Abstract
Dengue is a major public health issue in tropical and subtropical regions worldwide. The four serotypes of dengue virus (DENV1-4) are spread primarily by Aedes aegypti and Ae. albopictus mosquitoes, whose geographic range continues to expand. Humans are the only host for epidemic strains of DENV, and the virus has developed sophisticated mechanisms to evade human innate immune responses. The host cell's first line of defense begins with an intracellular signaling cascade resulting in production of interferon (IFN)-α/β, which promotes intracellular antiviral responses and helps initiates the adaptive response during the course of DENV infection. In response, DENV has developed numerous ways to subvert these intracellular antiviral responses and directly inhibit cellular signaling cascades. Specifically, DENV manipulates the unfolded protein response and autophagy to counter cellular stress and delay apoptosis. The DENV non-structural protein NS4B and subgenomic sfRNA interfere with the RNAi pathway by inhibiting the RNAse Dicer. During heterotypic secondary DENV infection, subneutralizing antibodies can enable viral uptake through Fcγ receptors and down-regulate signaling cascades initiated via the pattern recognition receptors TLR3 and MDA5/RIG-I, thus reducing the antiviral state of the cell. The DENV NS2B/3 protein cleaves human STING/MITA, interfering with induction of IFN-α/β. Finally, DENV NS2A, NS4A, and NS4B complex together to block STAT1 phosphorylation, while NS5 binds and promotes degradation of human STAT2, thus preventing formation of the STAT1/STAT2 heterodimer and its transcriptional induction of ISGs. Here we discuss the host innate immune response to DENV and the mechanisms of immune evasion DENV has developed to manipulate cellular antiviral responses.
Introduction
Four dengue virus serotypes (DENV-1,-2, -3, -4) cause dengue fever (DF) as well as more severe disease manifestations, traditionally referred to as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS)1. DF is an acute febrile illness with headache, retro-orbital pain, myalgia, arthralgia, rash, hemorrhagic manifestations, and/or leukopenia. The hallmark features of DHF consist of thrombocytopenia, hemorrhagic manifestations, and signs of plasma leakage, which can lead to hypotensive shock (DSS) and, without appropriate treatment, death. The disease was recently reclassified into dengue, with and without warning signs, and severe dengue2. Bhatt et al.3 recently estimated that up to 96 million dengue cases and a total of 390 million DENV infections occur each year worldwide3, leading to approximately 500,000 hospitalizations and 25,000 deaths, primarily among children4. Dengue occurs throughout tropical and subtropical regions around the world, with disease burden most well documented in Southeast Asia and Latin America3. DENV is transmitted by Aedes aegypti and Ae. albopictus mosquitoes, which continue to expand geographically, facilitated by increased global trade and travel, unplanned urbanization, poor waste and water management, as well as human behavior and ecology5. No commercial vaccine or specific antiviral treatment exists for dengue, though these are areas of substantial research and development efforts. Dengue is a human disease with no known animal reservoirs, and the virus has evolved successfully to evade human immune responses, especially innate antiviral immunity. This review focuses on mechanisms of the innate intracellular antiviral response and DENV evasion within infected cells.
The dengue virus life cycle
DENV is a positive-strand RNA enveloped flavivirus whose 10.7 kb genome contains a 5′ type I m7G cap structure and encodes a polyprotein that is post-translationally cleaved by host and viral proteases into three structural proteins (C, capsid; pr/M, membrane; E, envelope) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5). In humans, DENV primarily infects immune cells of the myeloid lineage, including monocytes, macrophages and dendritic cells as well as hepatocytes, as shown in situ in human autopsy tissues by immunohistochemistry6,7,8,9,10, in peripheral blood mononuclear cells (PBMC) during the acute phase of infection by flow cytometry11, and ex vivo in skin explants12. Though several reports exist of DENV infection of endothelial cells in vivo and in vitro, the role of viral infection of endothelial cells in dengue disease progression remains controversial7,8,9,10,13,14,15. Finally, only one autopsy report from a patient with fatal DHF showed DENV protein in myocytes, indicating that myocytes are likely not a primary target of DENV infection16.
Initial viral infection is mediated by clathrin-dependent receptor-mediated endocytosis17,18, and in a secondary infection, virions complexed with antibodies from a prior DENV infection(s) can enter cells via Fcγ receptors (FcγR) (Figure 1). Once inside the cell, the endosomal vesicle becomes acidified, and the virus undergoes conformational changes that enable fusion with the endosomal membrane, releasing the single-stranded RNA (ssRNA) into the cytosol19. The DENV ssRNA is then translated and replicated in the endoplasmic reticulum (ER), which undergoes hypertrophy after flavivirus infection20. Recent electron microscopy and electron tomography studies have shown that DENV RNA replication occurs in virus-induced membrane vesicles in the ER, with budding of DENV particles on ER membranes directly apposed to vesicle pores21. The newly replicated positive-strand viral RNA is packaged with capsid protein and assembled into an enveloped virion that is covered with 180 E and prM/M proteins, with the E proteins arranged in antiparallel dimers22. Vesicles containing the newly-formed virions pass through the Golgi, where the host protease furin cleaves prM to generate mature and partially mature virions that are secreted from the host cell (Figure 1). The non-structural viral proteins participate in the replication complex23 and have also been shown to play a role in disrupting cellular antiviral pathways to enhance viral replication, as discussed below.
Figure 1. Dengue virus life cycle.
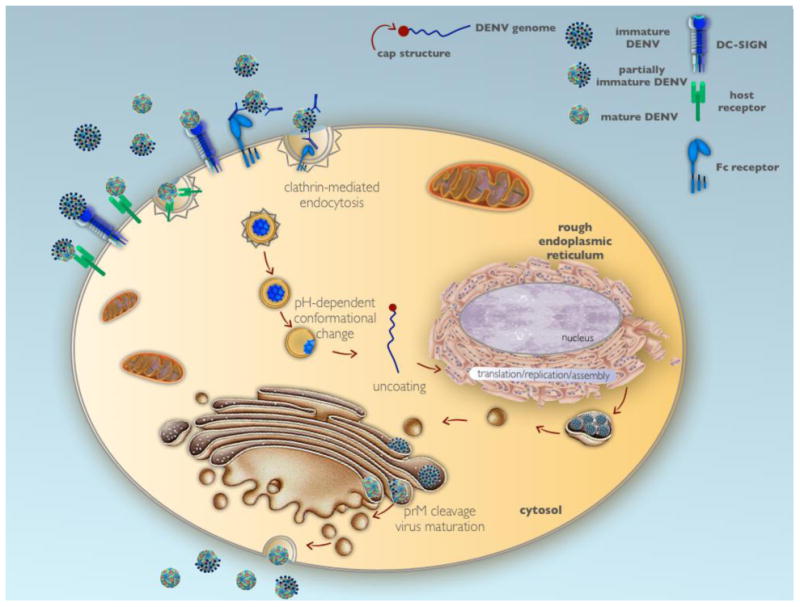
Dengue virus infects host cells via receptor-mediated endocytosis, and following vesicle acidification and conformational changes in the viral prM/M and E proteins, the viral RNA is uncoated and released into the cytosol. The viral RNA migrates to the endoplasmic reticulum where its translation, replication and assembly occur. Packaged virions are transported to the Golgi, where they undergo proteolytic cleavage that results in virus maturation and are then exported from the cell using the host secretory system.
The dengue virion
DENV replicates in two distinct host environments, human and mosquito. During replication, cleavage of prM/M on the virion surface is required for a mature virion to be generated. Mature and immature virions differ in size and structure. Immature virions prior to prM/M cleavage are ∼600Å in size and have a spiky appearance, whereas cleavage of prM allows the E protein to change conformation and form a smoother layer over the viral phospholipid membrane. During viral processing, prM has been shown to remain associated with mature virus at acidic pH, preventing membrane fusion24. Recent studies have shown that the maturation state of flaviviruses influences both the infectable cell type(s) and the interaction of the virion with particular antibodies, thus impacting the results of neutralization assays25 and potentially affecting the triggering of innate immune responses. However, the relative percentage of immature or mature DENV virions in humans and the role of maturation state in disease are currently unknown. Previous work has shown that during virus propagation in vitro, both mature and immature virions are produced26,27, although the relative amounts vary substantially by cell type28,29. Recent reports have shown that not only maturation state, but temperature as well plays a role in the structure of the virion. At temperatures above 33°C, the mature virion lacking prM is irreversibly altered from a “smooth marble-like” structure to a bumpy form that is 10% larger, which has been shown to be a stable prefusion intermediate30,31. Another important conceptual advance is that DENV virions are not static but rather dynamic, breathing structures, thus enabling antibodies with “cryptic” epitopes to bind and also exposing the membrane underneath the layer of viral E and M proteins32,33,34. As a result, both the temperature and time of incubation of antibodies and virus can dramatically alter the measurement of neutralizing antibody titers33.
Host receptors for dengue virus
DENV is transmitted to the human host during the mosquito's blood meal and infects human cells via several distinct receptors (Figure 1). Direct interaction between the glycosylated residues in Domain II of the DENV E protein and the carbohydrate recognition domain (CARD) of dendritic-cell-specific ICAM3-grabbing non-integrin (DC-SIGN) facilitates infection of dendritic cells27,35,36. However, as DENV infection of DCs does not require DC-SIGN internalization signals, DC-SIGN is considered an attachment factor37. Interestingly, polymorphisms in the human DC-SIGN gene CD209 have been suggested to correlate with an increased risk of dengue and disease severity38,39. DENV also utilizes the DC-SIGN homolog L-SIGN to infect liver endothelial cells40. Of note, virus produced in vitro in mosquito cells was found to utilize DC-SIGN, whereas virus propagated in human dendritic cells utilized L-SIGN to infect target cells36,40. In addition to DC-SIGN and L-SIGN, the mannose receptor expressed on human macrophages was found to bind the carbohydrate moieties on the DENV envelope protein41.
DENV has been shown to bind to a number of cell surface molecules. DENV is able to complex with heat shock protein (HSP) 90 and HSP70 on the surface of mammalian cells42,43 and p74 on the surface of mosquito cells44, among others. Following heat shock treatment, host cells were found to have increased HSP expression, viral uptake and virus output43,44. In cells lacking selectin-type receptors, recent studies have shown that DENV utilizes the transmembrane receptors TIM and TAM, two receptors involved in phosphatidylserine-dependent removal of cells undergoing apoptosis45. TIM binds DENV directly, whereas TAM interacts indirectly with DENV via two bridge proteins, Gas6 and ProS45. Finally, during secondary DENV infection with a heterotypic serotype, the adaptive immune response can act to enhance viral infection via antibody-dependent enhancement (ADE) of FcγRI- and FcγRII-bearing cells46,47. Cross-reactive antibodies from a previous infection with a different serotype bind to the infecting DENV serotype, forming an immune complex that is recognized by FcγRs, which then mediate uptake into the target cells of myeloid lineage48.
The host innate immune response against dengue virus
Pattern recognition receptors (PRRs) such as toll-like receptors (TLRs), particularly TLR-3, TLR-7, and TLR-8, and intracellular sensors (e.g., the DExE/H box RNA helicases) are some of the first lines of defense in the innate immune recognition of double-stranded RNA (dsRNA), ssRNA or modified RNA. The primary TLR involved in dengue viral recognition, TLR-3, recognizes DENV RNA after endosomal acidification (Figure 2A) and has been shown to induce strong IL-8 and interferon (IFN)-α/β responses in vitro49. Recognition of DENV RNA by TLR-3 results in phosphorylation of TIR domain-containing-adapter-inducing interferon β (TRIF). TRIF interacts with both TNF receptor associated factor (TRAF) 3 and 6. TRAF3 interacts with TANK binding kinase (TBK-1) and Ik kinase 1(Ikk1), resulting in the phosphorylation of IRF-3. TRAF6 signals through association with TAK1, activating AP-1 and initiating Ikk1/2 dephorphorylation of IkB, leading to NFkB activation. Nuclear translocation of IRF3, AP-1 and NFkB induces production of IFN-α/β, interferon stimulating genes (ISGs) and chemokines50. Experiments in non-human primates infected with DENV demonstrated that the administration of TLR-3 and TLR7/8 agonists resulted in significantly decreased viral replication and increased production of pro-inflammatory chemokines as well as increased anti-DENV serum antibody titers, indicating a protective role for TLRs during DENV infection51.
Figure 2. Host innate immune response to dengue virus infection.
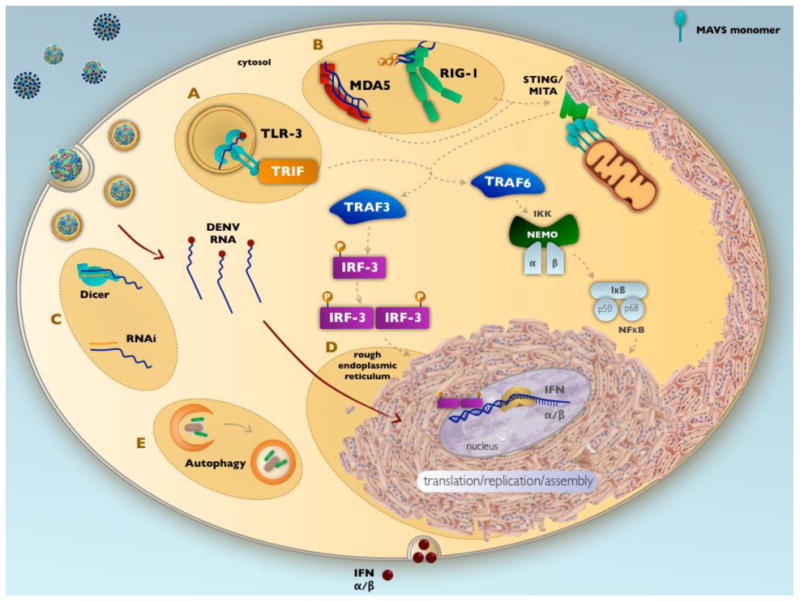
DENV RNA is recognized by pattern recognition receptors TLR-3 (A) and MDA5 and RIG-I (B), resulting in a signaling cascade that induces IFN-α/β production. Viral infection induces the host RNAi pathway (C) to control viral RNA replication; hypertrophy of the ER (D), where viral translation and replication occurs, and autophagy (E) to balance cellular stress and prevent apoptosis.
In conjunction with TLRs, cytoplasmic helicases sense RNA in the cytoplasm. Both retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which recognize dsRNA52, are induced during DENV infection and are involved in IFN-β induction52,53 (Figure 2B). Once bound to the dsRNA, RIG-I oligomerizes in an ATP-dependent fashion54. One important role of these cytoplasmic dsRNA sensors is to distinguish between viral and host cell RNA55. In particular, MDA5 is able to distinguish host RNA from viral RNA based on a ribose 2′-O methylation found on host mRNA.56 RIG-I and MDA5 signal through the mitochondrial antiviral signaling (MAVS) protein on mitochondria. MAVS interacts with the ER protein stimulator of interferon genes (STING, also known as MITA). Once activated, MAVS oligomerizes and attracts multiple ubiquitin E3 ligases such as TRAF3 and TRAF6 to activate the signaling cascade57. TRAF3 activation results in translocation of both IRF3 and IRF7 to the nucleus to induce transcription of IFN-α/β, as described above50. In addition, activation of TRAF6 results in NFkB translocation to the nucleus and production of IFN-α/β. Both RIG-I and MDA5 were found to synergize with TLR3 to limit DENV replication in vitro53.
IFN-α/β secreted by virus-infected cells triggers warning signals to adjacent cells that an infection is occurring as well as an autocrine induction of cellular antiviral responses. IFN-α/β is a powerful inhibitor of DENV infection. IFN-α/β signals through the IFN-α/β receptor (IFNAR) and activates the JAK/STAT pathway via phosphorylation of the adaptor molecules TYK2 and JAK158. The activation of these adaptor molecules results in the phosphorylation and dimerization of various signal transducer and activator of transcription (STAT) molecules, including STAT1, STAT2, STAT3, and STAT5. An important signaling complex formed by STAT1 and STAT2 along with IRF9 in response to IFNAR activation is the interferon stimulating gene factor 3 (ISGF3) complex that translocates to the nucleus and binds to IFN-stimulated response elements located in the promoter region of IFN-stimulated genes. ISGF3 complexes bind to these key elements and induce the production of numerous antiviral proteins and pro-inflammatory cytokines59. IFNAR signaling also induces alternative signaling cascades, including the mitogen-activated protein (MAP) kinase p38 cascade and the phosphatidylinositol 3 (PI3) kinase cascade59 that result in production of proinflammatory cytokines and chemokines.
In addition to IFN-α/β production, induction of NFkB translocation to the nucleus results in production of pro-interleukin 1 and activation of the inflammasome, which results in production of mature interleukin 1 and 18. Both cytokines are important for antiviral responses and result in recruitment of immune cells to the site of infection60. During infection, DENV interacts with the macrophage surface marker C-type lectin domain family 5, member A (CLEC5A) and induces inflammasome activation in human macrophages61. In addition, blockage of the CLEC5A/DENV interaction in vivo reduced mortality by 50% in a murine lethal challenge model62.
Small RNAs such as micro RNAs (miRNA) are utilized by the host to control cellular gene expression and in response to viral infections (Figure 2C). miRNAs are processed by and interact with proteins in the RNAi pathway, such as Dicer, Drosha, Argo1 and Argo2. Interestingly, knockdown of these proteins during DENV infection of mammalian cells resulted in increased viral replication, indicating the RNAi pathway may play an important role in the cellular anti-DENV response63. miRNAs not only regulate TLR and IL-1 signaling, but are also induced in response to these signals during viral infection, thus providing control of host innate immune responses. Approximately 22 nucleotides in length, miRNAs inhibit translation of target RNAs. Base-pair matching between miRNA and RNA does not need to be exact; thus, one miRNA can interact with many different RNAs.
Dengue virus subversion of the cellular antiviral response
DENV has evolved to evade the host's innate immune response in two distinct ways: sequestration and active subversion of innate immune responses. DENV infection of a host cell causes extensive rearrangements of cellular membranes, yet the virus must control the innate immune mechanisms such as stress that trigger cell death. DENV manipulates the cell to maintain host metabolism and protein production while sequestering itself in vesicles that are not degraded by host lysosomes. This complex process involves a delicate balance of activating cellular pathways for ER expansion and inducing lipid metabolism while preventing ER stress-induced death. DENV uses the cell's own pathways of survival such as autophagy to facilitate viral replication. Finally, DENV non-structural proteins act directly on components of the innate immune response's signaling cascade, inhibiting the RNAi pathway and IFN-α/β induction and signaling.
ER stress/Unfolded protein response (UPR)
DENV utilizes several mechanisms to hijack the host cell machinery to facilitate replication. DENV replication occurs in the ER of host cells, and early during infection (e.g., 12 hours post-infection), the ER undergoes rearrangement and expansion64 (Figure 2D). This ER expansion requires de novo synthesis of viral proteins but does not require host transcription and is independent of the unfolded protein response (UPR) and the sterol-regulatory-element-binding-protein-2 (SREBP-2) pathway, as shown using a series of mouse embryo fibroblasts deficient in key genes in the UPR and SREBP pathways64.
While early rearrangement of the ER has been shown to be independent of the UPR response, DENV infection induces the UPR and can manipulate the UPR to cope with ER stress (Figure 3A)65. In particular, DENV NS2B/3 was shown to induce X-box binding protein 1 (XBP1), leading to an expansion of the ER for viral replication66. This also results in reduction of the UPR response and decreased DENV-induced cytopathic effects, indicating that a reduced UPR may prevent or delay host cell death during viral replication66. It has also been reported that enhancing eIF2α phosphorylation using Salubrinal, a small molecule inhibitor of eIF2α phosphatases, increased translational inhibition, leading to reduced DENV production67. Overall, these data indicate that virus-mediated induction of the UPR is beneficial for viral production but must be controlled to prevent cell death.
Figure 3. Dengue virus subversion of host innate immune responses.
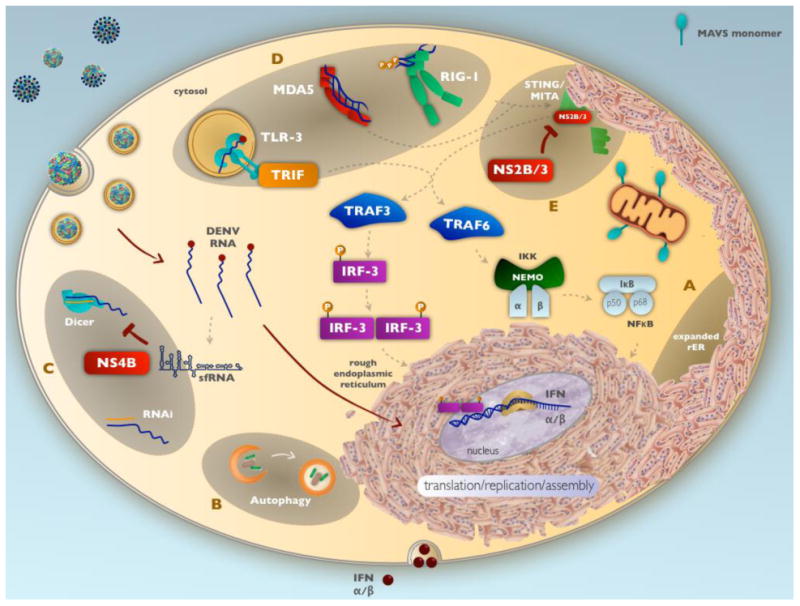
DENV manipulates host cellular processes and innate immune responses within infected cells by A) manipulating the unfolded protein response (UPR) to counter cellular stress and delay apoptosis. B) To offset ER stress and access fatty acid metabolism networks, DENV also initiates autophagy, which has shown to be necessary for production of infectious virions. C) The DENV non-structural protein NS4B and subgenomic sfRNA interfere with the RNAi pathway by inhibiting the RNAse Dicer. D) During heterotypic secondary DENV infection, subneutralizing antibodies can enable viral uptake through FcγR and down-regulate signaling cascades initiated via the pattern recognition receptors TLR3 and MDA5/RIG-I, thus reducing the antiviral state of the cell. E) The DENV NS2B/3 protein cleaves human STING/MITA, interfering with the signaling pathway leading to induction of IFNα/β.
Authophagy and lipid metabolism
Autophagy is lysosomal degradation of cytoplasmic contents that is part of the normal cellular recycling of macromolecules as well as a cellular host response to starvation or stress68. Autophagy has been implicated as an innate immune response to trigger the death of pathogen-infected cells. A critical aspect of the autophagic pathway is the formation of cytosolic double-membrane vesicles, autophagosomes, that engulf portions of cytoplasmic material and facilitate fusion with lysosomes69. However, many positive-strand RNA viruses have evolved to benefit from the autophagic pathway by subverting autophagosome formation and using the machinery to enhance viral replication70.
DENV-2 was first found to activate autophagic machinery (Figure 2E), resulting in increased autophagosome formation along with enhanced viral replication71 (Figure 3B). Further in vitro work found that both DENV-2 and -3 can inhibit autophagosome fusion with lysosomes72 and that the autophagosomal membrane-bound microtubule-associated protein light chain 3 (LC3) co-localized with DENV dsRNA and NS1 in infected HepG2 cells, indicating that replicating virus can be found in autophagic vacuoles73. More recent work tested an inhibitor of autophagy, spautin-1, in vitro and in the AG129 mouse model, concluding that autophagic capabilities are required for infectious viral particle production74. Thus, DENV manipulates the autophagy pathway to increase viral replication while avoiding lysosomal degradation.
Lipid droplets are cellular storage vesicles containing triglycerides and cholesterol. As a part of normal lipid metabolism, cells degrade lipid droplets as needed to obtain fatty acids for metabolism or vesicle formation75. Lipid metabolism is linked to autophagy, as during autophagy lipid droplets are degraded and engulfed into autophagosomes. Therefore, the regulation of lipid metabolism by DENV was suggested to result from the induction of autophagy that can increase the degradation of lipid droplets and produce more fatty acid material for viral replication75. In addition, several studies have implicated a direct role for DENV in controlling lipid metabolism for enhanced viral replication. Using an siRNA library, the DENV NS3 protein was found to recruit fatty acid synthase (FASN) and increase fatty acid biosynthesis76. In another study, the N-terminus of DENV capsid (C) protein was shown to contain a lipid droplet-binding motif77. The ability to target the DENV C protein to lipid droplets may provide a scaffolding for infectious viral particle formation78. Heaton et al.79 reported that DENV NS3 modulates host cellular fatty acid synthesis by recruiting fatty acid synthase to sites of DENV replication. The interaction between the NS3 and FASN is consistent with other studies in which DENV has been shown to modulate lipid metabolism by reabsorption of lipid droplets from host cells64,76. Thus, through careful manipulation of cellular processes, such as ER expansion, autophagy and lipid metabolism, DENV is able to sequester itself from the host's antiviral responses.
sfRNA
RNA interference is an important host defense against viral infections. Recent studies have shown that DENV is able to interfere with RNAi pathways via two distinct mechanisms, involving NS4B and subgenomic flavivirus RNA (sfRNA) (Figure 3C). DENV infection is able to suppress RNAi pathways by expression of the nonstructural protein NS4B, which directly interferes with Dicer's ability to process small RNAs in vitro80. Utilizing a GFP reporter that is silenced by miRNA, Kakumani et al80 showed that NS4B was sufficient to increase GFP expression in mammalian, mosquito, and plant cell lines, thus providing evidence that NS4B is able to inhibit RNAi. These findings were confirmed by the use of NS4B mutants that were unable to modulate the RNAi pathway. In addition, DENV infection results in production of sfRNA from the 3′ untranslated region of the viral genomic RNA. sfRNA is generated by stable secondary structures at the 5′ end of the 3′UTR blocking the host 5′-3′ exonuclease XRN181. Flavivirus sfRNA has been shown to inhibit cleavage of dsRNA by Dicer by binding to and saturating the RNase. In a dose-dependent manner, increasing concentrations of sfRNA inhibited Dicer cleavage of long dsRNA into siRNAs82. In addition, the sfRNA directly interacts with XRN1, increasing overall messenger RNA stability within the host cell83,84, which could also benefit the viral RNA.
Interference with innate signaling responses during ADE
During a secondary DENV infection, the virus can infect FcγR-bearing cells via ADE. A series of in vitro experiments have shown that when human monocytic cell lines are infected by DENV under conditions of ADE, as compared to high-dose infection in the absence of antibody, the innate antiviral state of the cell is compromised. Specifically, uptake of DENV in complex with anti-DENV antibodies through FcγRI and FcγRIIa on THP-1 monocytic cells yielded enhanced infection as a result of down-regulation of both TLR gene expression and negative regulators of the NFkB pathway, as well as disruption of RIG-I and MDA5 signaling cascades, leading to decreased innate immune responses and increased viral replication85,86 (Figure 3D). In addition, viral uptake through ADE enhanced anti-inflammatory cytokine production such as IL-10 from dendritic cells86,87. This phenomenon has been termed “intrinsic ADE”88 and is still somewhat controversial. For instance, Kou et al.86showed that viral replication was increased during ADE in primary human monocytes in the absence of increased IL-10 production, indicating that mechanisms of antibody-dependent enhancement of infection are not yet fully understood.
Interference with IFN-α/β induction
DENV non-structural proteins abrogate IFN-α/β production and signaling within infected cells via several distinct pathways. First, the 2′-O methylation of the viral mRNA cap structure by NS5 evades MDA5 recognition and host restriction via Ifit1-dependent and -independent mechanisms, blunting the IFN response against flaviviruses89,90,91. Second, DENV interferes with downstream signaling from RIGI and MDA5 by directly targeting human MITA/STING in this pathway to down-regulate the antiviral responses triggered upon viral infection. Recent reports have shown that expression of the DENV NS2B/3 protease directly cleaves MITA/STING, thereby disrupting IFN production92,93,94 (Figure 3E). DENV NS2B/3 only recognizes human STING and not the murine homologue MPYS92,93. When human STING was engineered to contain the mouse sequence in the NS2B/3 cleavage site, IFN production was restored in DENV-infected cells93. Thus, NS2B/3 inhibits IFN-α/β induction in a species-specific manner.
Interference with IFN-α/β signaling
Cells infected with DENV have been shown to be defective in responding to external treatment with IFN-α/β, indicating that DENV infection is able to prevent IFN-α/β signaling95,96,97. Signaling through the IFN-α/β receptor is mediated via the STAT1/2 signaling pathway and is an important component of the host cellular innate response to viral infection (Figure 4). DENV has evolved several mechanisms to interfere with this signaling pathway via inhibition of STAT1 phosphorylation as well as degradation of STAT2. DENV NS2A, NS4A and NS4B associate with cellular membranes and when expressed together have been shown to inhibit STAT1 phosphorylation and nuclear translocation in host cells95 (Figure 4A). Expression of these three DENV non-structural proteins enhanced replication of α/β-sensitive viruses in the presence of exogenous IFN-α/β95. In the related West Nile Virus (WNV), the sfRNA has been implicated in viral immune evasion of the IFN-α/β pathway. WNV deficient in sfRNA production was less virulent in vitro and in vivo when compared to wild-type virus. Interestingly, this phenotype was rescued when the mutant virus was grown under Type I IFN-deficient conditions82, suggesting that sfRNA may play a role in modulating IFN-α/β responses during flaviviral infection.
Figure 4. Dengue virus interference with interferon-α/β signaling.
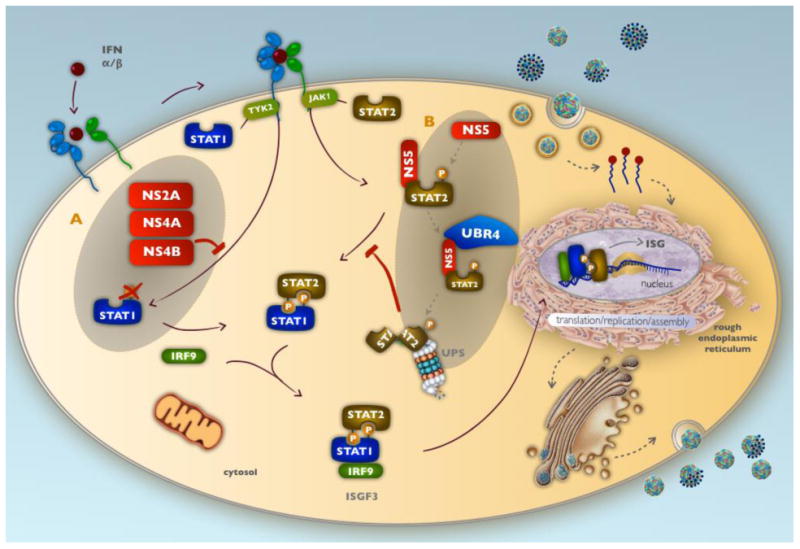
DENV non-structural proteins impede IFN-α/β signaling in virally infected cells. A) NS2A, NS4A, NS4B complex together to block STAT1 phosphorylation, nuclear translocation, and transcriptional activation of ISGs. B) DENV NS5 binds and degrades human STAT2, targeting it toward the proteasomal degradation pathway, thus preventing formation of the STAT1/STAT2 heterodimer and its transcriptional induction of ISGs.
Furthermore, DENV inhibits IFN-α/β signaling by triggering the degradation of STAT2. Initial studies observed reduced levels of STAT2 in cells expressing DENV replicons98. More recently, it has been shown that DENV NS5 mediates STAT2 degradation via the proteasome (Figure 4B). Mature NS5 was found to bind STAT2 but not to mediate protein degradation, as proteolytic cleavage of STAT2 required NS5 to be in the polyprotein form (Figure 4B). NS5 binds directly to the coiled-coil region in the first half of the human STAT2 protein99 acting as a bridge between UBR-4 and STAT2, thus directing STAT2 toward the ubiquitination and proteasomal degradation pathway99,100. Interestingly, DENV is only able to degrade human STAT2, but not mouse STAT2101. Thus, these DENV non-structural proteins enable specific evasion of human IFN-α/β receptor signal transduction.
Finally, the ability of DENV to perturb the host's innate immune response may impact the adaptive immune response and modulate disease outcome. Though it is difficult to determine the direct effect of the reduced innate responses, dengue patients from Vietnam and Nicaragua with more severe disease (DSS) had decreased expression levels of IFN-α/β-stimulated genes102,103,104. DENV infection of monocytes, macrophages and dendritic cells may also affect the quality and quantity of T cells, as T cells from patients infected with DENV exhibited reduced proliferation in vitro105.
Conclusions
DENV is an RNA virus that triggers innate antiviral immune mechanisms within infected cells that should protect the cell from viral replication and warn surrounding cells of the pathogen's presence. In response, DENV subverts the innate antiviral responses by sequestering itself and manipulating cellular machinery and signing pathways. DENV controls cellular processes in order to delay cell death, inhibit RNAi pathways, and provide a niche hidden from intracellular innate RNA sensors. In addition, non-structural DENV proteins inhibit the induction and signaling cascade of IFN-α/β, thus preventing cellular pathways that lead to the expression of interferon response genes and antiviral mechanisms. Recent and ongoing research continues to uncover a growing number of unique mechanisms used by DENV to outsmart the host innate immune response.
Highlights.
Dengue virus infection triggers numerous intracellular innate immune responses
DENV avoids intracellular detection via sequestration of viral replication in the ER
DENV manipulates cellular signaling cascades to inhibit innate antiviral responses
Acknowledgments
The figures in this review were illustrated by A. Hadjilaou.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Dengue haemorrhagic fever: Diagnosis, treatment, prevention, and control. 2nd. World Health Organization; Geneva: 1997. [Google Scholar]
- 2.World Health Organization. Dengue guidelines for diagnosis, treatment, prevention and control: new edition. World Health Organization; Geneva: 2009. [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 6.Balsitis SJ, Coloma J, Castro G, Alava A, Flores D, McKerrow JH, Beatty PR, Harris E. Tropism of dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunostaining. Am J Trop Med Hyg. 2009;80:416–24. [PubMed] [Google Scholar]
- 7.Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J infect Dis. 2004;189:1411–8. doi: 10.1086/383043. [DOI] [PubMed] [Google Scholar]
- 8.Bhoopat L, Bhamarapravati N, Attasiri C, Yoksarn S, Chaiwun B, Khunamornpong S, Sirisanthana V. Immunohistochemical characterization of a new monoclonal antibody reactive with dengue virus-infected cells in frozen tissue using immunoperoxidase technique. Asian Pac J Allergy Immunol. 1996;14:107–13. [PubMed] [Google Scholar]
- 9.Hall WC, Crowell TP, Watts DM, Barros VL, Kruger H, Pinheiro F, Peters CJ. Demonstration of yellow fever and dengue antigens in formalin-fixed paraffin-embedded human liver by immunohistochemical analysis. Am J Trop Med Hyg. 1991;45:408–17. doi: 10.4269/ajtmh.1991.45.408. [DOI] [PubMed] [Google Scholar]
- 10.Ramos C, Sanchez G, Pando RH, Baquera J, Hernandez D, Mota J, Ramos J, Flores A, Llausas E. Dengue virus in the brain of a fatal case of hemorrhagic dengue fever. J Neurovirol. 1998;4:465–8. doi: 10.3109/13550289809114548. [DOI] [PubMed] [Google Scholar]
- 11.Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376:429–35. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. Human skin Langerhans cells are targets of dengue virus infection. Nat Med. 2000;6:816–20. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- 13.Warke RV, Becerra A, Zawadzka A, Schmidt DJ, Martin KJ, Giaya K, Dinsmore JH, Woda M, Hendricks G, Levine T, Rothman AL, Bosch I. Efficient dengue virus (DENV) infection of human muscle satellite cells upregulates type I interferon response genes and differentially modulates MHC I expression on bystander and DENV-infected cells. J Gen Virol. 2008;89:1605–15. doi: 10.1099/vir.0.2008/000968-0. [DOI] [PubMed] [Google Scholar]
- 14.Balsitis SJ, Coloma J, Castro G, Alava A, Flores D, McKerrow JH, Beatty PR, Harris E. Tropism of dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunostaining. Am J Trop Med Hyg. 2009;80:416–424. [PubMed] [Google Scholar]
- 15.Halstead SB. Controversies in dengue pathogenesis. Paediatr Int Child Health. 2012;32(Suppl 1):5–9. doi: 10.1179/2046904712Z.00000000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salgado DM, Eltit JM, Mansfield K, Panqueba C, Castro D, Vega MR, Xhaja K, Schmidt D, Martin KJ, Allen PD, Rodriguez JA, Dinsmore JH, Lopez JR, Bosch I. Heart and skeletal muscle are targets of dengue virus infection. Ped Infect Dis J. 2010;29:238–42. doi: 10.1097/INF.0b013e3181bc3c5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acosta EG, Castilla V, Damonte EB. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J Gen Virol. 2008;89:474–84. doi: 10.1099/vir.0.83357-0. [DOI] [PubMed] [Google Scholar]
- 18.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS pathog. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaitseva E, Yang ST, Melikov K, Pourmal S, Chernomordik LV. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 2010;6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–15. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 21.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–75. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–7. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 24.Yu IM, Holdaway HA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J Virol. 2009;83:12101–7. doi: 10.1128/JVI.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 2012;8:e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zybert IA, van der Ende-Metselaar H, Wilschut J, Smit JM. Functional importance of dengue virus maturation: infectious properties of immature virions. J Gen Virol. 2008;89:3047–51. doi: 10.1099/vir.0.2008/002535-0. [DOI] [PubMed] [Google Scholar]
- 27.Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–93. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 28.Junjhon J, Edwards TJ, Utaipat U, Bowman VD, Holdaway HA, Zhang W, Keelapang P, Puttikhunt C, Perera R, Chipman PR, Kasinrerk W, Malasit P, Kuhn RJ, Sittisombut N. Influence of pr-M cleavage on the heterogeneity of extracellular dengue virus particles. J Virol. 2010;84:8353–8. doi: 10.1128/JVI.00696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junjhon J, Lausumpao M, Supasa S, Noisakran S, Songjaeng A, Saraithong P, Chaichoun K, Utaipat U, Keelapang P, Kanjanahaluethai A, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J Virol. 2008;82:10776–91. doi: 10.1128/JVI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci U S A. 2013;110:6795–9. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J Virol. 2013;87:7585–92. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–7. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 33.Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 2011;7:e1002111. doi: 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84:9227–39. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–8. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, Steinman RM, Schlesinger S, Marovich MA. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–9. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozach PY, Burleigh L, Staropoli I, Navarro-Sanchez E, Harriague J, Virelizier JL, Rey FA, Despres P, Arenzana-Seisdedos F, Amara A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J Biol Chem. 2005;280:23698–708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Chen RF, Liu JW, Lee IK, Lee CP, Kuo HC, Huang SK, Yang KD. DC-SIGN (CD209) Promoter -336 A/G polymorphism is associated with dengue hemorrhagic fever and correlated to DC-SIGN expression and immune augmentation. PLoS Negl Trop Dis. 2011;5:e934. doi: 10.1371/journal.pntd.0000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakuntabhai A, Turbpaiboon C, Casademont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM, Tangnararatchakit K, Tangthawornchaikul N, Vasanawathana S, Chaiyaratana W, Yenchitsomanus PT, Suriyaphol P, Avirutnan P, Chokephaibulkit K, Matsuda F, Yoksan S, Jacob Y, Lathrop GM, Malasit P, Despres P, Julier C. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–13. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dejnirattisai W, Webb AI, Chan V, Jumnainsong A, Davidson A, Mongkolsapaya J, Screaton G. Lectin switching during dengue virus infection. J Infect Dis. 2011;203:1775–83. doi: 10.1093/infdis/jir173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JL, de Wet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, Gordon S. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes-Del Valle J, Chavez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol. 2005;79:4557–67. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chavez-Salinas S, Ceballos-Olvera I, Reyes-Del Valle J, Medina F, Del Angel RM. Heat shock effect upon dengue virus replication into U937 cells. Virus Res. 2008;138:111–8. doi: 10.1016/j.virusres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Salas-Benito J, Reyes-Del Valle J, Salas-Benito M, Ceballos-Olvera I, Mosso C, del Angel RM. Evidence that the 45-kD glycoprotein, part of a putative dengue virus receptor complex in the mosquito cell line C6/36, is a heat-shock related protein. Am J Trop Med Hyg. 2007;77:283–90. [PubMed] [Google Scholar]
- 45.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12:544–57. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Littaua R, Kurane I, Ennis FA. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol. 1990;144:3183–3186. [PubMed] [Google Scholar]
- 47.Rodrigo WW, Block OK, Lane C, Sukupolvi-Petty S, Goncalvez AP, Johnson S, Diamond MS, Lai CJ, Rose RC, Jin X, Schlesinger JJ. Dengue virus neutralization is modulated by IgG antibody subclass and Fcgamma receptor subtype. Virology. 2009;394:175–82. doi: 10.1016/j.virol.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–69. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 49.Tsai YT, Chang SY, Lee CN, Kao CL. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol. 2009;11:604–15. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 50.Lee KG, Xu S, Kang ZH, Huo J, Huang M, Liu D, Takeuchi O, Akira S, Lam KP. Bruton′s tyrosine kinase phosphorylates Toll-like receptor 3 to initiate antiviral response. Proc Natl Acad Sci U S A. 2012;109:5791–6. doi: 10.1073/pnas.1119238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sariol CA, Martinez MI, Rivera F, Rodriguez IV, Pantoja P, Abel K, Arana T, Giavedoni L, Hodara V, White LJ, Anglero YI, Montaner LJ, Kraiselburd EN. Decreased dengue replication and an increased anti-viral humoral response with the use of combined Toll-like receptor 3 and 7/8 agonists in macaques. PLoS One. 2011;6:e19323. doi: 10.1371/journal.pone.0019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–7. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zinzula L, Tramontano E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: Hide, mask, hit. Antiviral Res. 2013;100:615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–43. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–22. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 59.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 60.van de Veerdonk FL, Netea MG. New Insights in the Immunobiology of IL-1 Family Members. Front Immunol. 2013;4:167. doi: 10.3389/fimmu.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu MF, Chen ST, Yang AH, Lin WW, Lin YL, Chen NJ, Tsai IS, Li L, Hsieh SL. CLEC5A is critical for dengue virus-induced inflammasome activation in human macrophages. Blood. 2013;121:95–106. doi: 10.1182/blood-2012-05-430090. [DOI] [PubMed] [Google Scholar]
- 62.Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, Lei HY, Lee CK, Chiou TW, Wong CH, Hsieh SL. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453:672–6. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- 63.Kakumani PK, Ponia SS, Rajgokul KS, Sood V, Chinnappan M, Banerjea AC, Medigeshi G, Malhotra P, Mukherjee SK, Bhatnagar RK. Role of RNAi in dengue viral replication and identification of NS4B as a RNAi suppressor. J Virol. 2013 doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pena J, Harris E. Early dengue virus protein synthesis induces extensive rearrangement of the endoplasmic reticulum independent of the UPR and SREBP-2 pathway. PLoS One. 2012;7:e38202. doi: 10.1371/journal.pone.0038202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pena J, Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem. 2011;286:14226–36. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu CY, Hsu YW, Liao CL, Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–80. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umareddy I, Pluquet O, Wang QY, Vasudevan SG, Chevet E, Gu F. Dengue virus serotype infection specifies the activation of the unfolded protein response. Virol J. 2007;4:91. doi: 10.1186/1743-422X-4-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 69.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 70.Richards AL, Jackson WT. How positive-strand RNA viruses benefit from autophagosome maturation. J Virol. 2013;87:9966–72. doi: 10.1128/JVI.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–8. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khakpoor A, Panyasrivanit M, Wikan N, Smith DR. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J Gen Virol. 2009;90:1093–103. doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- 73.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol. 2009;90:448–56. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- 74.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Jr, Yuan J, Kirkegaard K. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol. 2013;87:1312–21. doi: 10.1128/JVI.02177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saka HA, Valdivia R. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol. 2012;28:411–37. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- 76.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–32. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martins IC, Gomes-Neto F, Faustino AF, Carvalho FA, Carneiro FA, Bozza PT, Mohana-Borges R, Castanho MA, Almeida FC, Santos NC, Da Poian AT. The disordered N-terminal region of dengue virus capsid protein contains a lipid-droplet-binding motif. Biochem J. 2012;444:405–15. doi: 10.1042/BJ20112219. [DOI] [PubMed] [Google Scholar]
- 78.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–50. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kakumani PK, Ponia SS, S RK, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK. Role of RNA Interference (RNAi) in Dengue Virus Replication and Identification of NS4B as an RNAi Suppressor. J Virol. 2013;87:8870–83. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–91. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 82.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol. 2012;86:5708–18. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–40. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Modhiran N, Kalayanarooj S, Ubol S. Subversion of innate defenses by the interplay between DENV and pre-existing enhancing antibodies: TLRs signaling collapse. PLoS Negl Trop Dis. 2010;4:e924. doi: 10.1371/journal.pntd.0000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kou Z, Lim JY, Beltramello M, Quinn M, Chen H, Liu S, Martinez-Sobrido L, Diamond MS, Schlesinger JJ, de Silva A, Sallusto F, Jin X. Human antibodies against dengue enhance dengue viral infectivity without suppressing type I interferon secretion in primary human monocytes. Virology. 2011;410:240–7. doi: 10.1016/j.virol.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 87.Ubol S, Phuklia W, Kalayanarooj S, Modhiran N. Mechanisms of immune evasion induced by a complex of dengue virus and preexisting enhancing antibodies. J Infect Dis. 2010;201:923–35. doi: 10.1086/651018. [DOI] [PubMed] [Google Scholar]
- 88.Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis. 2010;10:712–22. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–6. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012;8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szretter KJ, Daffis S, Patel J, Suthar MS, Klein RS, Gale M, Jr, Diamond MS. The innate immune adaptor molecule MyD88 restricts West Nile virus replication and spread in neurons of the central nervous system. J Virol. 2010;84:12125–38. doi: 10.1128/JVI.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J Virol. 2010;84:9760–74. doi: 10.1128/JVI.01051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–13. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100:14333–8. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol. 2005;79:5414–20. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–18. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, Garcia-Sastre A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, Garcia-Sastre A. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8:410–21. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, Dung NT, Long TH, Hoang DM, Chau NV, Thao le TT, Hien TT, Relman DA, Farrar J. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Popper SJ, Gordon A, Liu M, Balmaseda A, Harris E, Relman DA. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLoS Negl Trop Dis. 2012;6:e1966. doi: 10.1371/journal.pntd.0001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Long HT, Hibberd ML, Hien TT, Dung NM, Van Ngoc T, Farrar J, Wills B, Simmons CP. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J Infect Dis. 2009;199:537–46. doi: 10.1086/596507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mathew A, Kurane I, Green S, Vaughn DW, Kalayanarooj S, Suntayakorn S, Ennis FA, Rothman AL. Impaired T cell proliferation in acute dengue infection. J Immunol. 1999;162:5609–15. [PubMed] [Google Scholar]


