Abstract
Colloidal dispersion of single-walled carbon nanotubes (SWCNTs) is often the first processing step to many of their unique applications. However, dispersed SWCNTs often exist in kinetically trapped states where aggregation can be of concern. Recent work revealed prominent DNA–SWCNT aggregation following intravascular injection. In this study, we performed detailed analysis of DNA–SWCNT aggregate formation, structure, and composition in the context of endothelial cell condition media. Interestingly, we found that aggregates formed within condition media from cells that have undergone a stress response differ in size and organization from that of the control. We also found that temperature increases also promote DNA–SWCNT associations. A mathematical model was developed to describe the kinetics of SWCNT extraction from solution. Through orthogonal optical analysis and imaging modalities, we verified that proteins form the bulk of the aggregate structure and dictate aggregate assembly at multiple levels of organization. Finally, physiochemical analysis indicated preferential extraction of low-abundance hydrophobic and charged proteins. The formed aggregates also remain relatively stable in solution, making them potential macroscopic indicators of solution content.
Keywords: single-walled carbon nanotubes, DNA, endothelial, secretome, aggregation, self-assembly
Protein adsorption to surfaces is a common but complex step of any interaction between biomaterials and the human body.1 While sometimes preferred in tissue engineering, such as in the case of scaffold vascularization,2 protein adsorption is often an unintended first step of complex biological responses that can result in thrombosis, inflammation, and fouling of bioengineered materials.3−6 In the case of therapeutics, proper delivery7 and bioavailability8 are intricately linked to drug or vehicle interactions with serum protein elements. Thus, understanding the mechanism of biomaterial and protein interactions is especially important.
Single-walled carbon nanotubes (SWCNTs) are a unique class of materials with unusual physical and chemical attributes.9 Recently, they have been explored for a variety of potential biomedical applications such as sensors,10−12 biological scaffolds,13 drug delivery vehicles,14 and imaging contrast.15 Since aqueous dispersion through noncovalent conjugation gives rise to many properties of SWCNTs, major classes of both natural and synthetic biomolecules have been explored as potential dispersion candidates.16−19 Specifically, single-stranded nucleotide conjugation allows for high SWCNT optical efficiency photoluminescence (PL).
While DNA–SWCNT have been implicated in many biologically relevant applications,20,21 recent work has found that they aggregate when injected in vivo.22 Although the aggregation process can be circumvented through improvement of colloidal stability with poly(ethylene glycol) (PEG) conjugation,22,23 the factors and molecular mechanisms surrounding the aggregation process of DNA–SWCNT remain unclear. The study of aggregation in therapeutics is especially important, as the phenomenon affects availability and targeting. Additionally, it is known that SWCNT cytotoxicity is concentration dependent,24 an instance where aggregation may play a role. There is a need to understand whether the unique properties of DNA–SWCNT are modified through aggregation and interactions with the local environment.
Although serum proteins are the major suspects, most current protein–SWCNT studies involve unconjugated SWCNTs,5,25,26 homogeneous protein species,27,28 or short peptides,25,29 which all are not applicable in explaining the observed in vivo phenomena. In this work, we emulate intravascular conditions by studying the changes in SWCNT conjugation and structure in the presence of endothelial cell-secreted factors. DNA–SWCNTs were found to interact with the cellularly secreted proteins, the secretome, as they aggregate. Additionally, we found that temperature changes also played a role in aggregate formation. Through novel modes of analysis, aggregation structures were studied at multiple levels of organization and found to be intricately linked to protein content. Protein analysis revealed that DNA–SWCNT aggregates possess a unique affinity for specific protein types, a property that may be applicable for future biomedical applications. On a fundamental level, we have improved the understanding of DNA–SWCNTs in the intravascular environment, a concept that potentially can be extended to other rod-shaped and DNA-assembled nanomaterials.
Results and Discussion
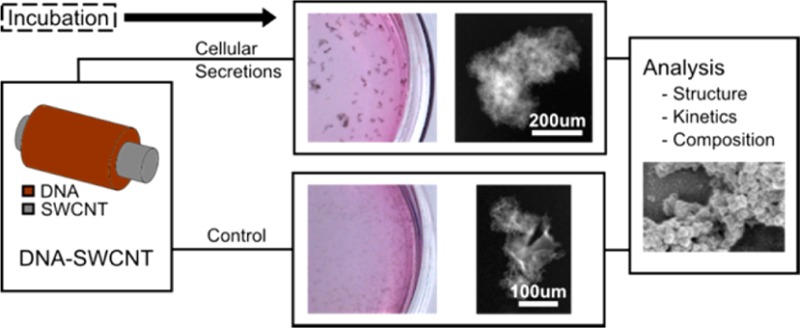
(AT)15-conjugated SWCNTs are being explored for biosensor applications22,30 due to their high fluorescence efficiency and response selectivity34 in a range of aqueous environments. We first observed in vitro aggregation of (AT)15–SWCNT following a 6 h incubation at 2 μg/mL with human umbilical vein endothelial cells (HUVECs). The effect was replicated in conditioned media to control for cellular responses. To elucidate potential factors responsible for these phenomena, we tested for various media components (Figure 1A). In congruence with previous studies,31 increases in fetal bovine serum (FBS) induced aggregate formation. However, aggregates were most prominently observed in conditioned media that had less FBS but also contained secreted proteins of HUVEC that have undergone stress responses such as hypoxia and starvation. This suggests that cellularly secreted components play a role in aggregate formation. Solution pH did not vary significantly during the course of the incubation, and factors such as CO2 levels, phenol red, and media supplements had no observable effect on the outcome. Thus, we hypothesize that aggregation is mainly protein mediated and that their size differences may be attributed to differential solution protein content. Formed aggregates remained stable in H2O at room temperature for 2 weeks postexperiment (Figure 1B), indicating that the interactions were near equilibrium and the associations between its individual components remain intact.
Figure 1.
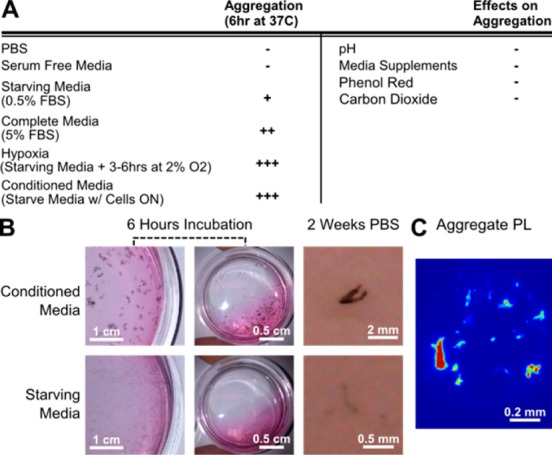
Aggregate formation conditions. (A) Different solutions exhibit different degrees of SWCNT aggregation. Media components having no effect were ruled out. (B) Visual comparison of aggregation formation of starving media and conditioned media. Aggregations formed are stable after 2 weeks. (C) Strong nIR signal from aggregates showed that SWCNT DNA wrapping is likely undisturbed.
To study the effect of the HUVEC secretome, the subsequent experiments involve the following solution types: (1) starving media containing 0.5% FBS, (2) complete media containing 5% FBS, and (3) conditioned media containing 0.5% FBS and incubated with HUVECs for 10 h. All media are filtered at 0.4 um to remove large debris prior to SWCNT addition. A typical aggregate experiment consists of incubating a test solution with SWCNTs for 6 h in a humidified incubator at 37 °C.
Structure Analysis
We studied the (AT)15–SWCNT associations and differences in aggregation macrostructure through a panel of orthogonal optical techniques.
Near-Infrared (nIR) Microscopy
nIR microscopy was used to image aggregates directly (Figure 1C). With excitation at 660 nm, aggregates showed intense PL, indicating that SWCNT single dispersion was not lost and that the aggregation process did not involve complete stripping of the nucleotide conjugation.
Photoluminescence
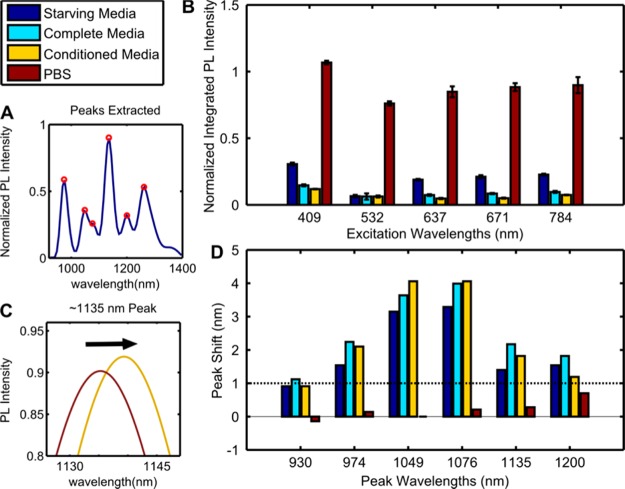
When SWCNTs aggregate, they are removed from solution, which can be interpreted as a loss of solution PL signal. Total PL was measured for supernatants of the aggregation media (Figure 2A,B). To study whether cells directly affected aggregate formation, both cell-containing and conditioned media were used. While SWCNTs in phosphate-buffered saline (PBS) experienced only minimal PL change postincubation, which will be further discussed in the single-particle tracking section, aggregate formation within other media types resulted in significant SWCNT removal from solution of up to 90%.
Figure 2.
Photoluminescence. (A) Example emission spectra from laser excitation. (B) Integrated PL emissions from five lasers normalized to preincubation PBS control (the PBS plotted is after experimental incubation). (C) Example peak-shift between control and conditioned media. (D) Peak-shifts from control. Dotted line indicating approximate instrument sensitivity.
We also detected significant SWCNT solvatochromism.32 The PL emission shifts reflect the SWCNT response to surface changes such as conjugation modification and protein adsorption (Figure 2C,D). In comparison to PBS control, media-incubated SWCNTs exhibited a ∼5 meV red shift, consistent with modification of noncovalent conjugation. This evidence suggests that the solution factors must have associated with the SWCNT and altered the nucleotide conjugation.
Raman Spectroscopy
The radial breathing mode (RBM) region of the SWCNT Raman spectra is indicative of SWCNT organization differences within aggregates.33 SWCNT arrangement differences shift the interband transition energy, changing RBM peak intensities as different chirality species move in and out of resonance with the excitation laser. The RBM of SWCNTs of conditioned media aggregates, as compared to that of the starting (AT)15–SWCNT (Figure 3), indicates significant differences in SWCNT arrangement. The ratios of the G peak to D peak of the Raman spectra has been correlated with aggregation state, with higher values indicative of increased aggregation.34 We found the G/D ratios of the condition media sample and control were 14.5 and 6.6, respectively, suggesting that intertube distances are greater for condition media formed aggregates.
Figure 3.
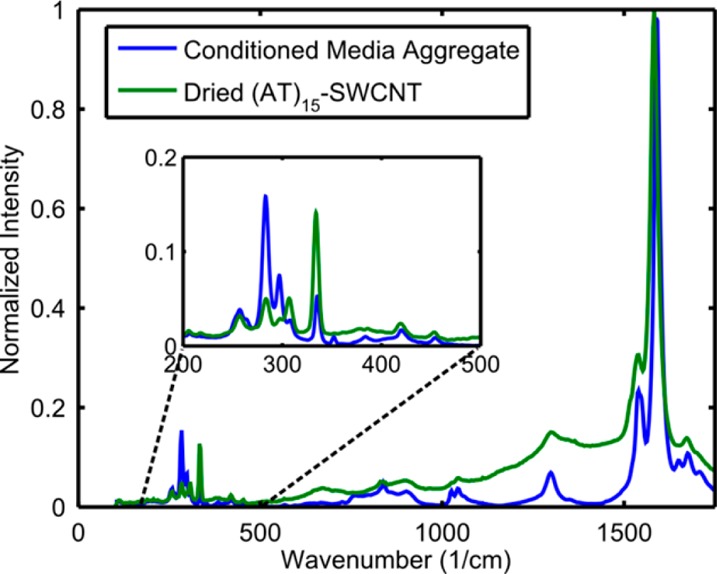
Raman spectroscopy. Radial breathing mode (RBM) region of the SWCNT Raman spectra shows changes in band gap energy consistent with differences in aggregation structure.
Fractal Analysis
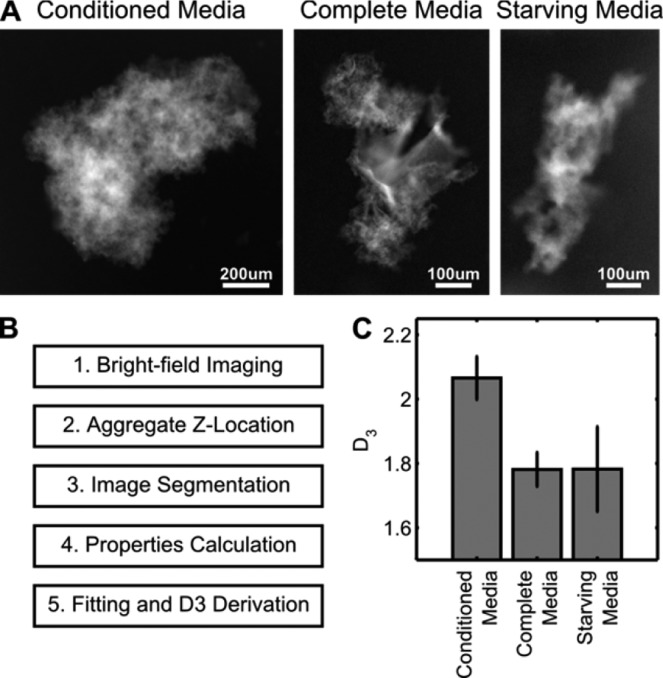
Bright-field images from different solutions were taken to study the aggregate macrostructure (Figure 4A). Starving media form aggregates of a clearly smaller size. While full media aggregates were larger, they were loosely packed as compared to conditioned media aggregates. Side-by-side comparisons reveal differences in topology, which is visual evidence of protein content dictating aggregation structure.
Figure 4.
Bright-field image analysis. (A) Inverted bright-field images of SWCNT aggregates formed from different media conditions, showing differences in size and structure. (B) Work flow of image analysis code. (C) Resulting D3 fractal coefficients from calculation show conditioned media aggregates with higher structural complexity.
To quantitatively compare the observed structural differences, we performed fractal analysis on the acquired images (Figure 4B). Fractal dimensions (FDs) are statistical indices that reflect how the detail of a structure changes with the scale with which it is measured and is often used to study protein aggregation.35 FD is a useful measure of organization complexity independent of overall size, and we expect a structure formed from more densely packed SWCNTs to exhibit a higher FD. Standard methods to derive the third fractal dimension (D3) requires complex instrumentation such as electrical sensing36 and angle-dependent static light scattering.37 We utilize a simpler method of first calculating the second fractal dimension (D2) through image analysis and then approximating D3 using an empirical relation.38 Following aggregate detection and segmentation (see the Supporting Information for Methods), we calculated the maximum projected length (LP), defined as the longest distance between any two perimeter points, and the area of occupation (AO), a normalized amount of SWCNTs occupying per pixel. An exponential fit of LPversusAO was used to solve for D2 (eq 1, Figure S2), and then an empirical relation was used to calculate D3 (eq 2).38
| 1 |
| 2 |
A higher D3 correlates with an aggregate population of higher three-dimensional structural complexity. We find that, despite protein concentration differences between complete and starving media, the aggregates formed had comparable D3 (Figure 4C). However, aggregates formed from conditioned media show a significant increase in D3 or structural complexity. This fractal image analysis provides quantitative evidence to support the observation that condition medial aggregates are more complex in nature and also implicates that protein composition of the corona phase, instead of concentration, dictates aggregate structure.
Scanning Electron Microscopy (SEM)
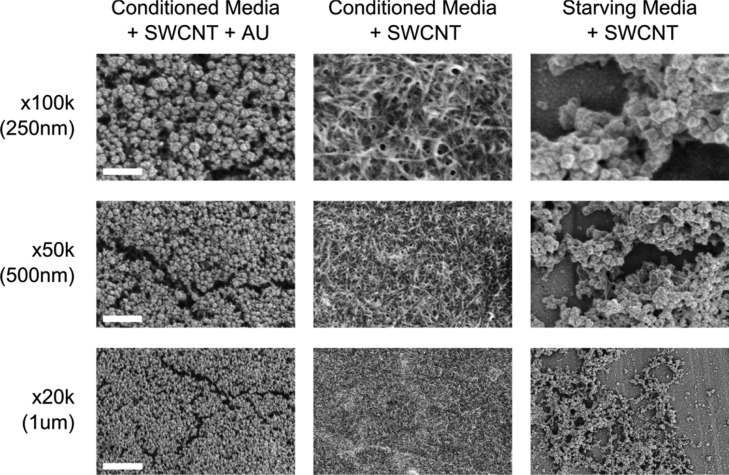
Traditionally, SEM has been used to study both protein deposition39 and SWCNT aggregation.33 In this study, we performed SEM on uncoated and Au sputter-coating aggregates (Figure 5). While SEM of Au-coated samples displayed a protein-dominant aggregate surface morphology, a low kV electron beam on uncoated samples gave contrast to the underlying SWCNT network. Although protein aggregates are anhydrous when imaged, we can still draw reasonable connections to the microstructure under hydration. The structure likely changes and collapses through drying, but large voids would still be preserved, as would some alignment and ordering. The protein to SWNT ratio also must necessarily be preserved, regardless of structure.
Figure 5.
SEM imaging. SEM of SWCNTs with or without gold coating showed aggregates consisted of both protein and an underlying network of SWCNTs. Starving media aggregates show consistently less density and interconnectedness.
From these images, we can reasonably conclude that proteins were responsible for SWCNT aggregation. Due to the density of the protein visualized, protein–protein interaction must have also played a major role in aggregate assembly. In agreement with the fractal analysis results, images from starving media show relatively loose packing. Control SEM showed SWNCTs with DNA coating only, which were more densely packed than the protein-containing samples (Figure S3). The packing difference of control versus media aggregates agrees with the band gap shifts observed in the Raman results.
Furthermore, proteins can alter electron scattering and charging characteristics on uncoated SEM. In addition to SWCNTs, contrast from proteins revealed larger protein networks within SWCNT aggregates (Figure S4). These observed structures suggest that proteins not only interact with (AT)15–SWCNT but also interact with each other within the aggregate structure.
Steps of Aggregation
Dynamic Light Scattering (DLS)
Aggregation was performed at 37 °C for 8 h with DLS measured every 15 min. Conditioned media samples after 6 h of incubation showed a marked increase in particle size, as indicated by a right-shifted correlation curve as compared to the serum-free media samples (Figure S5). Due to the high polydispersity, modeling the correlation curve to derive size distribution is not advised. However, time to half-maximum of the correlation curve showed that conditioned media aggregates increased in size significantly within the first 30 min and maintained a larger size distribution throughout the experiment as compared to control.
Single Particle Tracking (SPT) of Temperature Response
We used SPT to study the initial steps of aggregation. Image processing and particle tracking were performed on microscopy videorecordings of SWCNTs in solution. The diffusion coefficient was calculated from the mean-squared displacement, which was used to derive the hydrodynamic radius from the Stokes–Einstein equation.40 This technique has a sizing range from 10 nm to 10 μm and the advantage of sizing particles directly, which avoids over-representation of larger particles in the case of DLS. The Stokes–Einstein equation for the translational diffusion coefficient of spherical objects (eq 3) was used in this study to calculate an effective hydrodynamic diameter (HD).
| 3 |
| 4 |
Variables are D, the translational diffusion coefficient; kb, Boltzmann’s constant; T, temperature; and η0, solvent viscosity. While the spherical equation has radius, r, cylindrical objects (eq 4) include the translational hydrodynamic end correction coefficient, C, and the ratio of cylinder length to diameter, p.41 However, once (AT)15–SWCNT aggregates form, they are more likely noncylindrical in shape. We chose to use eq 3 for consistency throughout this study.
First, SPT analysis is performed on (AT)15–SWCNT in PBS without protein content to study its properties with temperature change. We observed that an increase of temperature from 24 °C to 37 °C induced associations between (AT)15–SWCNTs but not SDS–SWCNTs (Figure 6A). This is counterintuitive, as increased kinetic energy due to a temperature increase should prevent intermolecular associations. We propose that temperature-associated (AT)15–SWCNT interactions are DNA mediated. DNA conjugation occurs via π–π interactions of the nitrogenous bases with the SWCNT surface.42 This is a weak intermolecular interaction that can be overcome with slight temperature increases.43 The temperature change to 37 °C lifted the ends of the noncovalently conjugated DNA, which are then free to interact and hybridize with DNA attached to another SWCNT molecule. This is a process similar to a recent study on creating ordered SWCNT surfaces through DNA origami.44 Visualization of the scattering pattern from a single SWCNT dimer supported this hypothesis (Figure 6B). If two SWCNTs were indeed joined via DNA, we would observe a scattering pattern consisting of two particles interconnected via a narrow bridge and their movement resembling a tumbling dumbbell exhibiting both translational and rotational diffusion. If SWCNT associations involved direct surface contact, scattering would instead show a single spot. Our observations supported the DNA-linker model, with the centers of the SWCNT subunits within the dimer measuring 1 μm apart. Since the SWCNTs we used range in length from 0.1 to 1 μm, it is feasible for two SWCNT molecules to be joined lengthwise via DNA.
Figure 6.
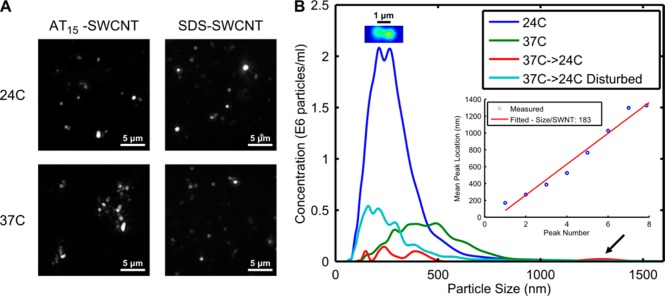
SPT temperature response. (A) SPT of (AT)15–SWCNT shows aggregation formation following a change from 24 °C to 37 °C, a phenomenon not present with SDS–SWCNTs. (B) SPT analysis of particle size distribution for temperature changes and solution disturbance. Imaging of a SWCNT dimer shows two connected but distinct elements. Linear fitting of size peaks shows each SWCNT contributes 183 nm in particle size to the aggregate.
SPT of (AT)15–SWCNT at 24 °C revealed multiple peaks (Figure 6B), showing that singly dispersed (AT)15–SWCNT can exist in solution in multimeric form. With a temperature increase to 37 °C, the distribution shifted right as smaller aggregates are combined to form larger ones. Subsequent cooling of this solution resulted in even larger aggregate formation, which can be attributed to thermodynamic effects. Lastly, physical disturbance, such as pipetting, of the solution containing large aggregates recovered a smaller particle size distribution, showing that the temperature-associated (AT)15–SWCNT aggregations were mediated via weak intermolecular forces such as nucleotide hybridization. Control SDS–SWCNT heating to 37 °C did not show size change (Figure S6), supporting the idea that DNA may be responsible for the temperature-induced effects. The observation of temperature-associated effects explains the slight decrease of PL in the PBS sample of Figure 2B.
SPT analysis of SWCNT populations resulted in distinct and broad peaks, illustrating different sized SWCNT species. We believe that the widths of each peak represent the distributions of SWCNT lengths, and locations of each peak represent a different sized SWCNT aggregate. Linearly fitting the position of the peaks revealed that each SWCNT contributed an average of 183 nm HD to aggregate size (Figure S6). A nearly linear increase in aggregate size indicated a loosely packed structure for initial aggregate formation. Using an approximate SWCNT diameter, an effective SWCNT length of 815 ± 73 nm is derived (see Supporting Information for calculations). Although reasonable, this is an overestimate of SWCNT length due to the rigid rod assumption in eq 4, which inflates the aspect ratio term.
Solution Component Extraction
The removal of SWCNT and protein from solution was measured to study the kinetics of aggregation.
SPT Kinetics
SPT was performed every 15 min for 6 h. SWCNT was extracted from solution over time, as shown by the left-ward shift of the size distribution (Figure 7A). A volume estimate calculated from HD was used to correlate the amount of SWCNT remaining in solution. As the experiment proceeded, the normalized SWCNT concentration decreased significantly (Figure 7B). A majority of SWCNT was extracted within the first hour of incubation, consistent with the DLS results. Control aggregation with starving media did not exhibit similar trends (Figure S7). We modeled SWCNT aggregation as a simple second-order reaction and solved for the concentration of SWCNT as a function of time (eqs 5 and 6, N is SWNCT concentration, t is time, k is the rate constant, and c is the integration constant).
| 5 |
| 6 |
Figure 7.
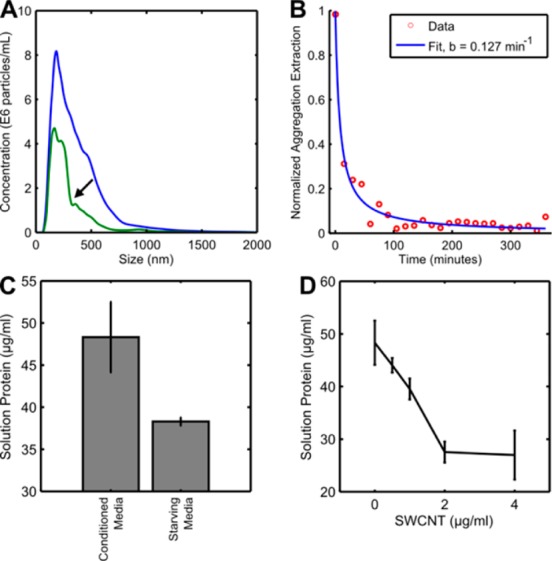
Aggregation kinetics. (A) SPT shows aggregation extractions of total SWCNTs from solution. (B) Approximating total SWCNT volume through 8 h of aggregation at 37 °C shows significant SWCNT extraction. (C) Comparison of protein concentration through BCA assay for two media types. (D) Amount of free protein postaggregation as a function of SWCNT concentration for conditioned media.
Fitting the aggregation model to the normalized SWCNT concentration, we found an association rate constant of 0.127 min–1 (R = 0.98).
Protein Extraction
The bicinchoninic acid (BCA) assay, which is a linearly calibrated colorimetric assay for protein concentration measurement, was used to quantify postaggregation protein concentration in solution. Conditioned media was found to contain ∼20% less total protein than starving media, possibly due to cellular metabolism (Figure 7C). This further reinforces the hypothesis that aggregation depended more on protein type then quantity. For each experiment, supernatants were measured following SWCNT aggregate removal through centrifugation. With increased concentration of SWCNT, the amount of total protein decreased (Figure 7D). Through a linear fit, we found the ratio of protein extraction to be 4.0 ± 0.9 (μg protein)/(μg SWCNT).
Protein Analysis
Gel Quantification
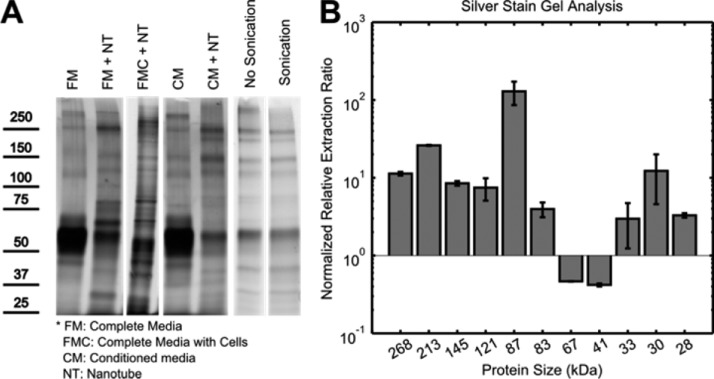
Protein was extracted from SWCNT aggregates via standard denaturing methods45 and run on SDS-PAGE for silver stain analysis (Figure 8A). Due to lysis buffer lipid displacement of DNA conjugation,46 postprocessing SWCNTs remained close to the starting wells and did not interfere with electrophoresis. Contrary to current belief,47 comparison of media and SWCNT aggregate proteins showed differential SWCNT preference against albumin. In aggregates formed from both starving media and conditioned media, we found increased relative concentrations (extraction ratio) of many proteins (Figure 8B). Gel image quantification of aggregate proteins from conditioned media showed that some secreted protein species have extraction ratios of >100. Introduction of cells to the aggregation solution significantly altered the extracted protein content, possibly a result of cellular–SWCNT interactions (Figure 8A). Also, large protein aggregates were observed at ∼500 kDa in the aggregate protein samples only and can be disrupted via sonication. This evidence suggests that protein aggregation is another aspect of SWCNT aggregate formation in biological settings.
Figure 8.
Gel quantification. (A) Silver stain of proteins within SWCNT aggregates from full media, conditioned media, and in the presence of cells are compared along with control. A ∼500 kDa protein aggregate band is also observed, which can be eliminated via sonication. (B) Quantification of band intensities normalized to albumin shows differential SWCNT preference for certain protein species.
Mass Spectroscopy (MS)
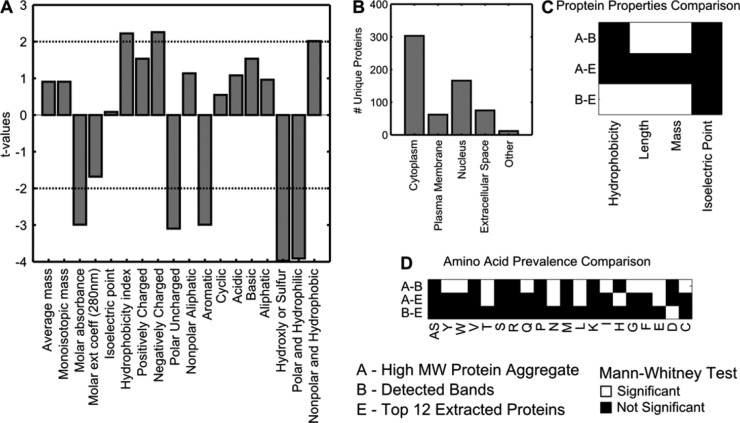
MS was performed on all non-albumin components of aggregate proteins. Physiochemical properties of the top 10 proteins found corresponding to bands of high extraction ratio were tested against bovine serum albumin (BSA) (Figure 9A). Aggregates preferred proteins with greater hydrophobicity and charged amino acids. These findings were reasonable, as hydrophobicity allows for increased interaction with the SWCNT surface, and charged subunits facilitate interactions between the DNA backbone and other protein species.
Figure 9.
Mass spectroscopy analysis. (A) Top 12 high-concentration proteins near the molecular weight of the gel bands are compared against BSA in a panel of material properties. Results show preferred proteins to be more hydrophobic and contain more charged amino acids. (B) Hydrophobicity versus length for detected protein species. (C) Detected protein cellular distribution, showing a majority to be either cytoplasmic or nuclear. Statistical tests for protein properties between three groups.
Our MS analysis of the proteome associated with SWCNT aggregates identified 618 statistically significant unique human proteins. Protein characterization showed preferences for different cellular species such as cytoplasmic and nuclear proteins (Figure 9B). As 20–25% of all cellular proteins are secreted, the detected distribution is not unexpected.48 The same preference for hydrophobicity was again shown when top extracted proteins were compared to the detected protein population (Figure 9C). Furthermore, the proteins that remain in aggregated form independent of SWCNT, within the high molecular weight band of the silver staining, were significantly different from those that were not (Figure 9D). On organizing detected proteins by functional groups, we found a preference for transmembrane regions, lipid moiety binding regions, and DNA binding regions (Figure S8). Taken together, we believe that the SWCNT aggregation phenomenon is a demonstration of preferential extraction of proteins from a complex mixture.
Analysis of the SWCNT-Associated Proteome
To gain molecular insight into proteins associated with SWCNT aggregates, our data were subjected to pathway and network analysis. Proteins participating in several canonical pathways are associated DNA–SWCNT aggregates, among which are protein ubiquitination, remodeling of epithelial adherens junctions, and glycolysis (Table S1). Protein ubiquitination is expected, as protein exocytosis from cellular turnover is often first processed through the ubiquitination and proteasome pathway. We also find that the highest functional group of extracted proteins involves nucleotide binding and processing, which suggests that the nucleotide conjugations on the SWCNT surface may provide the unique ability to attract DNA-binding proteins (Table S2).
On the basis of molecular and cellular function, major groups of detected proteins included cellular growth, proliferation, survival, cellular assembly, and organization (Table S3). Furthermore, upstream regulators of these networks were transcription regulators such as TP53 and MYC and growth factors TGF beta 1 (Figure S10). P53 is the master transcriptional regulator of cellular responses to stresses and DNA damage. Normally, p53 remains undetectable due to MDM2-mediated degradation.49 During the cellular stress response, P53 is stabilized to induce cell cycle arrest and DNA damage repair and promote cellular senescence or apoptosis.49 In our study, the DNA–SWCNT aggregates capture proteins associated with these important biological processes, a phenomenon that can be potentially engineered for sensor applications.
Conclusions
In the present study, we observed HUVEC secretome induced aggregation of DNA–SWCNTs in a cell-free system. (AT)15–SWCNTs form complexes with solution proteins and subsequently organize into macrostructures in the context of the extracellular environment. Through nanoparticle tracking, (AT)15–SWCNTs were found to form noncovalent associations upon temperature increases, which can be potentially responsible for the initial steps of SWCNT aggregation. Light scattering reveals a significant increase in particle polydispersity within the first 30 min of an experiment, corresponding to a loss of PL signal due to extraction of singly dispersed SWCNTs. Calculation of total hydrodynamic volume from tracking data over the course of aggregate formation indicates solution SWCNTs were extracted to form aggregates. A second-order reaction model was used to fit the kinetics to derive the rate of SWCNT recruitment, 0.127 min–1.
We studied the aggregate structure at multiple levels of organization and found that there was a greater dependence on protein content than concentration. Through SEM, light microscopy, and visual inspection, we found that the HUVEC secretome, in response to stresses such as starvation, facilitated the formation of larger and more densely packed aggregates than proteins from media supplements only. Lastly, we analyzed the protein components of aggregates formed and found the ratio of protein extraction from conditioned media to be 4.0 ± 0.9 μg protein/μg SWCNT. Silver staining revealed (AT)15–SWCNTs exhibited up to a 100× increased preference for certain protein species.
We used the shotgun proteomics approach to study the proteins associated with DNA–SWCNT aggregates; 618 human proteins were identified through MS. Aggregate proteins showed selective preference for increased hydrophobicity and charged amino acids. Network analyses reveal aggregate protein content is congruent with cellular stress responses.
From the experimental data, we propose a three-step model for the process of (AT)15–SWCNT aggregation formation in the intravascular setting. (1) Upon introduction, the change in temperature from 24 °C to 37 °C induces formation of small SWCNT oligomers. Concurrently, solution proteins are recruited to the SWCNT surface, possibly due to hydrophobic interactions. (2) The rod-shaped aspect of SWCNTs promotes larger aggregates50 and protein structure changes.51 The confinement of protein to the SWCNT surface and the close associations of SWCNT molecules with each other facilitate protein–protein interactions, stabilizing the aggregate structure. (3) Over time, smaller aggregates associate with each other through intermolecular forces, such as charge, and pack differentially due to differential protein content. Equilibrium is reached when repulsive forces cannot be overcome.
Aggregation is one of the major concerns of engineering nanomaterials for biomedical applications. It alters the effectiveness and biodistribution of the associated material and often can be correlated with resulting toxicity. SWCNT toxicity has also often been attributed to individual molecular interactions with cellular components.44 Demonstration of DNA–SWCNT aggregation suggests that their toxicity may also involve multimeric components. Here, we report one of the first works indicating that aggregation is affected by physiologic state. This finding is particularly important as we currently test the colloidal stability of novel materials and drugs in vitro using select proteins (i.e., HSA), human serum, and other blood components.52 However, this study shows that low concentrations of secreted proteins from altered physiologic states can disrupt colloidal stability. The type of protein and complex protein interactions are potentially responsible for why some drugs may not work as well in vivo as designed. Thus, strenuous in vitro biomaterial testing is necessary but may not be sufficient.
Our study of aggregation also opens the door to various potential applications. Compared to previous BSA–SWCNT studies, our EM showed much higher protein density per SWCNT.53 Combined with aggregate preferences for protein types, there is potential to utilize aggregation for the detection54,55 and extraction of low-concentration species. Lastly, the new understanding of temperature-dependent DNA–SWCNT associations can lead to novel methods of bioresponsive self-assembly.
Noncovalent conjugation of SWCNTs involves the dispersion of an intrinsically hydrophobic material into kinetically trapped states,47 often in the process of reaggregation. Our work is the first detailed study of DNA–SWCNT aggregation in a biological environment. Through structure and protein content analysis, we study the process of DNA–SWCNT aggregation. We demonstrated differential preference of DNA–SWCNT aggregates for proteins within the vascular secretome. This information could be useful for the application of SWCNTs in the in vivo setting. At the basic level, we have improved the understanding of the behavior of DNA-coated and high aspect ratio nanomaterials in the extracellular environment.
Methods
Materials
Chemicals, unless stated otherwise, were purchased from Sigma-Aldrich (USA). (AT)15 single-stranded DNA was purchased from Integrated DNA Technologies (USA). SWCNTs produced via the high-pressure carbon monoxide (HiPco) method were purchased from Unidym (USA) and processed via extraction of non-SWCNT material by phase separation in water/hexane.
SWCNT Preparation and Characterization: (AT)15–SWCNT Conjugation
(AT)15-SSDNA and SWCNT were mixed in a 1 mg to 0.5 mg weight ratio in a 1 mL volume of 0.1 M NaCl. The mixture was chilled on ice and probe-tip sonicated (Cole Parmer) with a 1/4 in. tip at 40% amplitude for 20 min. The dispersion was then centrifuged twice for 90 min at 16100g to remove large particulates, undispersed SWCNTs, and other residual impurities.
All samples are analyzed by UV–vis–nIR spectroscopy (Shimadzu UV-3101PC). To calculate SWCNT concentration, absorbance at ∼632 nm was multiplied by a previously found empirical coefficient, 27.8 μg/mL.56 A NanoSpectralyzer 3 (Applied NanoFluorescence) was used to study the fluorescence efficiency and Raman spectra of the dispersion. This instrument consists of five laser excitation wavelengths (409, 532, 637, 671, and 784 nm), a 512-element TE-cooled InGaAs array, and a 2048-element back-thinned CCD Raman detector. The singly dispersed (AT)15–SWCNTs exhibited high optical efficiency (Figure S1).
Cell Culture and Aggregation Experiments
HUVECs were used throughout this study and cultured as described previously.45 HUVECs, endothelial basal medium (EBM), and supplements (EGM-MV: 5% fetal bovine serum, 12 μg/mL bovine brain extract, 1 μg/mL hydrocortisone, and 1 μg/mL GA-1000) were purchased from Lonza (USA). Cells were grown on 30 μg/mL bovine collagen type-I (BD Biosciences)-coated plates in a humidified incubator with 5% CO2 at 37 °C. A hypoxic chamber (Billups-Rothenberg Inc.) was filled with 2% O2 and placed within the tissue culture incubator. HUVECs were grown to passage 4 at ∼80% confluence and used for all experiments. Complete media consisted of EBM, 5% FBS, and all EGM-MV supplements, while starving media had reduced FBS levels of 0.5%. Serum-free media contained no additives. Conditioned media was made by incubating ∼80% confluent cells with starving media for 10 h. The resulting supernatants were filtered with a 0.4 μm filter (Corning, USA) to remove debris. Unless otherwise stated, aggregates were formed via introduction of (AT)15–SWCNT in 0.1 M NaCl at 2 μg/mL to test solutions and incubated under culture conditions for 6 h.
Aggregate Structure Analysis
Near-Infrared Microscopy
Microscopy of aggregate nIR PL was done on an inverted microscope (Zeiss Axiovert 200) with a 20× objective (Zeiss, Plan-APOCHROM(AT) 20×) attached to a 2D InGaAs CCD array (OMA-V 2D, Princeton Instruments). Fluorescence was excited by a 660 nm 100 mW diode-pumped solid-state laser (CrystaLaser, CL660-100).
Photoluminescence
Photoluminescence spectra were collected with a NanoSpectralyzer 3 (Applied NanoFluorescence).
Raman Spectroscopy
Raman spectra were collected with a Horiba LabRAM instrument with a 633 nm laser excitation.
Bright-Field Microscopy and Image Processing
Bright-field images were taken with a Zeiss ApoTome.2. To acquire images of moving solution objects with maximum contrast, the system light source was set at 2.7 V with the camera (AxioCam MRm) at the minimum exposure of 1 ms. Additional detail of the supervised automated image processing can be found in the Supporting Information.
Scanning Electron Microscopy
SEM was performed on a Hitachi S-4700 scanning electron microscope using a field emission filament. The electron beam was set at 1 kV for all imaging. Au-coated samples were sputter-coated at 2.2 kV for 30 s.
Aggregation Formation Studies
Dynamic Light Scattering
DLS was performed on a Zetasizer ZS (Malvern). Sample cells were equilibrated for 30 s at 37 °C and then measured repeatedly via 173° backscatter for the duration of the experiment.
Single Particle Tracking
SPT was performed with a NanoSight NS300 (Malvern). It is equipped with a 532 nm, <60 mW laser, a 20× objective lens with a 3 mm working distance, and a sample chamber temperature controller. The camera readout was calibrated to 137 nm per pixel. The NanoSight software package performed the background subtraction, segmentation, tracking of each individual particle, and the calculation of diffusion coefficients.57 A minimum of 1000 particles was collected to construct each size distribution.
Solution Kinetics Studies
SPT Kinetics
The NanoSight device described above was programmed to collect 90 s of data at 37 °C every 15 min throughout the aggregation experiment.
BCA Protein Extraction
Total media protein concentration was measured via a standard BCA protein assay (Thermo Scientific/Pierce). Calibration was performed with an albumin standard, and samples were measured at 562 nm with a SpectraMax M5e microplate reader (Molecular Devices).
Protein Content Analysis
Silver Stain Analysis
Aggregates were formed and then centrifuged at 16100g for 10 min. Supernatants were removed, and pellets were washed and centrifuged three times to remove residual media. Protein was extracted via suspension in RIPA buffer (Boston Bioproducts) and heated to 95 °C for 10 min. SDS-PAGE was performed with 7.5% Mini-PROTEAN TGX precast gels (BioRad) at 100 V. A silver stain kit (Thermo Scientific/Pierce) was used to develop the result.
Mass Spectrometry
The silver-stained gel was cut into three parts, digested with trypsin, and subjected to mass spectrometry analyses. Mass spectrometry of the samples was performed by nanoflow liquid chromatography electrospray tandem mass spectrometry (nanoLC-ESI-MS/MS) using a Thermo Scientific Orbitrap Elite Hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled to a Thermo Ultimate 3000 RSLCnano HPLC system. The resulting tandem mass spectra were extracted by Msconvert (version 3.0.4019; ProteoWizard), and all MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.4.0), Sequest (Thermo Fisher Scientific, San Jose, CA, USA; version 27, rev. 12), and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1)) (see Supporting Information).
Protein Physiochemical and Bioinformatics Analysis
Physiochemical properties of proteins identified were extracted using PROMPT: Protein Mapping and Comparison Tool,58 which also performed the statistical tests between protein groups. Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA, USA) was used to obtain insight into potential cellular pathways, networks (molecular relationship between proteins), and their biological functions. Fisher’s exact test was used to calculate the p values.
Calculations and Statistics
All image analysis, modeling, and calculations were performed in M(AT)LAB (Natick, Massachusetts: The MathWorks Inc., 2010). Figures are constructed in M(AT)LAB and vectorized and assembled in Inkscape (The Inkscape Team, 2012).
Acknowledgments
This work was partly supported by grants from National Institutes of Health (NIH), HL70567 and CA150190, to D.M. X.G. acknowledges the Langan Scholarship and the National Institute of General Medical Sciences (T32 5T32CA148073). M.S.S. acknowledges support from the Juvenile Diabetes Research Foundation and a grant from the Institutes of Soldier Nanotechnology at MIT funded by the Army Research Office. The authors thank S. Shimizu for help with Raman spectroscopy, R. Eby and P. Hirsova for help with single-particle tracking analysis, B. J. Madden and the Mayo Proteomics Core for help with MS readings, S. I. Gamb, J. E. Charlesworth, and the Mayo Microscopy Core for help with SEM, S. Dutta for help with protein analysis, and J. Lau and H. Satyarthi for editing the manuscript.
Supporting Information Available
Supporting Information includes figures of control experiments and additional detailed methods. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Rabe M.; Verdes D.; Seeger S. Understanding Protein Adsorption Phenomena at Solid Surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. [DOI] [PubMed] [Google Scholar]
- Poldervaart M. T.; Gremmels H.; van Deventer K.; Fledderus J. O.; Oner F. C.; Verhaar M. C.; Dhert W. J.; Alblas J. Prolonged Presence of VEGF Promotes Vascularization in 3D Bioprinted Scaffolds with Defined Architecture. J. Controlled Release 2014, 184, 58–66. [DOI] [PubMed] [Google Scholar]
- Jung F.; Braune S.; Lendlein A. Haemocompatibility Testing of Biomaterials Using Human Platelets. Clin. Hemorheol. Microcirc. 2013, 53, 97–115. [DOI] [PubMed] [Google Scholar]
- Junge K.; Binnebosel M.; von Trotha K. T.; Rosch R.; Klinge U.; Neumann U. P.; Lynen Jansen P. Mesh Biocompatibility: Effects of Cellular Inflammation and Tissue Remodelling. Langenbecks Arch. Chir. 2012, 397, 255–270. [DOI] [PubMed] [Google Scholar]
- Salvador-Morales C.; Flahaut E.; Sim E.; Sloan J.; Green M. L.; Sim R. B. Complement Activation and Protein Adsorption by Carbon Nanotubes. Mol. Immunol. 2006, 43, 193–201. [DOI] [PubMed] [Google Scholar]
- Radomski A.; Jurasz P.; Alonso-Escolano D.; Drews M.; Morandi M.; Malinski T.; Radomski M. W. Nanoparticle-Induced Platelet Aggregation and Vascular Thrombosis. Br. J. Pharmacol. 2005, 146, 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvinte T.; Palais C.; Green-Trexler E.; Gregory S.; Mach H.; Narasimhan C.; Shameem M. Aggregation of Biopharmaceuticals in Human Plasma and Human Serum: Implications for Drug Research and Development. mAbs 2013, 5, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U.; Mukherjee S. Binding, Unfolding and Refolding Dynamics of Serum Albumins. Biochim. Biophys. Acta 2013, 1830, 5394–5404. [DOI] [PubMed] [Google Scholar]
- Saito R.; Dresselhaus M. S.; Dresselhaus G.. Physical Properties of Carbon Nanotubes; World Scientific Publishing Company: Singapore, 1998. [Google Scholar]
- Deacon R.; Chuang K.-C.; Doig J.; Mortimer I.; Nicholas R. Photoluminescence Study of Aqueous-Surfactant-Wrapped Single-Walled Carbon Nanotubes under Hydrostatic Pressure. Phys. Rev. B: Condens. Matter Mater. Phys. 2006, 74, 1–4. [Google Scholar]
- Zhu Z.; Yang R.; You M.; Zhang X.; Wu Y.; Tan W. Single-Walled Carbon Nanotube as an Effective Quencher. Anal. Bioanal. Chem. 2010, 396, 73–83. [DOI] [PubMed] [Google Scholar]
- Lefebvre J.; Finnie P.; Homma Y. Temperature-Dependent Photoluminescence from Single-Walled Carbon Nanotubes. Phys. Rev. B: Condens. Matter Mater. Phys. 2004, 70, 1–8. [Google Scholar]
- Chahine N. O.; Collette N.; Thomas C.; Genetos D. C.; Loots G. G.. Nanocomposite Scaffold for Chondrocyte Growth and Cartilage Tissue Engineering: Effects of Carbon Nanotube Surface Functionalization. Tissue Eng. Part A 2014, [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Bhatnagar I.; Venkatesan J.; Kiml S. K. Polymer Functionalized Single Walled Carbon Nanotubes Mediated Drug Delivery of Gliotoxin in Cancer Cells. J. Biomed. Nanotechnol. 2014, 10, 120–130. [DOI] [PubMed] [Google Scholar]
- Hong G.; Lee J. C.; Jha A.; Diao S.; Nakayama K. H.; Hou L.; Doyle T. C.; Robinson J. T.; Antaris A. L.; Dai H.; et al. Near-Infrared Ii Fluorescence for Imaging Hindlimb Vessel Regeneration with Dynamic Tissue Perfusion Measurement. Circ. Cardiovasc. Imaging 2014, 7, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C.; Balavoine F.; Schultz P.; Ebbesen T. W.; Mioskowski C. Supramolecular Self-Assembly of Lipid Derivatives on Carbon Nanotubes. Science 2003, 300, 775–778. [DOI] [PubMed] [Google Scholar]
- O’Connell M. J.; Bachilo S. M.; Huffman C. B.; Moore V. C.; Strano M. S.; Haroz E. H.; Rialon K. L.; Boul P. J.; Noon W. H.; Kittrell C.; et al. Band Gap Fluorescence from Individual Single-Walled Carbon Nanotubes. Science 2002, 297, 593–596. [DOI] [PubMed] [Google Scholar]
- Kim O.-K.; Je J.; Baldwin J. W.; Kooi S.; Pehrsson P. E.; Buckley L. J. Solubilization of Single-Wall Carbon Nanotubes by Supramolecular Encapsulation of Helical Amylose. J. Am. Chem. Soc. 2003, 125, 4426–4427. [DOI] [PubMed] [Google Scholar]
- Zorbas V.; Ortiz-Acevedo A.; Dalton A. B.; Yoshida M. M.; Dieckmann G. R.; Draper R. K.; Baughman R. H.; Jose-Yacaman M.; Musselman I. H. Preparation and Characterization of Individual Peptide-Wrapped Single-Walled Carbon Nanotubes. J. Am. Chem. Soc. 2004, 126, 7222–7227. [DOI] [PubMed] [Google Scholar]
- Barisci J. N.; Tahhan M.; Wallace G. G.; Badaire S.; Vaugien T.; Maugey M.; Poulin P. Properties of Carbon Nanotube Fibers Spun from DNA-Stabilized Dispersions. Adv. Funct. Mater. 2004, 14, 133–138. [Google Scholar]
- Cheung W.; Pontoriero F.; Taratula O.; Chen A. M.; He H. DNA and Carbon Nanotubes as Medicine. Adv. Drug Delivery Rev. 2010, 62, 633–649. [DOI] [PubMed] [Google Scholar]
- Iverson N. M.; Barone P. W.; Shandell M.; Trudel L. J.; Sen S.; Sen F.; Ivanov V.; Atolia E.; Farias E.; McNicholas T. P.; et al. In Vivo Biosensing via Tissue-Localizable near-Infrared-Fluorescent Single-Walled Carbon Nanotubes. Nat. Nanotechnol. 2013, 8, 873–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J.; Ge C.; Liu Y.; Bai R.; Li D.; Yang Y.; Liao L.; Chen C. The Interaction of Serum Proteins with Carbon Nanotubes Depend on the Physicochemical Properties of Nanotubes. J. Nanosci. Nanotechnol. 2011, 11, 10102–10110. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S.; Roxbury D.; Gong X.; Mukhopadhyay D.; Jagota A. DNA Conjugated Swcnts Enter Endothelial Cells via Rac1 Mediated Macropinocytosis. Nano Lett. 2012, 12, 1826–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz E.; Sreelatha A.; Garriga R.; Baughman R. H.; Goux W. J. Amyloidogenic Peptide/Single-Walled Carbon Nanotube Composites Based on Tau-Protein-Related Peptides Derived from Acphf6: Preparation and Dispersive Properties. J. Phys. Chem. B 2013, 117, 7593–7604. [DOI] [PubMed] [Google Scholar]
- Cai X.; Ramalingam R.; Wong H. S.; Cheng J.; Ajuh P.; Cheng S. H.; Lam Y. W. Characterization of Carbon Nanotube Protein Corona by Using Quantitative Proteomics. Nanomedicine (N. Y., NY, U. S.) 2013, 9, 583–593. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Liu R.; Chi Z.; Teng Y.; Qin P. New Insights into the Behavior of Bovine Serum Albumin Adsorbed onto Carbon Nanotubes: Comprehensive Spectroscopic Studies. J. Phys. Chem. B 2010, 114, 5625–5631. [DOI] [PubMed] [Google Scholar]
- Li L.; Lin R.; He H.; Jiang L.; Gao M. Interaction of Carboxylated Single-Walled Carbon Nanotubes with Bovine Serum Albumin. Spectrochim. Acta, Part A 2013, 105, 45–51. [DOI] [PubMed] [Google Scholar]
- Tsyboulski D. A.; Bakota E. L.; Witus L. S.; Rocha J. D.; Hartgerink J. D.; Weisman R. B. Self-Assembling Peptide Coatings Designed for Highly Luminescent Suspension of Single-Walled Carbon Nanotubes. J. Am. Chem. Soc. 2008, 130, 17134–17140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo J. P.; Landry M. P.; Faltermeier S. M.; McNicholas T. P.; Iverson N. M.; Boghossian A. A.; Reuel N. F.; Hilmer A. J.; Sen F.; Brew J. A.; et al. Plant Nanobionics Approach to Augment Photosynthesis and Biochemical Sensing. Nat. Mater. 2014, 13, 400–408. [DOI] [PubMed] [Google Scholar]
- Khan I. A.; Aich N.; Afrooz A. R.; Flora J. R.; Schierz P. A.; Ferguson P. L.; Sabo-Attwood T.; Saleh N. B. Fractal Structures of Single-Walled Carbon Nanotubes in Biologically Relevant Conditions: Role of Chirality vs. Media Conditions. Chemosphere 2013, 93, 1997–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. H.; Strano M. S. Solvatochromism in Single-Walled Carbon Nanotubes. Appl. Phys. Lett. 2007, 90, 223114–223116. [Google Scholar]
- Heller D. A.; Barone P. W.; Swanson J. P.; Mayrhofer R. M.; Strano M. S. Using Raman Spectroscopy to Elucidate the Aggregation State of Single-Walled Carbon Nanotubes. J. Phys. Chem. B 2004, 108, 6905–6909. [Google Scholar]
- Lopez-Lorente A. I.; Simonet B. M.; Valcarcel M. Raman Spectroscopic Characterization of Single Walled Carbon Nanotubes: Influence of the Sample Aggregation State. Analyst (Cambridge, U. K.) 2014, 139, 290–298. [DOI] [PubMed] [Google Scholar]
- Deyoung L. R.; Fink A. L.; Dill K. A. Aggregation of Globular-Proteins. Acc. Chem. Res. 1993, 26, 614–620. [Google Scholar]
- Qing J. A.; Logan B. E. Fractal Dimensions of Aggregates from Shear Devices. J. - Am. Water Works Assoc. 1996, 88, 100–113. [Google Scholar]
- Raper J. A.; Amal R. Measurement of Aggregate Fractal Dimensions Using Static Light-Scattering. Part. Part. Syst. Charact. 1993, 10, 239–245. [Google Scholar]
- Lee C.; Kramer T. A. Prediction of Three-Dimensional Fractal Dimensions Using the Two-Dimensional Properties of Fractal Aggregates. Adv. Colloid Interface Sci. 2004, 112, 49–57. [DOI] [PubMed] [Google Scholar]
- Uematsu I.; Matsumoto H.; Morota K.; Minagawa M.; Tanioka A.; Yamagata Y.; Inoue K. Surface Morphology and Biological Activity of Protein Thin Films Produced by Electrospray Deposition. J. Colloid Interface Sci. 2004, 269, 336–340. [DOI] [PubMed] [Google Scholar]
- Ortega A.; de la Torre J. G. Hydrodynamic Properties of Rodlike and Disklike Particles in Dilute Solution. J. Chem. Phys. 2003, 119, 9914–9919. [Google Scholar]
- Broersma S. Viscous Force and Torque Constants for a Cylinder. J. Chem. Phys. 1981, 74, 6989–6990. [Google Scholar]
- Johnson R. R.; Johnson A. T.; Klein M. L. Probing the Structure of DNA-Carbon Nanotube Hybrids with Molecular Dynamics. Nano Lett. 2008, 8, 69–75. [DOI] [PubMed] [Google Scholar]
- Li X.; Peng Y.; Qu X. Carbon Nanotubes Selective Destabilization of Duplex and Triplex DNA and Inducing B-A Transition in Solution. Nucleic Acids Res. 2006, 34, 3670–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo G. H.; Kang S. G.; Xiu P.; Zhao Y. L.; Zhou R. H. Interactions between Proteins and Carbon-Based Nanoparticles: Exploring the Origin of Nanotoxicity at the Molecular Level. Small 2013, 9, 1546–1556. [DOI] [PubMed] [Google Scholar]
- Zeng H.; Dvorak H. F.; Mukhopadhyay D. Vascular Permeability Factor (VPF)/Vascular Endothelial Growth Factor (VEGF) Peceptor-1 Down-Modulates Vpf/Vegf Receptor-2-Mediated Endothelial Cell Proliferation, but Not Migration, through Phosphatidylinositol 3-Kinase-Dependent Pathways. J. Biol. Chem. 2001, 276, 26969–26979. [DOI] [PubMed] [Google Scholar]
- Roxbury D.; Tu X.; Zheng M.; Jagota A. Recognition Ability of DNA for Carbon Nanotubes Correlates with Their Binding Affinity. Langmuir 2011, 27, 8282–8293. [DOI] [PubMed] [Google Scholar]
- Saleh N. B.; Pfefferle L. D.; Elimelech M. Influence of Biomacromolecules and Humic Acid on the Aggregation Kinetics of Single-Walled Carbon Nanotubes. Environ. Sci. Technol. 2010, 44, 2412–2418. [DOI] [PubMed] [Google Scholar]
- Kulasingam V.; Diamandis E. P. Proteomics Analysis of Conditioned Media from Three Breast Cancer Cell Lines: A Mine for Biomarkers and Therapeutic Targets. Mol. Cell Proteomics 2007, 6, 1997–2011. [DOI] [PubMed] [Google Scholar]
- Amaral J. D.; Xavier J. M.; Steer C. J.; Rodrigues C. M. The Role of P53 in Apoptosis. Discovery Med. 2010, 9, 145–152. [PubMed] [Google Scholar]
- Forstater J. H.; Kleinhammes A.; Wu Y. Self-Assembly of Protein-Based Biomaterials Initiated by Titania Nanotubes. Langmuir 2013, 29, 15013–15021. [DOI] [PubMed] [Google Scholar]
- Chakraborty S.; Joshi P.; Shanker V.; Ansari Z. A.; Singh S. P.; Chakrabarti P. Contrasting Effect of Gold Nanoparticles and Nanorods with Different Surface Modifications on the Structure and Activity of Bovine Serum Albumin. Langmuir 2011, 27, 7722–7731. [DOI] [PubMed] [Google Scholar]
- ASTM Standard F2027. Standard Guide for Characterization and Testing of Raw or Starting Biomaterials for Tissue-Engineered Medical Products; West Conshohocken, PA, 2008. [Google Scholar]
- Nithiyasri P.; Balaji K.; Brindha P.; Parthasarathy M. Programmable Self-Assembly of Carbon Nanotubes Assisted by Reversible Denaturation of a Protein. Nanotechnology 2012, 234656038. [DOI] [PubMed] [Google Scholar]
- Barone P. W.; Strano M. S. Reversible Control of Carbon Nanotube Aggregation for a Glucose Affinity Sensor. Angew. Chem. 2006, 45, 8138–8141. [DOI] [PubMed] [Google Scholar]
- Chen H.; Yu C.; Jiang C.; Zhang S.; Liu B.; Kong J. A Novel near-Infrared Protein Assay Based on the Dissolution and Aggregation of Aptamer-Wrapped Single-Walled Carbon Nanotubes. Chem. Commun. 2009, 5006–5008. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Boghossian A. A.; Barone P. W.; Rwei A.; Kim J. H.; Lin D.; Heller D. A.; Hilmer A. J.; Nair N.; Reuel N. F.; et al. Single Molecule Detection of Nitric Oxide Enabled by D(at)15 DNA Adsorbed to near Infrared Fluorescent Single-Walled Carbon Nanotubes. J. Am. Chem. Soc. 2011, 133, 567–581. [DOI] [PubMed] [Google Scholar]
- Carr B.; Hole P.; Malloy A.; Nelson P.; Wright M.; Smith J. Applications of Nanoparticle Tracking Analysis in Nanoparticle Research. Eur. J. Parenter. Pharm. Sci. 2009, 14, 45–50. [Google Scholar]
- Schmidt T.; Frishman D. Prompt: A Protein Mapping and Comparison Tool. BMC Bioinf. 2006, 7, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.