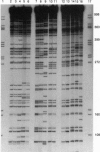
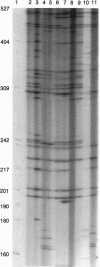
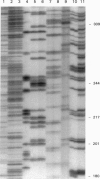
Abstract
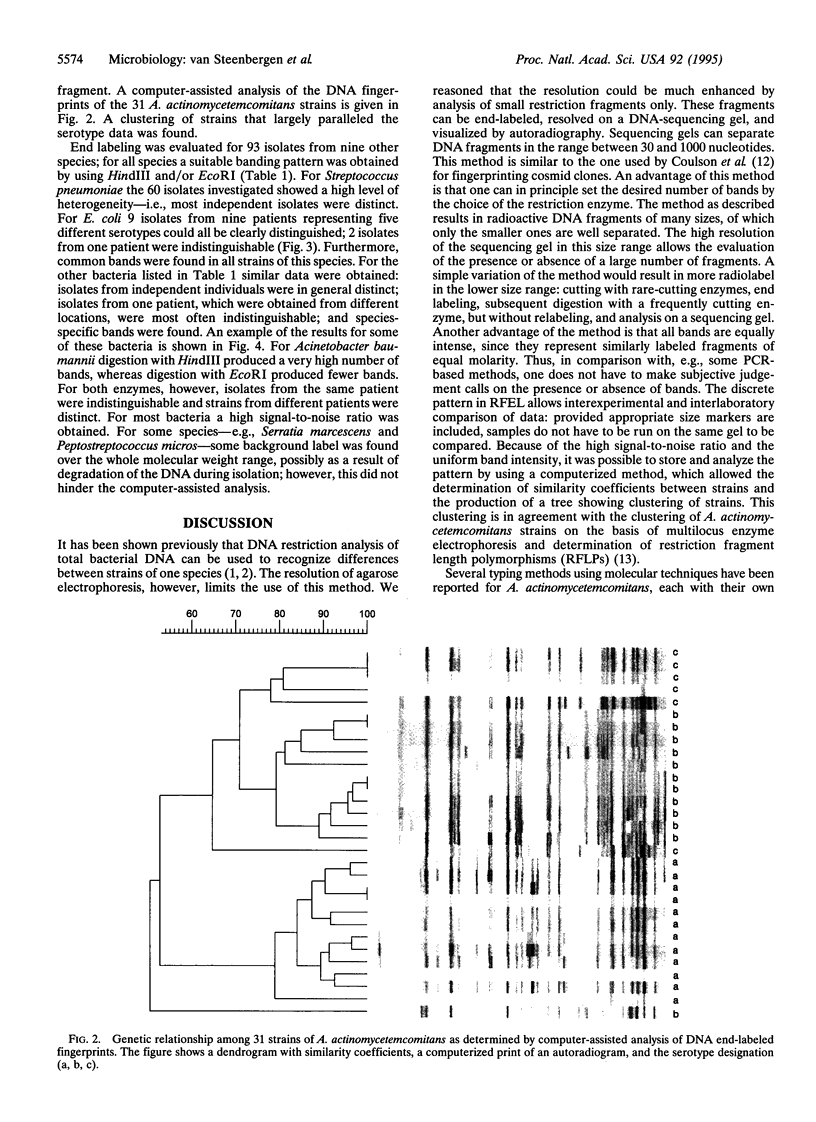
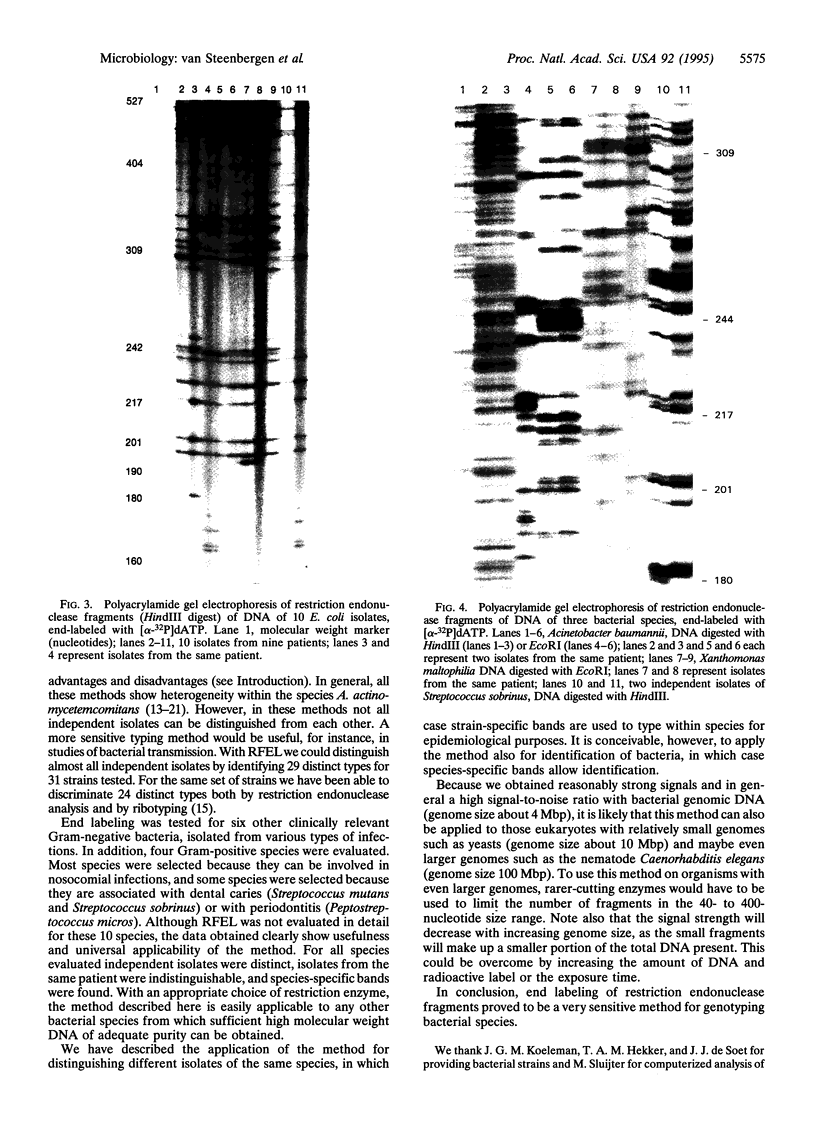
A typing method for bacteria was developed and applied to several species, including Escherichia coli and Actinobacillus actinomycetemcomitans. Total genomic DNA was digested with a restriction endonuclease, and fragments were enabled with [alpha-32P]dATP by using the Klenow fragment of DNA polymerase and separated by electrophoresis in 6% polyacrylamide/8 M urea (sequencing gel). Depending on the restriction endonuclease and the bacterium, the method produced approximately 30-50 well-separated fragments in the size range of 100-400 nucleotides. For A. actinomycetemcomitans, all strains had bands in common. Nevertheless, many polymorphisms could be observed, and the 31 strains tested could be classified into 29 distinct types. Furthermore, serotype-specific fragments could be assigned for the three serotypes investigated. The method described is very sensitive, allowing more distinct types to be distinguished than other commonly used typing methods. When the method was applied to 10 other clinically relevant bacterial species, both species-specific bands and strain-specific bands were found. Isolates from different locations of one patient showed indistinguishable patterns. Computer-assisted analysis of the DNA fingerprints allowed the determination of similarity coefficients. It is concluded that genomic fingerprinting by restriction fragment end labeling (RFEL) is a powerful and generally applicable technique to type bacterial species.
Full text
PDF
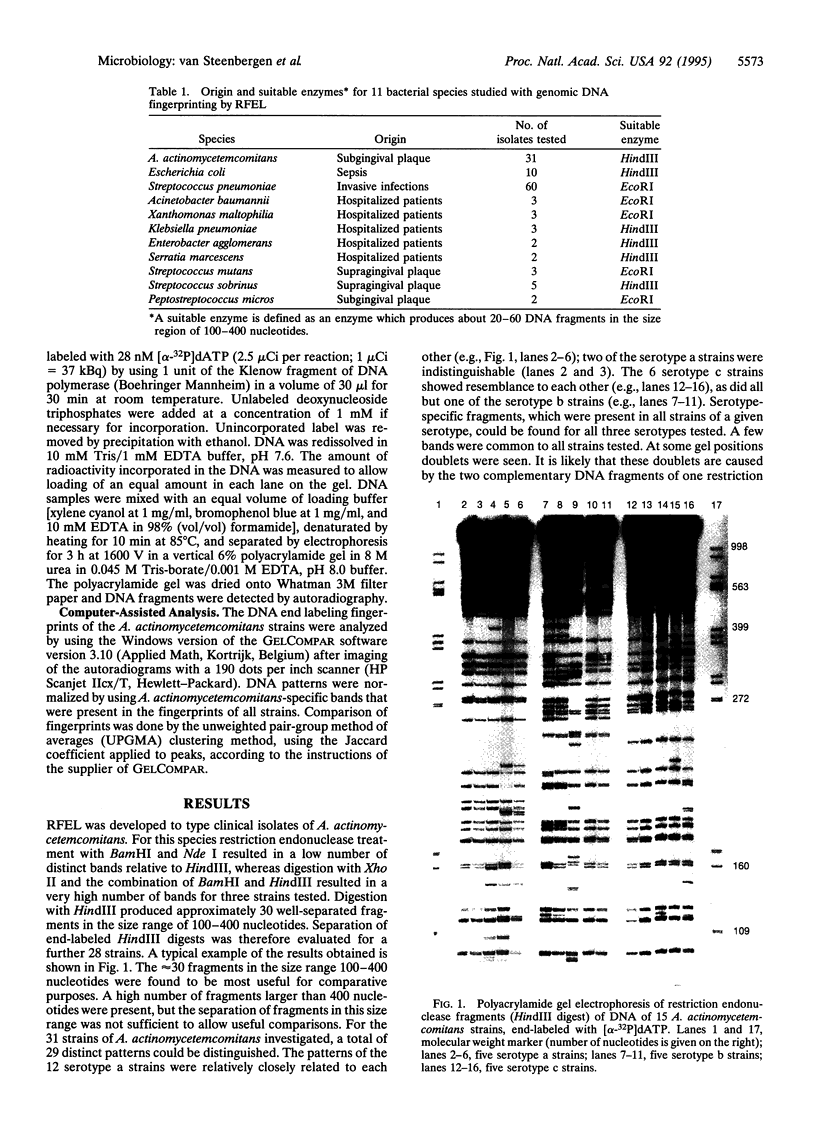

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alaluusua S., Saarela M., Jousimies-Somer H., Asikainen S. Ribotyping shows intrafamilial similarity in Actinobacillus actinomycetemcomitans isolates. Oral Microbiol Immunol. 1993 Aug;8(4):225–229. doi: 10.1111/j.1399-302x.1993.tb00564.x. [DOI] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmür R., McNabb H., van Steenbergen T. J., Baehni P., Mombelli A., van Winkelhoff A. J., Guggenheim B. Seroclassification of hitherto nontypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol Immunol. 1993 Apr;8(2):116–120. doi: 10.1111/j.1399-302x.1993.tb00556.x. [DOI] [PubMed] [Google Scholar]
- Griffen A. L., Leys E. J., Fuerst P. A. Strain identification of Actinobacillus actinomycetemcomitans using the polymerase chain reaction. Oral Microbiol Immunol. 1992 Aug;7(4):240–243. doi: 10.1111/j.1399-302x.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Guthmiller J. M., Kolodrubetz D., Kraig E. A panel of probes detects DNA polymorphisms in human and non-human primate isolates of a periodontal pathogen, Actinobacillus actinomycetemcomitans. Microb Pathog. 1993 Feb;14(2):103–115. doi: 10.1006/mpat.1993.1011. [DOI] [PubMed] [Google Scholar]
- Owen R. J. Chromosomal DNA fingerprinting--a new method of species and strain identification applicable to microbial pathogens. J Med Microbiol. 1989 Oct;30(2):89–99. doi: 10.1099/00222615-30-2-89. [DOI] [PubMed] [Google Scholar]
- Poulsen K., Theilade E., Lally E. T., Demuth D. R., Kilian M. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology. 1994 Aug;140(Pt 8):2049–2060. doi: 10.1099/13500872-140-8-2049. [DOI] [PubMed] [Google Scholar]
- Saarela M., Asikainen S., Jousimies-Somer H., Asikainen T., von Troil-Lindén B., Alaluusua S. Hybridization patterns of Actinobacillus actinomycetemcomitans serotypes a-e detected with an rRNA gene probe. Oral Microbiol Immunol. 1993 Apr;8(2):111–115. doi: 10.1111/j.1399-302x.1993.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Slots J., Listgarten M. A. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988 Feb;15(2):85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- Slots J., Liu Y. B., DiRienzo J. M., Chen C. Evaluating two methods for fingerprinting genomes of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1993 Dec;8(6):337–343. doi: 10.1111/j.1399-302x.1993.tb00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Whittam T. S., Berg C. M., Berg D. E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993 Dec 25;21(25):5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon J. J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985 Jan;12(1):1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Zambon J. J., Sunday G. J., Smutko J. S. Molecular genetic analysis of Actinobacillus actinomycetemcomitans epidemiology. J Periodontol. 1990 Feb;61(2):75–80. doi: 10.1902/jop.1990.61.2.75. [DOI] [PubMed] [Google Scholar]
- van Steenbergen T. J., Bosch-Tijhof C. J., van Winkelhoff A. J., Gmür R., de Graaff J. Comparison of six typing methods for Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1994 Nov;32(11):2769–2774. doi: 10.1128/jcm.32.11.2769-2774.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steenbergen T. J., Van der Velden U., Abbas F., de Graaff J. Microflora and bacterial DNA restriction enzyme analysis in young adults with periodontitis. J Periodontol. 1991 Apr;62(4):235–241. doi: 10.1902/jop.1991.62.4.235. [DOI] [PubMed] [Google Scholar]