Abstract
The major histocompatibility complex (MHC) region on chromosome 6p21.3 is suspected to host susceptibility loci for HIV-related Kaposi’s sarcoma (HIV-KS). A nested case-control study in the Multicenter AIDS Cohort Study was designed to conduct fine genetic association mapping across central MHC. Individuals co-infected with HIV-1 and HHV-8 who later developed KS were defined as cases (n=354) and were matched 1:1 with co-infected KS-free controls.
We report data for new independent MHC class II and III susceptibility loci. In particular, class II HLA-DMB emerged as a strong candidate, with the intronic variant rs6902982 A>G associated with a 4-fold increase of risk (OR= 4.09; 95% CI: 1.90–8.80; p= 0.0003). A striking multiplicative effect on the estimated risk was associated with further carriage of two non-synonymous variants, rs1800453 A>G (Asp697Gly) and rs4148880 A>G (Ile393Val), in the linked TAP1 gene (OR=10.5; 95% CI: 2.54–43.6; p=0.0012). The class III susceptibility variant is moderately associated with HIV-KS and lies within a 120 Kb-long haplotype (OR=1.52; 95% CI: 1.01–2.28; p=0.047) formed by rs7029 A>G (GPANK1 3’UTR), rs1065356 G>A (LY6G6C), rs3749953 A>G (MSH5-SAPCD1 readthrough) and rs707926 G>A (VARS). Our data suggest that antigen processing by MHC class II molecules is a target pathway in the pathogenesis of HIV-KS.
INTRODUCTION
Infection by the human immunodeficiency virus (HIV) greatly increases the risk of developing Kaposi’s sarcoma (1), an AIDS- (acquired immunodeficiency syndrome) defining malignancy (2). The development of KS within HIV-1 positive populations results from the uncontrolled expression of latency genes of human herpes virus-8 (HHV-8) (2), the etiologic agent of KS (3). While all cases of KS carry HHV-8, not all individuals with HHV-8 infection develop KS and among HIV-positive infected individuals, those who seroconvert to HHV-8 after HIV infection are at greater risk for KS than those who seroconvert prior to HIV infection (4).
Genetic susceptibility to HIV-related Kaposi’s sarcoma (HIV-KS) is poorly understood. Co-infection by HHV-8, also called KSHV (Kaposi’s sarcoma herpes virus), and the higher prevalence of KS in the setting of HIV infection suggested an important role for host immunity in the control of HIV progression to KS (1, 5–8). In particular, the observation that immunosuppressed transplant recipients have increased cumulative risks (8–30%) for classical KS (9, 10) furthermore suggested that immunosuppression is an important risk factor.
The advent of HAART (highly active anti-retroviral therapy) has significantly decreased the incidence of HIV-KS in the developed countries. Consequently interests in host genetics in HIV-KS have diminished and only a few studies have been reported in the last two decades. Early reports implicated variant genotypes of IL-6 (Interleukin-6) and Fc- γ receptor IIIA in the development of KS (11) (12). Further studies of HIV- and non-HIV KS that focused on the host factors encoded in central MHC (major histocompatibility complex), essentially those of the human leukocyte antigen (HLA) system, have reported positive associations with HLA-DR genes (1, 8, 13) corroborating early findings (14, 15).
As suggested by the higher incidence of classical KS among kidney transplant recipients relative to the general population, the risk associated with immunosuppression is apparently independent of HIV-1 infection. However, the highest risk of KS observed among HIV-infected individuals, notably among men who have sex with men who turned out to have the highest rate of HHV-8 infection (~ 40%), suggested that susceptibility to HIV-KS entails actions and/or interactions between HIV and HHV-8. However, which of the two aspects of immunosuppression -prolonged exposure to low CD4+ T-cell count (CD4+ count) or rate of decline of CD4+ count in the years preceding the diagnosis of KS- is the major determinant of risk is not known. Moreover, given the known implication of viral infection chronology on HIV-KS outcome, epidemiologic studies of HIV-KS may not have been adequately designed.
To overcome the limitations of previous studies, we have re-designed the initial case and control study of HIV-KS (8) nested within MACS (Multicenter AIDS Cohort Study) to allow for the control of infection chronology, immunosuppression and principal component analysis-based assessment of race/ethnicity in evaluating the independent effects of HLA and non-HLA loci across central MHC. Specifically, the degree of immunosuppression in the years preceding the diagnosis of KS was determined by the trapezoidal method (16) and was used together with the estimated slope of CD4+ count as time-dependent covariates.
In the present study, we evaluated the effects of the highly polymorphic class I HLA-B gene and a selection of 467 quality control-filtered SNPs encompassing about 5 Mb across the central MHC region on the natural history of HIV progression to KS. We report data for HLA-B-independent associations of HIV-KS with a variant HLA-DMB and linked TAP1 gene variants.
RESULTS
Single Locus Analysis
Exclusion of 29 SNPs (5.8%) that deviated from HWE in the controls led to a final set of 467 SNPs (Supplementary Table S1) available for further analyses. Based on the self-report of ancestry, 22 men were reclassified into other racial groups and re-matched based on the updated information from the PCA analysis. The majority of these individuals self-reported as Hispanic Europeans indicating that population admixture may explain the misclassification. Re-matching of the cases and controls within this subset of 22 individuals resulted in a loss of three additional individuals because no matched cases or controls could be identified. This resulted in samples of 348 and 318 pairs of matched cases and controls with available SNP and HLA-B typing data, respectively.
Among the modeled covariates, only age at baseline (age at the time of dual seroconversion) was significantly (p=0.004) associated with KS. The degree of immunosuppression (area under the curve of CD4+ count below 300) was not a significant predictor (p > 0.20); however, this covariate turned out to be marginally (0.05 < p < 0.10) associated with the risk for KS in models adjusted for HLA-B (see below). The rate of CD4+ count change (slope of CD4+ count since dual serocoversion) was not a significant predictor of the estimated risks (p > 0.20) regardless of the HLA-B effects; this indicated that the confounding effect of the declining immunity was accounted for by our case and control matching criteria.
The observed HLA-B allele and genotype frequencies for the entire case and control samples are concordant with those published for the Caucasian populations. Only two HLA-B alleles, the risk B*1401 (OR=4.2; 95% CI: 1.1–15.5; p=0.03) and the protection B*2705 (OR=0.37; 95% CI: 0.15–0.94; p=0.04) alleles passed the significance threshold of 5% (Supplementary Table S2).
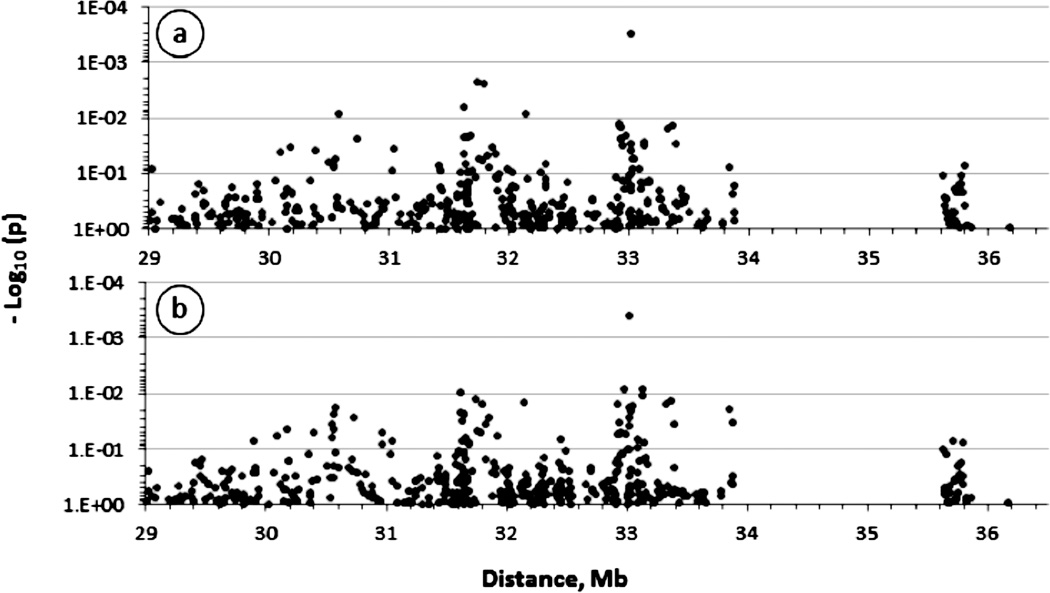
In single SNP analyses, significant associations (p < 0.05) were observed with risk alleles of several SNPs peaking essentially in two chromosomal locations, across a 120 Kb-long interval centromeric to HLA-B and spanning MHC class III GPANK1, LY6G6C, MSH5-SAPCD1 and VARS genes, and across a 95 Kb-long class II interval encompassing TAP1 and HLA-DMB loci (Fig. 2a). Vanishing class III signal but unchanged class II signal was observed after control for the associated HLA-B alleles (Fig. 2b); this indicated HLA-B-independent effects of target class II loci in contrast to those in the class III region, which appear to be confounded by the tightly linked HLA-B. Further sensitivity analyses restricted to the predominantly non-Hispanic European case and control group resulted in a similar pattern of association (Supplementary Fig. S1A and Table S1); thus false findings due to population structure was greatly minimized by the matched design and PCA-based assessment of race.
Figure 2.
Association of major histocompatibility complex (MHC) polymorphisms with HIV-related Kaposi’s sarcoma (HIV-KS) in men. The figure shows the strength of the association expressed as minus logarithm decimal of the p-value (−Log 10(p)) obtained from univariate analyses of 467 single nucleotide polymorphisms (SNP) spanning about 5 megabases across central and extended MHC. The association with the KS outcome was evaluated in conditional logistic models assuming additive variance and controlling for the effects of age at baseline, degree of immunosuppression (calculated as the area under the curve of CD4+ T-cell count below 300) and the rate of CD4+ T-cell count change at six month-intervals (slope of CD4+ T-cell count from the time of dual seroconversion to HIV-1 and HHV8). In panel B, the logistic models were further controlled for the effects of two HLA-B alleles (B*2705 and B*1401) significantly (p<5%) associated with KS. The large centromeric gap corresponds to a chromosome 6p21genomic interval where no genes of potential relevance to KS were found at the time this study was initiated.
Table 1 lists the SNPs associated with the risk of or protection from developing KS. A subset of these are non-synonymous or putative functional SNPs located in genes with relevance to cancer, including rs1116221 G>A, a missense (Glu421Lys) polymorphism in TRIM31 (OR=0.74; 95% CI: 0.56–0.96; p=0.033), rs909253 A>G in the 5’UTR of LT-α (OR=0.75; 95% CI: 0.58–0.96; p=0.022) and occurring in strong LD with non-synonymous SNP rs1041981 C>A (Thr60Asn) in the same gene (OR=0.75; 95% CI: 0.58–0.96; p=0.022), and rs3093665 A>C in the 3’UTR of TNF-α with a stronger effect but marginally associated (OR=2.1; 95% CI: 0.93–4.71; p=0.075) with the risk, possibly due to a low minor allele frequency (MAF).
Table 1.
Association of gene variants in central MHC with HIV-related Kaposi’s sarcoma in HIV-positive men enrolled in the Multicenter AIDS Cohort Study
| obs | polymorphism | position | gene/ closest gene | function | MAF*, count (%) | OR¶ (95% CI) | p | |
|---|---|---|---|---|---|---|---|---|
| cases | controls | |||||||
| 66 | rs356971 (A>C) | 30087776 | ZNRD1-AS1 | ncRNA | 88 (0.14) | 118 (0.19) | 0.73 (0.53–0.99) | 0.0422 |
| 72 | rs1116221 (G>A) | 30179309 | TRIM31 | Glu421Lys | 141 (0.23) | 177 (0.28) | 0.74 (0.56–0.96) | 0.0328 |
| 88 | rs9357097 (G>A) | 30393100 | HCG18 | intron | 185 (0.30) | 157 (0.25) | 1.32 (1.02–1.72) | 0.0394 |
| 100 | rs2429657 (A>G) | 30579499 | HLA-E | intergenic | 119 (0.19) | 152 (0.24) | 0.68 (0.51–0.90) | 0.0081 |
| 105 | rs1076829 (A>G) | 30735195 | DHX16 | intron | 194 (0.31) | 230 (0.37) | 0.76 (0.60–0.96) | 0.0229 |
| 169 | rs2071594 (G>C) | 31620699 | NFKBIL1 | 3'UTR | 188 (0.30) | 220 (0.35) | 0.74 (0.58–0.96) | 0.0220 |
| 170 | rs2071592 (T>A) | 31623319 | NFKBIL1 | intron | 174 (0.28) | 212 (0.34) | 0.70 (0.54–0.90) | 0.0063 |
| 172 | rs6929796 (G>A) | 31630648 | NFKBIL1 | intron | 89 (0.14) | 119 (0.19) | 0.74 (0.55–0.99) | 0.0444 |
| 174 | rs2239707 (A>G) | 31633298 | NFKBIL1 | intron | 217 (0.35) | 189 (0.30) | 1.26 (0.98–1.60) | 0.0683 |
| 185 | rs1800683 (G>A) | 31648050 | LTA | 5'UTR | 188 (0.30) | 220 (0.35) | 0.75 (0.58–0.96) | 0.0223 |
| 187 | rs909253 (A>G) | 31648292 | LTA | 5' region | 188 (0.30) | 220 (0.35) | 0.75 (0.58–0.96) | 0.0223 |
| 189 | rs1041981 (C>A) | 31648763 | LTA | Thr60Asn | 188 (0.30) | 220 (0.35) | 0.75 (0.58–0.96) | 0.0223 |
| 200 | rs3093665 (A>C) | 31653370 | TNF | 3'UTR | 19 (0.03) | 7 (0.01) | 2.00 (0.89–4.53) | 0.0965 |
| 214 | rs2844477 (A>G) | 31686751 | AIF-1, NCR3 | intergenic | 220 (0.35) | 263 (0.42) | 0.75 (0.59–0.96) | 0.0207 |
| 221 | rs7029 (A>G) | 31737932 | GPANK1 | 3'UTR | 167 (0.27) | 137 (0.22) | 1.55 (1.17–2.05) | 0.0022 |
| 224 | rs1065356 (G>A) | 31794987 | LY6G6C | cds-syn | 137 (0.22) | 109 (0.17) | 1.60 (1.18–2.16) | 0.0024 |
| 225 | rs3749953 (A>G) | 31821103 | MSH5-SAPCD1 | intronic | 92 (0.13) | 76 (0.11) | 1.45 (1.01–2.10) | 0.0459 |
| 227 | rs707926 (G>A) | 31856799 | VARS | cds-syn | 107 (0.17) | 84 (0.13) | 1.42 (1.03–1.97) | 0.0323 |
| 264 | rs3749962 (G>A) | 32144335 | TNXB | cds-syn | 41 (0.07) | 60 (0.10) | 0.57 (0.38–0.87) | 0.0084 |
| 349 | rs2071541 (A>G) | 32920836 | TAP1, TAPSAR1 | nearGene-3 | 100 (0.16) | 75 (0.12) | 1.60 (1.11–2.32) | 0.0124 |
| 350 | rs1800453 (A>G) | 32922953 | TAP1 | Asp697Gly | 116 (0.19) | 91 (0.14) | 1.54 (1.09–2.18) | 0.0136 |
| 352 | rs4148880 (A>G) | 32926752 | TAP1 | Ile393Val | 143 (0,21) | 117 (0.17) | 1.45 (1.05–1.99) | 0.0236 |
| 353 | rs4713600 (C>A) | 32930836 | TAP1 | nearGene-5 | 331 (0.48) | 293 (0.42) | 1.31 (1.04–1.66) | 0.0236 |
| 355 | rs9276820 (G>A) | 32937254 | PSMB9 | downstream | 305 (0.49) | 279 (0.44) | 1.33 (1.06–1.67) | 0.0140 |
| 356 | rs1383266 (G>A) | 32942710 | PSMB9, HLA-DMB | intergenic | 135 (0.22) | 169 (0.27) | 0.75 (0.58–0.97) | 0.0309 |
| 359 | rs2187688 (G>A) | 32979979 | PSMB9, HLA-DMB | intergenic | 274 (0.44) | 307 (0.49) | 0.75 (0.59–0.96) | 0.0199 |
| 367 | rs151719 (A>G) | 33011878 | HLA-DMB | intron | 161 (0.26) | 121 (0.19) | 1.39 (1.03–1.86) | 0.0291 |
| 370 | rs194675 (T>A) | 33013724 | HLA-DMB | intron | 272 (0.44) | 309 (0.49) | 0.77 (0.61–0.99) | 0.0383 |
| 372 | rs6902982 (A>G) | 33015859 | HLA-DMB | intron | 32 (0.05) | 10 (0.02) | 4.09 (1.90–8.80) | 0.0003 |
| 377 | rs209475 (G>A) | 33033563 | HLA-DMA | upstream | 215 (0.35) | 254 (0.41) | 0.78 (0.60–1.04) | 0.0533 |
| 379 | rs683208 (A>G) | 33045879 | BRD2 | nearGene-5 | 219 (0.35) | 257 (0.41) | 0.78 (0.60–1.00) | 0.0539 |
| 388 | rs12174395(A>G) | 33122296 | HLA-DPA1 | upstream | 191 (0.31) | 230 (0.37) | 0.75 (0.59–0.97) | 0.0270 |
| 389 | rs375912 (A>G) | 33124706 | HLA-DPA1 | upstream | 198 (0.32) | 236 (0.38) | 0.76 (0.60–0.98) | 0.0319 |
| 408 | rs461338 (A>G) | 33326158 | VPS52 | 3'UTR | 88 (0.14) | 115 (0.18) | 0.66 (0.47–0.92) | 0.0152 |
| 411 | rs466384 (A>G) | 33362643 | WDR46 | Val287Ala | 84 (0.13) | 110 (0.17) | 0.64 (0.45–0.91) | 0.0132 |
| 415 | rs1061801 (G>A) | 33390316 | ZBTB22 | UTR-3 | 123 (0.18) | 150 (0.22) | 0.72 (0.53–0.97) | 0.0288 |
Odds ratios (OR) and lower and upper 95% Wald confidence interval (CI) from conditional logistic models (assuming additive variance) with adjustment for baseline age, degree of immunosuppression (area under CD4+ count of 300) and rate of CD4+ count change (slope of CD4 counts).
Minor allele frequency (MAF).
The most elevated risk was observed with the MHC class II variant rs6902982 A>G, an intronic SNP in HLA-DMB associated with a four-fold increase of risk (OR=4.09; 95% CI: 1.90–8.80; p=0.0003). Within 95 Kb from HLA-DMB toward HLA-DR, significant but moderate associations with risk were observed with two non-synonymous SNPs in TAP1, rs1800453 A>G (Asp697Gly) (OR=1.54; 95%: 1.09–2.18; p=0.014) and rs4148880 A>G (Ile393Val) (OR=1.45; 95% CI: 1.05–1.99; p=0.024), and with rs2071541 A>G, a SNP located in the overlapping microRNA TAPSAR1 (OR=1.60; 95% CI: 1.11–2.32; p=0.012).
Significant associations were also observed with the 3’UTR SNP rs7029 A>G (OR=1.55; 95% CI: 1.17–2.05; p=0.002) in GPANK1 and with the synonymous SNP rs1065356 G>A (OR=1.60; 95% CI: 1.18–2.16; p=0.002) in LY6G6C located about 84 Kb centromeric to TNF-α.
Evaluation of SNP effects under non-additive genetic models suggested that the target class II gene, which is most likely HLA-DMB, acts co-dominantly (compare genetic models in Fig. 2b and Supplementary Figs. S1b and S1c).
Multiple Locus Analysis
The conservation of effect sizes across extended regions suggested that the associated SNPs occur in long-range haplotypes. To capture possible additive or multiplicative effects of two or more candidate MHC loci, we used the haplotype trend regression (HTR) approach separately for class III and II regions to assess the risk associated with MHC haplotypes formed by the risk SNPs. HTR-estimated posterior probabilities were included as explanatory variables in stepwise conditional logistic models with control for the appropriate covariates and additionally for the effects of HLA -B*1401 and -B*2705 for class II haplotypes.
Table 2 illustrates the estimated frequency of the reconstructed haplotypes in the case and control groups together with the magnitude and strength of their association with the KS outcome. Three unique haplotypes, one four-SNP class III haplotype and two seven-SNP class II haplotypes were significantly associated with the risk of KS. Class III G-A-G-A haplotype formed in the order by SNP221 (rs7029 A>G) in GPANK1 3’UTR, synonymous SNP224 (rs1065356 G>A) in LY6G6C, intronic SNP225 (rs3749953 A>G) in MSH5-SAPCD1 readthrough and synonymous SNP227 (rs707926 G>A) in VARS was associated with 50% increase of risk (OR=1.52; 95% CI: 1.01–2.28; p=0.047).
Table 2.
Distribution of the major histocompatibility complex haplotype at risk for HIV-related Kaposi’s sarcoma in a case and control study of HIV-positive men
| haplotype frequency, SE, 95% CL | conditional logistic regression | |||||||
|---|---|---|---|---|---|---|---|---|
| controls | cases | OR* (95% CI) | p | |||||
| SNP-221-224-225-227 | ||||||||
| A-A-G-A | 0.016 | 0.005 | 0.007–0.026 | 0.018 | 0.005 | 0.008–0.028 | - | ns |
| A-G-A-G | 0.738 | 0.016 | 0.705–0.771 | 0.685 | 0.018 | 0.650–0.719 | - | ns |
| G-A-A-A | 0.034 | 0.007 | 0.021–0.048 | 0.049 | 0.008 | 0.033–0.065 | - | ns |
| G-A-A-G | 0.049 | 0.008 | 0.033–0.065 | 0.065 | 0.009 | 0.047–0.084 | - | ns |
| G-A-G-A | 0.085 | 0.011 | 0.064–0.106 | 0.105 | 0.012 | 0.082–0.128 | 1.52 (1.01–2.28) | 0.047 |
| G-G-A-G | 0.064 | 0.009 | 0.046–0.083 | 0.063 | 0.009 | 0.045–0.081 | - | ns |
| SNP-349-350-352-353-355-367-372 | ||||||||
| A-A-A-A-A-A-A | 0.237 | 0.016 | 0.205–0.268 | 0.283 | 0.017 | 0.250–0.317 | 1.38 (1.02–1.86) | 0.035 |
| A-A-A-A-A-G-A | 0.036 | 0.007 | 0.022–0.049 | 0.022 | 0.007 | 0.011– 0.033 | - | ns |
| A-A-A-C-G-A-A | 0.412 | 0.019 | 0.375–0.449 | 0.314 | 0.018 | 0.279–0.349 | - | ns |
| A-A-A-C-G-G-A | 0.135 | 0.013 | 0.110–0.160 | 0.157 | 0.014 | 0.130–0.184 | - | ns |
| A-A-A-C-G-G-G | - | - | - | 0.013 | 0.004 | 0.004–0.021 | - | ns |
| A-A-G-A-A-A-A | 0.021 | 0.005 | 0.010–0.032 | 0.018 | 0.005 | 0.008–0.028 | - | ns |
| A-G-G-A-A-A-A | 0.020 | 0.005 | 0.010–0.031 | 0.019 | 0.005 | 0.009–0.029 | - | ns |
| A-G-G-A-A-G-A | 0.010 | 0.003 | 0.003–0.017 | 0.013 | 0.004 | 0.004–0.021 | - | ns |
| G-G-G-A-A-G-A | - | - | - | 0.019 | 0.005 | 0.009–0.029 | - | ns |
| G-G-G-A-A-A-A | 0.073 | 0.009 | 0.053–0.092 | 0.073 | 0.010 | 0.053–0.092 | - | ns |
| G-G-G-A-A-G-G | - | - | - | 0.026 | 0.006 | 0.014–0.039 | 10.5 (2.54–43.6) | 0.0012 |
| G-G-G-C-A-A-A | 0.018 | 0.005 | 0.008–0.028 | 0.020 | 0.005 | 0.009–0.030 | - | ns |
Odds ratios (OR) and lower and upper 95% confidence intervals (CI) were estimated by conditional logistic regression in haplotype trend regression models (HTR) adjusted for baseline age and for time-dependent CD4+ T-cell covariates. (ns)- not significant at the 5% threshold level to enter or to stay in the stepwise selection logistic models.
-Test of the distal haplotype was further adjusted for the effects of HLA -B. Note that the frequencies do not add to 100% because only haplotypes with frequencies greater than 1% were modeled; the remaining haplotypes were grouped together and included in the HTR models as a single term. (−) -not observed at the frequency cutoff of 1%.
The strongest effect was observed with class II G-G-G-A-A-G-G haplotype (OR=10.5; 95% CI: 2.54–43.6; p=0.0012), which carries among other risk alleles, the “G” allele of intronic SNP372 (rs6902982 A>G) in HLA-DMB shown to be at a 4-fold increase of risk in the single locus analysis (Table 1). This high-risk haplotype was formed strictly by the risk alleles at all the composite SNPs and occurred at a frequency of 2.6% in the cases (36 individuals) and at less than the HTR frequency cutoff of 1% in the controls. The second seven-SNP class II haplotype explained a small fraction of the risk (OR=1.38; 95% CI: 1.02–1.86; p=0.035) and unexpectedly involved both risk and non-risk alleles at the composite SNPs (A-A-A-A-A-A-A haplotype). With this haplotype occurring on average in 26% of the study sample, we reasoned that homozygous A-A-A-A-A-A-A diplotypes must also be common and may occur more frequently in the cases than in the controls. A closer examination of haplotype distributions showed that homozygous A-A-A-A-A-A-A diplotypes indeed occurred more frequently in the cases (n=27) than in the controls (n=16) and exclusion of these individuals resulted in a complete loss of the association with the A-A-A-A-A-A-A haplotype and a slight diminution of the association strength with the risk G-G-G-A-A-G-G haplotype (OR= 9.03; 95% CI: 2.17–37.7; p=0.0025).
DISCUSSION
We reported data from an extensive investigation of the MHC determinants of the natural history of HIV progression to KS. Using a carefully-designed matched case and control study nested within the MACS cohort and analytical models with appropriate time-dependent CD4+ count covariates, we reported data suggesting the implication of MHC class III and II susceptibility loci in the etiology of HIV-KS. Our most important finding indicated that a HLA-DMB variant tagged by intronic rs6902982 increased the risk for HIV-KS by four-fold in HIV- and HHV8-infected men. A significant increase of risk (adjusted OR=10.5) was associated with further carriage of non-synonymous rs1800453 (A>G) and rs4148880 (A>G) alleles encoding Asp697Gly and Ile393Val mutations in TAP1, respectively. Importantly, we have shown that the reported associations are controlled for the confounding effects of immunosuppression and are independent of HLA-B.
We also reported supportive data for a candidate class III susceptibility gene located within a 120 Kb interval flanked by the proximal VARS and distal GPANK1 genes, and spanning members of the leucocyte antigen-6 (LY6) gene superfamily and other class III genes including casein kinase 2B (CSNK2B) implicated in endometrial and esophageal carcinoma and colorectal cancer (17–19), von Willebrand factor A domain containing 7 (VWA7) in lung cancer susceptibility (20) and chloride intracellular channel 1 (CLIC1) in gliomas (21), gastric (22) and hepatic cancer (23).
The possibility that the positive association with HLA-B is an apparent association cannot be excluded in the present discovery stage of the study. HLA-B can influence the association in different ways, i) directly as a true etiologic factor, ii) mechanistically through confounding by linkage disequilibrium with one of the candidate class III susceptibility genes or iii) through joint carriage of specific HLA-B alleles with the risk HLA-DMB variant (locus and allelic genetic heterogeneities). While the protective effect of B*27 from HIV progression is well documented, that of B*14 is not known and surprisingly our E-M estimates of reconstructed HLA-B and SNP221 -224 -225 and -227 joint haplotypes did not reveal the B*1401-G-A-G-A haplotype in either cases or controls, even at a haplotype frequency cutoff as low as 0.3% (not shown). The bulk of the risk G-A-G-A class III haplotype occurred on B*1501 and B*3501 chromosomes in the controls and additionally on B*2705 and accessorily on B*1801 in the cases. The observation that the risk G-A-G-A haplotype is carried on B*2705 is consistent with the drop of the association strength seen at these four SNPs after adjustment for B*2705 and B*1401 (Fig. 2b and Table S1). Furthermore, the drop of the association strength was explained by the control for B*2705 and not for B*1401 (not shown); this excluded the hypothesis of confounding by linkage disequilibrium with B*1401 allele but not with B*2705. Thus given the low frequency of B*1401, replication studies are needed to confirm or reject the association with this allele.
Based on our previous haplotyping data for the present study population, the risk G-A-G-A haplotype is most likely carried on the TNF-α superhaplotype VI (Supplementary Table S3 (24)).
Several additional candidate gene loci were highlighted in this study, with some of them being tagged by potentially functional SNPs such as non-synonymous SNPs in TRIM31 and LTA, polymorphisms in non-coding RNA ZNRD1-AS1 and TAPSAR1, as well as polymorphisms in the 5’UTR of LTA and TAP1 or in the 3’UTR of LTA, TNF-a, GPANK1, VPS52 and ZBTB22. Most of these polymorphisms have protective effects and depending on the HLA background, they may occur or not in long-range linkage disequilibrium with each other or with the risk alleles.
Our data suggested a spurious association with class II A-A-A-A-A-A-A haplotype for SNPs-349-350-352-353-355-367-372 and the default assumption of additive models in the HTR approach is consistent with this finding. Alternatively, homozygosity for this haplotype may also be considered disadvantageous if it tags a unique HLA-DMB allele, thus multiple genetic etiologies cannot be excluded.
None of the listed candidate genes have previously been implicated in the development of KS; nonetheless, after Bonferroni corrections for multiple testing, only HLA-DMB rs6902982 SNP passed the significance threshold of 5%.
Non-classical MHC class II molecules encoded by HLA-DM have recently emerged as important molecules involved in the stabilization of classical MHC class II molecules (25). Specifically, HLA-DM exert their critical role in antigen presentation by MHC class II molecules to CD4+ T-lymphocytes by accelerating the removal of CLIP and by editing the peptide content of MHC class II molecules such that the display of high-affinity peptides is favored (26).
To date, HLA-DMB and the linked TAP-1 gene have not been implicated in the pathogenesis of HIV-related KS. However, TAP1 peptide transporter and the proteasome subunit beta type 9 (PSMB9), which are required for class I antigen presentation, were shown to be inactivated by IFN-γ-mediated down-modulation in response to an ectopic expression of LANA (HHV8-encoded latency-associated nuclear antigen) (27).
In a previous investigation of candidate non-MHC determinants of HIV-KS, which we have conducted on the present study sample, we reported that variants of human homologs of two latently expressed HHV-8 genes, cyclin D1 (CCND1) and interleukin-6 (IL-6), in conjunction with angiogenic gene variants (VEGF, EDN-1 and EDNRB) conferred significant risk for HIV-KS (OR=2.84–3.92; Bonferroni-adjusted p=9.9×10−3– 2.6×10−4) (28). Here, we ruled out possible long-range LD between markers in central MHC and the risk variants reported for the endothelin-1 (ET-1)-encoding EDN-1 gene, which maps few megabases away from central MHC (not shown).
It should be stressed that allelotyping of non-classical HLA -DMB and -DMA loci was not available at the time this study was conducted and only partial typing data were available for HLA-DR, the interaction partners of HLA-DM.
Our data lay foundation for a model in which prolonged exposure to low levels of CD4 count (< 300) may provide favorable conditions for HHV8 to down-regulate antigen processing by class II molecules in susceptible individuals (those carrying a combination of risk alleles in target HLA-DM and TAP1 loci).
METHODS
Study Participants
This study uses information on patients enrolled in the Multicenter AIDS Cohort Study (MACS) (29). MACS is a prospective longitudinal study of HIV-1 infection that recruited 5,622 homosexual men in 1984–1985 and 1987–1990. The MACS cohort began in 1984 in four US cities: Baltimore, MD; Chicago, IL; Pittsburgh, PA; and Los Angeles, CA. To be included in the study participants could not have a clinical AIDS diagnosis at baseline, they must be > 18 years at baseline, and participate in homosexual behavior within 5 years of study entry. The last date of follow-up for any of the study participants was January 1, 1996, when HAART became more widely available. Participants completed semiannual physical examinations and questionnaires that included information on treatments, medication utilization and information from routine blood draws.
Study Design
This study built on an initial selection of 360 matched cases and controls pairs nested within the MACS cohort. Men with dual HIV-1 and HHV-8 infections who later developed KS were defined as cases (n=360). Matched controls (n=360) were chosen based on HIV/HHV-8 serostatus, race, KS free time and CD4+ counts, and were defined as men who were free of KS. These pairs were matched for CD4+ counts at the visit within 1 year prior to the index diagnosis. If a control could not be matched by CD4+ count then it was matched by CD4+ slope (+/− 25 cells/year) up to the time of KS diagnosis. Due to the temporal relationship between HIV-1/ HHV-8 and their influence on progression time to KS, cases were matched to controls within each of the four different sero-status groups. These four sero-status groups are defined as HIV-1 seroprevalent (SP)/ HHV-8 SP; HIV-1 SP/ HHV-8 seroconverter (SC); HIV-1-SC/HHV-8-SP; and HIV-1-SC/HHV-8-SC. Follow-up time in the cases is defined as the date of KS diagnosis minus the baseline date or minus the date of seroconversion. The date of seroconversion is defined as the mid-point between the last HIV-1 or HHV-8-negative date and the first positive date.
Ascertainment of Seropositivity and Variable Definition
Sera were tested at the enrollment visit, or the subsequent study visit for a baseline measurement and the final measurement of sera was conducted at the most recent visit the participant was tested. HIV-1 seropositivity was defined as an immunoblot-confirmed positive ELISA. Standardized T-cell phenotyping was performed at each follow-up visit. Specimens of peripheral blood mononuclear cells, plasma, and serum from each participant have been stored in repositories. HHV-8 antibodies against HHV-8 lytic antigens were determined by use of an indirect immunofluorescence assay using 10-Q-tetradecanoyl phorbol 13-acetate induced body cavity B cell lymphoma-1 cells containing the HHV-8 genome. Known HHV-8 positive and negative sera were assayed for each batch of serum samples tested. Serum samples were tested twice in a blinded fashion and were assessed microscopically for the presence of whole cell immunofluorescence by the same reader. Positivity at either sample defined an HHV-8- infected man and HHV-8 negativity at both visits defined an uninfected man.
Laboratory Methods
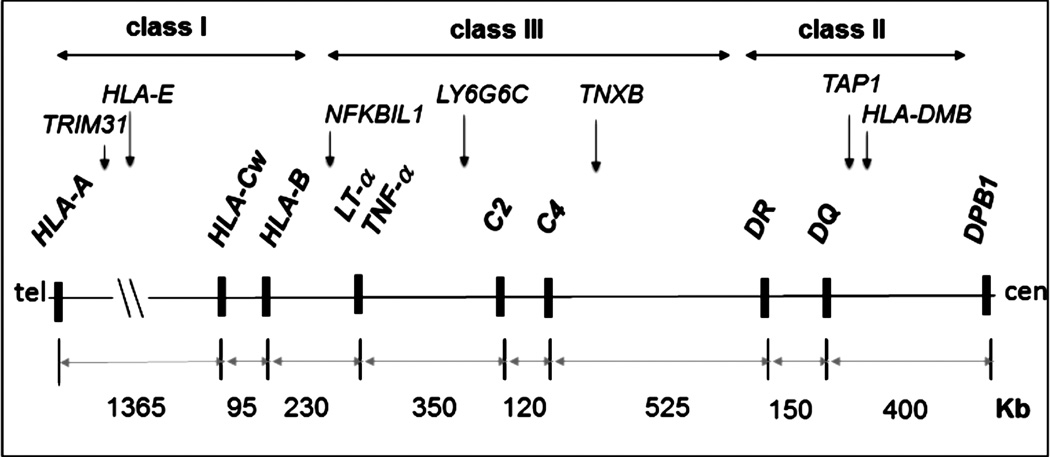
Single nucleotide polymorphisms were typed on a commercial genotyping platform (BeadArray®, Illumina Inc., San Diego, CA). A total of 496 SNPs were selected from candidate genes across 5 Mb of human chromosome 6p21 spanning central MHC and flanking regions (Figure 1).
Figure 1.
Genomic map of the target central MHC region. The genomic map of central major histocompatibility complex (MHC) on human chromosome 6p21.3 targeted for single nucleotide polymorphism association mapping is shown in the telomere (tel) to centromere (cen) orientation. Major human leukocyte antigen (HLA) and non-HLA class I, II and III landmark loci are shown in bold. A sample of MHC genes associated with HIV-related Kaposi’s sarcoma in the present study is shown above the landmark loci. The three known hot spots for recombination map between HLA-B and NFKBIL1, DR and DQ and between HLA-DMB and DPB1.
High-resolution HLA-B genotyping (four-digit) was carried out by PCR amplification followed by automated capillary electrophoresis-based sequencing with Cy5-labeled primers (Abbott Park, IL) in an ABI Prism 3130×l DNA Analyzer (Applied Biosystems).
Quality Control
Reliability in the typing data was assessed by a small set of intra- and inter-plates blind duplicates. SNP calls were checked for adherence to Hardy-Weinberg equilibrium (HWE) in each of the KS outcome categories and only SNPs showing no significant deviation (p>0.01) from HWE in KS-free controls were included for analyses.
Statistical Analysis
Covariates
Variables of interest within the demographic characteristics, clinical parameters, and co-morbidities that are known to be associated with KS were compared between cases and controls using chi-square and Student’s t-test. We controlled for the effects of age (age at the time of dual seroconversion to HIV-1 and HHV-8 infections), the degree of immunosuppression and the rate of immunity decline over the exposure time (time from dual infection to index diagnosis). We adopted the trapezoidal method (16) to the CD4+ count to estimate the degree of immunosuppression. Copy-years were defined as the CD4+ cell count per year below the cut-off of 300 CD4+ count and integrated over the number of years from study entry (for the seroprevalents) or one year after dual seroconversion (for the seroconverters). The slope of the CD4+ count from baseline to the study end captured the rate of immunity decline.
Population Structure
Principle component analysis (PCA) based on the current MHC SNPs and of an additional set of 284 non-MHC SNPs was reported in a previous study that used the present case and control sample (28). Rigorously checking the population of origin is very important especially for MHC-linked diseases because of the differential and long-range pattern of linkage disequilibrium (LD) across MHC in populations. The same SNP may capture different DNA variants depending on the population of origin, which usually has a distinct HLA distribution.
Single Locus Analysis
SNP markers were examined separately in case and control groups for adherence to HWE using Pearson’s chi-squared test. Conditional logistic regression under different genetic models (additive, dominant and over-dominant) was used to assess the association between the SNP genotypes and the odds of developing KS in models with adjustment for the two time-dependent CD4+ count covariates. All statistical tests were performed in SAS and adjusted odds ratios (OR), 95% confidence interval (95% CI) and two-sided p-values are reported.
For the multi-allelic HLA-B locus, we conducted the analyses by carrier status including only alleles with a frequency greater than 1.0 % in the entire study sample. Given the differentiation of MHC in highly diverse HLA-B-specific MHC haplotypes, we avoided grouping together related HLA-B alleles to increase specificity.
Multiple Locus Analysis
To evaluate the effects of MHC haplotypes, we estimated the haplotypes formed by the composite loci (non-HLA SNPs alone or joint HLA-B and non-HLA SNPs) that showed significant association with the outcome and assessed the overall differences in their distribution in cases and controls using the haplotype trend regression (HTR) approach (30). An additive model was assumed, estimating posterior probabilities for each subject for all expectation-maximization-inferred haplotypes. These posterior probabilities were treated as independent variables in the HTR model with the weights in the design matrix reflecting various alternative inferences about haplotypes. A logistic regression model containing the weighted haplotypes was used due to accommodate the case-control design. Confounding by differential CD4+ count was controlled for in multivariable logistic models, with the adjusted odds ratios representing the risk increase per haplotype copy. Haplotypes with a frequency < 1% were aggregated as a single term in the model. Haplotype associations were tested using the most prevalent haplotype in the controls as the referent haplotype for calculating the odds ratios.
Multiple Testing
Due to the high correlation (strong LD) among the tested SNPs and the conservative nature of the conventional methods used to correct for multiple testing, we present the results with no correction and discuss them in the specific context of correlated data.
Supplementary Material
ACKNOWLEDGEMENTS
Funding support for this study was provided by the National Institutes of Health/ National Cancer Institute grant R01-CA106168 (RAK) and administrative supplement (BA). The MACS cohort is supported by the National Institute of Allergy and Infectious Diseases, the National Cancer Institute and the National Heart, Lung and Blood Institute grant numbers: UO1-AI-35042, 5-MO1-RR-00722 (GCRC), UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613,UO1-AI-35041.
Data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centers (Principal Investigators) at The Johns Hopkins University Bloomberg School of Public Health (Joseph B. Margolick, Lisa Jacobson), Howard Brown Health Center and Northwestern University Medical School (John Phair), University of California, Los Angeles (Roger Detels), and University of Pittsburgh (Charles Rinaldo). Website located at http://www.statepi.jhsph.edu/macs/macs.html.
Footnotes
The authors of the present manuscript have no conflict of interest, financial or otherwise, to declare.
REFERENCES
- 1.Gaya A, Esteve A, Casabona J, McCarthy JJ, Martorell J, Schulz TF, et al. Amino acid residue at position 13 in HLA-DR beta chain plays a critical role in the development of Kaposi's sarcoma in AIDS patients. Aids. 2004 Jan 23;18(2):199–204. doi: 10.1097/00002030-200401230-00008. [DOI] [PubMed] [Google Scholar]
- 2.Goedert JJ. The epidemiology of acquired immunodeficiency syndrome malignancies. Seminars in oncology. 2000 Aug;27(4):390–401. [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994 Dec 16;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson LP, Jenkins FJ, Springer G, Munoz A, Shah KV, Phair J, et al. Interaction of human immunodeficiency virus type 1 and human herpesvirus type 8 infections on the incidence of Kaposi's sarcoma. The Journal of infectious diseases. 2000 Jun;181(6):1940–1949. doi: 10.1086/315503. [DOI] [PubMed] [Google Scholar]
- 5.Papasteriades C, Kaloterakis A, Filiotou A, Economidou J, Nicolis G, Trichopoulos D, et al. Histocompatibility antigens HLA-A, -B, -DR in Greek patients with Kaposi's sarcoma. Tissue antigens. 1984 Nov;24(5):313–315. doi: 10.1111/j.1399-0039.1984.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 6.Contu L, Cerimele D, Pintus A, Cottoni F, La Nasa G. HLA and Kaposi's sarcoma in Sardinia. Tissue antigens. 1984 Apr;23(4):240–245. doi: 10.1111/j.1399-0039.1984.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 7.Goedert JJ. Infectious and genetic factors in AIDS-associated Kaposi sarcoma. Lancet. 1990 Mar 3;335(8688):547. doi: 10.1016/0140-6736(90)90785-4. [DOI] [PubMed] [Google Scholar]
- 8.Dorak MT, Yee LJ, Tang J, Shao W, Lobashevsky ES, Jacobson LP, et al. HLA-B, -DRB1/3/4/5, and -DQB1 gene polymorphisms in human immunodeficiency virus-related Kaposi's sarcoma. Journal of medical virology. 2005 Jul;76(3):302–310. doi: 10.1002/jmv.20361. [DOI] [PubMed] [Google Scholar]
- 9.Frances C, Mouquet C, Marcelin AG, Barete S, Agher R, Charron D, et al. Outcome of kidney transplant recipients with previous human herpesvirus-8 infection. Transplantation. 2000 May 15;69(9):1776–1779. doi: 10.1097/00007890-200005150-00008. [DOI] [PubMed] [Google Scholar]
- 10.Regamey N, Cathomas G. HHV-8: a newly recognized pathogen in transplantation. Transplantation. 2000 May 15;69(9):1768–1769. doi: 10.1097/00007890-200005150-00003. [DOI] [PubMed] [Google Scholar]
- 11.Foster CB, Lehrnbecher T, Samuels S, Stein S, Mol F, Metcalf JA, et al. An IL6 promoter polymorphism is associated with a lifetime risk of development of Kaposi sarcoma in men infected with human immunodeficiency virus. Blood. 2000 Oct 1;96(7):2562–2567. [PubMed] [Google Scholar]
- 12.Lehrnbecher TL, Foster CB, Zhu S, Venzon D, Steinberg SM, Wyvill K, et al. Variant genotypes of FcgammaRIIIA influence the development of Kaposi's sarcoma in HIV-infected men. Blood. 2000 Apr 1;95(7):2386–2390. [PubMed] [Google Scholar]
- 13.Masala MV, Carcassi C, Cottoni F, Mulargia M, Contu L, Cerimele D. Classic Kaposi's sarcoma in Sardinia: HLA positive and negative associations. International journal of dermatology. 2005 Sep;44(9):743–745. doi: 10.1111/j.1365-4632.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 14.Mann DL, Murray C, O'Donnell M, Blattner WA, Goedert JJ. HLA antigen frequencies in HIV-1-related Kaposi's sarcoma. Journal of acquired immune deficiency syndromes. 1990;3(Suppl 1):S51–S55. [PubMed] [Google Scholar]
- 15.Scorza Smeraldi R, Fabio G, Lazzarin A, Eisera NB, Moroni M, Zanussi C. HLA-associated susceptibility to acquired immunodeficiency syndrome in Italian patients with human-immunodeficiency-virus infection. Lancet. 1986 Nov 22;2(8517):1187–1189. doi: 10.1016/s0140-6736(86)92197-5. [DOI] [PubMed] [Google Scholar]
- 16.Cole SR, Napravnik S, Mugavero MJ, Lau B, Eron JJ, Jr, Saag MS. Copy-years viremia as a measure of cumulative human immunodeficiency virus viral burden. American journal of epidemiology. 2010 Jan 15;171(2):198–205. doi: 10.1093/aje/kwp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Dong Z, Cao C, Wu D, Huang Z. Effect of casein kinase 2beta in esophageal carcinoma and its clinical significance. Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2012 Oct;32(10):1491–1494. [PubMed] [Google Scholar]
- 18.Zou JJ, Luo HS, Huang ZY, Dong ZY, Zeng Q, Wu DH. Correlation of casein kinase 2beta overexpression to the metastatic ability of colorectal cancer cells in vitro. Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2011 Apr;31(4):628–632. [PubMed] [Google Scholar]
- 19.Pallares J, Llobet D, Santacana M, Eritja N, Velasco A, Cuevas D, et al. CK2beta is expressed in endometrial carcinoma and has a role in apoptosis resistance and cell proliferation. The American journal of pathology. 2009 Jan;174(1):287–296. doi: 10.2353/ajpath.2009.080552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snoek M, Albertella MR, van Kooij M, Wixon J, van Vugt H, de Groot K, et al. G7c, a novel gene in the mouse and human major histocompatibility complex class III region, possibly controlling lung tumor susceptibility. Immunogenetics. 2000 Apr;51(4–5):383–386. doi: 10.1007/s002510050634. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, He S, Tu Y, Ji P, Zong J, Zhang J, et al. Elevated expression of chloride intracellular channel 1 is correlated with poor prognosis in human gliomas. Journal of experimental & clinical cancer research : CR. 2012;31:44. doi: 10.1186/1756-9966-31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma PF, Chen JQ, Wang Z, Liu JL, Li BP. Function of chloride intracellular channel 1 in gastric cancer cells. World journal of gastroenterology : WJG. 2012 Jun 28;18(24):3070–3080. doi: 10.3748/wjg.v18.i24.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Wang XM, Yin ZY, Zhao WX, Zhou JY, Zhao BX, et al. Chloride intracellular channel 1 is overexpression in hepatic tumor and correlates with a poor prognosis. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2013 Nov;121(11):1047–1053. doi: 10.1111/apm.12093. [DOI] [PubMed] [Google Scholar]
- 24.Merino AM, Zhang K, Kaslow RA, Aissani B. Structure of tumor necrosis factor-alpha haploblocks in European populations. Immunogenetics. 2013 Jul;65(7):543–552. doi: 10.1007/s00251-013-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze MS, Wucherpfennig KW. The mechanism of HLA-DM induced peptide exchange in the MHC class II antigen presentation pathway. Current opinion in immunology. 2012 Feb;24(1):105–111. doi: 10.1016/j.coi.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders AK, Call MJ, Schulze MS, Fowler KD, Schubert DA, Seth NP, et al. HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide. Nature immunology. 2011 Jan;12(1):54–61. doi: 10.1038/ni.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu F, Tsai K, Chen HS, Wikramasinghe P, Davuluri RV, Showe L, et al. Identification of host-chromosome binding sites and candidate gene targets for Kaposi's sarcoma-associated herpesvirus LANA. Journal of virology. 2012 May;86(10):5752–5762. doi: 10.1128/JVI.07216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aissani B, Wiener HW, Zhang K, Kaslow RA, Ogwaro KM, Shrestha S, et al. A candidate gene approach for virally induced cancer with application to HIV-related Kaposi's sarcoma. International journal of cancer Journal international du cancer. 2013 Jul 1; doi: 10.1002/ijc.28351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. American journal of epidemiology. 1987 Aug;126(2):310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 30.Aissani B, Ogwaro KM, Shrestha S, Tang J, Breen EC, Wong HL, et al. The major histocompatibility complex conserved extended haplotype 8.1 in AIDS-related non-Hodgkin lymphoma. Journal of acquired immune deficiency syndromes. 2009 Oct 1;52(2):170–179. doi: 10.1097/QAI.0b013e3181b017d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.