Abstract
The delivery of oligonucleotide antagonists to cytosolic RNA targets such as microRNA represents an avenue for the post-transcriptional control of cellular phenotype. In tumor cells, oncogenic miRNAs, termed oncomirs, are tightly linked to processes that ultimately determine cancer initiation, progression, and response to therapy. Therefore, the capacity to redirect tumor cell fate towards therapeutically beneficial phenotypes holds promise in a future clinical scenario. Previously, we have designed “nanodrugs” for the specific inhibition of oncogenic microRNAs in tumor cells. The basic design of these nanodrugs includes dextran coated iron oxide nanoparticles, conjugated to a tumor-targeting peptide, and a locked nucleic acid (LNA)-modified antisense oligonucleotide that stably binds and inhibits the complementary mature miRNA. Here, we focus on elucidating an optimal nanodrug design for effective miRNA inhibition in tumor cells. Specifically, we investigate the choice of chemical linker for the conjugation of the oligonucleotide to the nanoparticles and evaluate the contribution of tumor-cell targeting to nanodrug uptake and functionality. We find that short labile linkers (SPDP; N -Succinimidyl 3-(2-pyridyldithio)-propionate) are superior tonon-labile short linkers (GMBS; N-(γ-Maleimidobutyryloxy)succinimide ester) or non-labile long linkers (PEG24; Succinimidyl-([N-maleimidopropionamido]-24 ethyleneglycol)ester) in terms of their capacity to gain access to the cytosolic cellular compartment and to engage their cognate miRNA. Furthermore, using the nanodrug design that incorporates SPDP as a linker, we establish that the addition of tumor-cell targeting through functionalization of the nanodrug with the αvβ3-specific cyclic RGDfK-PEG peptide does not confer an advantage in vitro at long incubation times required for inhibition.
Keywords: gene silencing, microRNA, locked nucleic acids, integrin specific peptide, iron oxide nanoparticles, delivery vehicle
INTRODUCTION
MicroRNAs (miRNAs) are important post-transcriptional regulators of gene expression.1–4 The recent literature abounds in examples of the key role played by miRNAs in determining cell fate. These examples are particularly compelling with regard to cancer emergence, progression, and response to therapy.5–9 Consequently, miRNAs represent promising candidates as targets of therapeutic intervention. To specifically inhibit oncogenic miRNA, we designed nanodrugs capable of accumulatingin tumor cells and engaging their cognate miRNA. The basic design of these nanodrugs includes dextran coated iron oxide nanoparticles, conjugated to a tumor-targeting peptide, and a locked nucleic acid (LNA)-modified antisense oligonucleotide that stably binds and inhibits the complementary mature miRNA. The choice of oligonucleotide chemistry is supported by the literature, which has demonstrated that incorporating high binding affinity modifications, such as LNA, increases potency while maintaining specificity.10 Recently, using a nanodrug specific for the pro-metastatic miR-10b7, we demonstrated effective prevention and arrest of lymph node metastasis in a murine model of breast cancer. 11
Here, we focus on elucidating an optimal nanodrug design for effective miRNA inhibition in tumor cells. Specifically, we investigated the choice of chemical linker for the conjugation of the oligonucleotide to the nanoparticles. A variety of heterobifunctional cross-linkers are commercially available and can be classified as permanent (non-labile) or temporary (labile) with various lengths, which can be mainly adjusted by the introduction of polyethylene glycol.11–19 We compared three conjugation designs:
a labile short linker (SPDP; N -Succinimidyl 3-(2-pyridyldithio)-propionate) that ensures dissociation of the oligonucleotide in the reducing intracellular environment and minimal steric interference with the binding between the oligo and the target miRNA but may be unstable in circulation;
a non-labile short linker (GMBS; N-(γ-Maleimidobutyryloxy)succinimide ester) that maximizes the stability of the nanodrug in circulation but may reduce nanodrug functionality due to steric hindrance of the binding to the target miRNA;
a non-labile long linker (PEG24; Succinimidyl-([N-maleimidopropionamido]-24 ethyleneglycol)ester) that maximizes the stability of the nanodrug in circulation and may minimize the steric interference with the binding between the oligo and the target miRNA.
Finally, we compared a nanodrug design that incorporates a peptide-targeting moiety for enhanced intracellular delivery by receptor-mediated endocytosis and a simplified design that is devoid of a peptide-targeting moiety and is primarily taken up via phago/macropinocytosis.20
We find that short labile linkers (SPDP; N -Succinimidyl 3-(2-pyridyldithio)-propionate) are superior to non-labile short linkers (GMBS; N-(γ-Maleimidobutyryloxy)succinimide ester) or non-labile long linkers (PEG24; Succinimidyl-([N-maleimidopropionamido]-24 ethyleneglycol)ester) in terms of their capacity to gain access to the cytosolic cellular compartment and to engage their cognate miRNA. Furthermore, using the nanodrug design that incorporates SPDP as a linker, we establish that the addition of tumor-cell targeting through functionalization of the nanodrug with the αvβ3-specific cyclic RGDfK-PEG peptide, which is expressed by the test cell line does not confer an advantage in vitro at long incubation times required for inhibition.
RESULTS
Nanodrug testing to determine the optimal linker design for oligonucleotide conjugation
To compare nanodrug uptake and miRNA inhibition, as a function of linker design, amine derivatized dextran-coated magnetic nanoparticles (MN) were prepared and modified with the near-IR dye, Cy5.5-NHS. Three different heterobifunctional linkers (GMBS, SPDP and PEG24) were loaded onto MN-Cy5.5 and used for subsequent conjugation to the LNA-modified antisense oligonucleotides (anti-miR10b for the inhibition of miRNA-10b or an irrelevant oligonucleotide with no known miRNA specificity as a control; Fig. 1A and Fig. S1 and Table 1). The molar ratio of MN to Cy5.5 was determined spectrophotometrically as 1:3.8. The molar ratio of MN to oligonucleotide on MN-SPDP was estimated by electrophoresis as 1:10. The number of oligonucleotides on MN-GMBS and MN-PEG24 was assumed to be the same as on MN-SPDP. The change of nanoparticle size following each modification step is presented in Fig. 1B. GMBS and SPDP increased nanoparticle size by 21 and 24%, respectively. The final nanodrugs, following oligonucleotide conjugation, reflected a respective increase of 53 and 49% in nanoparticle diameter. The PEG24 linker doubled the diameter of the nanoparticles from 19.6 to 41.1nm. The final nanodrug represented a 290% (76nm) increase in diameter.
Figure 1.
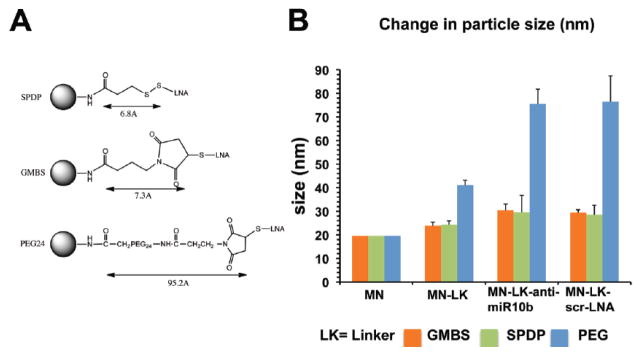
Nanoparticle synthesis and characterization. A. Three different heterobifunctional linkers (GMBS, SPDP and PEG24) were loaded onto MN-Cy5.5 and used for subsequent conjugation to the LNA-modified antisense oligonucleotides. B. Nanoparticle size following modification with SPDP, GMBS, or PEG24 and addition of anti-miR10b LNA-modified oligonucleotide. GMBS and SPDP increased nanoparticle size by 21 and 24%, respectively. The final nanodrugs, following oligonucleotide conjugation, reflected a respective increase of 53 and 49% in nanoparticle diameter. The PEG24 linker doubled the diameter of the nanoparticles from 19.6 to 41.1nm. The final nanodrug represented a 290% (76nm) increase in diameter. Z-average size is shown.
Table 1.
Abbreviations of probes used in the study
| Probes | Details |
|---|---|
| 1. MN | Amine derivatized dextran-coated magnetic nanoparticle |
| 2. MN-Cy5.5 | MN with Cy5.5 |
| 3. MN-SPDP, GMBS, or PEG24 | MN with Cy5.5 and SPDP, GMBS, or PEG24 |
| 4. MN-GMBS-anti-mir10b | MN with Cy5.5, GMBS linker, and anti-miR10b oligo |
| 5. MN-GMBS-scr-LNA | MN with Cy5.5, GMBS linker, and irrelevant oligo |
| 6. MN-PEG24-anti-miR10b | MN with Cy5.5, PEG24 linker, and anti-miR10b oligo |
| 7. MN-PEG24-scr-LNA | MN with Cy5.5, PEG24 linker, and irrelevant oligo |
| 8. MN-SPDP-anti-miR10b | MN with Cy5.5, SPDP linker, and anti-miR10b oligo |
| 9. MN-SPDP-scr-LNA | MN with Cy5.5, SPDP linker, and irrelevant oligo |
| 10. MN-cRGD-anti-miR10b | MN with Cy5.5, SPDP linker, anti-miR10b oligo, and cRGD peptide |
| 11. MN-cRGD-scr-LNA | MN with Cy5.5, SPDP linker, irrelevant oligo, and cRGD peptide |
| 12. MN-cRAD-anti-miR-10b | MN with Cy5.5, SPDP linker, anti-miR10b oligo, and cRAD peptide |
| 13. MN-cRAD-scr-LNA | MN with Cy5.5, SPDP linker, irrelevant oligo, and cRAD peptide |
The three different nanodrugs, MN-GMBS-anti-miR10b, MN-SPDP-anti-miR10b, and MN-PEG24-anti-miR10b, were compared to establish which design was associated with the most efficient uptake by the test cell line. Compared with PBS-treated cells, cells treated with MN-GMBS-anti-miR10b or MN-SPDP-anti-miR10b showed the most robust nanodrug uptake. By contrast, MN-PEG24-anti-miR10b demonstrated limited uptake (Fig. 2A). MN-GMBS-anti-miR10b and MN-SPDP-anti-miR10b showed a respective 14.6-(p = 0.003) and 9.8-fold (p = 0.005) higher uptake than MN-PEG24-anti-miR10b (Fig. 2B). This difference reflected true uptake and did not extend from a possible fluorescence quenching phenomenon, associated with the choice of linker, because the relative fluorescence/μmole of the three designs was comparable (Table 2).
Figure 2.
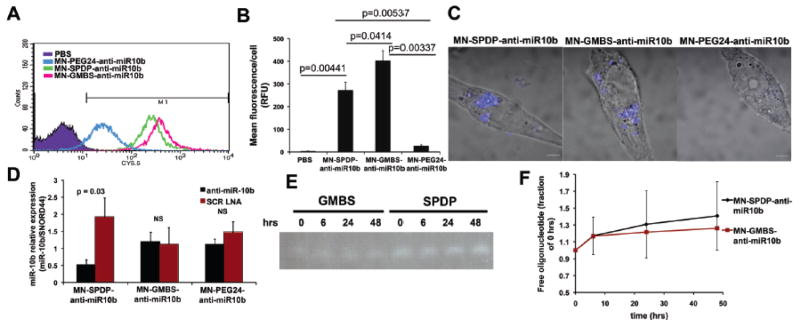
Cell uptake, target miRNA inhibition and serum stability as a function of linker design. A. Flow cytometry to determine which of the three nanodrug designs, MN-GMBS-anti-miR10b; MN-SPDP-anti-miR10b, or MN-PEG24-anti-miR10b, was associated with the most efficient uptake by the test cell line (MDA-MB-231). Compared with PBS-treated cells, cells treated with MN-GMBS-anti-miR10b or MN-SPDP-anti-miR10bshowed the most robust nanodrug uptake. By contrast, MN-PEG24-anti-miR10b demonstrated limited uptake. B. Quantification of the results from A. MN-GMBS-anti-miR10b (p = 0.00337, n = 2, t-test) and MN-SPDP-anti-miR10b (p = 0.00537, n = 2, t-test) showed a respective 14.6 and 9.8-fold higher uptake thanMN-PEG24-anti-miR10b. C. Confocal microscopy to investigate the subcellular localization of the nanodrugs. MN-GMBS-anti-miR10b and MN-SPDP-anti-miR10b localized in perinuclear regions of the cytoplasm. MN-PEG24-anti-miR10b showed negligible uptake. D. qRT-PCR to evaluate the capacity of the nanodrugs to inhibit the target miR-10b. MN-SPDP-anti-miR10b inhibited miRNA-10b expression by 63% compared to the inactive nanodrug, MN-SPDP-scr-LNA. By contrast, neither MN-GMBS-anti-miR10b nor MN-PEG24-anti-miR10b inhibited miR-10b (p = 0.03, n = 4, t-test). E. Stability of the nanodrugs in 10% fetal calf serum. F. Quantification of the results from E. There was no change in the amount of free oligo released either from MN-SPDP-anti-miR10b or MN-GMBS-anti-miR10b between 0 and 48 h. of incubation in serum. (p > 0.05, n = 2, repeated measures ANOVA).
Table 2.
Cy5.5 fluorescence (RFU μM−1 of nanodrug)
| UV Abs | Concentration (μM) | RFU | RFU μM−1 | P value | |
|---|---|---|---|---|---|
| MN-GMBS-anti-miR10b | 0.5638 | 2.2552 | 2249.287 | 997.26 | 0.168* |
| MN-SPDP-anti-miR10b | 0.6277 | 2.5108 | 2482.602 | 988.74 | 0.013** |
| MN-PEG24-anti-miR10b | 0.6106 | 2.4424 | 2828.087 | 1158.26 | 0.015*** |
MN-GMBS-anti-miR10b vs. MN-SPDP-anti-miR10b,
MN-SPDP-anti-miR10b vs. MN-PEG24-anti-miR10b,
MN-PEG24-anti-miR10b vs. MN-GMBS-anti-miR10b
To investigate a possible difference in the subcellular localization of the nanodrugs, we performed confocal microscopy. MN-GMBS-anti-miR10b and MN-SPDP-anti-miR10b localized in perinuclear regions of the cytoplasm. MN-PEG24-anti-miR10b showed negligible uptake (Fig. 2C).
Finally, we evaluated the capacity of the nanodrugs to inhibit the target miR-10b by qRT-PCR. MN-SPDP-anti-miR10b inhibited miRNA-10b expression by 74% (p = 0.03) compared to the inactive nanodrug, MN-SPDP-scr-LNA. By contrast, neither MN-GMBS-anti-miR10b nor MN-PEG24-anti-miR10b inhibited miR-10b (Fig. 2D).
Considering that the ultimate application of these nanodrugs would be in an in vivo setting, we analyzed their stability in serum. (Fig. 2E and F). In both the MN-GMBS-anti-miR10b and MN-SPDP-anti-miR10b designs, the change in the amount of free oligonucleotide between 0 hrs and 48 hrs of incubation in serum was negligible, suggesting limited degradation of the nanodrugs over time (Fig. 2F). MN-PEG24 demonstrated negligible uptake and was not analyzed.
Nanodrug testing to determine the contribution of peptide targeting to nanodrug uptake and functionality
For the purpose of enhanced oligonucleotide delivery into cancer cells, we used an integrin (ανβ3) receptor targeting peptide, which is expressed by cancer cells. MN was labeled with either cyclic RGDfK-PEG or control cyclic RADfK-PEG and then conjugated to anti-miR10b or scr-LNA oligonucleotides. The ratio of Cy5.5/peptide/oligonucleotide/MN was determined as 3.8/15/10/1.
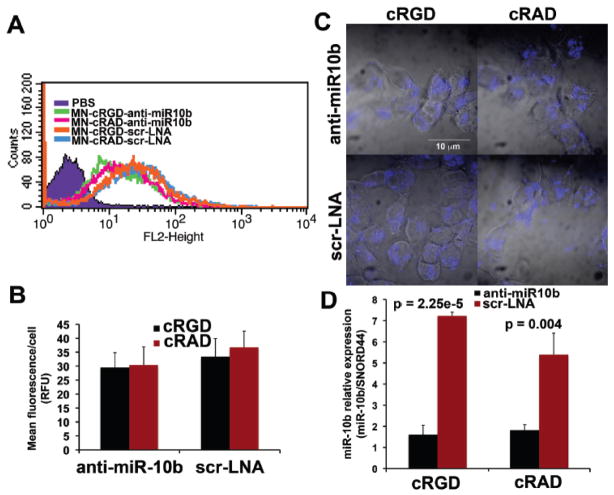
To evaluate the contribution of targeting through the cRGD peptide to nanodrug uptake, cells were incubated with MN-cRGD-anti-miR10b, MN-cRGD-scr-LNA, MN-cRAD-anti-miR10b, or MN-cRAD-scr-LNA for 48 hrs, which is necessary for achieving miRNA inhibition. All four designs showed comparable uptake (Fig. 3A). The mean fluorescence per cell was also not significantly different between active or inactive nanodrugs with and without a targeting peptide (Fig. 3B).
Figure 3.
Cell uptake, target miRNA inhibition and serum stability as a function of peptide targeting. A. Flow cytometry to evaluate the contribution of targeting through the cRGD peptide to nanodrug uptake. Cells were incubated with MN-cRGD-anti-miR10b, MN-cRGD-scr-LNA, MN-cRAD-anti-miR10b, or MN-cRAD-scr-LNA and analyzed based on fluorescence from the Cy5.5 dye. All four designs showed comparable uptake. B. Quantification of the results from A. The mean fluorescence per cell was not significantly different between active or inactive nanodrugs with and without a targeting peptide. C. Confocal microscopy to assess the subcellular localization of the nanodrugs. All four designs were associated with abundant perinuclear fluorescence. D. qRT-PCR to determine if peptide targeting through cRGD contributes to the capacity of the nanodrugs to inhibit miR-10b in the test cell line. MN-cRGD-anti-miR10b andMN-cRAD-anti-miR-10b suppressed miRNA-10b expression by a significant 78% (p = 2.25e-5, n = 3, t-test) and 66% (p = 0.004, n = 3, t-test), respectively, compared to the MN-cRGD-scr-LNA and MN-cRAD-scr-LNA controls. Also, there was no significant difference in miRNA-10b expression betweenMN-cRGD-anti-miR10b andMN-cRAD-anti-miR-10b.
These results were confirmed by confocal microscopy. All four designs were associated with abundant perinuclear fluorescence (Fig. 3C).
Finally, to determine if peptide targeting through cRGD contributes to the capacity of the nanodrugs to inhibit miR-10b in the test cell line, following a 48-hr in vitro incubation, we analyzed miR-10b expression by qRT-PCR. We found that MN-cRGD-anti-miR10b and MN-cRAD-anti-miR10b suppressed miRNA-10b expression by a respective 78% (p = 2.25e-5) and 66% (p = 0.004) compared to the MN-cRGD-scr-LNA and MN-cRAD-scr-LNA controls. Also, there was no significant difference in miRNA-10b expression between MN-cRGD-anti-miR10b and MN-cRAD-anti-miR10b (Fig. 3D), indicating that in this model system, targeting through cRGD did not contribute to the nanodrug’s ability to inhibit the target miRNA.
These findings likely do not reflect a low expression of the target ανβ3 receptor by the test cell line. MDA-MB-231 cells express a high level of ανβ3 integrin (Fig. S2A). The antigen provided a basis for differential cell uptake of the free cRGD and cRAD peptides at concentrations of the peptide above 1 μM, as shown by confocal microscopy, following peptide labeling with FITC (Fig. S2B).
DISCUSSION
The current studies aim to arrive at an optimized design for nanodrug inhibitors of tumor microRNAs. The model used here is based on dextran-coated iron oxide nanoparticles(MN) as delivery vehicles and LNA-modified antisense oligonucleotides as direct miRNA inhibitors.11
We investigated three different heterobifunctional linkers, GMBS, SPDP and PEG24. GMBS and PEG24 linkers form permanent linkages with an amine through an N-hydroxy succinimidyl group and a thiol through a maleimide group. SPDP forms a permanent linkage with an amine through an N-hydroxy succinimidyl group and a labile linkage with a thiol through a disulfide bond on the other end. The disulfide bond is breakable under reducing conditions, including in the cytosolic intracellular environment and ensures release of the oligo from the nanoparticles. These designs allowed us to mimic three scenarios: one in which the oligo remains tethered to the nanoparticle and is in close proximity to the nanoparticle surface (GMBS); one in which the oligo remains tethered to the nanoparticle but is distant from the nanoparticle surface (PEG24); and one in which the oligo is released from the nanoparticle surface in the reducing intracellular environment (SPDP).
With respect to the biological function of the nanodrugs, after conjugation to MN, PEG24 mediated a large increase in the hydrodynamic diameter of the nanodrugs, which was associated with negligible cellular uptake. This finding is not surprising. The positive surface charge of MN is likely neutralized by a thick boundary layer of PEG24 that would interfere with the interaction between the nanodrug and the negatively charged cellular membrane. Consequently, while in vivo, PEG 24 could be helpful in enhancing circulation time, the increase in nanodrug size could restrict the permeation of the nanodrug within the tumor tissue, and, importantly, inhibit nanodrug uptake in the absence of specific targeting. 21
The MN-SPDP and MN-GMBS nanodrugs were associated with very high uptake by the tumor cells. In the absence of targeting, the nanodrugs were taken up by nonspecific phagocytosis/macropinocytosis. 20 Despite the similar uptake, the nanodrugs were significantly different in their capacity to inhibit the target miRNA. Whereas MN-SPDP was efficient at miRNA inhibition, MN-GMBS was not effective. The absence of miRNA inhibition by the MN-GMBS design could be the effect of steric interference between the binding of the antisense oligo to the target miRNA or from ineffective subcellular distribution of the oligo that would occur if the intact nanodrug cannot successfully relocate from endosomes to the cytosol. By contrast in the MN-SPDP design, dissociation of the oligo from the nanoparticles in the cytosol can ensure more efficient cytosolic escape of the free oligo22.
With an outlook towards in vivo translation of these findings, it is reasonable to avoid the use of the labile SPDP linker because of possible degradation of the nanodrug in serum. However, our studies imply that the nanodrug is relatively stable in serum irrespective of whether GMBS or SPDP is used as a linker. This observation differs from prior studies,13 which have found limited stability of the SPDP linker in serum. However, the prior research used quantum dots as nanoparticle carriers of oligonucleotides (small interfering RNA). In our design, it is possible that the uneven dextran coat on the MN nanoparticles provides improved steric protection of the linker from serum enzymes.
To enhance delivery of nanodrugs into cancer cells, nanodrugs are often modified with targeting peptides. In our studies, to investigate the contribution of peptide targeting to nanodrug uptake and functionality, MN-SPDP was modified with the cyclic RGD peptide (cRGD), which targets integrin ανβ3 on the surface of the test cell line. We found no enhancement of nanodrug uptake or capacity to inhibit the target miRNA in the presence of cRGD, as compared to the non-targeting cRAD peptide. This finding is not unexpected considering that the cells were incubated with the nanodrugs in vitro and for a prolonged period of time (48 hrs). It suggests that in this scenario, the predominant uptake mechanism of these positively charged nanoparticles by cancer cells is phagocytosis/macropinocytosis and not receptor mediated internalization. It also indicates that, when the design permits oligo release from the nanoparticle, nonspecific endocytosis is an adequate mechanism for delivery of the oligonucleotide to the cytosol.
While not automatically relevant to an in vivo system, these findings could provide important information for the in vivo application of similar nanodrugs. There have been numerous reports that “active targeting” did not lead to increased tumor accumulation over a non-targeted control.23–26 These results were explained by suggesting that “passive targeting” through the enhanced permeability and retention effect (EPR) is the predominant mechanism of tumor accumulation. In fact, when the goal is to deliver antisense oligonucleotides to the intracellular compartment, the major difference between “passive” and “active” targeting would extend from the cellular uptake mechanism (phago/macropinocytosis vs. receptor-mediated endocytosis) as suggested by the prior literature. 26 Despite differences in the retention or wash-out times between active targeting and passive targeting of nanoparticles, a simplified non-targeted design may be optimal, provided that the nanoparticles are optimized to ensure efficient access to the target molecule (in this case miRNA).
CONCLUSION
Our results suggest a simplified nanodrug design that ensures oligonucleotide release from the surface of the nanoparticle carrier inside the cell and is devoid of peptide targeting. This design is narrowly based on the application of 20–30-nm dextran-coated iron oxide nanoparticles, as delivery vehicles, and LNA-modified antisense oligonucleotides, as effectors, for the inhibition of tumor cell miRNA. The relevance of these findings to other nanoparticles and applications or for in vivo delivery is not immediately obvious and remains to be determined.
EXPERIMENTAL PROCEDURES
Locked nucleic acid antisense oligonucleotides
The short locked nucleic acid (LNA) antisense oligonucleotides (ASO) were synthesized and provided by Exiqon (Woburn, MA). The sequences were: anti-miR10b- 5′-ThioMC6-D/GTGTAACACGTC TATACGCCCA-3′; mismatch control scr-LNA- 5′-ThioMC6-D/CACAAATTCGGTTCTACAGGGTA-3′). A 5′-Thiol modification was inserted into both sequences for conjugation to magnetic nanoparticles (MN). The oligonucleotides were described previously 11.
Synthesis of cRGD peptides
Peptide synthesis was performed as described in 11. Briefly, the synthesis of cyclic RGDfK-PEG-Cys-(Boc) as a targeting peptide (cRGD) was carried out as follows. Cyclic RGDfK-PEG was added to a solution of Boc-Cys(Tris)-OSu (11.2 mg, 0.02 mmol) in DMF (1 mL) with diisopropyl ethylamine (DIEA, 348μL, 0.2mmol) and the reaction mixture was allowed to stir at room temperature overnight. The progress of the reaction was monitored by thin layer chromatography (TLC). The final product was purified by high performance liquid chromatography with PDA detector (HPLC/PDA, column: C-18 semi-preparative column eluent; 0.1% TFA in water (A) /0.1% TFA in acetonitrile (B), gradient from 100% (A) to 100% (B) for 30 min and continues 100% (B) for 15 min, flow rate; 5mL min−1). Collected fractions were lyophilized and obtained as a white powder, which was analyzed by MALDI-TOF Mass spectrometry (expected; 1339.56, measured; 1340.57 [MH]+).
Cyclic RGDfK-PEG-Cys-(Boc) was treated with 2–3 mL of cleavage cocktail (94% trifluoroacetic acid (TFA)/1% triisopropylsilane (TIS)/2.5% H2O/2.5% 1,2 ethanedithiol (EDT)) at room temperature for 30 min. The resulting volatile mixture was completely removed under vacuum. Afterwards, the residue was dissolved in 100 mM NH4OAc buffer with 10mM EDTA (3 mL). The resulting solution was filtered, and the filtrate was purified by HPLC. The final product was analyzed by MALDI-TOF Mass spectrometry (expected: 997.13, measured: 998.14 [MH]+).
Following the same procedures, a control peptide, cyclic RADfK-PEG (cRAD), was conjugated with Boc-Cys(Trt)-OSu and deprotected by use of cleavage cocktail. The final product was purified by HPLC and characterized by MALDI-TOF Mass spectrometry (expected: 1011.15, measured: 1016.16 [MH]+).
Synthesis of FITC labeled peptides
Cyclic RGDfK-PEG-Cys (1.11mg, 1.1μmol) was conjugated with FITC-maleimide (4.7mg, 11μmol in 50μL DLSO) overnight. The final product was purified by HPLC-PDA and characterized by MALDI-TOF Mass spectrometry (expected: 1359.48, measured: 1360.49 [MH]+). Cyclic RADfK-PEG-Cys was conjugated with FITC following the same procedures to generate cRGDfK-PEG-Cys-FITC (expected: 1373.51, measured: 1374.51 [MH]+).
Synthesis of dextran coated magnetic nanoparticles (MN)
The magnetic nanoparticle (MN) synthesis was performed as described in 11. Briefly, 9 g of Dextan-T10 (Pharmacosmos, Denmark) was dissolved in 30 mL of double-distilled water and stirred in a round bottom flask on ice. FeCl3.6H2O (0.65 g) was added while flushing Argon gas into the reaction mixture for an hour. FeCl2.4H2O (0.4 g) was added into the mixture and then 15 mL of concentrated cold NH4OH (~28%) was added dropwise to the stirring mixture. The temperature increased to 85 °C for an hour to induce the formation of a nanoparticulate colloidal mixture, cooled to room temperature and concentrated to 20 mL using Amicon Ultra centrifugal units (MWCO 30kDa; Millipore). The resulting 20 mL dextran coated magnetic nanoparticles were cross-linked and aminated with subsequent addition of 35 mL of 5M NaOH, 14 mL of concentrated epichlorohydrin (8 h) and 60 mL of concentrated NH4OH. The nanoparticle solution was purified using a dialysis bag (MWCO 14kDa) against water and 20 mM citrate buffer (pH 8.0) and then concentrated to 20 mL by Amicon Ultra centrifugal units. The nanoparticle concentration was determined by iron assay 27 and the size of the nanoparticles was determined by dynamic light scattering (Zetasizer Nano ZS, Malvern Instruments Ltd)
Nanodrug synthesis and characterization for the linker optimization studies
Cy5.5 mono-reactive NHS ester (1mg, GE Healthcare) was dissolved in 100 μL of anhydrous DMSO and incubated with MN (10 mg Fe) in 20 mM citrate buffer (pH 8.0) overnight. The nanoparticles were purified using Sephadex PD-10 column (GE Healthcare) with PBS eluent. The number of Cy5.5 molecules per nanoparticle was determined as four Cy5.5 molecules per MN by spectrophotometry. Cy5.5-labeled MN (MN-Cy5.5) was loaded with hetero-bifunctional crosslinkers, N-γ-Maleimidobutyryl-oxysuccinimide ester (GMBS), N-Succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) or NHS-PEG24-Maleimide (PEG24), respectively and conjugated with each oligonucleotide (anti-miR10b and scr-LNA), separately following a previous report.11 Briefly, 2.4 mg (20.7nmol) of MN-Cy5.5 was reacted with 0.625 mg (100 equiv.) of SPDP in PBS (pH 8.0) and purified by PD-10 column after overnight reaction at 4°C. After determining the concentration of MN by iron assay and the amount of SPDP on MN (SPDP/MN; 56±6), 1.11 mg (9.6nmol), SPDP-MN-Cy5.5 was conjugated with activated oligo (96nmol, 15 equiv.), which was treated by TCEP to deprotect the thiol at the 5′-end disulfide protecting group. All other oligo-loaded MN with different linkers were prepared following the same procedures. Quantification of oligonucleotides per MN was described previously 11 and determined as ten oligos per MN.
Nanodrug synthesis and characterization for the peptide optimization studies
Nanodrug synthesis was carried out as described in 11. Cy5.5-labeled MN (MN-Cy5.5) was conjugated to heterofunctional linker N-Succinimidyl 3-[2-pyridyldithio]-propionate (SPDP, Pierce Biotechnology) in order to provide a thiol reactive terminus for LNA and cRGD peptide conjugation. MN-SPDP was conjugated to cyclic RGDfK-PEG-Cys-(Boc) or the control cyclic RADfK-PEG-Cys-(Boc) peptide through the peptide’s cysteine terminus in PBS and purified with a Sephadex PD-10 column. The LNA oligos were then conjugated to the resultant probes. Briefly the thiolated 5′ terminus of the oligo was activated via 3% TCEP treatment in nuclease free PBS. The LNA oligos were purified using ammonium acetate/ethanol precipitation method. After TCEP-activation and purification, the oligos were resuspended in PBS and 50 mM EDTA and incubated with the nanoparticles overnight. The resulting probe was purified using a G-50 Sephadex disposable quick spin columns (Roche Applied Science) followed by incubation with each LNA oligonucleotide to produce the final probes (anti-miR10b-MN-cRGD, scr-LNA-MN-cRGD, anti-mirR10b-MN-cRAD and scr-LNA-MN-cRAD). The number of peptides and LNA oligos per MN were determined as fifteen cRGD (or cRAD) and ten LNA oligos per MN. The MN probes are summarized in Table 1. The flow-chart of probe synthesis is described in Fig. S1.
Fluorescence confocal microscopy
Fluorescence confocal microscopy was performed on MDA-MB-231 (ATCC, Manassas, VA) or MDA-MB-231-GFP breast cancer cells (kindly provided by Dr. Danny R. Welch, University of Alabama at Birmingham), as indicated in the figure legends. Cells (2 × 106) were incubated with nanodrugs (17.35 nmol mL−1 oligo; 200 μg mL−1 iron; 26 nmol mL−1 of peptide) or FITC labeled peptides (0.25 to 50 nmol mL−1 of cRGDfK-PEG-Cys-FITC or cRADfK-PEG-Cys-FITC) for 48 h at 37 °C on a cover slip in an 8-well plate. The cells were then washed three times with HBBS buffer and fixed with 2% formaldehyde for 10 mins. The cover slip was placed on a glass slide with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). The slides were dried for 30 minutes in a dark room. The cells finally were imaged by confocal microscopy under phase contrast and in the Cy5.5 channel (MN-Cy5.5 detection) using a Zeiss LSM 5 Pascal laser confocal microscope. Image analysis was performed using Zeiss LSM 5 Pascal Confocal Microscopy Software (Release 3.2). The final images were color-coded blue for Cy5.5 (MN) and represented as overlays with phase contrast.
Flow cytometry
Nanodrug uptake was analyzed by flow cytometry. The cells were incubated with the nanodrugs (17.35 nmol mL−1 oligo; 200 μg mL−1 Fe) for 48 h at 37 °C in an 8-well plate, washed twice with HBBS buffer and removed from the plate using Hank’s based enzyme free cell dissociation buffer. The cells were then fixed in 2% paraformaldehyde for 1 h at 4°C and diluted in sheath solution for flow cytometry measurements. Nanodrug uptake was analyzed in the FL4 channel (Cy5.5, MN) using FACS Calibur (Becton Dickinson, Franklin Lakes, NJ) equipped with the CellQuest software package.
Real-time quantitative reverse transcription-PCR
To measure the extent of miR-10b inhibition by the nanodrugs, the cells were incubated with the nanodrugs (17.35 nmol mL−1 oligo; 200 μg mL−1 Fe) for 48 h at 37°C. The miRNA-enriched fraction from total extracted RNA was harvested using the miRNeasy mini kit, according to the manufacturer’s protocol (Qiagen Inc, Duesseldorf, Germany). Relative levels of miR-10b were determined by real-time quantitative reverse transcription-PCR (qRT-PCR; Taqman protocol) and compared to the internal housekeeping gene SNORD44. Taqman analysis was carried out using an ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, CA). The primers were provided by the manufacturer (RT2 miRNA First Strand Kit, miRNA qPCR assay for hsa-miR-10b and SNORD44 as endogenous control; SABiosciences, Valencia, CA).
In vitro stability of the nanodrugs in serum
The in vitro stability of the nanodrugs was studied in a solution of bovine serum (12.5% in PBS, pH 7.4). The solution was incubated at 37°C for 10 min before use. The reaction was initiated at 37°C by adding 20 μL MN-anti-miR10b (2.5 nmol of oligonucleotide) to 80 μL of preheated serum solution. 20 μL of aliquot was sampled at 0, 6, 24, and 48 h and added to an eppendorf tube containing 5 μL of nucleic acid loading buffer (5x). All collected samples were stored at 4°C (without freezing). Five μL of each sample was heated at 70°C for 3 min. and loaded on a 15% TBE urea gel, stained with ethidium bromide (1 μg mL−1) for 20 min., and washed twice with water. Free oligonucleotide, anti-miR10b (0–150 pmol), was run as a control to locate and quantify the released anti-miR10b oligonucleotide from the nanodrugs. The gels were visualized under UV light.
Western blot to determine αvβ3 expression by MDA-MB-231 cells
Cells were lysed in cell extraction buffer (Invitrogen, Camarillo, CA) containing 1mM PMSF and proteinase inhibitor cocktails (Sigma, St. Louis, MO). Protein content was determined with the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Total protein (20 or 40μg) was applied onto an SDS-PAGE (4 – 20%) gel under reducing conditions. Resolved proteins were transferred electrophoretically to nitrocellulose membranes and pre-blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) for 1h at room temperature. After blocking, the membrane was incubated overnight at 4°C in 1% nonfat dry milk/TBS containing either anti-human integrin αvβ3 (23C6) monoclonal antibody (1.0 μg mL−1) (Santa Cruz Biotech, Santa Cruz, CA) or anti-β-actin (1μg mL−1, (Applied Biosystems, Grand Island, NY) monoclonal antibody. The membrane was then washed 3 times with 0.05% Tween TBS (TBST) and incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Invitrogen, Camarillo, CA) for 60 min at room temperature, followed by washing 3 times with TBST and one time with TBS. Membranes were developed using ECL Plus Western Blotting detection reagents kit (GE Healthcare, Piscataway, NJ) according to the manufacturer’s specifications.
Statistical Analysis
Data were expressed as either mean ± SD. Statistical significance was determined by using Student’s t test. Time-course data were analyzed by one-way repeated-measures analysis of variance. A p value ≤ 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants NCIR00CA129070 and NCI-1R01CA163461-01A1 to ZM by the NIH and the Young Investigator Award to ZM by the Breast Cancer Alliance.
Contributor Information
Mehmet V. Yigit, Email: myigit@albany.edu.
Zdravka Medarova, Email: zmedarova@partners.org.
References
- 1.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs-the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 8.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 11.Yigit M, Ghosh S, Kumar M, Petkova V, Kavishwar A, Moore A, Medarova Z. Context-dependent differences in miR-10b breast oncogenesis can be targeted for the prevention and arrest of lymph node metastasis. Oncogene. 2012;32:1530–1538. doi: 10.1038/onc.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 13.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjugate Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 14.Singh N, Agrawal A, Leung AK, Sharp PA, Bhatia SN. Effect of nanoparticle conjugation on gene silencing by RNA interference. J Am Chem Soc. 2010;132:8241–8243. doi: 10.1021/ja102132e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derfus AM, von Maltzahn G, Harris TJ, Duza T, Vecchio KS, Ruoslahti E, Bhatia SN. Remotely triggered release from magnetic nanoparticles. Adv Mater. 2007;19:3932–3936. [Google Scholar]
- 16.Veiseh O, Kievit FM, Fang C, Mu N, Jana S, Leung MC, Mok H, Ellenbogen RG, Park JO, Zhang M. Chlorotoxin bound magnetic nanovector tailored for cancer cell targeting, imaging, and siRNA delivery. Biomaterials. 2010;31:8032–8042. doi: 10.1016/j.biomaterials.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiefenauer LX, Kuehne G, Andres RY. Antibody-magnetite nanoparticles: in vitro characterization of a potential tumor-specific contrast agent for magnetic resonance imaging. Bioconjugate Chem. 1993;4:347–352. doi: 10.1021/bc00023a007. [DOI] [PubMed] [Google Scholar]
- 18.Lee J-H, Huh Y-M, Jun Y-W, Seo J-W, Jang J-T, Song H-T, Kim S, Cho E-J, Yoon H-G, Suh J-S. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2006;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 19.Park J-H, Von Maltzahn G, Xu MJ, Fogal V, Kotamraju VR, Ruoslahti E, Bhatia SN, Sailor MJ. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc Natl Acad Sci USA. 2010;107:981–986. doi: 10.1073/pnas.0909565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canete M, Soriano J, Villanueva A, Roca AG, Veintemillas S, Serna CJ, Miranda R, Del Puerto Morales M. The endocytic penetration mechanism of iron oxide magnetic nanoparticles with positively charged cover: a morphological approach. Int J Mol Med. 2010;26:533–539. doi: 10.3892/ijmm_00000496. [DOI] [PubMed] [Google Scholar]
- 21.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Mediating tumor targeting efficiency of nanoparticles through design. Nano Letters. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 22.Song E, Lee SK, Dykxhoorn DM, Novina C, Zhang D, Crawford K, Cerny J, Sharp PA, Lieberman J, Manjunath N, Shankar P. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol. 2003;77:7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonvico F, Mornet SP, Vasseur SB, Dubernet C, Jaillard D, Degrouard J, Hoebeke J, Duguet E, Colombo P, Couvreur P. Folate-conjugated iron oxide nanoparticles for solid tumor targeting as potential specific magnetic hyperthermia mediators: synthesis, physicochemical characterization, and in vitro experiments. Bioconjugate Chem. 2005;16:1181–1188. doi: 10.1021/bc050050z. [DOI] [PubMed] [Google Scholar]
- 24.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Peng X, Wang Y, Wang Y, Shin DM, El-Sayed MA, Nie S. A reexamination of active and passive tumor targeting by using rod-shaped gold nanocrystals and covalently conjugated peptide ligands. ACS Nano. 2010;4:5887–5896. doi: 10.1021/nn102055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci USA. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar M, Yigit M, Dai G, Moore A, Medarova Z. Image-guided breast tumor therapy using a small interfering RNA nanodrug. Cancer Res. 2010;70:7553–7561. doi: 10.1158/0008-5472.CAN-10-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.