ABSTRACT
How the architecture of DNA binding sites dictates the extent of repression of promoters is not well understood. Here, we addressed the importance of the number and information content of the three direct repeats (DRs) in the binding and repression of the icdA promoter by the phosphorylated form of the global Escherichia coli repressor ArcA (ArcA-P). We show that decreasing the information content of the two sites with the highest information (DR1 and DR2) eliminated ArcA binding to all three DRs and ArcA repression of icdA. Unexpectedly, we also found that DR3 occupancy functions principally in repression, since mutation of this low-information-content site both eliminated DNA binding to DR3 and significantly weakened icdA repression, despite the fact that binding to DR1 and DR2 was intact. In addition, increasing the information content of any one of the three DRs or addition of a fourth DR increased ArcA-dependent repression but perturbed signal-dependent regulation of repression. Thus, our data show that the information content and number of DR elements are critical architectural features for maintaining a balance between high-affinity binding and signal-dependent regulation of icdA promoter function in response to changes in ArcA-P levels. Optimization of such architectural features may be a common strategy to either dampen or enhance the sensitivity of DNA binding among the members of the large OmpR/PhoB family of regulators as well as other transcription factors.
IMPORTANCE
In Escherichia coli, the response regulator ArcA maintains homeostasis of redox carriers under O2-limiting conditions through a comprehensive repression of carbon oxidation pathways that require aerobic respiration to recycle redox carriers. Although a binding site architecture comprised of a variable number of sequence recognition elements has been identified within the promoter regions of ArcA-repressed operons, it is unclear how this variable architecture dictates transcriptional regulation. By dissecting the role of multiple sequence elements within the icdA promoter, we provide insight into the design principles that allow ArcA to repress transcription within diverse promoter contexts. Our data suggest that the arrangement of recognition elements is tailored to achieve sufficient repression of a given promoter while maintaining appropriate signal-dependent regulation of repression, providing insight into how diverse binding site architectures link changes in O2 with the fine-tuning of carbon oxidation pathway levels.
INTRODUCTION
In Escherichia coli, the ArcAB two-component system, comprised of the membrane-bound sensor kinase ArcB and the response regulator ArcA, couples changes in the respiratory state of cells to a global transcriptional response (1). Under aerobic conditions, ArcB kinase activity is silenced, maintaining ArcA largely in the inactive, unphosphorylated state (1, 2). As O2 levels decrease, the proportion of phosphorylated ArcA increases accordingly, with maximal phosphorylation occurring under anaerobic conditions (3). Upon phosphorylation, ArcA binds extensively across the genome, regulating the expression of ~100 operons and acting predominantly as a global repressor of nonfermentative carbon oxidation pathways (4). Although the mechanism of repression has not been well studied, ArcA binding sites within the promoters of repressed operons contain a variable number of direct repeat (DR) sequence elements while almost exclusively overlapping the σ70 RNA polymerase (σ70-RNAP) DNA recognition elements (4). Defining how these ArcA cis-regulatory elements contribute to ArcA DNA binding and repression is critical to understanding how the ArcAB system coordinates this global reprogramming of transcription.
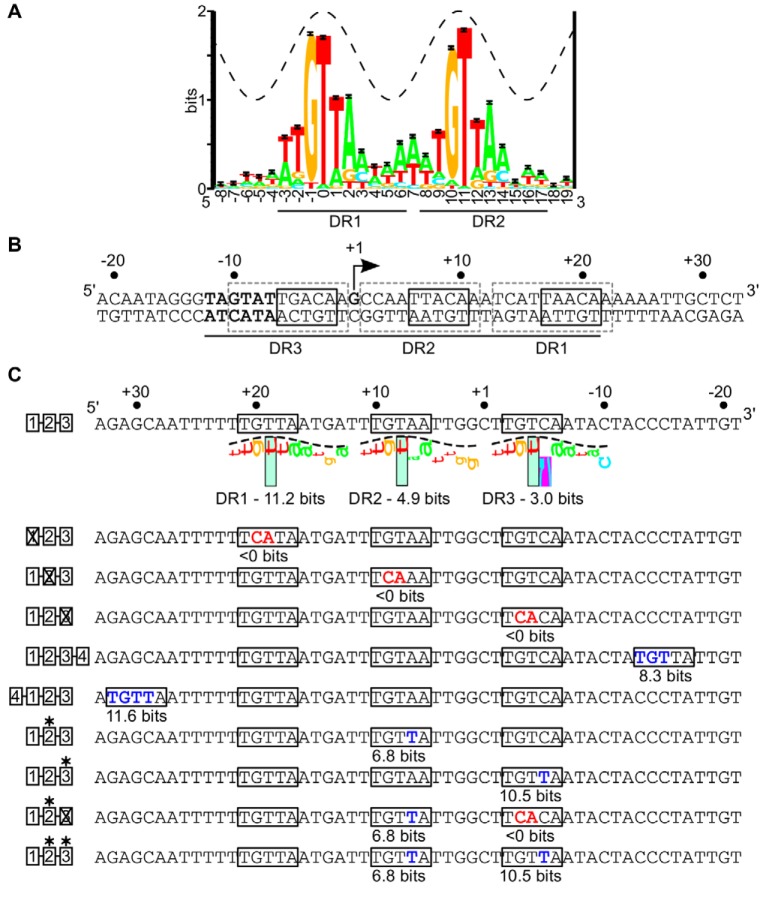
The DNA sequence determinants for ArcA binding have been obscured by the long, degenerate DNA elements bound by ArcA in vitro (5–10). Previous analyses of these footprinted regions have proposed a 15-bp DNA site (5′-GTTAATTAAATGTTA-3′) consisting of two adjacent direct repeats (underlined) as the minimal ArcA recognition site (11–14). However, recent analysis of the chromosomal binding regions of ArcA identified by chromatin immunoprecipitation-DNA sequencing (ChIP-seq) suggested that the minimal ArcA binding site is composed of two 10-bp direct repeat elements (5′-ATGTTAAAAA-1-ATGTTAAAAA-3′) (Fig. 1A) separated by a single nucleotide spacer (11 bp, center to center [CTC]). Furthermore, the majority of ArcA binding sites contain an additional one to three DR elements spaced by approximately one to two turns of the B-form DNA helix (11-bp or 22-bp CTC spacing) from the minimal two DR sites (4). DNase I footprinting assays suggest that these additional DR elements dictate the length of the ArcA binding site (4), providing an explanation for the long ArcA-P footprints.
FIG 1 .
(A) Sequence logo for the minimal ArcA binding site consisting of two 10-bp direct repeat elements (5′-ATGTTAAAAA-1-ATGTTAAAAA-3′) (4). The total sequence conservation is 15.6 ± 0.07 bits in the range from positions −3 to +14. The crest of the sine wave represents the major groove of B-form DNA. (B) Regulatory region of the icdA P1 promoter from E. coli. The arrow indicates the position of the previously mapped transcription start site (5), with the σ70-RNAP −10 promoter element in bold. Each of three 10-bp DR elements is indicated by dashed-line gray boxes, with the most conserved 5-bp 5′-TGTTA-3′ region within each DR element indicated with a solid-line black box. The ArcA-P footprint region is indicated underneath the sequence by the black line (4). (C) Noncoding strand of the icdA-lacZ promoter, depicting the ArcA binding site mutations used in this study. The degree of match of each DR element to the 10-bp ArcA DR element PWM (4) is indicated in bits and visualized using sequence walkers (40). The purple box surrounding the C at position 6 indicates a contact that is more unfavorable than −4 bits and, thus, off the scale. The boxes to the left of the binding sites are the key used to indicate mutations in subsequent figures. Mutations away from the consensus in each DR element (5′-TGTTA-3′ to 5′-TCATA-3′) are indicated in red and labeled with a × in the cartoon, while mutations toward the consensus are indicated in blue and are labeled with an asterisk. The information content for all 10 bp of each mutated DR element is listed below the DR element.
The abundance of ArcA binding sites with three DR elements (4) raises the question of how ArcA dimers bind to a DNA site with an odd number of DR elements. Tandem direct repeat element recognition by an ArcA dimer is supported by the cocrystal structure of the C-terminal DNA binding domain of the closely related response regulator PhoB bound to its tandem direct repeat site as a head-to-tail dimer (15). However, the structure of the activated N-terminal regulatory domain of ArcA bound to a phosphate analog is also dimeric but with a symmetric mode of dimerization (16). These data have led to a model for ArcA and other OmpR/PhoB response regulators that consists of the C-terminal DNA binding domains of the dimer bound to two DRs in a head-to-tail orientation, connected via a flexible linker to the N-terminal regulatory domains that are oriented symmetrically (head to head) along a common interface (16, 17). The recent structural characterization of full-length KdpE, another OmpR/PhoB family member, bound to its direct repeat site confirmed these different domain symmetries while identifying an additional level of asymmetry resulting from intramolecular contacts between the receiver and DNA binding domains within one KdpE subunit (18). Nevertheless, full-length ArcA-P has been reported to form oligomers (19), as have both the isolated N-terminal and C-terminal domains (16), suggesting that although the minimal DNA binding unit is likely a dimer, as demonstrated for PhoB and OmpR (20–22), oligomerization beyond a dimer may explain binding to multiple direct repeats.
To gain insight into the physiological function of multiple DR element binding sites, we evaluated the role of each of the three predicted 10-bp DR elements (DR1-1, DR2-1, DR3) in ArcA-P DNA binding and repression of icdA, encoding isocitrate dehydrogenase of the tricarboxylic acid (TCA) cycle. These repeats are directionally oriented on the noncoding strand and are numbered on the basis of their order in the 5′-to-3′ direction (Fig. 1B). This particular three-DR binding site was chosen because all three DR elements were protected from DNase I cleavage when ArcA is bound (4, 5) despite both strong (DR1 [11.2-bit]) and weak (DR2 [4.9-bit] and DR3 [3.0-bit]) matches to the position weight matrix (PWM) (Fig. 1) for a single DR element (4) and because ArcA-P is the only annotated regulator of the primary icdA promoter (P1) (23). Thus, changes in ArcA DNA binding should change P1 repression. In addition, the positions of DR3 adjacent to the −10 promoter element and of DR1 and DR2 downstream of the transcription start site (TSS) (5) (Fig. 1B) provided an opportunity to determine if there were any specific effects of DR element positioning on ArcA DNA binding and transcriptional regulation. To understand the contribution of each DR in ArcA repression of icdA, they were mutated toward or away from 5′-TGTTA-3′ (Fig. 1C), the most conserved sequence within each ArcA DR element (hereinafter referred to as the consensus) and, based on the PhoB DNA cocrystal structure (15), the region likely contacted by ArcA in the major groove. Mutant arcA alleles were used to determine the phosphorylation dependence of this regulation. Our data reveal that all three DR elements are important for full anaerobic repression of icdA and that degeneracy in these DR elements is important for preserving O2-dependent regulation.
RESULTS
All three DR elements within the icdA promoter contribute to ArcA-P DNA binding in vitro.
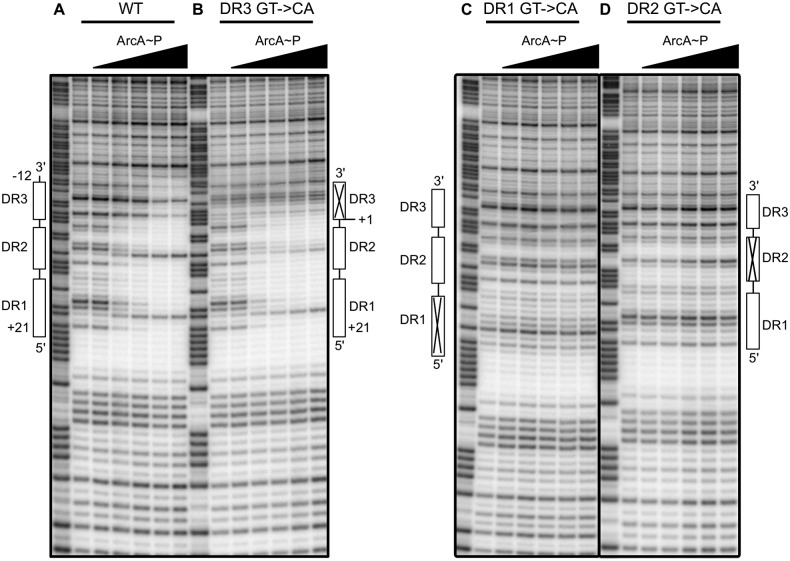
To test the role of each of the three DR elements in ArcA DNA binding to P1icdA, DNase I footprinting assays were performed using ArcA-P and either the wild type (wt) icdA promoter fragment or those in which each DR element was individually disrupted through mutation of highly conserved GT to CA (5′-TGTTA-3′ to 5′-TCATA-3′), reducing the information content of each DR element below the theoretical lowest limit of binding (0 bits) (24) (Fig. 1C). As previously observed (4), ArcA-P protected the three DR elements of the wt promoter region from −12 to +21 relative to the TSS (Fig. 2A). As expected from previous results (4, 5), more ArcA-P (600 nM) was required to observe maximum occupancy of the lower-information-content site, DR3 (3.0 bits), than the higher-information-content sites, DR1(11.2 bits) and DR2 (4.9 bits) (300 nM). Disruption of either DR1 or DR2 eliminated ArcA-P protection of all three DR elements, even at the highest ArcA-P levels tested (Fig. 2C and D). In contrast, when DR3 was mutated, ArcA-P binding to only DR3 was eliminated (Fig. 2B). Furthermore, the amount of ArcA-P required for maximal binding of either DR1 or DR2 was not affected by disruption of DR3, suggesting that ArcA binding to DR1 and DR2 is not enhanced by ArcA-P interactions with DR3 despite the dependence of DR3 binding on ArcA-P interactions with DR1 and DR2.
FIG 2 .
DNase I footprinting of ArcA binding to the wt or mutated icdA promoter region. (A) wt ArcA binding site containing all three DR elements; (B) elimination of DR3 by a 5′-TGTCA-3′-to-5′-TCACA-3′ mutation; (C) elimination of DR1 by a 5′-TGTTA-3′-to-5′-TCATA-3′ mutation; (D) elimination of DR2 by a 5′-TGTAA-3′-to-5′-TCAAA-3′ mutation. The regions protected by ArcA-P are indicated with vertical lines and are numbered to indicate the position relative to the previously determined transcription start site (5). The 10-bp DR elements are indicated by open boxes, with a × representing a DR element that has been eliminated through mutation (Fig. 1C). Samples were electrophoresed with Maxam-Gilbert ladders (A + G) made using the same DNA (lane 1). ArcA-P protein concentrations are given from left to right in nM total ArcA-P protein as follows: 0, 50, 150, 300, 600, and 1,000 nM.
The mechanisms governing the occupancy of DR3 may be complex, since we found that an N-terminal His tag variant of ArcA-P also eliminated binding to DR3, but not DR1 and DR2 (data not shown), suggesting that protein-protein interactions may be important for stabilizing ArcA-P binding to DR3. We also found that disruption of DR3 weakened a hypersensitive band at position +8 within DR2 (Fig. 2A and B). Because DNase I is sensitive to the minor groove width (25), this change in hypersensitivity may suggest that ArcA-P bends or kinks the DNA to a greater degree when bound to all three DR elements than when bound to just DR1 and DR2. Thus, an ArcA-P dimer bound to DR1 and DR2 may also stabilize the binding of ArcA-P to DR3 by bending the DNA.
All three DR elements are required for repression of icdA in vivo.
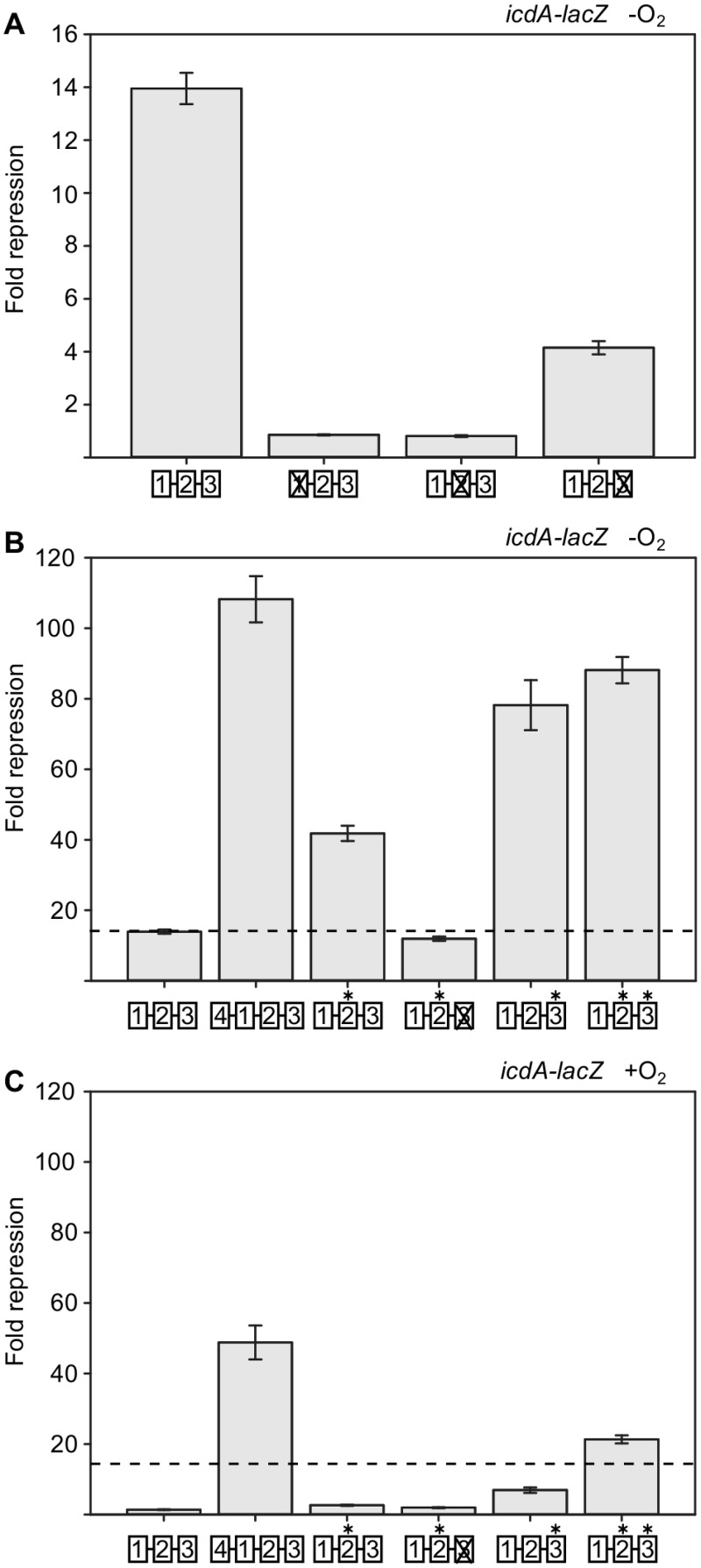
How ArcA binding to each DR element contributes to icdA repression was determined by measuring β-galactosidase activity produced from P1icdA-lacZ transcriptional fusions containing the GT-to-CA (5′-TGTTA-3′ to 5′-TCATA-3′) mutations within each DR element under anaerobic conditions. Basal promoter activity was not altered by any binding site mutation, as all variants exhibited the same activity as the wt promoter in the absence of ArcA repression (data not shown). P1icdA was repressed 14-fold by ArcA (Fig. 3A). However, disruption of either DR1 or DR2 completely abolished ArcA-dependent repression (Fig. 3A), consistent with the loss of DNA binding to all three DRs observed by DNase I footprinting (Fig. 2C and D). In contrast, disruption of DR3, which did not perturb ArcA binding to DR1 and DR2 (Fig. 2B), showed an ~3.5-fold loss in repression (Fig. 3A). This result suggests that in vivo occupancy of DR1 and DR2 is sufficient to direct a moderate amount of P1icdA repression but that additional occupancy of DR3 is required for maximal repression, perhaps because it overlaps the −10 promoter element.
FIG 3 .
Effects of mutations on ArcA-dependent repression of P1icdA. Strains containing P1icdA-lacZ were grown in minimal medium with 0.2% glucose, and fold repression was calculated by dividing the β-galactosidase activity of a ΔarcA strain (e.g., 803 Miller units for wt P1icdA without O2) by the activity of an arcA+ strain (e.g., 57 Miller units for wt P1icdA without O2). The 10-bp DR elements are indicated by open boxes, with a × representing a DR element that has been eliminated through mutation and an asterisk denoting DR elements that have been mutated toward the consensus (Fig. 1C). (A) Effects of mutations away from the consensus within each DR element assayed under anaerobic conditions (−O2). (B) Effects of mutations toward the consensus within each DR element assayed under anaerobic conditions. The dotted line represents anaerobic ArcA-dependent repression of wt P1icdA. (C) Effects of mutations toward the consensus within each DR element assayed under aerobic conditions. Error bars represent the standard errors of results from at least three independent replicates. We note that P1icdA expression in the construct with a fourth DR element was about 18% higher in a ΔarcA background than in the other strains tested (data not shown).
The three DRs of P1icdA are suboptimal for maximal repression.
Since DR2 and DR3 contain a lower information content than DR1, we tested whether mutations that improve the information content affect repression under anaerobic conditions. Mutation of DR2 toward the consensus (5′-TGTAA-3′ to 5′-TGTTA-3′) resulted in a 3-fold increase in anaerobic repression of P1icdA (Fig. 3B). This repression still depends on DR3 function, since the additional disruption of DR3 (5′-TGTCA-3′ to 5′-TCACA-3′) caused the same 3-fold reduction in repression as observed when DR3 was disrupted in an otherwise wt icdA sequence (Fig. 3A and B). When just DR3 was mutated toward the consensus (5′-TGTCA-3′ to 5′-TGTTA-3′), repression was increased 6-fold (Fig. 3B). Improving both DR2 and DR3 toward the consensus resulted in a level of repression similar to that observed with a consensus DR3 element alone, suggesting that maximal P1icdA repression by ArcA had been achieved (Fig. 3B). Assuming that these nucleotide changes simply improve DNA binding affinity, the enhanced anaerobic repression suggests that the three DRs of wt icdA are not completely occupied by ArcA under our standard anaerobic growth conditions.
A fourth DR element enhances ArcA binding affinity.
Although our DNase I footprinting analysis suggests that an ArcA-P dimer bound to DR1 and DR2 stabilizes the binding of ArcA-P to DR3, whether ArcA-P binds as a dimer or as a monomer to DR3 is an open question. The lack of DNase I protection of the DNA sequence adjacent to DR3 suggests that if a dimer is bound, then this sequence either contributes only weakly or not at all to stabilizing the binding of the second dimer. To determine whether adding a fourth DR element facilitates ArcA-P binding and increases the footprint length, a consensus DR element (5′-TGTTA-3′) was added at the same spacing (11 bp CTC) to either the 3′ or the 5′ end of the three-DR ArcA binding site within the icdA promoter region and DNase I footprinting experiments were performed. For both variants, the ArcA-P footprint encompassed all four DR elements, and the apparent ArcA-P DNA binding affinity was noticeably increased compared to that with the wt binding site (Fig. 4A and B). In addition, protection of the entire four-DR site occurred over a very narrow increase in ArcA-P levels (<4-fold), suggesting that cooperativity was also enhanced. Notably, the hypersensitive sites at positions +8 and +19 were unaffected by binding to a fourth repeat, suggesting bending or kinking similar to that with the wt binding site. Finally, as with the wt icdA fragment, binding depended on phosphorylation, since no binding was observed with unphosphorylated ArcA at protein concentrations up to 1 µM (data not shown).
FIG 4 .
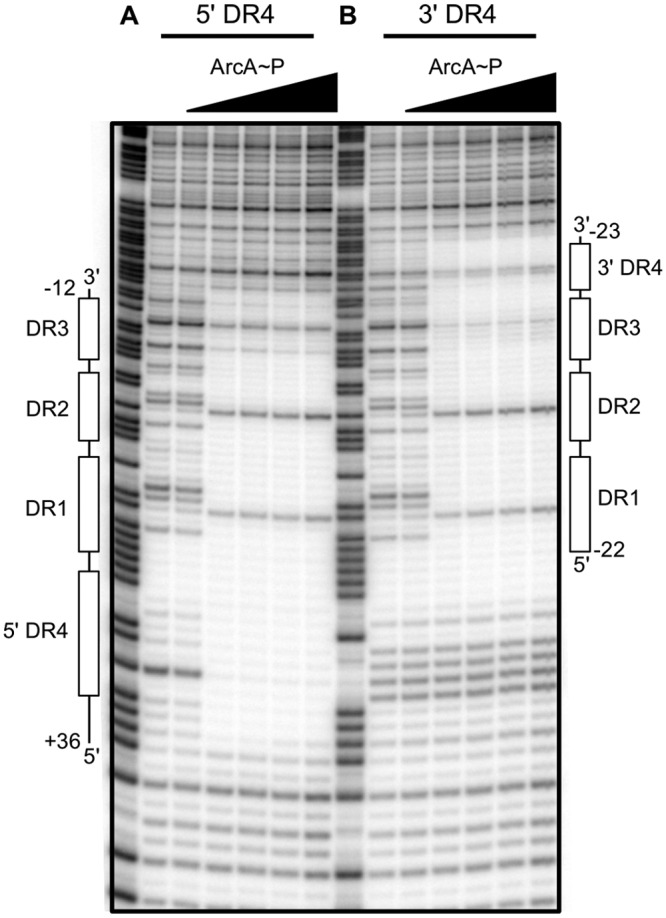
DNase I footprinting of ArcA binding to the icdA promoter region containing four DR elements. (A) Fourth DR element (5′-TGTTA-3′) located 5′ of DR1; (B) fourth DR element (5′-TGTTA-3′) located 3′ of DR3. The regions protected by ArcA-P are indicated with vertical lines, with DR elements indicated by open boxes. The numbers indicate positions relative to the previously determined transcription start site. Samples were electrophoresed with Maxam-Gilbert ladders (A + G) made using the same DNA (lane 1). ArcA-P protein concentrations are given from left to right in nM total ArcA-P protein as follows: 0, 50, 150, 300, 600, and 1,000 nM.
Adding a fourth DR element or improving DR2 or DR3 disrupts O2-dependent regulation of ArcA DNA binding.
Despite the potential for enhancement of DNA binding, multiple consecutive high-information-content DR sites are relatively rare in the E. coli genome (4), raising the question of whether there is a tradeoff between DNA binding and the ability to respond to the regulatory signal. To test whether the P1icdA variant with four consecutive DR binding sites still retains O2-dependent regulation, we measured β-galactosidase activity produced from a P1icdA-lacZ transcriptional fusion containing either the 3′ or the 5′ DR4 element and compared it to that produced with the wt promoter under anaerobic or aerobic conditions. As expected, ArcA-dependent repression of wt P1icdA was largely relieved in the presence of O2 (Fig. 3C), consistent with the known reduction in ArcA-P levels under aerobic conditions (3). However, addition of DR4 to the 5′ end not only resulted in an 8-fold increase in repression compared to the repression with the wt binding site under anaerobic conditions (Fig. 3B) but also increased repression by ArcA under aerobic conditions to nearly the same magnitude observed under anaerobic conditions, indicating that ArcA repression of this variant site was no longer O2 sensitive (Fig. 3C). The addition of DR4 to the 3′ end disrupted promoter function, preventing assessment of ArcA repression (data not shown). The simplest interpretation of these results is that strengthening binding affinity disrupts O2-dependent regulation of ArcA DNA binding.
We also tested whether the degeneracy of DR2 and DR3 (Fig. 1C) is important for maintaining O2-dependent regulation of P1icdA by assaying the variants where the sites were mutated toward the consensus. Improving DR2 or DR3 toward the consensus also increased aerobic P1icdA repression compared to that with the wild-type binding site, but the effect was more pronounced with a consensus DR3 element (~2.5-fold versus 7-fold repression) (Fig. 3C). P1icdA with both consensus DR2 and DR3 elements was even more repressed by ArcA under aerobic conditions (21-fold) than with either consensus site alone, suggesting that there was an additive effect (Fig. 3C). Together, these results suggest that improving binding affinity through the use of consensus DR elements disrupts the signal-dependent regulation of ArcA DNA binding, suggesting that the degeneracy of DR2 and DR3 is important for balancing anaerobic repression with O2-dependent relief of repression.
Enhanced ArcA repression is still dependent on phosphorylation.
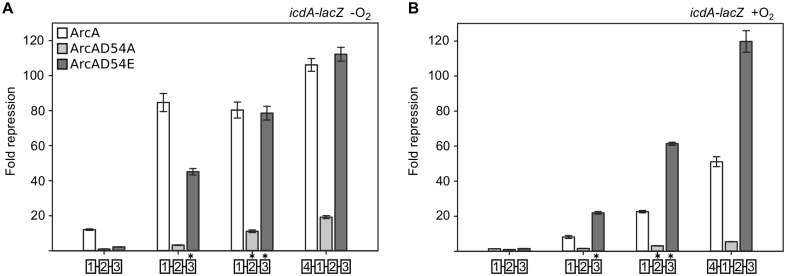
To test whether the enhanced repression of P1icdA with mutant ArcA binding sites is still dependent on phosphorylation, the aspartate residue at position 54 (site of phosphorylation [19]) in the chromosomal copy of arcA was mutated to yield either alanine or glutamate, preventing phosphorylation from ArcB (19). The D54A variant reduced the repression of all P1icdA-lacZ constructs compared to that with the wt protein under both aerobic and anaerobic growth conditions (Fig. 5A). This suggests that, independent of the strength of the binding site, repression is largely dependent on the phosphorylated form of ArcA. This result is consistent with the failure of unphosphorylated ArcA to bind to the four DR sites in vitro (data not shown). Thus, the elevated aerobic repression with the strengthened ArcA binding sites appears to result from increased occupancy of the small amount of ArcA-P likely present during aerobic conditions.
FIG 5 .
Phosphorylation dependence of ArcA repression of P1icdA in strains with strengthened ArcA binding sites. The fold repression of P1icdA-lacZ in strains containing arcA-FRT-cat-FRT (white bars), arcA(D54A)-FRT-cat-FRT (light-gray bars), or arcA(D54E)-FRT-cat-FRT (dark-gray bars) was determined from cells grown under anaerobic (A) or aerobic (B) conditions and calculated by dividing the β-galactosidase activity of a ΔarcA strain by the activity with each of the arcA alleles. Asterisks denote DR elements that have been mutated toward the consensus. Error bars represent the standard errors of results from at least three independent replicates.
We expected ArcA(D54E) to similarly reduce the repression of P1icdA, since this substitution has previously been shown to prevent both phosphorylation from ArcB and binding to the pfl promoter (19). Surprisingly, ArcA(D54E) still strongly repressed P1icdA constructs with strengthened binding sites even though repression of wt P1icdA was largely eliminated; repression of the construct with a consensus DR3 element was reduced by only 2-fold, while repression of constructs with consensus DR2 and DR3 elements or a fourth DR element was indistinguishable from that observed with the wt protein under anaerobic conditions (Fig. 5B). Furthermore, under aerobic conditions, ArcA(D54E) repression of P1icdA was increased compared to that of wt ArcA for all binding sites tested (Fig. 5B). Thus, D54E ArcA appears to partially mimic phosphorylated ArcA. An aspartate-to-glutamate substitution has previously been shown to elicit constitutive activity in some response regulators (26).
DISCUSSION
The results presented here provide new insight into the plasticity of the DNA elements that can control transcriptional repression. Our data suggest that for icdA, the arrangement of multiple DNA binding elements appears to be tailored to achieve both sufficient DNA binding affinity and repression by ArcA while maintaining O2-dependent regulation. We propose that the distribution of DNA binding information across several DR elements may be a design principle to achieve the appropriate level of repression and to tune the signal-dependent regulation of target genes for both ArcA and other repressors.
Interaction of ArcA with three DR elements of icdA.
Our analysis of the three DR elements of the icdA promoter indicate that ArcA-P binding to the lowest-information-content site, DR3, is stabilized by ArcA-P bound to DR1 and DR2, suggestive of a cooperative DNA binding mechanism. The lack of an observable defect in binding to DR1 or DR2 when DR3 was eliminated suggests that the cooperative energy is predominantly partitioned toward binding of DR3, as expected for sites with large differences in intrinsic levels of binding energy (27). Since ~67 genomic sites have an odd number of DR elements (4), cooperativity is likely an important determinant for ArcA binding genome-wide.
An unanswered question is what the stoichiometry of ArcA-P binding to DR1, -2, and -3 is. It is possible that ArcA-P binds to the icdA promoter as a dimer of dimers; one dimer binds DR1 and DR2, as depicted in the PhoB and KdpE DNA cocrystal structures (15, 18), and the second dimer binds DR3 but only weakly to adjacent DNA sequence, such that no footprint is observed (Fig. 6). This model is supported by the requirement for phosphorylation of ArcA to bind to DR3, which is also known to promote dimer formation among OmpR/PhoB response regulators (20, 21). However, phosphorylation may simply eliminate an interaction between the regulatory and DNA-binding domains, allowing ArcA-P to bind as a monomer to DR3 (Fig. 6). Thus, additional studies are necessary to determine the stoichiometry of ArcA binding to the icdA promoter and whether this stoichiometry is shared among other ArcA sites with three DR elements.
FIG 6 .
Model for ArcA-P binding to a three-DR binding site. The orientation and protein-protein contacts between the N- and C-terminal domains within an ArcA-P dimer are based on crystallographic data from ArcA and PhoB, respectively (15, 16). Energetically favorable contacts are indicated in blue, while contacts likely to be less favorable are indicated in red. We propose that two ArcA-P dimers bind to a three-DR site in a cooperative manner; the first dimer binds to DR1 and DR2, and a second dimer binds to DR3 and adjacent nonspecific sequences. A favorable energetic contribution from the interaction between ArcA-P dimers is likely required to overcome the poor binding affinity of an ArcA-P dimer to DR3 and adjacent nonspecific sequence. Alternatively, it is possible that dimerization is not required for binding to DR3; ArcA-P may bind to DR3 as a monomer. Potential regions of interaction between ArcA-P molecules in both scenarios are marked with question marks
In either scenario, the predominance of three DR sites with 11-bp CTC spacing between each DR in the E. coli genome, together with our previous finding that ArcA-P did not bind to a predicted DR3 element in which the CTC spacing was separated by an additional bp (4), suggests that protein-protein interactions between correctly spaced subunits is important for cooperative ArcA binding to multiple DRs. Because the C-terminal domain of ArcA binds as a dimer to two adjacent DRs, one can envision that binding of an ArcA-P dimer to DR1 and -2 stabilizes a second dimer or a monomer via protein interactions with DR3. Additionally, the hypersensitive site observed when all three (or four) DR elements of icdA were occupied may indicate a requirement for DNA bending to facilitate these protein-protein interactions. Our finding that an N-terminally His-tagged variant of ArcA failed to stabilize binding to DR3 suggests that the His tag specifically disrupts the mechanism needed to enhance the energetics of DR3 binding site occupancy. Additional work is needed to define the molecular interactions that stabilize ArcA-P binding to DR3 elements, but tagged protein variants may not recapitulate this important property of response regulators.
Maximizing repression by binding DR3.
The analysis of the effects of mutations eliminating individual DR elements suggests that DR1 and DR2 determine the overall strength of ArcA binding and that all three DR elements contribute to repression. However, the fact that DR3 overlaps the −10 hexamer as opposed to DR1 and DR2, which are located between positions +2 and +22 (Fig. 1B), suggests that ArcA binding to DR3 may interfere with the initial binding of RNA polymerase to form the closed complex, as has been shown for the Lac repressor bound to the Lac operator that overlaps the TSS (28). Furthermore, more-effective repression was observed when the Lac and Tet operators overlapped the −10 and −35 promoter elements than when they overlapped those placed downstream of the TSS (29–31). We do not expect this particular role of DR3 in icdA to be broadly applicable to all multiple-DR-element ArcA binding sites because of differences in both the strengths and the locations of DR3 elements relative to the TSS (4). Furthermore, because ArcA DNA elements are direct repeats, they can be found either in the same or in the opposite orientation from the promoter elements, providing additional flexibility for coding repressor information within a constrained sequence space. There are several instances where all three ArcA DR elements overlap the promoter elements or where DR3 is found downstream of the TSS and may thus play a role more akin to those of DR1 and DR2 of icdA. This flexible property of response regulators may also be confined to repressors, since activators are likely to be located in specific positions because of the typical requirement to interact with RNA polymerase.
The combinatorial effect of weak versus strong DR elements can create a range of responses to ArcA levels.
At icdA, the differences in binding affinity of an ArcA-P dimer for DR1/DR2 versus DR3 increases the amount of ArcA-P required for full occupancy in vitro. Assuming that ArcA-P binds the same way in vivo, this binding site architecture would extend the sensitivity to ArcA-P levels by increasing the amount of ArcA-P required for maximal repression. This property may be a feature shared with other response regulators, since in the case of OmpR, the binding of an OmpR-P dimer to box 1 (two DR elements) at the ompF and ompC promoters occurs at a lower concentration of OmpR-P than does binding of OmpR-P to adjacent OmpR boxes (32, 33). Similarly, binding of PhoB-P to the upstream PhoB box at the pstS promoter occurs at a lower concentration of PhoB-P than when it binds to the adjacent, downstream box (34).
On the other hand, promoters with three or more DR elements of high information content appear to result in ArcA-P occupancy over a very narrow range of protein concentrations. For example, when the disparity in ArcA-P binding affinities at icdA was reduced by replacing the nonspecific sequence adjacent to DR3 with a fourth DR element, the increase in binding affinity resulted in the promoter bound by ArcA-P in a highly cooperative manner. A similar switch-like occupancy of ArcA-P was also observed for the four DR elements at the astC promoter (4). Data obtained using an icdA-lacZ reporter fusion indicate that strengthening DR3 toward the consensus likely also enhances binding affinity. Indeed, all three DR elements at the acs promoter are bound over a narrow range of ArcA-P levels, likely due to a greater energetic contribution to ArcA-P binding provided by a stronger DR3 element (4). Thus, the combinatorial effect of strong or weak DR elements may be used to either dampen or enhance the concentration-sensitive occupancy by ArcA-P compared to that of a site with only two DRs.
Physiological significance of multiple DR binding sites.
The configuration of the ArcA DR elements may also provide a mechanism for achieving a stepwise response to changes in O2, as suggested for OmpR-P dimer binding to the ompF and ompC promoters in response to changes in osmolarity (32). For example, under aerobic conditions, ArcA-P levels are likely insufficient for appreciable binding to the icdA promoter; thus, icdA expression is high, consistent with the need for isocitrate dehydrogenase for carbon oxidation in the TCA cycle. However, as O2 becomes limiting, ArcA-P levels likely increase (3), perhaps allowing an ArcA-P dimer to bind DR1 and DR2, reducing icdA expression to an intermediate level. As O2 is further depleted, ArcA-P levels likely increase more, and we expect binding to all three DRs, reducing icdA expression to levels optimal for anaerobic metabolism. Experimental support for this model came from showing that O2-dependent regulation was disrupted at icdA either by adding a fourth DR element or by improving DR2 and DR3. Thus, these results suggest that the degeneracy in DR2 and DR3 and the absence of a recognizable fourth DR element is important for maintaining the balance between strong, but not complete, anaerobic repression and O2-dependent relief of repression.
Given the function of the majority of ArcA-repressed operons in aerobic respiratory metabolism, this balance between high-affinity ArcA-P binding and maintenance of O2-dependent regulation is likely widely applicable to genomic ArcA binding sites. Furthermore, it may explain why there are many three-DR sites without identifiable fourth DR elements in the E. coli genome and, additionally, why the average strength of DR elements decreases as the number of DR elements in the binding site increases (4). Nevertheless, both the strength of the promoter and the incorporation of other regulator binding sites should at least partially dictate the specific ArcA binding site architecture required to achieve optimal regulation, with four DR sites apparently necessary at some promoters.
It will also be informative to determine how expression of other ArcA-dependent promoters (e.g., acs and astC) with a strong DR3 and/or DR4 respond to changes in O2. The saturation of ArcA-P binding to these sites over a narrow range of ArcA-P concentrations in vitro (4) suggests that these promoters may respond to ArcA-P with a switch-like behavior as cells become limited for O2. For the engineered icdA promoter containing a four-DR site, it seems likely that the affinity of ArcA-P for this site is so strong that the concentration of ArcA-P present under aerobic conditions is sufficient to occupy this site so that an O2-dependent change in repression cannot be observed. Nevertheless, our data provide a model for how the ArcA binding site architecture may be optimized to achieve regulatory logic schemes not possible with a canonical two-DR binding site. This plasticity in the promoter architecture likely plays an important role in linking the redox-sensing properties of the ArcAB two-component system with the fine-tuning of expression of carbon oxidation pathway levels.
The incorporation of plasticity in the binding site architectures that we observed for ArcA may be a common regulatory strategy for other global transcriptional repressors (e.g., Fur, LexA). Like ArcA, Fur binding sites are variable in length (30 to 103 bp) and contain multiple Fur recognition elements of differing predicted strengths and locations with respect to the promoter elements (35). Although the physiological basis for this plasticity is unknown, it may similarly impose a differential sensitivity of regulatory target expression to changes in Fe-Fur concentrations. Furthermore, although LexA-regulated genes typically have only one LexA binding site, differences in the strengths and locations of these sites alter the absolute level and sensitivity of expression (36). In a few cases, adjacent LexA sites are bound in a cooperative manner, further enhancing the sensitivity to changes in signal (36), as hypothesized for the ArcA binding sites located upstream of acs and astC. Given the conserved dimerization mode and binding of direct repeat DNA sites among response regulators within the OmpR/PhoB family (16), this architectural plasticity may be a common regulatory strategy, particularly for regulators that act as repressors at many targets.
MATERIALS AND METHODS
Strain construction.
An icdA promoter-lacZ fusion was constructed as described previously (37) by amplifying the region from −50 to −330 with respect to the start of translation using primers flanked by XhoI or BamHI restriction sites. The icdA fragment contains two promoters: one whose expression is dependent on ArcA (P1) and a second whose expression is dependent on FruR (P2) (5, 38). To examine icdA expression from only P1, transcription from P2 was eliminated using QuikChange site-directed mutagenesis (Stratagene) as described previously (39) to mutate the −10 site from 5′-CATTAT-3′ to 5′-CGGTGA-3′, generating pPK9476. Mutations within the ArcA binding site of the icdA promoter were similarly generated using pPK9476 as a template (mutations are numbered with respect to P1 in Table 1). These lacZ promoter constructs were then recombined into the chromosomal lac operon as previously described (37) and then transduced using P1 vir into MG1655 and PK9416 to form the strain derivatives listed in Table 1.
TABLE 1 .
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | F− λ− rph-1 | This laboratory |
| PK9416 | MG1655 ΔarcA | 4 |
| PK9483 | MG1655 PicdA(-58GGTGA-54)-lacZ | 4 |
| PK9484 | PK9416 PicdA(-58GGTGA-54)-lacZ | 4 |
| PK9494 | MG1655 PicdA(-58GGTGA-54, 19TG20)-lacZ | This study |
| PK9495 | PK9416 PicdA(-58GGTGA-54, 19TG20)-lacZ | This study |
| PK9486 | MG1655 PicdA(-58GGTGA-54, 8TG9)-lacZ | This study |
| PK9487 | PK9416 PicdA(-58GGTGA-54, 8TG9)-lacZ | This study |
| PK9496 | MG1655 PicdA(-58GGTGA-54, -4TG-3)-lacZ | This study |
| PK9497 | PK9416 PicdA(-58GGTGA-54, -4TG-3)-lacZ | This study |
| PK9915 | MG1655 PicdA(-58GGTGA-54, 29AACA32)-lacZ | This study |
| PK9916 | PK9416 PicdA(-58GGTGA-54, 29AACA32)-lacZ | This study |
| PK9917 | MG1655 PicdA(-58GGTGA-54, -15ACA-13)-lacZ | This study |
| PK9918 | PK9416 PicdA(-58GGTGA-54, -15ACA-13)-lacZ | This study |
| PK9924 | MG1655 PicdA(-58GGTGA-54, -5A)-lacZ | This study |
| PK9925 | PK9416 PicdA(-58GGTGA-54, -5A)-lacZ | This study |
| PK9941 | MG1655 PicdA(-58GGTGA-54, 7A)-lacZ | This study |
| PK9942 | PK9416 PicdA(-58GGTGA-54, 7A)-lacZ | This study |
| PK9943 | MG1655 PicdA(-58GGTGA-54, 7A, -5A)-lacZ | This study |
| PK9944 | PK9416 PicdA(-58GGTGA-54, 7A, -5A)-lacZ | This study |
| PK10967 | MG1655 PicdA(-58GGTGA-54, 7A, -4TG-3)-lacZ | This study |
| PK10968 | PK9416 PicdA(-58GGTGA-54, 7A, -4TG-3)-lacZ | This study |
| BW25993 | lacIq ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 45 |
| PK9970 | PK9483 arcA::cat | This study |
| PK9973 | PK9915 arcA::cat | This study |
| PK9971 | PK9924 arcA::cat | This study |
| PK9972 | PK9943 arcA::cat | This study |
| PK9980 | PK9483 arcA-D54A::cat | This study |
| PK9983 | PK9915 arcA-D54A::cat | This study |
| PK9981 | PK9924 arcA-D54A::cat | This study |
| PK9982 | PK9943 arcA-D54A::cat | This study |
| PK9975 | PK9483 arcA-D54E::cat | This study |
| PK9978 | PK9915 arcA-D54E::cat | This study |
| PK9976 | PK9924 arcA-D54E::cat | This study |
| PK9977 | PK9943 arcA-D54E::cat | This study |
| Plasmids | ||
| pKD46 | Phage λ gam-bet-exo genes under ParaB control | B. L. Wanner |
| pKD13 | FRT-kan-FRT | K. A. Datsenko and B. L. Wanner |
| pKD32 | FRT-cat-FRT | B. L. Wanner |
| pPK7035 | kan gene from pHP45Ω and BamHI-NdeI fragment from pRS1553 into pBR322 | 37 |
| pPK9476 | pPK7035 PicdA(-58GGTGA-54)-lacZ | 4 |
| pPK9477 | pPK7035 PicdA(-58GGTGA-54 19TG20)-lacZ | This study |
| pPK9908 | pPK7035 PicdA(-58GGTGA-54 8TG9)-lacZ | This study |
| pPK9909 | pPK7035 PicdA(-58GGTGA-54 -4TG-3)-lacZ | This study |
| pPK9913 | pPK7035 PicdA(-58GGTGA-54 29AACA32)-lacZ | This study |
| pPK9914 | pPK7035 PicdA(-58GGTGA-54 -15ACA-13)-lacZ | This study |
| pPK15001 | pPK7035 PicdA(-58GGTGA-54, 7A, -4TG-3)-lacZ | This study |
| pPK9965 | arcA in pBR322 | This study |
| pPK9966 | BamHI FRT-cat-FRT in pPK9965 | This study |
| pPK9431 | Apr; His6-arcA cloned into the NheI and XhoI sites of pET-21d | 4 |
Chromosomally encoded arcA mutants in which aspartate at position 54 was replaced with glutamate or alanine were constructed in several steps. First, the arcA open reading frame (codons 1 to 238) was amplified using primers flanked by HindIII and BamHI and cloned into pBR322, generating pPK9965. The cat cassette from pKD32, which has flanking FRT (FLP recognition target) sites, was then cloned into the BamHI site, 6 bp after the arcA termination codon. The arcA gene on the resulting plasmid, pPK9966, was then mutated using QuikChange (Stratagene) site-directed mutagenesis to create the D54A and D54E mutants. The arcA-cat fragments were PCR amplified using a primer with homology to the region upstream of arcA (5′-GGTAGCAAACATGCAGACCCCGCACATTCTTATCG-3′) and a primer with homology to the region downstream of arcA (5′-GCGCCGTTTTTTTTGACGGTGGTAAAGCCGATTAGTGTAGGCTGGAGCTGCTTC-3′), and the DNA was electroporated into BW25993/pKD46. The correct recombinants were selected for chloramphenicol (Cm) resistance, confirmed with DNA sequencing, and then transduced with P1 vir into the desired icdA promoter-lacZ fusion strains (Table 1). Placement of the cat cassette downstream of arcA did not alter ArcA activity, as icdA promoter-lacZ activity was comparable to that of the wt arcA+ strain for all binding sites tested (Fig. 3B and C and 5A and B).
Determination of the information content of DR elements.
A 10-bp ArcA DR element, PWM, derived from the conservation of bases within aligned DR1 and DR2 elements from 128 sequences bound by ArcA in vivo (4) was used to guide the design of binding site mutations. The information content of each mutant DR element was determined by the scan program (24) and is indicated in bits (Fig. 1C). Greater information content should reflect stronger ArcA binding (24). Sequence walkers (40) were used to visualize how DR elements were evaluated by the PWM. Nucleotides extending upwards represent favorable DNA contacts, while letters extending downward represent unfavorable contacts.
β-Galactosidase assays.
All strains were grown in MOPS minimal medium (41) with 0.2% glucose at 37°C and sparged with a gas mix of 95% N2 and 5% CO2 (anaerobic) or 70% N2, 5% CO2, and 25% O2 (aerobic). Cells were harvested during mid-log growth (optical density at 600 nm [OD600] of ~0.3 on a PerkinElmer Lambda 25 UV/visible-light spectrophotometer). To terminate cell growth and any further protein synthesis, chloramphenicol (final concentration, 20 µg/ml) or tetracycline (final concentration, 10 µg/ml) was added, and cells were placed on ice until assayed for β-galactosidase activity (42). β-Galactosidase assays were repeated at least three times, and fold repression was calculated by dividing the β-galactosidase activity of a ΔarcA strain by the activity of an arcA+ strain. Standard errors for data plotted as “fold repression” were calculated using a formula for propagation of standard error (43).
Overexpression and purification of His6-ArcA.
E. coli BL21(DE3) plysS, containing the PK9431 gene, was grown at 37°C until an OD600 of ~0.4 was reached. A final concentration of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added, and cells were incubated at 30°C. Cells were harvested, suspended in 5 mM imidazole buffer containing 20 mM Tris-Cl (pH 7.9) and 0.5 M NaCl, and lysed by sonication. His6-ArcA was isolated from cell lysates by passing them over a Ni-nitrilotriacetic acid (NTA) column preequilibrated with 5 mM imidazole, washing the column extensively with the same buffer and then with 20 and 50 mM imidazole, and then eluting with 100 mM imidazole. Fractions containing the overexpressed His6-ArcA, determined by electrophoresis, were dialyzed against 50 mM Tris-Cl, pH 7.5, 0.1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 0.2 M NaCl. The His6 tag was removed from ArcA by overnight incubation with tobacco etch virus (TEV) protease at 4°C and passage over a Ni-NTA-agarose column (Qiagen). The protein concentration of ArcA (reported here as monomers) was determined as previously described (4).
DNase I footprinting.
icdA promoter fragments were isolated from pPK9476, pPK9477, pPK9908, pPK9909, pPK9913, pPK9914, and pPK15001 (Table 1) after digestion with XhoI and BamHI. Sequenase version 2.0 (USB Scientific) was used to 3′-end radiolabel the BamHI end of the fragment with [α-32P]dGTP (PerkinElmer). Labeled DNA fragments were isolated from a nondenaturing 5% acrylamide gel and were subsequently purified with Elutip-d columns (Schleicher and Schuell). ArcA was phosphorylated by incubating it with 50 mM disodium carbamyl phosphate (Sigma-Aldrich) in 50 mM Tris, pH 7.9, 150 mM NaCl, and 10 mM MgCl2 for 1 h at 30°C (6) and immediately used in the binding assays. Footprinting assays were performed by incubating phosphorylated ArcA with labeled DNA (~5 nM) for 10 min at 30°C in 40 mM Tris (pH 7.9), 30 mM KCl, 100 µg/ml bovine serum albumin (BSA), and 1 mM DTT followed by the addition of 2 µg/ml DNase I (Worthington) for 30 s. The DNase I reaction was terminated by the addition of sodium acetate and EDTA to final concentrations of 300 mM and 20 mM, respectively. The reaction mix was ethanol precipitated, resuspended in urea loading dye, heated for 60 s at 90°C, and loaded onto a 7 M urea–8% polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) buffer. An A+G ladder was made by formic acid modification of the radiolabeled DNA, followed by piperidine cleavage (44). The reaction products were visualized by phosphorimaging.
ACKNOWLEDGMENTS
We thank Wilma Ross for assistance with DNase I footprinting experiments and Erin Mettert for critically reading the manuscript.
This work was supported by NIH grant GM45844 to P.J.K. D.M.P. was supported in part by the DOE BACTER Program (grant DE-FG02-04ER25627).
Footnotes
Citation Park DM, Kiley PJ. 2014. The influence of repressor DNA binding site architecture on transcriptional control. mBio 5(5):e01684-14. doi:10.1128/mBio.01684-14.
REFERENCES
- 1. Malpica R, Sandoval GR, Rodríguez C, Franco B, Georgellis D. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8:781–795. 10.1089/ars.2006.8.781 [DOI] [PubMed] [Google Scholar]
- 2. Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101:13318–13323. 10.1073/pnas.0403064101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HM, Sanguinetti G, de Mattos JT, Poole RK, Green J. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J. Biol. Chem. 286:10147–10154. 10.1074/jbc.M110.211144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. 2013. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 9:e1003839. 10.1371/journal.pgen.1003839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chao G, Shen J, Tseng CP, Park SJ, Gunsalus RP. 1997. Aerobic regulation of isocitrate dehydrogenase gene (icd) expression in Escherichia coli by the arcA and fnr gene products. J. Bacteriol. 179:4299–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lynch AS, Lin EC. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178:6238–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cunningham L, Georgellis D, Green J, Guest JR. 1998. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: characterisation of an ArcA binding site in the lpd promoter. FEMS Microbiol. Lett. 169:403–408. 10.1111/j.1574-6968.1998.tb13347.x [DOI] [PubMed] [Google Scholar]
- 8. Pellicer MT, Lynch AS, De Wulf P, Boyd D, Aguilar J, Lin EC. 1999. A mutational study of the ArcA-P binding sequences in the aldA promoter of Escherichia coli. Mol. Gen. Genet. 261:170–176. 10.1007/s004380050954 [DOI] [PubMed] [Google Scholar]
- 9. Pellicer MT, Fernandez C, Badía J, Aguilar J, Lin EC, Baldom L. 1999. Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection. Characterization of the glc promoter. J. Biol. Chem. 274:1745–1752. 10.1074/jbc.274.3.1745 [DOI] [PubMed] [Google Scholar]
- 10. Drapal N, Sawers G. 1995. Purification of ArcA and analysis of its specific interaction with the pfl promoter-regulatory region. Mol. Microbiol. 16:597–607. 10.1111/j.1365-2958.1995.tb02422.x [DOI] [PubMed] [Google Scholar]
- 11. Gerasimova AV, Gelfand MS, Makeev VY, Mironov AA, Favorov AV. 2003. ArcA regulator of gamma-proteobacteria identification of the binding signal and description of the regulon. Biophysics 48:S21–S25 [Google Scholar]
- 12. McGuire AM, De Wulf P, Church GM, Lin EC. 1999. A weight matrix for binding recognition by the redox-response regulator ArcA-P of Escherichia coli. Mol. Microbiol. 32:219–221. 10.1046/j.1365-2958.1999.01347.x [DOI] [PubMed] [Google Scholar]
- 13. Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588–12597. 10.1074/jbc.M313454200 [DOI] [PubMed] [Google Scholar]
- 14. Wang X, Gao H, Shen Y, Weinstock GM, Zhou J, Palzkill T. 2008. A high-throughput percentage-of-binding strategy to measure binding energies in DNA-protein interactions: application to genome-scale site discovery. Nucleic Acids Res. 36:4863–4871. 10.1093/nar/gkn477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanco AG, Sola M, Gomis-Rüth FX, Coll M. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701–713. 10.1016/S0969-2126(02)00761-X [DOI] [PubMed] [Google Scholar]
- 16. Toro-Roman A, Mack TR, Stock AM. 2005. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face. J. Mol. Biol. 349:11–26. 10.1016/j.jmb.2005.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao R, Mack TR, Stock AM. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 32:225–234. 10.1016/j.tibs.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Narayanan A, Kumar S, Evrard AN, Paul LN, Yernool DA. 2014. An asymmetric heterodomain interface stabilizes a response regulator-DNA complex. Nat. Commun 5:3282. 10.1038/ncomms4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 276:40873–40879. 10.1074/jbc.M104855200 [DOI] [PubMed] [Google Scholar]
- 20. Mack TR, Gao R, Stock AM. 2009. Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB. J. Mol. Biol. 389:349–364. 10.1016/j.jmb.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barbieri CM, Wu T, Stock AM. 2013. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J. Mol. Biol. 425:1612–1626. 10.1016/j.jmb.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ritzefeld M, Walhorn V, Kleineberg C, Bieker A, Kock K, Herrmann C, Anselmetti D, Sewald N. 2013. Cooperative binding of PhoB(DBD) to its cognate DNA sequence—a combined application of single-molecule and ensemble methods. Biochemistry 52:8177–8186. 10.1021/bi400718r [DOI] [PubMed] [Google Scholar]
- 23. Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–D590. 10.1093/nar/gkq1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider TD. 1997. Information content of individual genetic sequences. J. Theor. Biol. 189:427–441. 10.1006/jtbi.1997.0540 [DOI] [PubMed] [Google Scholar]
- 25. Drew HR, Travers AA. 1984. DNA structural variations in the E. coli tyrT promoter. Cell 37:491–502. 10.1016/0092-8674(84)90379-9 [DOI] [PubMed] [Google Scholar]
- 26. Smith JG, Latiolais JA, Guanga GP, Pennington JD, Silversmith RE, Bourret RB. 2004. A search for amino acid substitutions that universally activate response regulators. Mol. Microbiol. 51:887–901. 10.1046/j.1365-2958.2003.03882.x [DOI] [PubMed] [Google Scholar]
- 27. Ackers GK, Shea MA, Smith FR. 1983. Free energy coupling within macromolecules. The chemical work of ligand binding at the individual sites in co-operative systems. J. Mol. Biol. 170:223–242. 10.1016/S0022-2836(83)80234-4 [DOI] [PubMed] [Google Scholar]
- 28. Schlax PJ, Capp MW, Record MT., Jr. 1995. Inhibition of transcription initiation by lac repressor. J. Mol. Biol. 245:331–350. 10.1006/jmbi.1994.0028 [DOI] [PubMed] [Google Scholar]
- 29. Elledge SJ, Davis RW. 1989. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3:185–197. 10.1101/gad.3.2.185 [DOI] [PubMed] [Google Scholar]
- 30. Lanzer M, Bujard H. 1988. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. U. S. A. 85:8973–8977. 10.1073/pnas.85.23.8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cox RS, III, Surette MG, Elowitz MB. 2007. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3:145. 10.1038/msb4100187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshida T, Qin L, Egger LA, Inouye M. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114–17123. 10.1074/jbc.M602112200 [DOI] [PubMed] [Google Scholar]
- 33. Rampersaud A, Harlocker SL, Inouye M. 1994. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J. Biol. Chem. 269:12559–12566 [PubMed] [Google Scholar]
- 34. Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15–26. 10.1006/jmbi.1996.0298 [DOI] [PubMed] [Google Scholar]
- 35. Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. 2007. Discovery of fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 35:6762–6777. 10.1093/nar/gkm631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Butala M, Zgur-Bertok D, Busby SJ. 2009. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 66:82–93. 10.1007/s00018-008-8378-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135–1160. 10.1128/JB.187.3.1135-1160.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prost JF, Nègre D, Oudot C, Murakami K, Ishihama A, Cozzone AJ, Cortay JC. 1999. Cra-dependent transcriptional activation of the icd gene of Escherichia coli. J. Bacteriol. 181:893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nesbit AD, Giel JL, Rose JC, Kiley PJ. 2009. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J. Mol. Biol. 387:28–41. 10.1016/j.jmb.2009.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schneider TD. 1997. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 25:4408–4415. 10.1093/nar/25.21.4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 43. Ku HH. 1966. Notes on the use of propagation of error formulas. J. Res. Natl. Bureau Stand. 70C:263–273 [Google Scholar]
- 44. Maxam AM, Gilbert W. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499–560. 10.1016/S0076-6879(80)65059-9 [DOI] [PubMed] [Google Scholar]
- 45. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]