becker A, Soreau P, Arlat M, Lauber E. 2014. Transcriptional reprogramming and phenotypical changes associated with growth of Xanthomonas campestris pv. campestris in cabbage xylem sap. FEMS Microbiol. Ecol. 89:527–541. 10.1111/1574-6941.12345 [DOI] [PubMed] [Google Scholar]
- 23. Erbs G, Silipo A, Aslam S, De Castro C, Liparoti V, Flagiello A, Pucci P, Lanzetta R, Parrilli M, Molinaro A, Newman MA, Cooper RM. 2008. Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chem. Biol. 15:438–448. 10.1016/j.chembiol.2008.03.017 [DOI] [PubMed] [Google Scholar]
- 24. Goodell EW. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng Q, Park JT. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434–6436. 10.1128/JB.184.23.6434-6436.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211–227. 10.1128/MMBR.00027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keyhani NO, Roseman S. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108–122. 10.1016/S0304-4165(99)00172-5 [DOI] [PubMed] [Google Scholar]
- 29. Grimont PA, Grimont F. 1978. The genus Serratia. Annu. Rev. Microbiol. 32:221–248. 10.1146/annurev.mi.32.100178.001253 [DOI] [PubMed] [Google Scholar]
- 30. Yang TC, Hu RM, Hsiao YM, Weng SF, Tseng YH. 2003. Molecular genetic analyses of potential beta-galactosidase genes in Xanthomonas campestris. J. Mol. Microbiol. Biotechnol. 6:145–154. 10.1159/000077245 [DOI] [PubMed] [Google Scholar]
- 31. da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Sluys MA, Almeida NF, Alves LM, do Amaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EG, Lemos MV, Locali EC, Machado MA, Madeira AM, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CFM, Miyaki CY, Moon DH, Moreira LM, Novo MTM, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, Rossi A, Sena JAD, Silva C, de Souza RF, Spinola LAF, Takita MA, Tamura RE, Teixeira EC, Tezza RID, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459–463. 10.1038/417459a [DOI] [PubMed] [Google Scholar]
- 32. Dow JM, Daniels MJ. 1994. Pathogenicity determinants and global regulation of pathogenicity of Xanthomonas campestris pv. campestris. Curr. Top. Microbiol. Immunol. 192:29–41 [DOI] [PubMed] [Google Scholar]
- 33. Slauch JM, Camilli A. 2000. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 326:73–96. 10.1016/S0076-6879(00)26047-3 [DOI] [PubMed] [Google Scholar]
- 34. Katta S, Ankati S, Podile AR. 2013. Chitooligosaccharides are converted to N-acetylglucosamine by N-acetyl-β-hexosaminidase from Stenotrophomonas maltophilia. FEMS Microbiol. Lett. 348:19–25. 10.1111/1574-6968.12237 [DOI] [PubMed] [Google Scholar]
- 35. Minic Z, Jamet E, Négroni L, Arsene der Garabedian P, Zivy M, Jouanin L. 2007. A sub-proteome of Arabidopsis thaliana mature stems trapped on concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 58:2503–2512. 10.1093/jxb/erm082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ligat L, Lauber E, Albenne C, Clemente San, Valot B, Zivy M, Pont-Lezica R, Arlat M, Jamet E. 2011. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11:1798–1813. 10.1002/pmic.201000781 [DOI] [PubMed] [Google Scholar]
- 37. Séveno M, Cabrera G, Triguero A, Burel C, Leprince J, Rihouey C, Vézina LP, D’Aoust MA, Rudd PM, Royle L, Dwek RA, Harvey DJ, Lerouge P, Cremata JA, Bardor M. 2008. Plant N-glycan profiling of minute amounts of material. Anal. Biochem. 379:66–72. 10.1016/j.ab.2008.04.034 [DOI] [PubMed] [Google Scholar]
- 38. Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, Cociancich S, Couloux A, Darrasse A, Gouzy J, Jacques MA, Lauber E, Manceau C, Mangenot S, Poussier S, Segurens B, Szurek B, Verdier V, Arlat M, Rott P. 2009. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616. 10.1186/1471-2164-10-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burnaugh AM, Frantz LJ, King SJ. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190:221–230. 10.1128/JB.01251-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Byers HL, Tarelli E, Homer KA, Beighton D. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469–479. 10.1093/glycob/9.5.469 [DOI] [PubMed] [Google Scholar]
- 41. Berger S, Menudier A, Julien R, Karamanos Y. 1995. Do de-N-glycosylation enzymes have an important role in plant cells? Biochimie 77:751–760. 10.1016/0300-9084(96)88193-4 [DOI] [PubMed] [Google Scholar]
- 42. Lanctot PM, Leclerc PC, Clément M, Auger-Messier M, Escher E, Leduc R, Guillemette G. 2005. Importance of N-glycosylation positioning for cell-surface expression, targeting, affinity and quality control of the human AT1 receptor. Biochem. J. 390:367–376. 10.1042/BJ20050189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao H, Sun L, Wang L, Xu Z, Zhou F, Su J, Jin J, Yang Y, Hu Y, Zha X. 2008. N-Glycosylation at Asn residues 554 and 566 of E-cadherin affects cell cycle progression through extracellular signal-regulated protein kinase signaling pathway. Acta Biochim. Biophys. Sin. (Shanghai) 40:140–148. 10.1111/j.1745-7270.2008.00380.x [DOI] [PubMed] [Google Scholar]
- 44. Dennis JW, Granovsky M, Warren CE. 1999. Glycoprotein glycosylation and cancer progression. Biochim. Biophys. Acta 1473:21–34. 10.1016/S0304-4165(99)00167-1 [DOI] [PubMed] [Google Scholar]
- 45. Zhao YY, Takahashi M, Gu JG, Miyoshi E, Matsumoto A, Kitazume S, Taniguchi N. 2008. Functional roles of N-glycans in cell signaling and cell adhesion in cancer. Cancer Sci. 99:1304–1310. 10.1111/j.1349-7006.2008.00839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boisson M, Gomord V, Audran C, Berger N, Dubreucq B, Granier F, Lerouge P, Faye L, Caboche M, Lepiniec L. 2001. Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20:1010–1019. 10.1093/emboj/20.5.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR. 2001. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 98:2262–2267. 10.1073/pnas.051625798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C. 2002. Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156:1003–1013. 10.1083/jcb.200111093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fumagalli O, Tall BD, Schipper C, Oelschlaeger TA. 1997. N-glycosylated proteins are involved in efficient internalization of Klebsiella pneumoniae by cultured human epithelial cells. Infect. Immun. 65:4445–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, Robatzek S, von Schaewen A. 2010. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285:4629–4636. 10.1074/jbc.M109.063073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arlat M, Gough CL, Barber CE, Boucher C, Daniels MJ. 1991. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 4:593–601 [DOI] [PubMed] [Google Scholar]
- 52. Oke V, Long SR. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837–849. 10.1046/j.1365-2958.1999.01402.x [DOI] [PubMed] [Google Scholar]
- 53. Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- 54. Turner P, Barber CE, Daniels MJ. 1985. Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 199:338–343. 10.1007/BF00330277 [DOI] [Google Scholar]
- 55. Morales CQ, Posada J, Macneale E, Franklin D, Rivas I, Bravo M, Minsavage J, Stall RE, Whalen MC. 2005. Functional analysis of the early chlorosis factor gene. Mol. Plant Microbe Interact. 18:477–486 [DOI] [PubMed] [Google Scholar]
- 56. Dugé de Bernonville T, Albenne C, Arlat M, Hoffmann L, Lauber E, Jamet E. 2014. Xylem sap proteomics. Methods Mol. Biol. 1072:391–405. 10.1007/978-1-62703-631-3_28 [DOI] [PubMed] [Google Scholar]
- 57. Meyer D, Lauber E, Roby D, Arlat M, Kroj T. 2005. Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant Pathol. 6:327–333. 10.1111/j.1364-3703.2005.00287 [DOI] [PubMed] [Google Scholar]
- 58. Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. 10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
X. campestris pv. campestris nixD to nixL genes form an operon with a putative additional promoter upstream of nixG. (A) Schematic representation of the nixE-nixL cluster with arrowheads indicating pVO155 insertion sites. Light gray, insertion in nixD (nixD*I mutant); white, insertion in nixE (nixE*I mutant); dark gray, insertion in nixG (nixG*I mutant); black, insertion in nixI (nixI*I mutant). NagR boxes are represented by black circles. Black arrows indicate transcription start sites. (B to E) Expression of nix genes relative to the wt strain in nixD*I (B), nixE*I (C), nixG*I (D), and nixI*I (E) insertion mutants. Expressions were analyzed by qRT-PCR with at least three independent experiments on RNA extracted from strains grown in MME minimal medium supplemented with 10 mM N-acetylglucosamine. Calculation of relative expression includes normalization against the endogenous control gene 16S RNA. Download
Neighbor-joining phylogenetic tree of X. campestris pv. campestris NixA, NixB, NixC, and NixD and Caulobacter crescentus CC0446 TonB-dependent transporters. This phylogenetic analysis was performed by using the Phylogeny.fr web server (http://www.phylogeny.fr/). The significance of each node was assessed by approximate-likelihood-ratio tests. Download
Relative black rot symptom intensity, obtained for different strains of X. campestris pv. campestris on Arabidopsis thaliana ecotype Sf-2. Disease index was scored at 8 dpi and is presented as a percentage of the value for the wt strain, except in panel D, where disease index was given as percentage of the value for the ampG*I mutant. (A) Disease index of single mutants and complemented strains. (B) Epistasis of the ΔnagA mutant. (C and D) Epistasis of the ΔnagA ampG*I mutant.
Epistasis during growth of X. campestris pv. campestris in MMC minimal medium containing chitohexaose (CHI-6) or in xylem sap (XS) of Brassica oleracea cv. Bartolo F1 cabbages. Results are given as maximal specific growth rates (µmax [h−1]) relative to the wild-type (wt) strain. Specific μmax were calculated from CFU counts during the log phase (between 3 and 9 h of growth). Standard deviations (SD) were obtained from at least 3 independent experiments. In red are values significantly different from those for the wt strain (sensitivity), in green are values significantly different from those for the ΔnagA strain (total epistasis), and in brown are values significantly different from those for both the wt and ΔnagA strains (partial epistasis), all with a P value of <0.0001. nd, not determined.
Epistasis during growth of X. campestris pv. campestris strains in MMC minimal medium containing N-acetylglucosamine (GlcNAc) or chitotriose (CHI-3) at 100 µM. Results are given as maximal specific growth rates (µmax [h−1]) relative to the wild-type (wt) strain. Specific µmax were calculated from OD600 values during the log phase of the wt strain in MMC minimal medium containing GlcNAc (between 7 and 12 h of growth). Standard deviations (SD) were obtained from at least 3 independent experiments. In red are values significantly different from those for the wt strain (sensitivity), in green are values significantly different from those for the ΔnagA mutant (total epistasis), and in brown are values significantly different from those for both the wt and ΔnagA strains (partial epistasis), all with a P value of <0.0001.
Growth of X. campestris pv. campestris strains MME minimal medium containing chitooligosaccharides. Results are given as maximal specific growth rates (µmax in h−1) relative to the wild-type (wt) strain. Specific µmax were calculated from OD600 values during the log phase of the wt in medium containing N-acetylglucosamine (GlcNAc) (between 7 and 12 h [A] or between 5 and 10 h [B] depending on the experiments). Substrates were added at a final concentration of 0.1% (wt/vol). Standard deviations (SD) were obtained from at least 3 independent experiments. Values underlined in gray indicate strains affected in growth compared to the wt strain, with a P value of <0.0001. CHI-2, chitobiose; CHI-3, chitotriose; CHI-4, chitotetraose; CHI-5, chitopentaose; CHI-6, chitohexaose; CHI-n, chitin; nd, not determined.
Conservation of genes belonging the X. campestris pv. campestris strain ATCC 33913 N-acetylglucosamine (GlcNAc) CUT system in sequenced and assembled genomes of Xanthomonas and Xylella strains and representative plant nitrogen-fixing, plant-associated, and plant-pathogenic bacteria.
List of X. campestris pv. campestris strains and plasmids used or generated in this study.
List of primers used in this study. Sequences corresponding to restriction sites are underlined and sequences nonhomologous to X. campestris pv. campestris are in italics.