Abstract
We suggest a bacteriophage genus, “Viunalikevirus”, as a new genus within the family Myoviridae. To date, this genus includes seven sequenced members: Salmonella phages ViI, SFP10 and ΦSH19; Escherichia phages CBA120 and PhaxI; Shigella phage phiSboM-AG3; and Dickeya phage LIMEstone1. Their shared myovirus morphology, with comparable head sizes and tail dimensions, and genome organization are considered distinguishing features. They appear to have conserved regulatory sequences, a horizontally acquired tRNA set and the probable substitution of an alternate base for thymine in the DNA. A close examination of the tail spike region in the DNA revealed four distinct tail spike proteins, an arrangement which might lead to the umbrella-like structures of the tails visible on electron micrographs. These properties set the suggested genus apart from the recently ratified subfamily Tevenvirinae, although a significant evolutionary relationship can be observed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00705-012-1360-5) contains supplementary material, which is available to authorized users.
Introduction
Bacteriophage classification is an evolving field that has traditionally been focused on morphology but has shifted more to genomics as DNA sequencing has become significantly less expensive. Accessible sequencing has resulted in a logarithmic increase in sequences deposited to databases and the need to classify them into new species, genera, subfamilies and families [1]. The subfamily category in the order Caudovirales is relatively new and was first introduced into the family Podoviridae with the creation of the subfamily Autographivirinae and the subfamily Picovirinae [2]. Similarly, within the family Myoviridae, the subfamily Tevenvirinae has been described, containing the genera T4likevirus and SchizoT4likevirus (http://www.ictvonline.org). Members of the first genus, with T4 as the type phage, are morphologically indistinguishable and comprise the former members of the T-even and pseudo-T-even type phages [3]. The genus Schizot4likevirus, named after the historical schizo-T-even phage group, has as its type virus Vibrio phage KVP40 [4]. The phages belonging to this genus have larger head sizes than T4, an identical tail structure and over 20 % of their coding sequences in common [4–6].
Many of the phages recognized as being T4-related by the marine ecology community, particularly a group of cyanomyoviruses, fall outside of these genera because their proteomes were considered to be too distinct in comparison to other clades within the viral domain. In a recent review of the “T4 supergroup of viruses,” Petrov and coworkers [7] defined T4-related bacteriophages as possessing a Core Genome encoding approximately 37 proteins. Appropriately, we feel this definition not only includes the members of the genus T4likevirus and the genus SchizoT4likevirus but also various cyanomyoviruses and such phages as Campylobacter phages Cpt10 (FN667788) and CP220 (FN667789), Delftia phage ΦW-14 (NC_013697), and Salmonella phage ViI (NC_015296), each of which seems to be a prototype for a different genus within this supergroup.
In this paper, we suggest the creation of a new genus “Viunalikevirus”, after phage ViI, the first phage to be identified and studied of this viral genus [8]. This bacteriophage was first described in the 1930s as Typhoid phage Q151, which later became known as Vi1 [9–11] and Salmonella Typhi phage ViI [12], specific for the Vi antigen of Salmonella enterica subsp. enterica serovar Typhi. Genome analysis of six other phages, Salmonella phages SFP10 and ΦSH19, Escherichia phages CBA120 and PhaxI, Shigella phage phiSboM-AG3 and Dickeya phage LIMEstone1, showed a DNA similarity of over 50 % with each other and a distant relationship to T4 [13–17]. The close relatedness of these seven phages is examined in this paper, and the features of members of the newly suggested genus are described.
Bacteriophages belonging to the genus “Viunalikevirus”
The properties of the seven phages with fully sequenced and annotated genomes belonging to the genus “Viunalikevirus” are summarized in Table 1. The distinguishing features of this genus include the genome size and organization, gene synteny, T4-like regulation of late transcription, the use of tail spikes for host recognition rather than T4-like tail fibers, and the use of a modified form of uracil (putatively HMdU) rather than thymine. The tail spikes of these phages were examined particularly closely, as this is the region in the genome that is most diverged. Morphologically near-identical phages occur in Acinetobacter, Bordetella, and Sinorhizobium, but their genomes have not yet been sequenced, and these phages are still awaiting classification.
Table 1.
Features of the phages belonging to the genus “Viunalikevirus”
| Phage name | Host | Head dimensions (nm) | Tail dimensions (nm) | Genome size (bp) | Mol%G + C | Number of ORFs annotated | Number of tRNAs | Number of HEases | Reference | Accession number |
|---|---|---|---|---|---|---|---|---|---|---|
| Phages with sequenced genomes | ||||||||||
| ViI (syn. Vi1, Vi01) | Salmonella Typhimurium | 90–95* | 105–115 × 17* | 157,061 | 45 | 208 | 6 | [8, 11, 12, 29] | NC_015296 | |
| SFP10 | S. Typhimurium | 86* | 109 × 18 | 157,950 | 44.5 | 201 | 4 | [17] | HQ259103 | |
| ΦSH19 | S. Typhimurium | 83 | 110 × 14 | 157,785 | 44.7 | 166 | 5 | 2 | [16] | JN126049 |
| CBA120 | Escherichia coli | 90* | 105 × 17* | 157,304 | 44.5 | 202 | 4 | [15] | JN593240 | |
| PhaxI | E. coli | 80 | 115 × 15 | 156,628 | 44.5 | 209 | 4 | 1 | Shahrbabak et al, unpublished | JN673056 |
| LIMEstone1 | “Dickeya solani” | 91* | 114 × 17* | 152,427 | 44.9 | 201 | 1 | 14 | [13] | HE600015 |
| phiSboM-AG3 | Shigella boydii | 83* | 110 × 14* | 158,006 | 50.4 | 216 | 4 | [14] | NC_013693 | |
| Phages with Vi1 morphology | ||||||||||
| 15, 16 | S. Typhimurium | 88* | 110–114 × 17* | – | – | – | – | [27] | ||
| 10 | S. Typhimurium | 88* | 112 × 19* | – | – | – | – | [30] | ||
| FGCSSa1 | S. Typhimurium | 107 | 123 × 17 | – | – | – | – | [31] | ||
| Det7 | S. Typhimurium | 90 | 120 × 17 | – | – | – | – | [32] | ||
| 344 | Serratia plymuthica | 85 | 117 × 17 | – | – | – | – | Grimont, PhD thesis | ||
| E14, E1, E2, E7 | Acinetobacter calcoaceticus | 91* | 106 × 17* | – | – | – | – | [33] | ||
| Ba5.2 | Bordetella avium | 80* | 107 × 17* | – | – | – | – | Ackermann, unpublished | ||
| CM1 to CM9, CM20, CM21 |
Sinorhizobium meliloti | 98* | 96–103 × 16* | – | – | – | – | [34, 35] | ||
* Measured after calibration with catalase crystals or T4 phage tails
Morphology
The phages of the genus “Viunalikevirus” have contractile tails and are thus members of the family Myoviridae. They are nearly indistinguishable in morphology, with similar head and tail dimensions, namely an isometric head of about 90 nm and a contractile tail of about 110 × 18 nm. The type phage ViI is depicted in Fig. 1. Based on this morphology, a number of other phages from the literature can be classified as probable viunalikeviruses (Table 1; Supplementary Figure 1). Heads are icosahedral, as indicated by the simultaneous presence of capsids with hexagonal and pentagonal outlines. Tails consist of a T4-like neck with a collar, a sheath surrounding a tail tube or core, a thin base plate, and an adsorption structure. The tail sheath has 24 transverse striations and resembles that of phage T4. The head and tail dimensions shown vary slightly, even when measured after magnification control by means of catalase crystals or T4 phage tails. The reasons are that micrographs were taken over a period of over 40 years using different electron microscopes and that uranyl-acetate-stained capsids tend to be smaller than those of phosphotungstate-stained phages (for references see Table 1). The collar is found in ViI-like phages of enterobacteria, Bordetella, and Sinorhizobium, but not of Acinetobacter.
Fig. 1.
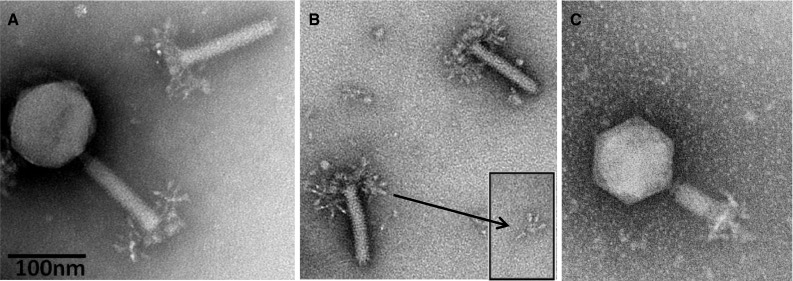
Electron micrographs of Salmonella phage ViI. (A) Complete phage ViI with quiescent tail and severed tail; (B) ViI tails with unfolded tail entities, displaying an umbrella-like structure; (C) ViI with contracted tail. The phage was applied to a glow-discharged carbon/Formavar-coated 200-mesh copper grid and then stained with 5 % aqueous ammonium molybdate plus 1 % trehalose. The grid was finally examined on a 120 KV Philips Tecnai Spirit BioTwin transmission electron microscope fitted with a Tietz F415 charge-coupled-device (CCD) TemCam camera
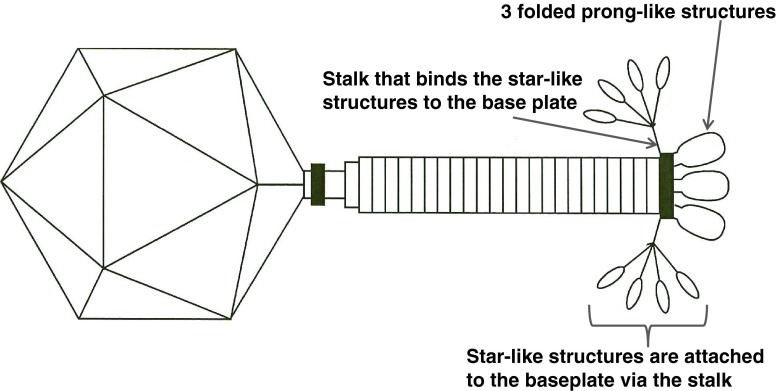
The adsorption organelle is complex and undergoes conformational changes. Quiescent tails show in profile 3-4 thick “prongs” with rounded ends (schematic representation in Fig. 2). In other instances, one observes a broad, umbrella-like, fibrous structure measuring 83-90 × 70-50 nm with star-like elements. The latter consists of 4 or 5 short filaments with bulbous tips (rays). Intermediate structures with both “prongs” and “stars” have been observed. Still other tails show, especially after tail contraction, an entangled mass of indistinct filaments. No similar tail structures have been reported in other phages. It is concluded that the tail has a six-folded symmetry and 6 “prongs” which unfold into as many “stars” and finally individual filaments. The most probable dimensions of the various ViI-like phages are indicated in Table 2.
Fig. 2.
Scale drawing of phage ViI with partially unfolded tail spikes. The sixth spike is behind the tail shaft and not shown. At present, the gene product responsible for the stalk, which attaches the star-like structures (and prongs) to the base-plate, has not been identified
Table 2.
Probable dimensions of the viunalikeviruses
| Head/tail | Structure | Dimensions |
|---|---|---|
| Head | Diameter | 90 nm between opposite apices |
| Tail | Overall | 108 × 18 nm in the extended state |
| Neck | 10 × 8 nm | |
| Collar | 12 × 3 nm | |
| Contracted sheath | 40 × 25 nm | |
| Core | 8 nm wide, channel of 3 nm | |
| Base plate | 23 × 3 nm | |
| Prongs | 17 × 6 nm | |
| Stars | 30 nm wide | |
| Rays | 20 nm long |
Comparative genomics
Genome organization
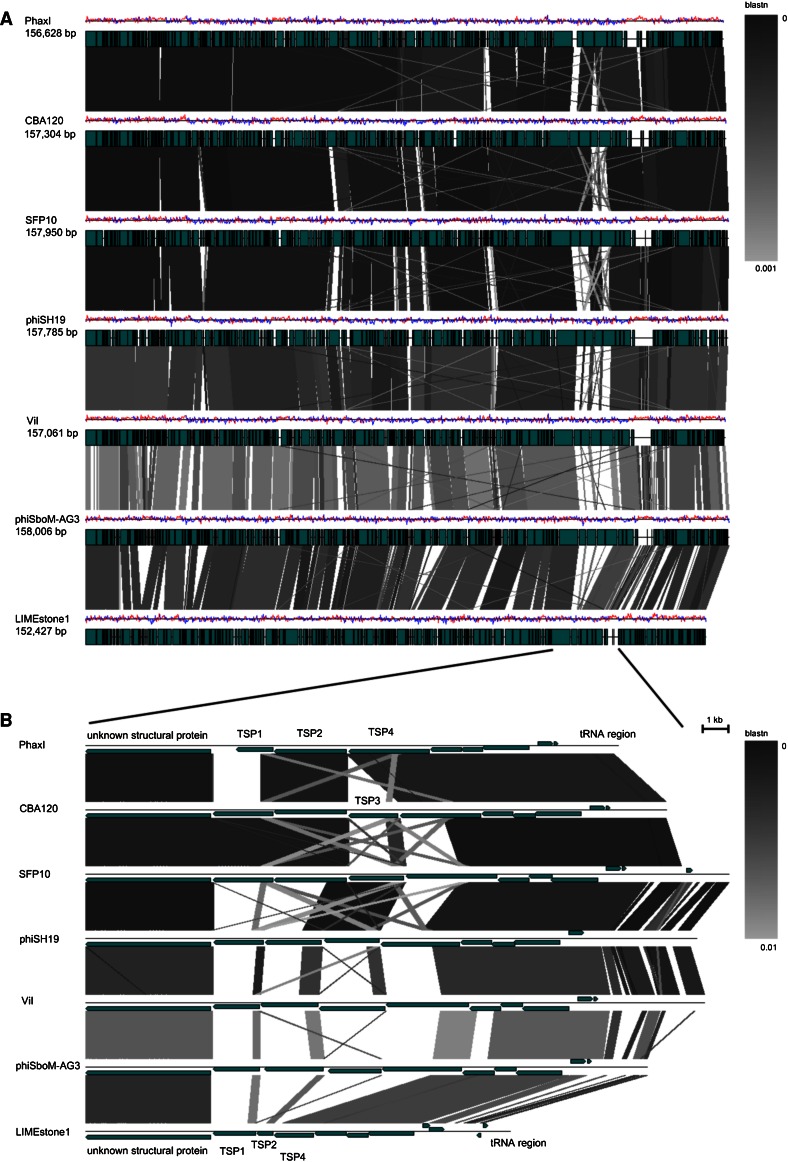
The seven sequenced phages of the genus “Viunalikevirus” have genomes of similar size and share a high degree of sequence similarity. Sizes range from 152 kb for phage LIMEstone1 to 158 kb for phiSboM-AG3. Using the EMBOSS Stretcher algorithm (http://www.ebi.ac.uk/Tools/psa/emboss_stretcher/), nucleotide similarities with the type phage ViI were computed. The Salmonella and Escherichia phages all shared 81 % DNA identity; the similarity of ViI with phiSboM-AG3 and LIMEstone1 was 66 % and 59 %, respectively. This high degree of similarity is clearly visible in Fig. 3, which shows a pair-wise BLASTN comparison between the different genomes, each phage compared with its nearest neighbour on the figure [18].
Fig. 3.
BLASTN comparison of the phages belonging to the suggested genus “Viunalikevirus”. Phages are compared pairwise with their nearest neighbours in the figure. (A) Genomes are represented by a GC skew plot (top line for each phage), and annotated ORFs are depicted as boxes (bottom line for each phage). (B) Subregion pairwise comparison. ORFs are depicted as arrows according to frame
At the protein level, similarity was determined using CoreGenes [19] at its default settings. Relative to bacteriophage ViI, phage SFP10 has 189 proteins in common (91.0 % similarity), PhaxI (90.4 %), CBA120 (88.5 %), phiSboM-AG3 (80.1 %), ΦSH19 (76.9 %), and LIMEstone1 (77.4 %).
The gene order is strongly conserved in all seven phages, even though various functional regions are randomly distributed throughout the genome, in contrast to the fairly extensive functional clustering seen in tevenviruses [7]. This particular genome organization appears to be a distinguishing feature of this genus.
Regulatory elements
To investigate the existence of unique regulatory regions within the new genus, 100 bp of 5’ upstream sequence data was extracted using extractUpStreamDNA (http://lfz.corefacility.ca/extractUpStreamDNA/ ) and submitted for MEME analysis [20] at http://meme.sdsc.edu/meme/cgi-bin/meme.cgi. Two motifs, with sequences TTCAAT[N14]TATAAT and CTAAATAcCcc, were found in all of the phages (Supplementary Figure 2). The core of the latter motif closely resembles the coliphage T4 late promoter core—TATAAATA [21, 22]; moreover, a number of these are located upstream of proven morphogenesis genes. Late transcription in T4 is dependent upon host polymerase and the products of genes 45 (RNA polymerase recruitment), 33 (co-activator of late transcription) and 55 (late promoter recognition protein). All of the phages in this genus possess homologs of these proteins; in the case of ViI, they are ViO1_135c, ViO1_069c, and ViO1_103c, respectively. In contrast, the sequence TTCAAT[N14]TATAAT is found upstream of many small conserved phage genes, reminiscent of the many immediate-early genes responsible for the transition from host to phage metabolism in phage T4 [23]. It is also found before various genes involved in DNA synthesis, including DNA polymerase, nrdA and B, the gp45 sliding clamp and DNA ligase, suggesting that these sequences are also active in delayed early transcription [23]. The fact that they resemble but are not identical to the RpoD-dependent host promoters (TTGACA[N15-17]TATAAT) suggests that an unknown phage factor may be involved in their recognition.
tRNAs and codon usage
The ViI-related phages possess a collection of tRNA genes (Table 3) identified using tRNAscan-SE [24] and ARAGORN [25]. These tRNA genes are located at comparable positions within the ViI-like genomes (indicated in the bottom half of Fig. 3). For phages ViI, ΦSH19, SFP10, CBA120, and PhaxI, the tRNA genes occupy regions that are associated with a perceptible increase over the average GC content (which peaks at around 60 %). A similar but less overt GC bias is observed in phiSboM-AG3, correlating with the higher mean GC content of this phage. In contrast, LIMEstone1 possesses a solitary tRNAMet, found in a region that is slightly below the mean GC content. Based on the regional nucleotide sequence conservation and a common tendency to exhibit a localized increase in GC content, we hypothesize that a tRNA gene set comprising tRNAMet, tRNAAsn, and tRNASer was horizontally acquired by an ancestral ViI-like phage at some stage during its evolutionary history. The region also appears to be more amenable to the acquisition of further tRNA genes such as the tRNAGln in ViI, tRNAIle in CBA120, and tRNASer (TCA) in SboM-AG3.
Table 3.
tRNAs and their codons of the phages of the genus “Viunalikevirus”
| tRNAa (Codon) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phage | Met (ATG) | Ser 1 (TCA) | Asn (AAC) | Ile (ATC) | Gln (CAA) | Tyr (TAC) | Ser 2 (AGC) | Undetermined (???) |
| ViI | + (+) | − (−) | + (+) | − (−) | + (+) | + (+) | + (+) | + (−) |
| phiSboM-AG3 | − (−) | + (+) | + (+) | − (−) | − (−) | + (+) | + (+) | − (−) |
| CBA120 | + (+) | − (−) | + (+) | − (+) | − (−) | − (−) | + (+) | + (−) |
| ΦSH19 | + (+) | − (−) | + (+) | − (−) | − (−) | + (+) | + (+) | + (−) |
| SFP10 | + (+) | − (−) | + (+) | − (−) | − (−) | − (−) | + (+) | + (−) |
| LIMEstone1 | + (+) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) |
| PhaxI | + (+) | − (−) | + (+) | − (−) | − (−) | − (−) | + (+) | + (−) |
a + present; − absent, non-bracketed—identified with tRNAscan-SE, brackets—identified with ARAGORN
To examine the relationship between tRNA carriage and codon usage, relative synonymous codon usage and codon adaptation indices were generated for each phage using E-CAI [26]. Examination of the codon usage of the ViI-like phages does not reveal any apparent link between codon preference and the presence of their cognate tRNAs. A wider examination of the ViI-like phages and host bacteria codon usage tables indicated that the codons specified by the ViI-like phage tRNAs are actually amongst the most frequently occurring in the host genomes. These data suggest that the requirements of the phage could be served by sufficient host tRNAs.
DNA modification
None of the seven sequenced viunalikeviruses carry any of the genes responsible for substituting C for hydroxymethylcytosine (HMdC) in the DNA of T4 as well as the more closely related T4-like phages, implying that these phages all use C in their DNA. However, while most of the genes involved in nucleotide metabolism appear homologous to corresponding genes in tevenviruses, the ORF originally annotated as a thymidylate synthase in most viunalikeviruses is much more closely related to the hydroxymethyluracil (HMdU) transferases found in a few other phages, such as Delftia phage ΦW-14 and Bacillus subtilis phage SP01 (Fig. 4). This appears to be a primary distinguishing feature of the new genus “Viunalikevirus”.
Fig. 4.
Dendogram of hydroxymethyluridylate transferase sequences of members of the genus “Viunalikevirus” compared with those of known hydroxymethyluridylate transferases as well as thymidylate synthase sequences from other phages. Proteins were aligned in MAFFT, with BLOSUM 30 used as the scoring matrix. The topology of the tree is based on NJ bootstrap values, shown above, which were calculated using the Poisson distance between sequences over 100 pseudoreplicates
The incorporation of a non-canonical nucleotide in place of thymine was first suspected when Kutter et al. [15] found that CBA120 could not incorporate tritiated thymidine into its DNA, while still making hundreds of phage per cell. To investigate the nature of this probable substitution, an assay was constructed that took advantage of the characterized methylation sensitivities of five commercially available restriction endonucleases according to the manufacturer’s instructions. Testing showed that both CBA120 and LIMEstone1 genomic DNA are sensitive to cleavage by restriction endonucleases that have been previously shown to be active against fully substituted HMdU-containing DNA but not endonucleases that are inhibited by that modification. Using this assay, we were able to distinguish HMdU-containing DNA from DNA containing canonical nucleotides as well as other non-canonical substitutions such as methylcytosine, hydroxymethylcytosine, glycosylated hydroxymethylcytosine, uracil, and phosphoglucuronated 5-(4’,5’-dihydroxypentyl) uracil (Blasdel, Brabban and Kutter, unpublished observations).
Tail spikes
Bacteriophages within the genus “Viunalikevirus” are closely related to each other in terms of their DNA sequence and the remarkable synteny of the vast majority of genes along their entire genomes (see Fig. 3). The sole region of significant divergence lies within the tail spike region of each phage (bottom half of Fig. 3), yet even within these regions, there are significant zones of homology at both the DNA and protein level. This arrangement of tail spike genes within their genomes is a unique signature of this group of phages.
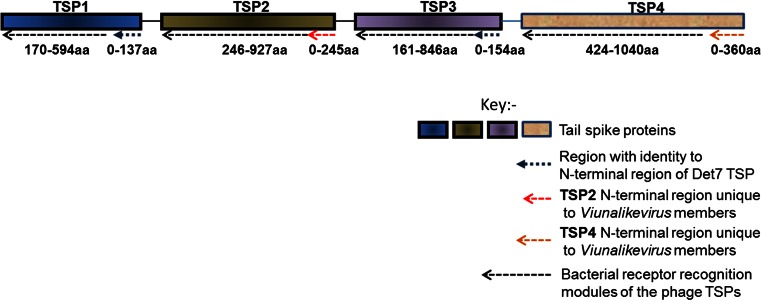
As more representatives of this phage genus have been identified, variants in the number of tail spike proteins (TSPs) have been found. For example, LIMEstone1 and PhaxI have one and three likely tail spike proteins, respectively. Most of the identified ViI-like phages to date are now believed to encode four tail spike proteins, as illustrated in Fig. 3. Many of the different modules and domains we have identified in TSP1-4 have been discussed in detail previously [8, 15, 16]. Briefly, tail spike proteins 1 to 3 (TSPs1-3) have a number of salient shared features and modules, while TSP4 is significantly longer and will be analyzed separately. Figure 5 provides an overview of the TSPs illustrating the modules present and their conservation with respect to most members of the genus “Viunalikevirus”.
Fig. 5.
A generalised depiction of the various domains and regions identified within the tail spike proteins (TSPs) of members of the genus “Viunalikevirus”
TSPs 1 to 3 are characterized by two main regions or modules. The N-terminal regions of the TSPs are highly conserved and may represent the domain for binding to the phage base-plate, reminiscent of those of podoviruses, although this has not yet been confirmed experimentally. TSP1 and TSP3 share a common N-terminal region that shares extensive homology with a tail spike from phage Det7 [27], a phage that most likely belongs to the genus “Viunalikevirus”. The order of TSP1 and TSP3 with respect to TSP2 is conserved within this variable region of the ViI-group; only TSP1 and TSP3 have homology to the one Det7 spike whose sequence has been published to date [27], while no TSP2 genes have shown any homologies to that spike. The TSP2 genes of the ViI-like phages (when present) share a highly homologous N-terminal domain, which to date has been found exclusively in viunalikeviruses. Figure 3 also illustrates this conservation.
The second, C-terminal region can again be divided into two domains in at least some of the spikes, as seen most clearly in the crystal structure determined for this portion of the published Det7 spike [27]. The proximal beta-helical region has an enzymatic function, exemplified in Det7 by an O-antigen binding and hydrolysis domain, in ViI by an acetyl-esterase, which catalyzes the acetyl-modification of the Vi capsule, and in CBA120 by a section related to the N-acetylneuraminidase domain in the spike of coliphage K1F. The C-terminal regions then each encode some sort of bacterial receptor-recognition proteins to elicit phage attachment.
The pairwise comparisons shown in Fig. 3, in conjunction with the features described in Fig. 5, underline a possible role for TSP4 when it is intact, as it is in the majority of the ViI-like phages. The N-terminal region of TSP4 encodes a novel base-plate-binding protein found only in the ViI-like phages, while the rest of this gene encodes one of a further range of bacterial recognition protein modules in a manner similar to TSP1-3.
The sequence data clearly point to up to four distinct TSPs, and the EM morphology, as shown in Fig. 1B with its small inserted image, appears to support this. There are four spikes surrounding a less dense central core, reminiscent of tail structures associated with podoviruses when seen separated from the phage head [8]. This central core could well be the tail stalk that binds the spikes to the base plate itself. The overall phage morphology is summarized in Fig. 2, but aspects of the tail spike structure require further investigation for each phage.
Table 4 summarizes the known and putative targets of the TSPs. The range typically encompasses both plant and animal pathogens of the family Enterobacteriaceae. Many of the TSP targets have been identified experimentally, but a number were putatively identified using BLASTP when the criteria included both very good E values and coverage of at least 70 % of the length of the TSP. In most cases, the putative homologies to the ViI-like TSPs were to tail spike genes from temperate phages. For example, the unique TSP1 of phiSboM-AG3 is very similar to a gene found in an uncharacterized temperate phage of E. coli UMNK88 with the accession number AEE57668 (266/428aa identity; 325/428aa positives). A further interesting example is TSP4 from phages CBA120 and PhaXI that is similar to a further uncharacterized lysogenic phage gene (accession number CBJ02120), this time located in E.coli H10407 (247/375aa identity; 299/375aa positives). It may not be a coincidence that E. coli H10407 is an O78 antigenic type strain, as this was also the serotype of the only member of the ECOR collection that CBA120 infected [15].
Table 4.
The known and putative targets of the tail spike proteins of the sequenced bacteriophages of the genus “Viunalikevirus”
| TSP | PhaXI | CBA120 | SFP10 | ΦSH19 | ViI | phiSboM-AG3 | LIMEstone1 |
|---|---|---|---|---|---|---|---|
| TSP1 | Unique TSP | Synteny with TSP1 of SFP10. Target-unknown |
Synteny with TSP1 of CBA120. Target-unknown |
Unique TSP | Unique TSP | Unique TSP. Target: putative ETEC (E. coli UMNK88; uncharacterised lysogenic phage AEE57668) |
Unique TSP. Target: “Dickeya solani” |
| TSP2 | Synteny with TSP2 of CBA120 and SFP10. Target: E. coli O157 | Synteny with TSP2 of PhaxI and SFP10. Target: E. coli O157 (E. coli phage phiV10 YP_512279) | Synteny with TSP2 of PhaxI and SFP10. Target: E. coli O157 |
Unique TSP | Unique TSP that targets the acetyl group of the S. Typhi Vi capsule | Unique TSP | Truncated TSP |
| TSP3 | Not present | Unique TSP | Synteny with TSP3 of ΦSH19. Target—Salmonella Typhimurium (S. Typhimurium phage 9NA AET41722 P22 tail spike) | Synteny with TSP3 of SFP10. Target—Salmonella Typhimurium | Unique TSP. Target: putatively Escherichia fergusonii (Uncharacterised lysogenic phage YP_002383156) | Unique TSP. Target: putatively Enterobacter hormaechei
ATCC 49162 (Uncharacterised lysogenic phage ZP_08496176) |
Not present |
| TSP4 | Synteny with TSP4 of CBA120. Target: putatively ETEC or Citrobacter freundii | Synteny with TSP4 of PhaX1. Target: putatively ETEC or Citrobacter freundii (E. coli H10407-Uncharacterised lysogenic phage CBJ02120) | Unique TSP | Unique TSP | Unique TSP | Unique TSP | Truncated TSP |
The putative targets of some of the TSPs are based on BLASTP scores and significant E values with tail fibers/spikes found in lysogenic phage infecting the named bacterium
Given the remarkable synteny observed in these phages, other genes may be conserved that have a pan-global inhibitory effect on their enteric hosts. These will be a target in future research.
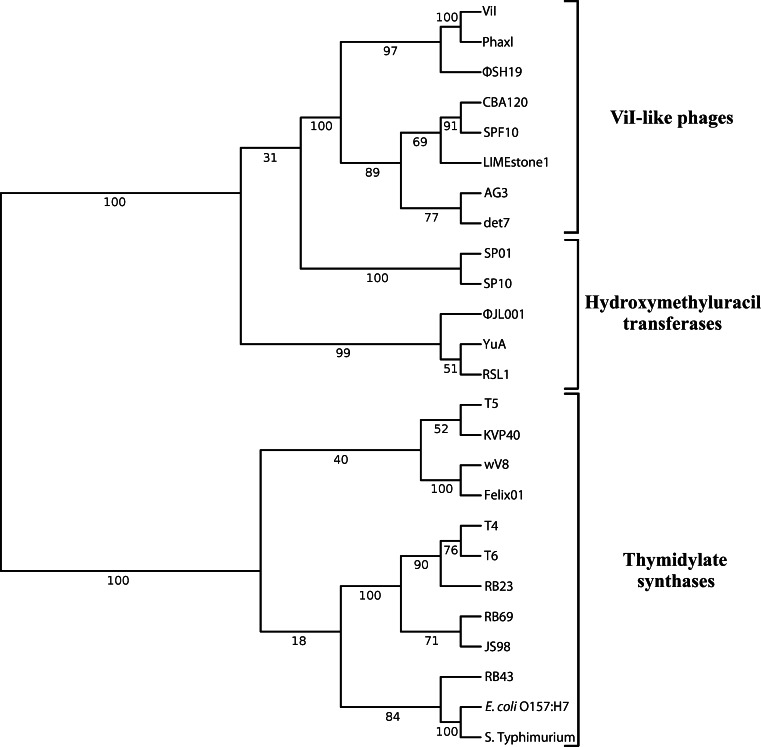
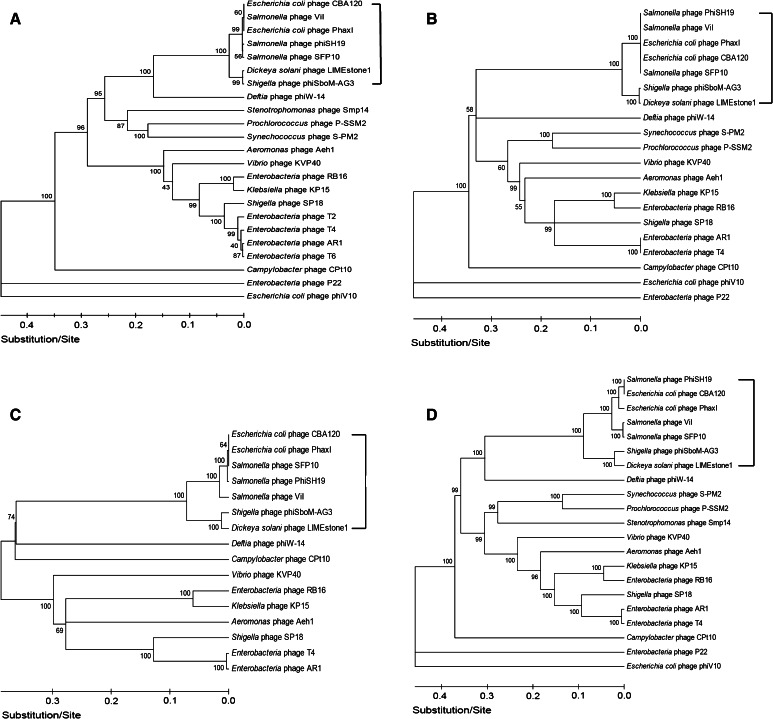
Phylogeny
To further explore this new group of ViI-like phages and to differentiate them from other phages, phylogenetic analysis of highly conserved proteins such as the major capsid proteins (T4 gp23 homologues), DNA polymerases (T4 gp43 homologues), DNA ligases (T4 gp30 homologues), and terminase large subunits (T4 gp17 homologues) was carried out (Fig. 6). All of these analyses substantiated the establishment of the new phage genus “Viunalikevirus” and established the same pattern of their relationship to the tevenviruses and other T4-related phages (S-PM2, P-SSM2). Interestingly, while Delftia acidovorans phage ΦW-14 is not a member of this new phage group, it is most closely related to the ViI-like phages in all of these phylogenetic analyses. Also of interest, this has led to the somewhat surprising finding that Shigella phage phiSboM-AG3 is much more closely related to Dickeya phage LIMEstone1 than it is to the other enteric phages.
Fig. 6.
Phylogenetic trees. Comparisons of (A) major capsid proteins (T4 gp23 homologues), (B) DNA polymerases (T4 gp43 homologues), (C) DNA ligases (T4 gp30 homologues), and (D) terminase large subunits (T4 gp17 homologues). Square brackets indicate the suggested genus “Viunalikevirus”
These findings could ultimately lead to a reclassification of the higher orders of bacterial viruses, but at present, the genus “Viunalikevirus” will be an independent genus in the family Myoviridae, despite the relationship of its members to the phages of the subfamily Tevenvirinae.
Discussion
The group of ViI-like phages differs from any established phage genus by its characteristic morphology and genomic properties. It stands virtually alone in the phage world in a number of ways despite its distant relationships to T4-like phages. The phages share a large degree of DNA homology, a similar genome organization, the presence of a modified base (suspected to be HMdU), and conserved patterns of early and late promoter sequences. Furthermore, there seems to be a horizontally acquired tRNA set conserved among most ViI-like phages. We suggest the term “Viunalikevirus” as the taxonomic name for this new genus.
Combining the morphology information of the EMs and the sequence data of the tail spike regions of these phages, new hypotheses can be made about the morphology of the ViI-like tail structure. Figure 1A shows the general morphology of the ViI-like bacteriophages, with six apparent tail spike entities. Some of these can be seen to be unfolded, while others are not, creating “stars” and “prongs”, respectively. Figure 1b shows the arrangement of the tail spikes in more detail with respect to most of the ViI-like family, where we now hypothesize that up to four spikes are present on each of the six tail entities, encoded by four tail spike genes (TSP1-4) (as represented schematically in Fig. 2). In this respect, comparisons to the EM of LIMEstone1 are illuminating (Supplementary Figure 3), since it encodes just one complete tail spike protein (TSP1) and a truncated TSP4. The overall structure is less elaborate, and the tail spikes are less complex, showing no splitting into the umbrella-like arrangement characteristic of ViI and several of the others, as seen in Fig. 1A and B, for example.
Figure 3 provides extensive evidence that regions of DNA are shared by the various TSPs, both within the same phage and among the other viunalikeviruses. These homologous regions are most likely involved in the domain exchange evident in this phage family within the tail spike repertoire. Such re-assortment of tail spike receptor recognition ‘modules’ would be further enhanced by having so many TSPs and shared enteric hosts. For example, a substantial region of DNA (278 bases over a range of 386 bases—72 % match) from TSP1 of PhaXI is also located in TSP4, and regions like these can serve as recombination hot spots for domain exchange.
A result of the broad tail spike repertoire and putative targets (Table 4) might be that the host range of some of the ViI-like phages is more extensive than previously considered. This was already witnessed for SFP10, which infects both Salmonella Typhimurium and E. coli O157. For phages PhaXI and CBA120, a shared TSP2 was found with ETEC or Citrobacter freundii as potential targets, and TSP1 of CBA120 and SFP10 showed synteny, with the target of the tail spike unknown. So far, for all the sequenced ViI-like phages, unique TSPs are present for which the targets have not yet been identified. At the same time, their specificity can be highly targeted. For example, CBA120 does not target any of the common laboratory E. coli strains, and only hits one member of the ECOR collection (ECOR70) in addition to hitting most tested E. coli O157 strains [15]. It also does not target most Salmonella strains; only one (very weak) hit has been found to date, in subspecies indica [15]. Determining the roots of this host specificity will be an interesting area for future research.
If these ViI-like phages have a capacity to infect such diverse enteric hosts, they will need to have mechanisms for overcoming an equally diverse range of bacterial restriction and modification systems. The probable replacement of thymidine (dThd) with HMdU appears to be one such method employed by this phage genus to circumvent the host’s restriction and modification pathways. Aside from defense against some restriction enzymes, it is not clear what, if any, evolutionary advantages the incorporation of HMdU provides for viunalikeviruses. However, for Bacillus phage SPO1, the middle-mode genes are preferentially transcribed from DNA that has HMdU rather than dThd [28]. Many members of the related genus T4likevirus also use the presence of HMdC in their DNA to rapidly block all transcription of cytosine-containing DNA and to much more gradually degrade all cytosine-containing DNA without damaging their own DNA [23].
We conclude that the creation of the genus “Viunalikevirus” is another appropriate step towards phage classification.
Electronic supplementary material
Supplementary Figure 1: Electron micrographs of ViI-like phages outside the enterobacteria. Phage 7 of Sinorhizobium meliloti with quiescent tail, uranyl acetate (left) and phage E14 of Acinetobacter calcoaceticus, with unfolded tail spikes, phosphotungstate (right). The bar indicates 100 nm.
Supplementary Figure 2: Consensus promoter sequences of ViI. A) Putative early promoter. B) Putative late promoter.
Supplementary Figure 3: Electron micrograph of LIMEstone1. Image was prepared as described for Figure 1 except no trehalose was used.
Acknowledgments
AMK would like to thank Andre Villegas for developing “extractUpStreamDNA”. BB and EMK would like to thank NIH grant 2R15GM63637-3A1 for support, along with the many members of the Evergreen Phage lab who participated in these studies. DG, DP and ALT are very grateful for funding from the Wellcome Trust. RL and AMK are respectively the chair and vice-chair of the ICTV Prokaryotic Virus Subcommittee.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.King AMQ, Lefkowitz E, Adams MJ, Carstens EB. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. London: Academic Press; 2012. [Google Scholar]
- 2.Lavigne R, Seto D, Mahadevan P, Ackermann H-W, Kropinski AM (2008) Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res Microbiol 159: 406–414. doi:10.1016/j.resmic.2008.03.005 [DOI] [PubMed]
- 3.Desplats C, Krisch HM (2003) The diversity and evolution of the T4-type bacteriophages. Res Microbiol 154: 259–267. doi:10.1016/S0923-2508(03)00069-X [DOI] [PubMed]
- 4.Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol. 2003;185:5220–5233. doi: 10.1128/JB.185.17.5220-5233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ackermann H-W, Krisch HM. A catalogue of T4-type bacteriophages. Arch Virol. 1997;142:2329–2345. doi: 10.1007/s007050050246. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki S, Tanaka S, Koga T, Kawata T. A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol Immunol. 1992;36:93–97. doi: 10.1111/j.1348-0421.1992.tb01645.x. [DOI] [PubMed] [Google Scholar]
- 7.Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD (2010) Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J 7. doi:10.1186/1743-422X-7-292 [DOI] [PMC free article] [PubMed]
- 8.Pickard D, Toribio AL, Petty NK, van TA, Yu L, Goulding D, Barrell B, Rance R, Harris D, Wetter M, Wain J, Choudhary J, Thomson N, Dougan G (2010) A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J Bacteriol 192: 5746–5754. doi:10.1128/JB.00659-10 [DOI] [PMC free article] [PubMed]
- 9.Craigie J, Brandon KF. Bacteriophage specific for the O-resistant V form of B. typhosus. J Pathol. 1936;43:233–248. doi: 10.1002/path.1700430202. [DOI] [Google Scholar]
- 10.Craigie J, Yen CH. V Bacteriophages for B. typhosus. Trans Royal Soc Can. 1937;5:79–87. [Google Scholar]
- 11.Bradley DE, Kay D. The fine structure of bacteriophages. J General Microbiol. 1960;23:553–563. doi: 10.1099/00221287-23-3-553. [DOI] [Google Scholar]
- 12.Ackermann H-W, Berthiaume L, Kasatiya SS. Ultrastructure of Vi phages I to VII of Salmonella typhi. Can J Microbiol. 1970;16:411–413. doi: 10.1139/m70-070. [DOI] [PubMed] [Google Scholar]
- 13.Adriaenssens EM, Van Vaerenbergh J, Vandenheuvel D, Dunon V, Ceyssens PJ, De Proft M, Kropinksi AM, Noben JP, Maes M, Lavigne R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by ‘Dickeya solani’. PLoS ONE. 2012;7:e33227. doi: 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anany H, Lingohr E, Villegas A, Ackermann HW, She YM, Griffiths M, Kropinski A. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol J. 2011;8:242. doi: 10.1186/1743-422X-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutter EM, Skutt-Kakaria K, Blasdel B, El-Shibiny A, Castano A, Bryan D, Kropinski AM, Villegas A, Ackermann HW, Toribio AL, Pickard D, Anany H, Callaway T, Brabban AD (2011) Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J 8:430. doi:10.1186/1743-422X-8-430 [DOI] [PMC free article] [PubMed]
- 16.Hooton SP, Timms AR, Rowsell J, Wilson R, Connerton IF (2011) Salmonella Typhimurium-specific bacteriophage PhiSH19 and the origins of species specificity in the Vi01-like phage family. Virol J 8:498. doi:10.1186/1743-422X-8-498 [DOI] [PMC free article] [PubMed]
- 17.Park M, Lee JH, Shin H, Kim M, Choi J, Kang DH, Heu S, Ryu S (2011) Characterization and comparative genomic analysis of a novel bacteriophage SFP10 simultaneously inhibiting both Salmonella and Escherichia coli O157:H7. Appl Environ Microbiol. doi:10.1128/AEM.06231-11 [DOI] [PMC free article] [PubMed]
- 18.Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27. doi:10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed]
- 19.Zafar N, Mazumder R, Seto D. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinform. 2002;3:12. doi: 10.1186/1471-2105-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Menlo Park: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 21.Nechaev S, Geiduschek EP. Dissection of the bacteriophage T4 late promoter complex. J Mol Biol. 2008;379:402–413. doi: 10.1016/j.jmb.2008.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiduschek EP, Kassavetis GA. Transcription of the T4 late genes. Virol J. 2010;7:288. doi: 10.1186/1743-422X-7-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W (2003) Bacteriophage T4 genome. Microbiol Mol Biol Rev 67:86–156 [DOI] [PMC free article] [PubMed]
- 24.Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–W689. doi:10.1093/nar/gki366 [DOI] [PMC free article] [PubMed]
- 25.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puigbo P, Bravo IG, Garcia-Vallve S (2008) E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI). BMC Bioinform 9:65. doi:10.1186/1471-2105-9-65 [DOI] [PMC free article] [PubMed]
- 27.Ackermann H-W, Gershman M. Morphology of phages of a general Salmonella typing set. Res Virol. 1992;143:303–310. doi: 10.1016/S0923-2516(06)80118-4. [DOI] [PubMed] [Google Scholar]
- 28.Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, Pedulla ML (2009) The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 388:48–70. doi:10.1016/j.jmb.2009.03.009 [DOI] [PMC free article] [PubMed]
- 29.Ackermann H-W, DuBow MS. Viruses of prokaryotes. Boca Raton: CRC Press; 1987. [Google Scholar]
- 30.Demczuk W, Ahmed R, Ackermann H-W (2004) Morphology of Salmonella enterica serovar Heidelberg typing phages. Can J Microbiol 50:873–875. doi:10.1139/w04-075 [DOI] [PubMed]
- 31.Carey-Smith GV, Billington C, Cornelius AJ, Hudson JA, Heinemann JA (2006) Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol Lett 258: 182–186. doi:10.1111/j.1574-6968.2006.00217.x [DOI] [PubMed]
- 32.Walter M, Fiedler C, Grassl R, Biebl M, Rachel R, Hermo-Parrado XL, Llamas-Saiz AL, Seckler R, Miller S, van Raaij MJ (2008) Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J Virol 82:2265–2273. doi:10.1128/JVI.01641-07 [DOI] [PMC free article] [PubMed]
- 33.Ackermann H-W, Brochu G, Emadi Konjin HP. Classification of Acinetobacter phages. Arch Virol. 1994;135:345–354. doi: 10.1007/BF01310019. [DOI] [PubMed] [Google Scholar]
- 34.Krsmanovic-Simic D, Werquin M. Study of bacteriophages of Rhizobium meliloti. C R Acad Sci Hebd Seances Acad Sci D. 1973;276:2745–2748. [PubMed] [Google Scholar]
- 35.Werquin M, Ackermann H-W, Levesque RC. A Study of 33 Bacteriophages of Rhizobium meliloti. Appl Environ Microbiol. 1988;54:188–196. doi: 10.1128/aem.54.1.188-196.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.