Abstract
Study Objectives:
To address whether changes in gene expression in blood cells with sleep loss are different in individuals resistant and sensitive to sleep deprivation.
Design:
Blood draws every 4 h during a 3-day study: 24-h normal baseline, 38 h of continuous wakefulness and subsequent recovery sleep, for a total of 19 time-points per subject, with every 2-h psychomotor vigilance task (PVT) assessment when awake.
Setting:
Sleep laboratory.
Participants:
Fourteen subjects who were previously identified as behaviorally resistant (n = 7) or sensitive (n = 7) to sleep deprivation by PVT.
Intervention:
Thirty-eight hours of continuous wakefulness.
Measurements and Results:
We found 4,481 unique genes with a significant 24-h diurnal rhythm during a normal sleep-wake cycle in blood (false discovery rate [FDR] < 5%). Biological pathways were enriched for biosynthetic processes during sleep. After accounting for circadian effects, two genes (SREBF1 and CPT1A, both involved in lipid metabolism) exhibited small, but significant, linear changes in expression with the duration of sleep deprivation (FDR < 5%). The main change with sleep deprivation was a reduction in the amplitude of the diurnal rhythm of expression of normally cycling probe sets. This reduction was noticeably higher in behaviorally resistant subjects than sensitive subjects, at any given P value. Furthermore, blood cell type enrichment analysis showed that the expression pattern difference between sensitive and resistant subjects is mainly found in cells of myeloid origin, such as monocytes.
Conclusion:
Individual differences in behavioral effects of sleep deprivation are associated with differences in diurnal amplitude of gene expression for genes that show circadian rhythmicity.
Citation:
Arnardottir ES, Nikonova EV, Shockley KR, Podtelezhnikov AA, Anafi RC, Tanis KQ, Maislin G, Stone DJ, Renger JJ, Winrow CJ, Pack AI. Blood-gene expression reveals reduced circadian rhythmicity in individuals resistant to sleep deprivation. SLEEP 2014;37(10):1589-1600.
Keywords: sleep deprivation, gene expression, microarray analysis, circadian rhythm, psychomotor vigilance test
INTRODUCTION
Insufficient sleep length is increasingly common1,2 and has been shown to have serious behavioral consequences such as increased human error-related accidents and decreased cognitive performance.3 Moreover, sleep loss has been shown to accelerate neurodegeneration in mouse models of Alzheimer disease.4
Although sleep deprivation is common, there are substantial interindividual differences in the resulting behavioral impairment,5,6 as reflected in differences in brain activation and cognitive performance.7,8 These differences appear “trait-like” because they are highly reproducible over time in the same individual, are very different between individuals,5,6 and are highly heritable.9 Sleep loss leads to extensive changes in gene expression in different brain regions in mice10 as well as in other organs, including liver,11 heart, and lung.12 Thus, it is conceivable that changes in gene expression produced by sleep loss might be different in those individuals who are behaviorally resistant to sleep loss compared with those who are particularly sensitive.
One approach to assess this hypothesis is to determine whether sleep loss produces changes in gene expression in peripheral blood from humans with different susceptibility to sleep deprivation.13 Surprisingly, in a recent study, acute sleep loss following normal sleep resulted in few genes showing increased (n = 46) or decreased (n = 76) trends in expression as a function of time awake, and the magnitude of expression change of these genes was small.14 Another study identified more genes with altered expression following sleep deprivation, although the level of these changes was extremely small.15 A more robust effect of sleep loss that was observed was a decrease in amplitude of the diurnal rhythmicity of gene expression in peripheral blood during acute sleep deprivation.14 This may be the result of the known effect of sleep loss on binding of the CLOCK/BMAL transcription factor complex to E-boxes of clock genes.16
To address whether changes in gene expression with sleep loss are different in those individuals who are resistant to, or sensitive to, the effects of sleep loss, we assessed subjects who had been previously identified as having widely different neurobehavioral responses to sleep deprivation.9 We reassessed these subjects in this study by psychomotor vigilance reaction task (PVT) every 2 h while awake to assess performance lapses17 with parallel blood draws every 4 h during 24-h normal baseline (sleep and wakefulness), 38 h of continuous wakefulness, and during subsequent recovery sleep, for a total of 19 time-points per subject. The aims of this study were to assess whether these two groups of subjects demonstrate different diurnal transcriptional rhythms at baseline, different changes in these rhythms as a function of sleep state, or different transcriptional patterns as a function of time awake.
We found that subjects who were behaviorally sensitive to sleep deprivation had a less adaptable circadian rhythm of gene expression in blood than those behaviorally resistant to sleep deprivation, who showed a blunting of circadian rhythm in blood while sleep deprived.
MATERIALS AND METHODS
Participants
Based on PVT analysis of 200 twin-pair participants in a previous sleep deprivation study,9 subjects were selected for this study as being resistant and sensitive to sleep deprivation (discussed in Supplemental Methods). Only a single member of a twin pair was recruited. The consent of the Institutional Review Board of the University of Pennsylvania and the Clinical and Translational Research Center of the University of Pennsylvania was granted for this study and informed consent obtained from all research subjects.
Study Procedure
Following actigraphy for 2 w, subjects arrived at the Clinical and Translational Research Center at the University of Pennsylvania, (Philadelphia, PA) in the evening of Day 0 and had 1 night of undisturbed sleep for adjustment. The study started with a baseline assessment starting at 08:00 the next morning (Day 1) with blood sampling every 4 h and PVTs every 2 h (when awake) (see Figure 1). On Day 2 the sleep deprivation started at 08:00 and lasted until 22:00 on Day 3, a total of 38 h. Thereafter the subjects were allowed one night of recovery sleep, from 22:00 to 08:00 on Day 4 (final blood draw and PVT) (see Figure 1). For more detailed information on the sleep deprivation visit, the initial telephone screen and the baseline visit, see Supplemental Methods.
Figure 1.
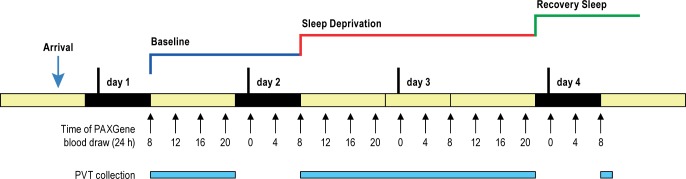
Experimental design. Time points for blood draws (every 4 h across wake [yellow] and sleep periods [black]) and psychomotor vigilance test (PVT) data collection (every 2 h while awake). A total of 27 PVTs and 19 blood draws per subject were performed throughout the study period.
Blood Sampling and Microarray
Blood was sampled every 4 h from 08:00 on the baseline day, throughout 38 h of sleep deprivation and 10 h of recovery sleep, for a total of 19 time points (see Figure 1). 2.5 mL of blood were collected into PAXgene” Blood RNA Tubes (PreAnalytiX GmbH, Hombrechtikon, Switzerland) at room temperature. After blood draw, the tubes were gently inverted and kept at room temperature for less than 10 min prior to freezing at -20°C. Samples were moved to -80°C freezer 24 h after completion of all sample collection in accordance with the instructions from PreAnalytiX (www.preanalytix.com). Two of 266 total blood samples (19 time points per 14 subjects) could not be collected because of faulty intravenous devices.
Messenger RNA Isolation, Microarray Profiling and Processing
Messenger RNA (mRNA) isolation and analysis was performed by the Covance Gene Expression Laboratory (Seattle, WA) using standard protocols. Briefly, Paxgene tubes were processed according to manufacturer's protocol. Total RNA was isolated using TRIzol water solution (4:1 ratio). Chloroform (100%) was added to the TRIzol/GITC lysate (1:5 ratio) to facilitate separation of the organic and aqueous components using the phaselock (Eppendorf) system. The aqueous supernatant was further purified using the Promega SV-96 total RNA kit, incorporating a deoxyribonuclease treatment during the procedure. Isolated total RNA samples were then assayed for quality (Agilent Bioanalyzer, Agilent Technologies, Inc., Santa Clara, CA) and yield (Quant-iT” RiboGreen®, Life Technologies, Carlsbad, CA) metrics prior to amplification. Fifty ng total RNA samples were amplified and labeled using a custom automated version of the NuGEN Ovation WB protocol (NuGEN Technologies, Inc., San Carlos, CA) before hybridization to custom human Affymetrix GeneChip® microarrays arrays (Affymetrix, Inc., Santa Clara, CA) containing 52,378 probe sets (representing 18,983 unique gene symbols) designed to monitor additional annotated transcript variants and poly A sites compared to commercially available microarrays (GEO Accession #GPL10379, www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL10379). All analyses in this paper are done at probe set level and not gene level because of the custom design of the microarray. Hybridization, labeling, and scanning were conducted using Affymetrix ovens, fluidics stations, and scanners following recommended protocols (NuGEN Technologies, Inc., San Carlos, CA). Two hundred sixty-four of the 266 planned samples were collected successfully (14 subjects × 19 time points). Upon extraction, 13 of the 264 samples failed Covance RNA quality control metrics. Two additional samples did not pass Covance amplification quality control metrics. Therefore, a total of 249 samples were hybridized to micro-arrays. All 249 microarray profiles passed standard quality control metrics and were included in downstream analysis. A principal component analysis showed no obvious outliers after hybridization and that the data clustered mainly by subject.
Probe intensity data were processed using RMAExpress 1.0.4 (http://rmaexpress.bmbolstad.com/).18 Data quality was assessed using boxplots and histograms of raw signal intensities. Normalization was performed using the robust multiarray average (RMA) method19 to form one expression measure per probe set per array. Briefly, the RMA method is a three-step process that adjusts the background of perfect match (PM) probes, applies a quantile normalization of the corrected PM values, and calculates final expression measures using the Tukey median polish algorithm.
Agglomerative hierarchical bottom-up clustering was performed, starting with every single object (gene or sample) in a single cluster. Then, in each successive iteration, it agglomerated the closest pair of clusters according to our selected distance metric, until all of the data were in one cluster. We used Pearson correlation distance (1 - correlation) as the distance metric. The hierarchy within the final cluster has the following properties: clusters generated in early stages are nested in those generated in later stages and clusters with different sizes in the tree can be valuable for discovery. Matlab statistical toolbox R2010b (7.11.0) was used to perform clustering (The Math-Works, Inc., Natick, MA).
The data discussed in this paper have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus20 and are accessible through GEO Series accession number GSE56931 study (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56931).
Statistical Analyses
Statistical analysis for demographics, PVT measurements, and subjective reporting were performed using STATA 11.0 and SAS 9.2 for intraclass correlation coefficient (ICC) analysis. For bivariate analysis, the chi-square test and t-test were used for nominal and continuous variables respectively. All microarray statistical tests were based on the Fs statistic21 implemented within the R/maanova software package.22 P values were adjusted according to the false discovery rate (FDR) procedure of Benjamini and Hochberg23 using the p.adjust() function in R. A 5% FDR threshold was used for all analyses unless otherwise noted.
We used a linear mixed-model approach to detect probe sets with circadian patterns. Gene expression measures (yij) for each probe set are fit to the model:
where μ is the overall mean, β1 and β2 are the coefficients of regression for the circadian effect terms, tij for the jth observation on the ith patient, si is the fixed effect of sex (female or male) for the ith patient, Pi is the random patient effect and εij refers to unknown random errors. Collecting repeated measurements on the same subjects required the random effects parameter (Pi) to avoid violating the assumption of independent error terms. Random effects are assumed to have a normal distribution with a mean of 0 and a variance of σ2, where σ2 is estimated using the restricted maximum likelihood method.24 Probe sets with significant circadian effects were found by testing the hypothesis β1 = β2 = 0 using data subset for the baseline condition (Day 1, 08:00 until Day 2, 20:00). Amplitude was calculated as:
and phase was calculated as the angle between the positive x-axis and the vector from the origin to the point given by (β1, β2) using the two-argument arctangent function in R. Linear trends during sleep deprivation (Day 3, 12:00 until Day 3, 20:00) were found by adding the term (β3t) to the model and testing the hypothesis β3 = 0.
Expression differences between baseline and sleep deprivation states (analysis 2) or baseline, sleep deprivation and recovery (analysis 3) were identified using the linear mixed-model:
where STATE is incorporated as a fixed effect and the other terms are described previously. For analysis 2, the data were subset for “24:00, 04:00, 08:00, 12:00, 16:00, and 20:00” from each state. For analysis 3, the data was subset for “24:00, 04:00, 08:00” from each state. A number of other models were evaluated that incorporated interactions between sex (female or male) or subject status (sensitive or resistant) and other model terms. However, no sex- or subject-dependent interactions were found in any scenario and therefore these models are not described here. Random effects were assumed to be independently normally distributed with a mean of 0.
Additionally, the results from the linear modeling approach to assess circadian genes were compared to results obtained from an analysis involving cosinor analysis25 as well as the non-parametric JTK_Cycle algorithm.26 All analyses were reported with FDR < 5% and were only based on data prior to the actual sleep deprivation.
The cosinor analysis was applied to within-patient normalized expression data. To remove baseline interpatient variability, all data for each subject were ratioed to the average of the six baseline time points covering the first 24 h of the study, so that the average of all genes over this period is zero for each subject. The expression Yi, was modeled using multiple regression:
The regression coefficients ai and bi were used to determine the amplitude:
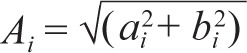 |
and the phase in hours, 24atan2(bi,ai) / 2π. The latter uses two-argument arctangent that correctly assigns the phase quadrant over the entire 24-h period.
The JTK_cycle algorithm uses the Jonckheere-Terpstra (JT) statistical test to assess the relative ordering of time course expression data and then applies Kendall's tau test to assess the rank correlation between this ordering and a set of oscillatory reference curves.26 Given the wide intrasubject variation in baseline expression values, the time course data for each gene and subject was normalized by the median expression of that gene in that subject prior to application of the JTK_Cycle algorithm.
Significant probe sets were identified by a FDR < 5% threshold and were cross-referenced for enrichment of biological processes and pathways against multiple public and commercial databases, specifically GeneGo (www.genego.com), Gene Ontology (www.geneontology.org), KEGG (www.genime.jp/kegg/), and Ingenuity (www.ingenuity.com). The hypergeometric distribution (probability distribution after sampling without replacement) was used to calculate over-representation of P values in the combined databases. Bonferroni corrected P values (expectation, or E-values) were calculated to control for multiple comparisons across all resources. E-values below 0.1 were considered significant. Lights on and lights off periods were defined as times 8:00 – 22:00 and 22:00 – 8:00, respectively. Probe sets with peak expression during these time periods were grouped for blood cell type enrichment analyses.
Additionally, the Gene Set Enrichment Analysis Tool from the Broad Institute (GSEA v 2.08, Cambridge, MA),27 was used to identify functionally related gene sets that showed overall expression changes in any of the employed statistical tests. GSEA identifies gene sets that are enriched toward the top or bottom of a ranked list of genes.28 Core genes accounting for the enrichment signal were defined by the leading-edge subset as defined by the GSEA program.27 The probes were ranked by the value of the F-statistic for the relevant statistical test and the “preranked” tool in the GSEA package was used with default settings. Gene sets describing KEGG pathways29 were obtained from the GSEA molecular signatures databases (c2.cp.kegg. v3.1). Pathways that met a FDR threshold of 10% were considered significant. We followed the recommended guidelines for each application described above for selecting thresholds for statistical significance in order to obtain a reasonably sized list of genes or pathways to explore.
For blood cell type enrichment analysis, we used previously published blood gene expression Ramilo modules30 to identify the cell types and/or biological processes that showed significant enrichment within the identified circadian signature, during the lights on and lights off periods. Hypergeometric P values were calculated for the overlap between genes that belonged to a Ramilo module and all circadian genes (8,064 probe sets, FDR < 5%). FDRs were calculated using the Benjamini-Hochberg method for the 28 assessed modules.
RESULTS
Demographic Data and Behavioral Analysis
The 14 subjects studied were five males and nine females. Eleven were Caucasian and 3 African-American, with an average ± standard deviation age 29.7 ± 8.9 y and mean body mass index of 25.4 ± 3.6 kg/m2. There were no significant demographic differences between the seven sensitive and seven resistant subjects to sleep deprivation (Table S1, supplemental material).
Subjects were defined as neurobehaviorally sensitive and resistant to sleep deprivation based on performance in a PVT17 during sleep deprivation in a prior study9 as measured by the number of performance lapses (see Methods). We specifically recruited seven of the most resistant and seven of the most sensitive subjects to the effects of sleep loss (see Supplemental Methods). During the extended wakefulness period of the present study, subjects performed PVTs every 2 h (Figure 1). Both resistant and sensitive subjects had an increase in the number of performance lapses during the progression of sleep deprivation but the increase was significantly higher for the sensitive group (Figure S1, supplemental material). No significant differences were found between sensitive and resistant subjects in subjective sleepiness, mood, and exhaustion measures during sleep deprivation (Table S2, supplemental material). To confirm that the neurobiological response to sleep deprivation demonstrates trait-like stability, we compared the PVT and subjective sleepiness scores from the previous and current trials (these trials were separated by a mean ± standard deviation of 885.6 ± 507 days). The highest ICC found was for the mean PVT lapses in the last 24 h of sleep deprivation and in the linear slope of PVT lapses during sleep deprivation after adjustment for circadian effects (ICC 0.61 and 0.56, respectively; see Table S3, supplemental material). Thus, despite the large time difference between the repeated tests, there was substantial reproducibility of behavioral response to sleep loss.
During the baseline period of the normal sleep/wake cycle and during sleep deprivation, as well as recovery sleep, blood was repeatedly obtained every 4 h and collected in PAXgene tubes (Figure 1).
Circadian Signature of Gene Expression Under Normal Sleep-Wake Conditions
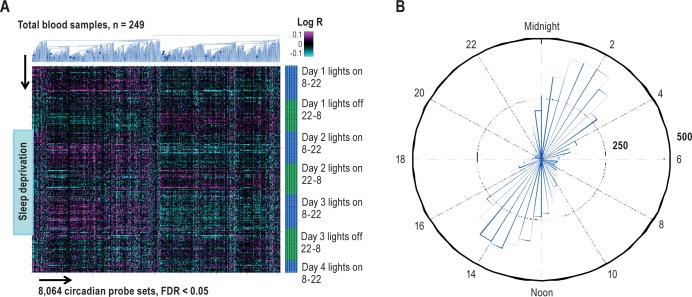
A mixed-effects linear regression model incorporating effects for sex and random effects for subjects was used to identify genes that cycled with a 24-h period during the baseline protocol preceding sleep deprivation (see Methods). All subjects and all time points prior to actual sleep deprivation (Day 1, 08:00 – Day 2, 20:00) were included in the analysis. We found 4,481 unique cycling genes (8,064 probe sets) out of a total of 18,983 genes on the array (52,378 probe sets) using a FDR threshold below 5%. This corresponds to 23.6% of all unique genes and 15.4% of all tested probe sets on the array used in this study. Diurnal changes in gene expression are shown in Figure 2A.
Figure 2.
A robust circadian signature identified in human blood. (A) Agglomerative hierarchical clustering (blue dendrogram) of the mixed-model 8,064 probes with significant 24-h circadian changes (false discovery rate [FDR] < 5%, x-axis) ordered by collection time on y-axis, mixed subjects (y-axis, total blood samples n = 249). The total sleep deprivation period is shown by blue block (start: day 2, 08:00, end: day 3, 10:00). Values shown are the log-transformed ratios (Log R) of observed expression values to the corresponding average across the initial baseline 24-h period for each subject. (B) The phase distribution of peak expression of the 8,064 probe sets (or 4,481 genes) with significant 24-h circadian cycling during baseline sleep and wake: 48-bin histogram (30 min each across 24 h) shows the number of probe sets that reach their maximum expression in a particular half-hour period. The peak time of day is calculated from the sine and cosine regression coefficients in the statistical model.
We compared the mixed-model 8,064 probe sets identified with those obtained by 2 other published approaches to identify genes with cycling expression, i.e., cosinor analysis25 (8,909 probe sets) and the JTK_cycle algorithm31 (7,924 probe sets). The results from all three approaches were highly overlapping with 6,465 (80%) of probe sets being identified as significantly cycling by all three analytical approaches (FDR < 5%). No significant interactive effect on diurnal changes in gene expression involving subject status (sensitive or resistant) was found during the normal baseline condition (data not shown).
Most of the mixed-model 8,064 circadian probe sets (4,481 unique genes) peaked between 01:00-03:00 or 13:00-15:00 as shown in Figure 2B. Overall, a much higher number of biological functional categories were significantly enriched among genes that peaked during the day, 08:00-22:00 (266 pathways) than during the night, 22:00 – 08:00 (71 pathways) (Table S4, supplemental material). The biggest portion of “day pathways” (195 in total) were identified between 10:00 and 14:00. Immune system biological functional categories dominated during the day, whereas RNA processing and protein synthesis were top functions identified during the night.
We also assessed the diurnal rhythm of known core clock genes32–34 in blood and found that most of them showed a robust diurnal rhythm. Specifically, several of the clock genes analyzed, including NPAS2, ARNTL (BMAL1), CSNK1D, NR1D2(REVERB), and DBP peaked near the same time as the majority of cycling genes identified (around 02:00 and 14:00). But others such as PER1, PER2, PER3, and CSNK1E peaked 3 - 6 h prior to the 14:00 peak (Figure S2, supplemental material). Other canonical circadian genes, including CRY1, CRY2, RORA, RORB, RORC, DEC1, BHLHE41 (DEC2), and CLOCK, showed some variation in expression but did not pass the FDR < 5% threshold. No differences in the diurnal rhythm of clock genes were observed in sensitive and resistant subjects (Figure S2).
Effects of Sleep Deprivation on Gene Expression
We tested for changes in gene expression with prolonged sleep deprivation after accounting for circadian effects and sex. We used three different analysis strategies to find: (1) genes with a significant linear change (up or down) in expression as a function of time awake during sleep deprivation after accounting for circadian effects; (2) genes with significant mean differences in expression between the baseline and sleep deprivation days comparing the same time of day; and (3) genes with differences in expression during habitual sleep hours (12:00, 04:00, 08:00) among the three “states,” i.e., normal sleep, sleep deprivation and recovery sleep. These final two comparisons mitigated circadian effects by doing the analysis at the same time of day.
Temporal Analysis: Linear Changes in Gene Expression
At the FDR threshold of 5%, there were only two significant unique genes (three probe sets), SREBF1 and CPT1A, across all subjects that showed a linear trend with time awake. On average, SREBF1 had decreased expression while CPT1A showed increased expression during sleep deprivation. CPT1A also had a significant circadian rhythm in expression but SREBF1 did not based on the FDR < 5% threshold. The observed effect of sleep deprivation on the expression of these genes was very minor (linear slope of -0.03 and 0.06 Ig/h, respectively) (Figure 3). Both of these genes are involved in the regulation of lipid metabolism, specifically regulation of acetyl-CoA carboxylase 1 activity in lipogenic tissue. To compliment this analysis of individual probe sets, we did a gene enrichment analysis (GSEA)27 on the same normalized data using the KEGG Pathway.29 We found one significant biological class; fatty acid metabolism (FDR = 0.08) with CPT1A being the top core enrichment gene. We examined nine of 13 core enriched genes in fatty acid metabolism in our study and found that they had a significant but small linear effect with the same trend (four positive and five negative linear slopes ranging from -0.02 to 0.06). There was no difference between sensitive and resistant subjects.
Figure 3.
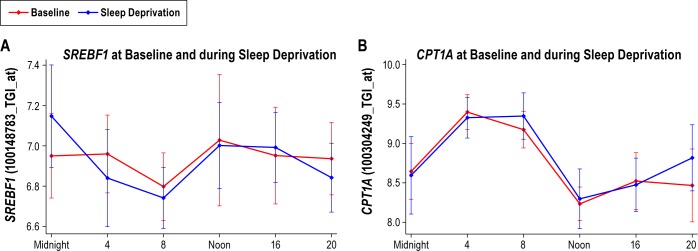
The two genes with a significant linear trend in expression with sleep deprivation (false discovery rate < 5%); (A) SREBF1 and (B) CPT1A. Data are shown as mean ± standard deviation in normalized intensity on y-axis. The linear effects of sleep deprivation were very small for both genes: -0.03 and 0.06, respectively. No difference was found between sensitive and resistant subjects.
State Analysis: Baseline Versus Sleep Deprivation
A second analysis was used to assess whether significant changes in gene expression were found for a pairwise 24-h comparison of baseline sleep/wake and sleep deprivation. No significant differences in gene expression between baseline and sleep deprivation were found (FDR < 5%) and no significant biological functional classes were found by GSEA on the same normalized data (KEGG Pathway, FDR 5%)29.
Sleep Time Analysis: Baseline, Sleep Deprivation, and Recovery Sleep
A third approach was used to maximize our ability to detect the possible effect of state on gene expression by analyzing all three different states (normal sleep, sleep deprivation, and recovery sleep) simultaneously, using the habitual “sleep hours” only (12:00, 04:00, 08:00). Forty-eight unique genes (67 probe sets) were found to differ significantly between the three different states (FDR < 5%, Table S5, supplemental material).
Post hoc analysis was performed to assess the significance of differences between normal sleep, sleep deprivation, and recovery sleep. Out of 48 “state” responsive genes, comparing normal sleep to sleep deprivation, 12 genes decreased expression from normal sleep to sleep deprivation (fold change range -1.05 to -1.16), 18 genes increased expression (fold change range 1.06 to 1.39) and 18 did not change significantly. Comparing sleep deprivation to recovery sleep, 8, 29, and 11 genes decreased, increased expression, or did not change significantly, respectively. Similarly, the fold changes were small between enforced wakefulness and recovery sleep with a range of 1.23 to -1.42. There was also no difference between sensitive and resistant subjects between the three different states and we found no significant biological enrichment for those 48 genes by either multiple database or GSEA approaches.
Effect of Sleep Loss on Diurnal Rhythm of Gene Expression
Because previous studies have shown that sleep loss affects the amplitude of diurnal rhythm of gene expression,14 we questioned whether sleep loss affected the diurnal amplitude differently in resistant and sensitive subjects.
We first compared the distribution of cycling gene probes using different P values across the 24 h at baseline and during the sleep deprivation period in resistant and sensitive subjects for all 52,378 probes (Figure 4). At baseline, sensitive subjects with a high number of PVT lapses and resistant subjects with a low number of PVT lapses had similar numbers of significantly cycling probe sets. However, during sleep deprivation, resistant subjects had far fewer differentially cycling probe sets than sensitive subjects at any given P value (Figure 4).
Figure 4.
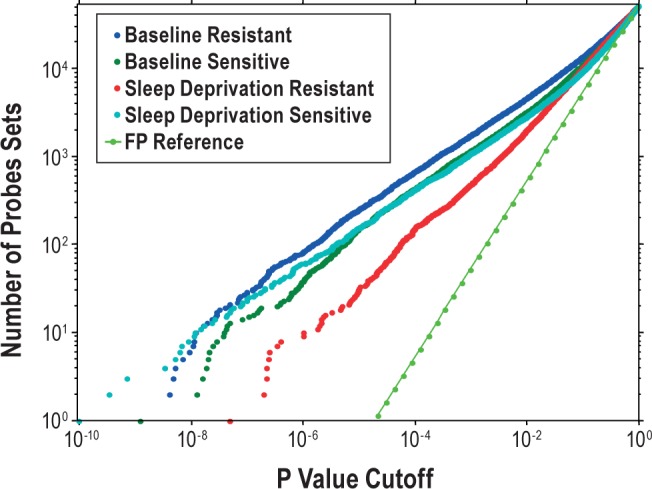
Decreased number of cycling probe sets in behaviorally resistant subjects during sleep deprivation. The graph shows the cumulative distribution of the number of cycling probe sets across 24 h (y-axis) below a specified P value (x-axis) during baseline and sleep deprivation. Data are shown for behaviorally sensitive subjects and resistant subjects separately. Although both groups are almost identical at baseline, the resistant subjects show markedly less cycling probe sets during sleep deprivation than sensitive subjects. The reference line represents the expected number of false-positives (FP) calculated based on the uniform P value distribution that is expected under the null hypothesis.
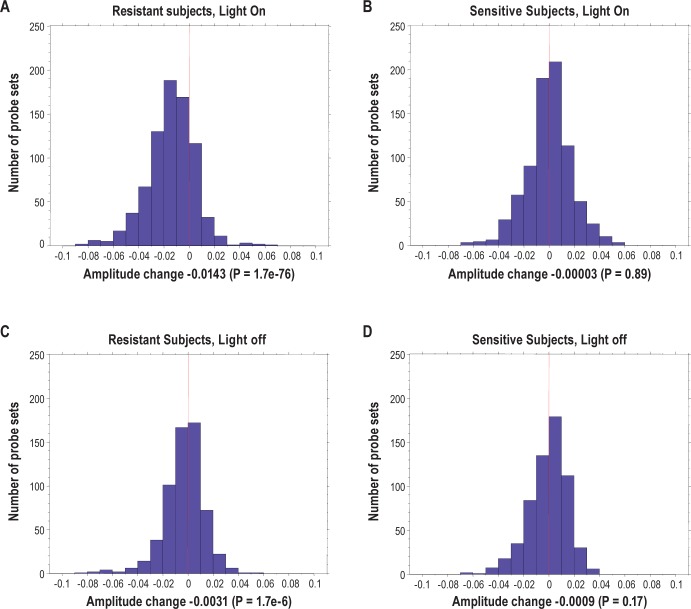
To further investigate the difference between sensitive and resistant subjects, we looked at the amplitude changes in the diurnal rhythm of gene expression going from baseline to sleep deprivation in the two groups. We specifically examined the most robust circadian genes by assessing the 1,397 probe sets that cycled in both sensitive and resistant subjects at an FDR < 5% during baseline (when groups were analyzed separately), broken down into sets that peaked during the light-on and light-off periods, 788 and 609 probe sets, respectively.
We found that in sensitive subjects with a high number of lapses during sleep deprivation, there were no significant changes in amplitude for diurnal cycling between baseline and sleep deprivation either in the lights-on (Figure 5B, paired t-test P = 0.89) or lights-off periods (Figure 5D, paired t-test P = 0.17). In contrast, in subjects resistant to sleep deprivation there was a highly significant decrease in circadian probe set amplitude in the lights-on period (Figure 5A, paired t-test P = 1.7e-76) and a smaller but significant effect in the lights- off period (Figure 5C, paired t-test P = 1.7e-6).
Figure 5.
Distribution of amplitude changes in behaviourally resistant (A and C) and sensitive (B and D) subjects from baseline to sleep deprivation using 1,397 most robust circadian probe sets (false discovery rate < 5%). Probe sets are divided into lights on (A and B) and lights off (C and D) based on their peak expression time. No significant changes are found in behaviorally sensitive subjects but a highly significant decrease in amplitude is found in resistant subjects, especially during lights on. The amplitude change is shown as sleep deprivation minus baseline.
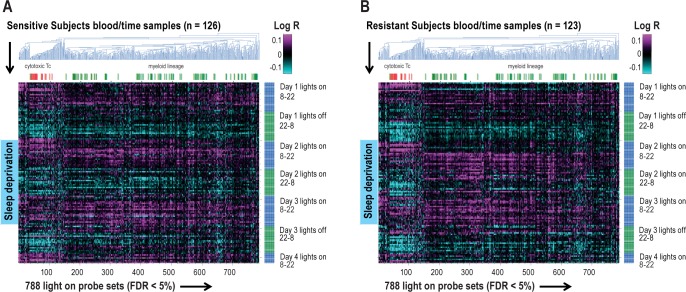
To better understand the biological meaning of this observation, we compared both the primary circadian signature (8,064 probe sets, FDR < 5%) and the subset most effected in resistant subjects (the 788 probe sets with peak expression during the lights-on period) to published expression modules of specific blood cell types.30 For the primary circadian signature across all subjects, we found that probe sets expressed higher during the day than night were significantly enriched for myeloid lineage (M1.5, M2.6, e.g., monocytes) and cytotoxic T cells (M2.1, see Table S6, supplemental material). During the night, the higher expressed probe sets were enriched for T cells (M2.8), and B cells (M1.3, see Table S6). Of note, platelet-, plasma cell-, neutrophil-, and hemoglobin-associated gene sets had no significant day-night oscillations (not shown). The signature 788 probe sets that demonstrated expression pattern differences between sensitive and resistant subjects were also enriched with genes from myeloid lineage and cytotoxic T cell modules (Figure 6A and 6B, respectively). Agglomerative clustering of these 788 probe sets suggested that the myeloid lineage was associated with the largest pattern of amplitude change among these genes (Figure 6).
Figure 6.
Interindividual circadian differences in response to sleep deprivation. A reduction in the pattern of diurnal cycling during sleep deprivation is not found in behaviourally sensitive subjects (A), only in resistant subjects (B). The heat maps represent agglomerative hierarchical clustering of the 788 probe sets that peak during light on in both (A) sensitive and (B) resistant subjects, respectively (false discovery rate [FDR] < 5%), ordered by time (y-axis) in (A) sensitive and (B) resistant subjects, respectively. The log-transformed ratios (Log R) of observed expression values relative to the corresponding average across the initial baseline 24-h period for each subject are shown. There is a much larger reduction in the amplitude of diurnal cycling (amplitude) of the 788 probe sets in resistant subjects (B) than in sensitive subjects (A) when comparing the baseline and sleep deprivation periods, which predominantly was represented by myeloid lineage cells (green labeled). Cytotoxic T cell gene expression is shown in red d.
DISCUSSION
We found that a large percentage of genes expressed in blood had a diurnal rhythm. The major difference between subjects who are resistant to sleep loss and those who are not is in the effect of sleep deprivation on diurnal rhythmicity of gene expression. Those who are resistant to the effects of sleep loss showed a significant reduction in the amplitude of the diurnal cycling of gene expression, whereas those who are sensitive did not, a potential molecular mechanism to the individual differential response to sleep deprivation.
Circadian Changes in Gene Expression
Earlier studies have examined circadian signatures in the mouse brain35 and mouse peripheral tissues such as liver, heart, and fat tissue.35–37 These studies have found that circa-dian oscillations occur in 8-21% of the gene transcripts.35,36,38 The percentage of transcripts cycling in these mice tissues is comparable to the 23.6% of the unique genes and 15.4% of probe sets cycling in blood in humans that we observed, despite species differences between studies. Two recently published studies on the circadian human metabolome have also shown cycling of 15% of the metabolome in plasma and saliva39 and that constructing a blood metabolome timetable to determine internal clock time is possible.40 A recently published study by Möller-Levet et al.14 have also shown cycling of 9% of genes in human blood cells under constant routine conditions. Similar to our study, they found a biphasic peak in the expression of circadian genes where most genes peak at the same time of day.14 However, the peaks found by Möller-Levet et al.14 were delayed by approximately 2 h compared to our results. This phase difference may be because of interference caused by the inclusion of a linear regression term related to time awake in their model in addition to the circadian terms. When assessed in our data, the linear term interfered with phase determination if the assessment period was 24 h. The phase difference may also reflect the use of melatonin rhythms to establish a baseline time scale in the study by Möller-Levet et al.
The oscillations of known core clock genes32–34 were assessed specifically in our study. We found that ARNTL (BMAL1), NPAS2, NR1D1(REV-ERB), PER1, PER2, and PER3, as well as CSNK1D and CSNK1E, had a circadian rhythm in blood cells. This is in agreement with previous studies looking at circadian gene expression in several peripheral tissues.36,37,41–44 However, a circadian rhythm for CRY1, CRY2, RORA, RORB, RORC, DEC1, BHLHE41 (DEC2), and CLOCK was not observed. CRY1 and CRY2 have been found to have a circadian rhythm in some tissues43,44 but not others.35–37 RORA has been shown to have only a slight oscillation in many peripheral tissues41,42 and RORC to have various rhythms in different peripheral tissues.42 CLOCK was not found to have a circadian expression pattern in agreement with earlier studies in blood and other peripheral tissues.35–37,43,45 As mentioned in the results, the circadian genes PER1, PER2, and PER3 preceded the peak expression of most cycling genes, suggesting they are upstream of the later changes. James et al.46 similarly found that PER1 and PER2 expression in blood peaked around the time of awakening. Similar to our findings, they also observed that ARNTL(BMAL1) peaked around 14:00.
Immune processes were the most significantly enriched biological functions among the genes peaking in expression during the morning and afternoon time blocks (06:00-10:00, 13:00-14:00, and 14:00-18:00). A few hours prior to lights off/normal sleep (18:00-22:00 pm), CD40 antigen signaling through tumor necrosis factor receptor superfamily member 5 was most significant. During the nighttime hours, specifically, between 22:00-02:00 and 02:00-06:00, protein biosynthesis and RNA processing were the most enriched biological functions, respectively. Interestingly, these same biological pathways were found previously to peak during sleep in the mouse brain.47
Sleep Deprivation Changes in Gene Expression
Our study shows very stable interindividual differences in neurobehavioral impairment to sleep deprivation, for a longer time period than has previously been shown (average 2.4 y between our studies), confirming and extending the findings of Van Dongen et al.5 and in agreement with Kuna et al.,9 which showed a large effect of heritability on performance during sleep deprivation.
Our results show, however, a very limited transcriptional signature for sleep loss. Only two genes, SREBF1 and CPT1A, were found to have significantly altered linear expression (FDR < 5%) during sleep deprivation compared to a normal sleep/wake cycle. SREBF1, involved in lipid synthesis,48 decreased expression whereas CPT1A, also involved in lipid metabolism,49 increased expression as sleep deprivation progressed. These changes were very modest in scale. SREBF1 has previously been found to be decreased in the brain of mice (cerebral cortex and hypothalamus) during sleep deprivation47 but no significant changes were found for CPT1A in the brain in the same study.47 Macromolecule biosynthesis, including cholesterol synthesis and lipid transport, was found to be a key function of sleep in mouse brain47 in accordance with our results as SREBF1 and CPT1A are involved in lipid synthesis48 and lipid metabolism,49 respectively. Interestingly, studies in mouse brain found significant linear changes in thousands of genes in the brain. Moreover, changes in expression of a large number of genes in specific biological pathways between sleeping and sleep deprived animals have been found in liver, heart, and lung.11,12 These studies were conducted, however, in mice with the same genotype, thereby reducing the effect of interindividual differences. We did find 48 genes with a sleep signature when comparing normal sleep to sleep deprivation and recovery sleep during the night hours only. However, all changes were very modest and no enriched biological categories were found among the differentially expressed genes. Therefore, based on our data, assessment of individual genes in blood will not show whether a person is sensitive or resistant to sleep deprivation.
Our results can be compared to other studies of changes in blood-gene expression with sleep deprivation. A recent study by Pellegrino et al.15 found approximately 500 genes changing expression when they compared a normal sleep cycle following 2 nights of sleep deprivation and following recovery sleep, at the same diurnal time. The study design is different in that Pellegrino et al. compared expression at one time of day only (08:00) in the three conditions. However, similar to our study, all changes are extremely small with fold changes ranging from -1.18 to 1.18. The difference in number of genes found between these two studies may lie in the variations in study design and in stringency of statistical thresholds. Only three of the 50 genes found significant in our study are also found in the study by Pellegrino et al. (PSMF1, SLC25A37 and TUBB2A) and all three genes are downregulated after recovery sleep compared to sleep deprivation in their study but upregulated in ours.
Another recent study by Möller-Levet et al.14 compared the gene expression profiles of 26 subjects during sleep deprivation both after 1 w of normal sleep and 1 w of insufficient sleep (≤ 6 h per night) in a crossover design. This study found 122 genes with either a linear increase (46 genes) or decrease (76 genes) in expression during sleep deprivation following a normal sleep/ wake cycle. The number of genes increased to 856 genes when sleep deprivation was performed following 1 w of insufficient sleep (368 up and 488 down). Surprisingly, even though they used a within-subject design, only 53 of the original 122 genes that showed a linear change in expression in sleep deprivation following a normal sleep/wake cycle continued to do so after 1 w of insufficient sleep, indicating that the expression changes found in sleep deprivation can be quite varied even within individuals. As with our study and the Pellegrino study, all changes were small. Möller-Levet et al.14 used a different statistical approach than that used here. They first assessed within individual subjects genes that changed expression with sleep deprivation and then looked at the number of subjects in which the same gene was found. They did not assess change averaged across all subjects. Of the large number of genes found to change in any way in the study by Möller-Levet et al. because of sleep deprivation, only eight were also found significant in our study (ALPL, ANXA3, CD59, EML4, FOSB, NRG1, RIOK3, TMOD1). It is, however, difficult to assess whether the changes are in the same direction because the study by Möller-Levet et al. did not measure gene expression during a normal wake/sleep cycle or recovery sleep for reference.
Another recent study50 assessed gene expression changes with sleep deprivation in human saliva for 96 inflammation-related genes using a low-density array. They compared expression in the same subjects at the same time of day (14:00) after normal sleep and 30 h of extended wakefulness. Two genes were found to have elevated expression, ANXA3 and 17GAM. The changes in expression of these genes in saliva are much greater (1.5-2.5 fold changes) than that found in peripheral blood. It is of interest that ANXA3 was identified in our study, in the study by Möller-Levet et al.14 and also in saliva.
Taken together, the results of studies looking at changes in the blood transcriptome with sleep loss suggest the following: (1) whereas the diurnal changes in gene expression are robust and affect a large percentage of total transcripts, changes with sleep deprivation are very limited; (2) the very small changes in gene expression with sleep deprivation, the lack of reproducibility across studies and within the same individual raises concerns whether the changes observed represent a true biological signal or potentially statistical noise.
Effect of Sleep Loss on Diurnal Rhythm of Gene Expression
An important finding of the study by Möller-Levet et al.14 is that a large number of genes that have a circadian expression pattern in baseline conditions lose circadian rhythmicity during sleep deprivation. We replicated this finding and extended it. Specifically, we find a major difference in the suppression of circadian rhythmicity of gene expression between sensitive and resistant subjects to sleep loss. Resistant subjects showed robust suppression of circadian rhythmicity whereas sensitive subjects did not. This was observed both when assessing the number of cycling probe sets at different P value thresholds to define cycling and when looking at changes in amplitude of the most robust circadian cycling probe sets. The decrease in circadian rhythm amplitude was highly significant in resistant subjects but not in sensitive subjects. Blood cell type module analysis showed that genes associated with cytotoxic T cells and cells of myeloid origin peak in expression during daytime, as has been previously published.51 Our data additionally suggest that the decrease in circadian rhythm amplitude found in resistant subjects, is mostly found in cells of myeloid origin, such as monocytes.
The mechanism behind the blunting of the circadian rhythm in blood may be caused by the decreased binding of certain core clock gene transcription factors to the promoter regions of clock genes during sleep deprivation as shown by Mongrain et al.16 in the mouse cerebral cortex. They found, using chromatin-immunoprecipitation, decreased binding of CLOCK and BMAL1 to the DBP promoter, as well as decreased binding of NPAS2 and BMAL1 to PER2 during sleep deprivation. It is possible, although not yet proven, that our observed association between changes in circadian rhythmicity with sleep deprivation and the resistant versus sensitive subject trait in our study is causative. Recent studies suggest that at a molecular level the processes underlying sleep homeostasis and circadian regulation are not independent52 as was originally proposed in the two-process model.53 In support of this is the demonstration that a mutation in the clock-associated gene DEC2 in humans results in short sleep and resistance to the effects of sleep loss.54 The causality of this mutation was demonstrated by expressing the human mutation in Drosophila and mice resulting in shorter sleep and less recovery sleep following sleep deprivation, i.e., reduced sleep homeostasis.54 DEC2 represses the binding of Clock1-BMAL1 heterodimer to E boxes in the transcriptional regulatory area of cycling genes.33,55 The mutation that alters sleep duration and confers resistance to sleep loss reduces the ability of DEC2 to suppress CLOCK-BMAL1 transactivation of clock gene expression.54 Thus, there is clearly a link between regulation of clock gene expression and response to sleep loss. The mechanisms by which sleep loss alters the binding of CLOCK-BMAL1 to regulatory sites for clock genes is unknown, as is the reason why this mechanism appears to differ between individuals. These topics will require further study.
Implications for Identifying a Biomarker for Sleep Loss
One important goal of the current research is to address whether identifying a biomarker/s that can be used to assess chronically reduced sleep is feasible.56 One approach would be to assess changes in gene expression in peripheral blood, which is readily accessible and cost effective. Our data and other recently published studies14,15 do not provide evidence that this is a viable strategy. The changes in expression are small and there are very robust time-of-day effects that would need to be accounted for in applying this approach to individuals. It is conceivable, however, that the small signal might, in part, be a consequence of the cellular heterogeneity from which the RNA was derived. Studying only a homogeneous population of circulating cells such as monocytes, based on our study results, is of future interest. A similar approach is being used for studies of biomarkers in cardiovascular disease reviewed in a previous study57 and cancer (solid tumors),58 but the fold changes are still relatively small.
Although challenging, there have been assertions with respect to biomarkers for sleep drive in humans.59 There is an increase in salivary amylase mRNA in humans with extended wakefulness,59 but this is quite variable between individuals. In considering any biomarker, it is important to address sensitivity and specificity. Salivary amylase activity has been proposed as a marker of sympathetic nervous system activity.60,61 As such, levels are altered by psychological stress62 and acute increases may reflect levels of emotional arousal.63 Thus, it is highly unlikely that salivary amylase will be a specific enough biomarker of sleep loss to be of value in this regard.
In conclusion, we find a robust circadian oscillation for nearly a quarter of assessed unique genes in peripheral blood in humans. The effects of sleep deprivation on gene expression independent of circadian processes are small. They affect few genes and are small in magnitude. The major effect of sleep loss on the blood transcriptome is suppression of the circadian rhythm of gene expression. We observed differences in the degree of suppression of circadian rhythmicity of gene expression between those who are sensitive and those who are resistant to the effects of sleep loss. Specifically, resistant individuals have more marked suppression of circadian rhythmicity than sensitive individuals and likely have more robust compensatory mechanisms to sustain wakefulness. Based on other data, it is conceivable that this finding could be the basis for differential vulnerability to sleep loss, although determining this will require additional studies.
DISCLOSURE STATEMENT
This work was supported by the NIH grant HL94307 and the Eimskip Fund of the University of Iceland, Merck Research Laboratories (Merck & Co., Inc.) as well as by the National Center for Research Resources grant UL1RR024134. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ZIA ES103166-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health. Dr. Arnardottir is a part-time consultant for Nox Medical (Reykjavik, Iceland) and has received grant support from the ResMed foundation as well as paid speaking engagement from ResMed, Sweden and Pfizer, Iceland. Dr. Nikonova was employed by Merck & Co during the study period. Drs. Podtelezhnikov, Tanis, Stone, Renger, and Winrow are employees of Merck & Co. Drs. Tanis, Stone, Renger, and Winrow are also shareholders of Merck & Co. Mr. Maislin is Principal Biostatistician of Biomedical Statistical Consulting (BSC). The other authors have indicated no financial conflicts of interest. The study was performed at University of Pennsylvania Perelman School of Medicine, Philadelphia, PA.
ACKNOWLEDGMENTS
Drs. Arnardottir and Nikonova are co-first authors. Drs. Winrow and Pack are co-senior authors. The authors thank Brendan Keenan, a statistician at the Sleep Center of the University of Pennsylvania for his assistance with the figures and graphic designer Arni Collett for his assistance with tables. We also thank Melissa Fernando, Frances Pack, Bethany Staley and the other staff at the Sleep Center of the University of Pennsylvania. We would also like to thank Merck & Co., Inc. employees Terrence McDonald and Eva Finney and the staff at the Clinical & Translational Research Center (CTRC) at the University of Pennsylvania for their help. Finally, we thank Shyamal Peddada and Gregg Dinse from the Biostatistics Branch, NIEHS, for their useful suggestions.
Footnotes
A commentary on this article appears in this issue on page 1581.
SUPPLEMENTAL MATERIAL
SUPPLEMENTAL METHODS
Participants
Based on psychomotor vigilance task (PVT) analysis of 200 twin-pair participants in a previous sleep deprivation study,1 a single member of the twin pair studied previously was invited to participate in this study. The number of PVT performance lapses, i.e., a reaction > 500 msec, across the previous sleep deprivation was used as selection criterion, inviting only subjects below the 33rd (resistant subjects) and above the 67th (sensitive subjects) percentiles to participate in the study. The slope of the increase in PVT lapses was calculated using the following regression model:
where ΔYit is change in the value of the PVT parameter from time = 0 to time = t for subject i. The parameters of interest are subject-specific linear changes in performance measures during sleep deprivation, βi3 after adjustment for circadian effects (cos and sin terms). The use of change scores allows removal of the intercept parameter, therefore only assessing the change in performance, not the actual baseline score. The PVT lapses prior to sleep deprivation were not different between sensitive and resistant subjects. Subjects in the sensitive (slope of PVT lapses > 0.286) and resistant groups (slope of PVT lapses < 0.121), were contacted for participation in this study representing the 33rd and 67th percentiles of PVT performance, respectively. The slopes of the change in subjective sleepiness (Karolinska Sleepiness Scale [KSS],2 visual analog scales (VAS) of mood and exhaustion (scale 0-10, with the higher number referring to worse condition)3,4 across the sleep deprivation trial were calculated in the same manner.
Study Design: Initial Telephone Screen
Exclusion criteria included shift and night work, major medical illnesses in the past year (such as diabetes or cancer), pregnancy, hot flashes caused by menopause, diagnosis of a sleep disorder, excessive daytime sleepiness (Epworth Sleepiness Scale score > 10),5 depression in the past 6 mo, kidney problems, anemia or low blood count, excessive caffeine use (more than eight cups per day), alcohol problems (two or more drinks per day or two or more positive answers on the CAGE questionnaire,6 and drug addiction. Subjects were excluded from the study if they were taking the following medications: central nervous system stimulants, sleeping pills, sedatives, hypnotics, beta-blockers, and cholesterol-lowering medication. Subjects were screened for sleep disorders before the previous study performed on average 2.4 y earlier. If the weight of subjects had increased more than 10% since the previous study or subjects reported habitual snoring and witnessed apneas, we planned that they would be asked to come in for a repeat sleep study to exclude sleep apnea. No subject met these criteria. Forty subjects were contacted for participation in the study. Twelve subjects were ineligible for the study because of use of antidepressants (n = 1), not interested (n = 4), time constraints (n = 5), and moving to a different time zone (n = 2). Therefore, 28 subjects were enrolled into the study. No subject needed a repeat sleep study based on the aforementioned criteria. Of 28 enrolled subjects, 22 subjects completed the study, six withdrew from study because of time constraints or other reasons, one had a positive pregnancy test, and one did not follow up. The first seven sensitive and seven resistant subjects to complete the study were included in the microarray analysis reported here.
Neurobehavioral impairment of sensitive versus resistant subjects during sleep deprivation. The average number of performance lapses throughout the sleep deprivation period (psychomotor vigilance test [PVT] every 2 h). Behaviorally sensitive subjects had a significantly higher increase in PVT lapses during 38 h of sleep deprivation than behaviorally resistant subjects (mean ± standard deviation slope of resistant versus sensitive subjects; 0.59 ± 0.37 versus 0.18 ± 0.14, P = 0.02) after adjustment for circadian effects, see Supplemental Methods.
Demographic data for sensitive and resistant subjects to the behavioral impairment of sleep deprivation.
The subjective behavioral impairment of sensitive and resistant subjects during sleep deprivation.
Trait-like interindividual difference in psychomotor vigilance task as a measure of impairment to sleep loss.
Enriched biological pathways by 4-h bins in human blood during normal sleep/wake
Study Design: Baseline Visit
A physical examination was performed and medical history obtained. Subjects were screened for the presence of drugs and alcohol in urine. A pregnancy test was performed in the females. Blood was collected for a complete blood count (CBC) and blood urea nitrogen (BUN) measurement. Subjects with CBC or BUN outside the normal range were to be excluded from the study. No subject was excluded based on these criteria. Subjects also completed the Center for Epidemiological Studies Depression Scale (CES-D)7 and the 36-Item Short Form Health Survey (SF-36).8
Study Design: Sleep Deprivation Visit
Subjects were asked to maintain a regular sleep-wake cycle for 2 w prior to the study, going to bed between 21:30 and midnight every night for 2 w and a minimum sleep of 7 h at night without naps. This was confirmed by actigraphy (Actigraphy devices AWLP and AW64, Philips Respironics, Bend, OR) and sleep-wake diary for 2 w prior to the sleep deprivation visit. Subjects were asked to abstain from nicotine (one smoker with fewer than 10 cigarettes per day in the study) and caffeine use for 24 h before and during the study. Subjects drinking more than eight cups of caffeine-containing beverages were excluded from the study to minimize stress caused by abstinence from caffeine.
Two subjects were assessed simultaneously in the sleep laboratory and their blood draws were staggered by 5 min, always in the same order.
The caloric content of meals was standardized using the Harris-Benedict equation (an activity factor of 1.3 was assumed for all subjects).9 The meals were composed of 50% carbohydrate, 20% protein, and 30% fat. The meals were served to the subjects at fixed intervals during the day at 08:00, 12:00, 16:00, and 20:00 after the blood draws.
The PVT was administered for 10 min every 2 h when the subjects were awake (at 09:15, 11:15, 13:15, etc.).10 Additionally, subjects answered three questionnaires before and after each PVT: the KSS,2 and VAS-F and VAS-M (for fatigue and mood, respectively).3,4 The KSS is a scale from 1-9 from “very alert” to “very sleepy, an effort to stay awake, fighting sleep” and the VAS-F and VAS-M are 10-point scales from “fresh to exhausted” and “elated to depressed”, respectively. The PVT measurements were missing for one sensitive subject because of a faulty PVT device.
Circadian changes in expression of four clock genes during normal sleep/wake in baseline for subjects sensitive and resistant to sleep deprivation. Data shown as mean ± standard deviation in normalized probe intensity on the y-axis; (A) PER2, (B) PER3, (C) DBP, and (D) NPAS2. No significant differences were found between the two groups. The PER genes peak between 04:00 and 08:00, DBP peaks around 03:00 and NPAS2 around 01:30.
Probe sets with differential mean expression during three states: normal sleep/wake versus sleep deprivation versus recovery sleep (12:00, 04:00, and 08:00) using a false discovery rate < 5%.
During the study, subjects were on 14-h lights on:10 h lights off regimen with lights on during the entire sleep deprivation period. Subjects were not allowed to leave the research center, use the telephone, read the newspaper, watch television, or perform vigorous exercise during the study. A trained staff member was present at all times to make sure that all requirements for the study were fulfilled and to keep subjects awake during the sleep deprivation period.
Diurnal differences in blood cell types using baseline combined sensitive and resistant subjects circadian signature (8,064 probe sets, false discovery rate < 5%).
REFERENCES
- 1.Kuna ST, Maislin G, Pack FM, et al. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35:1223–33. doi: 10.5665/sleep.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillberg M, Kecklund G, Akerstedt T. Relations between performance and subjective ratings of sleepiness during a night awake. Sleep. 1994;17:236–41. doi: 10.1093/sleep/17.3.236. [DOI] [PubMed] [Google Scholar]
- 3.Lee KA, Hicks G, Nino-Murcia G. Validity and reliability of a scale to assess fatigue. Psychiatry Res. 1991;36:291–8. doi: 10.1016/0165-1781(91)90027-m. [DOI] [PubMed] [Google Scholar]
- 4.Monk TH. A visual analogue scale technique to measure global vigor and affect. Psychiatry Res. 1989;27:89–99. doi: 10.1016/0165-1781(89)90013-9. [DOI] [PubMed] [Google Scholar]
- 5.Johns MW. Reliability and factor analysis of the Epworth Sleepiness Scale. Sleep. 1992;15:376–81. doi: 10.1093/sleep/15.4.376. [DOI] [PubMed] [Google Scholar]
- 6.Mayfield D, McLeod G, Hall P. The CAGE questionnaire: validation of a new alcoholism screening instrument. Am J Psychiatry. 1974;131:1121–3. doi: 10.1176/ajp.131.10.1121. [DOI] [PubMed] [Google Scholar]
- 7.Radloff LS. The CES-D scale: a self-report depression scale for research in the general population. App Psychol Meas. 1977;1:385–401. [Google Scholar]
- 8.Brazier JE, Harper R, Jones NM, et al. Validating the SF-36 health survey questionnaire: new outcome measure for primary care. BMJ. 1992;305:160–4. doi: 10.1136/bmj.305.6846.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris JA, Benedict FG. Washington, DC: Carnegie Institute of Washington (Publication no. 279.); 1919. A biometric study of basal metabolism in man. [Google Scholar]
- 10.Dinges DF, Powell JW. Microcomputer analyses of performance on a portable, simple visual RT task during sustained operations. Behav Res Meth Instr Comp. 1985;17:652–5. [Google Scholar]
REFERENCES
- 1.Luckhaupt SE, Tak S, Calvert GM. The prevalence of short sleep duration by industry and occupation in the National Health Interview Survey. Sleep. 2010;33:149–59. doi: 10.1093/sleep/33.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leger D, Roscoat E, Bayon V, Guignard R, Paquereau J, Beck F. Short sleep in young adults: Insomnia or sleep debt? Prevalence and clinical description of short sleep in a representative sample of 1004 young adults from France. Sleep Med. 2011;12:454–62. doi: 10.1016/j.sleep.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Goel N, Rao H, Durmer JS, Dinges DF. Neurocognitive consequences of sleep deprivation. Seminars in Neurology. 2009;29:320–39. doi: 10.1055/s-0029-1237117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang JE, Lim MM, Bateman RJ, et al. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science. 2009;326:1005–7. doi: 10.1126/science.1180962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Dongen HP, Baynard MD, Maislin G, Dinges DF. Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004;27:423–33. [PubMed] [Google Scholar]
- 6.Leproult R, Colecchia EF, Berardi AM, Stickgold R, Kosslyn SM, Van Cauter E. Individual differences in subjective and objective alertness during sleep deprivation are stable and unrelated. Am J Physiol Regul Integr Comp Physiol. 2003;284:R280–90. doi: 10.1152/ajpregu.00197.2002. [DOI] [PubMed] [Google Scholar]
- 7.Mu Q, Mishory A, Johnson KA, et al. Decreased brain activation during a working memory task at rested baseline is associated with vulnerability to sleep deprivation. Sleep. 2005;28:433–46. doi: 10.1093/sleep/28.4.433. [DOI] [PubMed] [Google Scholar]
- 8.Chee MW, Chuah LY, Venkatraman V, Chan WY, Philip P, Dinges DF. Functional imaging of working memory following normal sleep and after 24 and 35 h of sleep deprivation: Correlations of fronto-parietal activation with performance. Neuroimage. 2006;31:419–28. doi: 10.1016/j.neuroimage.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Kuna ST, Maislin G, Pack FM, et al. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35:1223–33. doi: 10.5665/sleep.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackiewicz M, Zimmerman JE, Shockley KR, Churchill GA, Pack AI. What are microarrays teaching us about sleep? Trends Mol Med. 2009;15:79–87. doi: 10.1016/j.molmed.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maret S, Dorsaz S, Gurcel L, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–5. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anafi RC, Pellegrino R, Shockley KR, Romer M, Tufik S, Pack AI. Sleep is not just for the brain: transcriptional responses to sleep in peripheral tissues. BMC Genomics. 2013;14:362. doi: 10.1186/1471-2164-14-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohane IS, Valtchinov VI. Quantifying the white blood cell transcriptome as an accessible window to the multiorgan transcriptome. Bioinformatics. 2012;28:538–45. doi: 10.1093/bioinformatics/btr713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Möller-Levet CS, Archer SN, Bucca G, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci U S A. 2013;110:E1132–41. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrino R, Sunaga DY, Guindalini C, et al. Whole blood genome-wide gene expression profile in males after prolonged wakefulness and sleep recovery. Physiol Genomics. 2012;44:1003–12. doi: 10.1152/physiolgenomics.00058.2012. [DOI] [PubMed] [Google Scholar]
- 16.Mongrain V, La Spada F, Curie T, Franken P. Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PloS One. 2011;6:e26622. doi: 10.1371/journal.pone.0026622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinges DF, Powell JW. Microcomputer analyses of performance on a portable, simple visual RT task during sustained operations. Behav Res Meth Instr Comp. 1985;17:652–5. [Google Scholar]
- 18.Bolstad B. RMAExpress. [cited 2009 Aug 24]; Available from: http://rmaexpress.bmbolstad.com/
- 19.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, Kerr MK, Cui XQ, Churchill GA. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments. In: Parmgiani G, Garrett ES, Irizarry RA, Zeger SL, editors. The analysis of gene expression data: an overview of methods and software. New York: Springer; 2003. pp. 313–431. [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 24.Searle SR, Casella G, McCulloch CE. Variance components. Hoboken, NJ: John Wiley & Sons, Inc.; 2008. Frontmatter. [Google Scholar]
- 25.Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinorrhythmometry. Chronobiologia. 1979;6:305–23. [PubMed] [Google Scholar]
- 26.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–80. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark NR, Ma'ayan A. Introduction to statistical methods for analyzing large data sets: gene-set enrichment analysis. Sci Signal. 4:tr4. doi: 10.1126/scisignal.2001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–60. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaussabel D, Quinn C, Shen J, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 25:372–80. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–7. doi: 10.1093/hmg/ddl207. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 33.Honma S, Kawamoto T, Takagi Y, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–4. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 34.Crumbley C, Burris TP. Direct regulation of CLOCK expression by REVERB. PloS one. 2011;6:e17290. doi: 10.1371/journal.pone.0017290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 36.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 37.Zvonic S, Ptitsyn AA, Conrad SA, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–70. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 38.Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasukawa T, Sugimoto M, Hida A, et al. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci U S A. 2012;109:15036–41. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–8. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 42.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 43.Zieker D, Jenne I, Koenigsrainer I, et al. Circadian expression of clock-and tumor suppressor genes in human oral mucosa. Cell Physiol Biochem. 2010;26:155–66. doi: 10.1159/000320547. [DOI] [PubMed] [Google Scholar]
- 44.Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Leonardson AS, Zhu J, Chen Y, et al. The effect of food intake on gene expression in human peripheral blood. Hum Mol Genet. 2010;19:159–69. doi: 10.1093/hmg/ddp476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James FO, Boivin DB, Charbonneau S, Bélanger V, Cermakian N. Expression of clock genes in human peripheral blood mononuclear cells throughout the sleep/wake and circadian cycles. Chronobiol Int. 2007;24:1009–34. doi: 10.1080/07420520701800736. [DOI] [PubMed] [Google Scholar]
- 47.Mackiewicz M, Shockley KR, Romer MA, et al. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–57. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 48.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–9. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramsay RR, Gandour RD, van der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim Biophys Acta. 2001;1546:21–43. doi: 10.1016/s0167-4838(01)00147-9. [DOI] [PubMed] [Google Scholar]
- 50.Thimgan MS, Gottschalk L, Toedebusch C, et al. Cross-translational studies in human and Drosophila identify markers of sleep loss. PloS One. 2013;8:e61016. doi: 10.1371/journal.pone.0061016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–8. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franken P, Dijk DJ. Circadian clock genes and sleep homeostasis. Eur J Neurosci. 2009;29:1820–9. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 53.Borbely AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 54.He Y, Jones CR, Fujiki N, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–70. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azmi S, Sun H, Ozog A, Taneja R. mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity and Stra13 expression. J Biol Chem. 2003;278:20098–109. doi: 10.1074/jbc.M210427200. [DOI] [PubMed] [Google Scholar]
- 56.Quan SF, Shaw PJ, Naidoo N, Haeggström E, Krueger JM, Church GM. Panel discussion: can there be a biomarker for sleepiness? J Clin Sleep Med. 2011;7:S45–8. doi: 10.5664/JCSM.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnardottir ES, Mackiewicz M, Gislason T, Teff KL, Pack AI. Molecular signatures in obstructive sleep apnea in adults: a review and perspective. Sleep. 2009;32:447–70. doi: 10.1093/sleep/32.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Showe MK, Vachani A, Kossenkov AV, et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 2009;69:9202–10. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seugnet L, Boero J, Gottschalk L, Duntley SP, Shaw PJ. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci U S A. 2006;103:19913–8. doi: 10.1073/pnas.0609463104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nater UM, Rohleder N. Salivary alpha-amylase as a non-invasive biomarker for the sympathetic nervous system: current state of research. Psychoneuroendocrinology. 2009;34:486–96. doi: 10.1016/j.psyneuen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 61.Granger DA, Kivlighan KT, el-Sheikh M, Gordis EB, Stroud LR. Salivary alpha-amylase in biobehavioral research: recent developments and applications. Ann N Y Acad Sci. 2007;1098:122–44. doi: 10.1196/annals.1384.008. [DOI] [PubMed] [Google Scholar]
- 62.Robles TF, Shetty V, Zigler CM, et al. The feasibility of ambulatory biosensor measurement of salivary alpha amylase: Relationships with self-reported and naturalistic psychological stress. Biol Psychol. 2011;86:50–6. doi: 10.1016/j.biopsycho.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adam EK, Till Hoyt L, Granger DA. Diurnal alpha amylase patterns in adolescents: associations with puberty and momentary mood states. Biol Psychol. 2011;88:170–3. doi: 10.1016/j.biopsycho.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neurobehavioral impairment of sensitive versus resistant subjects during sleep deprivation. The average number of performance lapses throughout the sleep deprivation period (psychomotor vigilance test [PVT] every 2 h). Behaviorally sensitive subjects had a significantly higher increase in PVT lapses during 38 h of sleep deprivation than behaviorally resistant subjects (mean ± standard deviation slope of resistant versus sensitive subjects; 0.59 ± 0.37 versus 0.18 ± 0.14, P = 0.02) after adjustment for circadian effects, see Supplemental Methods.
Demographic data for sensitive and resistant subjects to the behavioral impairment of sleep deprivation.
The subjective behavioral impairment of sensitive and resistant subjects during sleep deprivation.
Trait-like interindividual difference in psychomotor vigilance task as a measure of impairment to sleep loss.
Enriched biological pathways by 4-h bins in human blood during normal sleep/wake
Circadian changes in expression of four clock genes during normal sleep/wake in baseline for subjects sensitive and resistant to sleep deprivation. Data shown as mean ± standard deviation in normalized probe intensity on the y-axis; (A) PER2, (B) PER3, (C) DBP, and (D) NPAS2. No significant differences were found between the two groups. The PER genes peak between 04:00 and 08:00, DBP peaks around 03:00 and NPAS2 around 01:30.
Probe sets with differential mean expression during three states: normal sleep/wake versus sleep deprivation versus recovery sleep (12:00, 04:00, and 08:00) using a false discovery rate < 5%.
Diurnal differences in blood cell types using baseline combined sensitive and resistant subjects circadian signature (8,064 probe sets, false discovery rate < 5%).