Abstract
Background
Cancers of the bile duct and the pancreas are virtually indistinguishable using conventional histopathological and clinical characteristics. We sought to use microRNA (miR) profiling to differentiate these two cancers.
Methods
RNA was harvested from the tumors of patients undergoing curative resection for cholangiocarcinoma or pancreatic adenocarcinoma and compared with adjacent normal bile duct or pancreas, respectively. There were 31 pairs of cholangiocarcinoma with matched tumor and adjacent bile duct and nine pairs of pancreatic cancer with matched tumor and adjacent uninvolved pancreas that had sufficient quantity of RNA that were included in the final analysis. Differential microRNA expression profiles were determined using the nCounter System from nanoString Technologies (Seattle, WA, USA).
Results
A total of 41 differentially expressed miRs were identified in cholangiocarcinoma (25 overexpressed, 16 underexpressed) and 52 differentially expressed miRs were found in pancreatic adenocarcinoma (30 overexpressed, 22 underexpressed) relative to adjacent normal tissue. Of these two profiles, 15 miRs were commonly dysregulated between tumor types. Also, eight miRs were similarly overexpressed or underexpressed in cholangiocarcinoma and pancreatic adenocarcinoma, whereas the other seven miRs had inverse expression levels.
Conclusions
Cholangiocarcinoma has a distinct miR profile from pancreatic adenocarcinoma. Discrimination between these two tumor types may be possible with as few as seven miRs.
Cholangiocarcinoma arises from the malignant transformation of cholangiocytes in the biliary epithelium.1 Cholangiocarcinoma is the second most common primary hepatic malignancy and its incidence is increasing in western countries.2 Tumors are often separated into intrahepatic or extrahepatic classifications based on their anatomic location despite similar histology because of differences in their clinical presentations.3,4 Bile duct tumors have a tendency to develop desmoplastic reactions and have early perineural invasion.3,5,6 Another gastrointestinal cancer, pancreatic adenocarcinoma, shares many similar characteristics with cholangiocarcinoma. Pancreatic adenocarcinomas also are characterized by dense desmoplastic reaction where stromal elements include lymphocytes, fibroblasts, and stellate cells.7 Cholangiocarcinoma and pancreatic adenocarcinoma have similarly poor prognoses with nearly all of the estimated 43,000 patients diagnosed with pancreatic adenocarcinoma dying within 1 year, while patients with cholangiocarcinoma fare slightly better with a median survival of less than 24 months.8–11 With few effective medical therapies for either cancer, surgical resection offers patients the only hope for cure. Cholangiocarcinoma and pancreatic adenocarcinoma are virtually indistinguishable histologically and have overlapping immunohistochemical profiles, but molecular differentiation may help explain their pathophysiologies leading to different treatment approaches or inclusion in clinical trials.
MicroRNAs, or miRs, are small (~20–22 nt) noncoding RNAs known to function through their regulatory effect on various genes involved in apoptosis, cell-cycle progression, and as transcription factors in normal and neoplastic cells.12 MicroRNAs may also function as oncogenes or tumor suppressor genes.13 These oncomirs have been described in both hematologic and solid organ malignancies, and their effects can be tissue-specific.14,15 For example, microRNA-21 (miR-21) is one of the most commonly deregulated microRNAs in cancer. In pancreatic cancer, miR-21 upregulation has been linked to cell proliferation, invasion, and chemoresistance in vitro and predicts poor survival in patients with node-negative disease.16,17 In cholangiocarcinoma cell lines, the upregulation of miR-21 decreased gemcitabine-induced apoptosis by blocking expression of the tumor-suppressor gene PTEN.18 However, factors that induce miR-21 overexpression are less understood. MicroRNA profiles can also distinguish between normal and cancerous tissues and have differentiated pancreatic adenocarcinoma from adjacent normal pancreas and chronic pancreatitis.19,20
Given the histopathological and clinical similarities between cancers of the bile duct and pancreas we sought to develop a miR profile that can objectively differentiate cholangiocarcinoma from pancreatic adenocarcinoma. Such an objective tool may have implications clinically as the systemic treatment of these two diseases is beginning to diverge and scientifically as similarities and differences in their pathogenesis become evident.
METHODS
After approval by the institutional review board at the Ohio State University, specimens from 38 patients who had undergone resection for cholangiocarcinoma and 10 specimens from patients who had undergone resection for pancreatic adenocarcinoma between 1993 and 2007 were identified from the archival files of the Department of Pathology. All samples were reviewed by a single senior pathologist (WLF), and site of origin (bile duct vs pancreas) confirmed. As well, clinical information (e.g., preoperative imaging) was used to separate pancreatic cancers from distal cholangiocarcinomas. The pancreas specimens were selected from the same patients who had undergone miR profiling previously using a microarray platform, and all pancreatic samples that had adequate remaining volume of RNA for analysis were used.19 Three 2-mm cores were obtained from macrodissected paraffin blocks for both tumor and matched adjacent benign tissue. Tissue cores were deparaffinized using 100 % xylene at 50 °C for 3 min. Total RNA was extracted according to the manufacturer’s instructions using RecoverAll Total Nucleic Acid Isolation Kit (Ambion by Life Technologies). Digital quantification of the microRNAs was calculated using the nCounter System from nanoString Technologies at our Nucleic Acid Shared Resource facility as per manufacturer’s protocol (nanoString Technologies). Briefly, the nanoString nCounter platform involved mixing total RNA with pairs of capture and reporter probes tailored to each miRNA, hybridizing, washing away excess probes, immobilizing probe-bound miRNAs on a surface and scanning color-coded bar tags on the reporter probes. The reporter probes representing individual copies of miRNA were then tabulated.21
Statistical Analysis
For nanoString data, positive spike-in controls were used for a direct assessment of sample preparation conditions. A linear model was fit on the log-transformed positive control data for each chip. Then the linear models from each chip were normalized to the average linear model. Negative controls were used to access background hybridization and for filtering out low-expression probes. A quantile normalization method was used to normalize samples. Linear mixed models for correlated data was then used to detect differentially expressed miRNAs. Variance smoothing method was used for improving variance estimates in testing.22 Significance was adjusted by controlling the mean number of false positives.23 Significance was accepted at p < 0.01 to correct for repeated measures. Statistical software SAS 9.2 and R was used for analysis.
RESULTS
There were 31 pairs of cholangiocarcinoma with matched tumor and adjacent bile duct and nine pairs of pancreatic cancer with matched tumor and adjacent uninvolved pancreas that had sufficient quantity of RNA that were included in the final analysis. Cholangiocarcinomas were intrahepatic in 11 and extrahepatic in 20. All cholangiocarcinomas and pancreatic cancers had been resected with curative intent. None of the patients had received preoperative chemotherapy or radiation. A total of 41 differentially expressed miRs were identified by nanoString nCounter platform in cholangiocarcinoma relative to adjacent normal bile duct epithelium. Of these, 25 miRs were overexpressed and 16 were underexpressed; p < 0.01 (Table 1). Also, 52 miRs were identified as being significantly dysregulated in pancreatic adenocarcinoma relative to adjacent normal tissue with upregulation of 30 miRs and downregulation of 22 miRs; p < 0.01 (Table 2). Comparison of the miR expression profiles established from cholangiocarcinoma and pancreatic adenocarcinoma versus their adjacent normal tissue revealed 15 miRs that were commonly dysregulated between tumor types (miRs -221, -10a, -135b, -21, -151-3p, -181a, -106b, -139-5p, -100, -145, -125b, -127-3p, -30e, -96, and miR-30b). There were 8 miRs that were similarly overexpressed or underexpressed in cholangiocarcinoma and pancreatic adenocarcinoma, whereas the other seven miRs had inverse expression levels (Fig. 1). For example, miR-21 was similarly overexpressed in both cancers while miR-100 had inverse expression levels; i.e., it was underexpressed in cholangiocarcinoma but overexpressed in pancreatic cancer.
TABLE 1.
Differentially expressed microRNAs for cholangiocarcinoma vs normal bile duct
| MicroRNA | Fold change | p value |
|---|---|---|
| Overexpressed microRNAs | ||
| hsa-miR-21 | 2.57 | 3.65E – 07 |
| hsa-miR-135b | 1.98 | 3.63E – 06 |
| hsa-miR-122 | 1.94 | 2.30E – 03 |
| hsa-miR-27a | 1.87 | 3.76E – 05 |
| hsa-miR-29a | 1.60 | 1.35E – 06 |
| hsa-miR-429 | 1.49 | 5.82E – 03 |
| hsa-miR-24 | 1.47 | 4.25E – 05 |
| hsa-miR-203 | 1.45 | 1.63E – 04 |
| hsa-miR-106b | 1.45 | 2.48E – 05 |
| hsa-miR-29b | 1.41 | 7.29E – 05 |
| hsa-miR-20a + hsa-miR-20b | 1.40 | 1.61E – 03 |
| hsa-miR-93 | 1.40 | 6.45E – 05 |
| hsa-miR-30e | 1.40 | 5.93E – 03 |
| hsa-miR-30b | 1.40 | 1.25E – 03 |
| hsa-miR-151-3p | 1.39 | 4.09E – 05 |
| hsa-miR-10a | 1.37 | 9.99E – 04 |
| hsa-miR-181a | 1.33 | 6.81E – 05 |
| hsa-miR-96 | 1.32 | 3.24E – 04 |
| hsa-miR-663b | 1.31 | 5.01E – 03 |
| hsa-miR-103 | 1.31 | 3.01E – 03 |
| hsa-miR-221 | 1.29 | 3.52E – 03 |
| hsa-miR-22 | 1.28 | 1.68E – 03 |
| hsa-miR-107 | 1.28 | 2.35E – 03 |
| hsa-miR-424 | 1.23 | 2.51E – 03 |
| hsa-miR-340 | 1.19 | 2.29E – 04 |
| Underexpressed microRNAs | ||
| hsa-miR-451 | 0.38 | 6.94E – 05 |
| hsa-miR-145 | 0.49 | 1.88E – 05 |
| hsa-miR-99a | 0.52 | 3.69E – 06 |
| hsa-miR-125b | 0.53 | 1.55E – 05 |
| hsa-miR-630 | 0.56 | 2.66E – 03 |
| hsa-let-7c | 0.63 | 1.50E – 03 |
| hsa-miR-144 | 0.64 | 3.99E – 04 |
| hsa-miR-100 | 0.67 | 6.36E – 04 |
| hsa-miR-127-3p | 0.69 | 8.89E – 04 |
| hsa-miR-139-5p | 0.72 | 2.75E – 03 |
| hsa-miR-337-3p | 0.74 | 3.21E – 04 |
| hsa-miR-1 | 0.74 | 4.93E – 03 |
| hsa-miR-126 | 0.76 | 4.69E – 03 |
| hsa-miR-376c | 0.80 | 8.33E – 03 |
| hsa-miR-517c + hsa-miR-519a | 0.80 | 4.74E – 04 |
| hsa-miR-520e | 0.82 | 7.83E – 03 |
TABLE 2.
Differentially expressed microRNAs for pancreatic adenocarcinoma vs normal pancreas
| MicroRNA | Fold change | p value |
|---|---|---|
| Overexpressed microRNAs | ||
| hsa-miR-205 | 7.54 | 1.01E – 03 |
| hsa-miR-222 | 5.80 | 4.49E – 08 |
| hsa-miR-100 | 4.07 | 1.68E – 04 |
| hsa-miR-210 | 3.36 | 8.41E – 05 |
| hsa-miR-31 | 3.36 | 1.35E – 04 |
| hsa-miR-221 | 3.22 | 2.32E – 05 |
| hsa-miR-10a | 3.13 | 1.46E – 04 |
| hsa-miR-135b | 3.08 | 4.42E – 04 |
| hsa-miR-145 | 2.88 | 9.78E – 04 |
| hsa-miR-21 | 2.82 | 1.80E – 03 |
| hsa-miR-151-3p | 2.68 | 2.45E – 05 |
| hsa-let-7i | 2.53 | 3.95E – 04 |
| hsa-miR-143 | 2.45 | 5.95E – 03 |
| hsa-miR-181a | 2.44 | 3.39E – 04 |
| hsa-miR-125a-5p | 2.41 | 4.60E – 05 |
| hsa-miR-125b | 2.33 | 4.07E – 03 |
| hsa-miR-484 | 2.20 | 3.49E – 04 |
| hsa-miR-1274a | 2.16 | 3.19E – 03 |
| hsa-miR-155 | 2.02 | 6.40E – 03 |
| hsa-miR-633 | 2.00 | 6.40E – 03 |
| hsa-miR-331-3p | 1.89 | 1.85E – 04 |
| hsa-miR-146a | 1.89 | 3.35E – 03 |
| hsa-miR-191 | 1.69 | 1.77E – 03 |
| hsa-miR-181b + hsa-miR-181d | 1.68 | 2.27E – 03 |
| hsa-miR-127-3p | 1.65 | 9.60E – 03 |
| hsa-miR-1274b | 1.63 | 6.34E – 03 |
| hsa-miR-320a | 1.50 | 6.76E – 03 |
| hsa-miR-10b | 1.49 | 4.01E – 03 |
| hsa-miR-106b | 1.48 | 5.09E – 03 |
| hsa-miR-874 | 1.45 | 8.43E – 03 |
| Underexpressed microRNAs | ||
| hsa-miR-217 | 0.05 | 5.00E – 06 |
| hsa-miR-148a | 0.07 | 5.42E – 06 |
| hsa-miR-216a | 0.08 | 1.10E – 05 |
| hsa-miR-375 | 0.11 | 8.76E – 04 |
| hsa-miR-141 | 0.19 | 1.96E – 05 |
| hsa-miR-220a | 0.21 | 4.20E – 06 |
| hsa-miR-29c | 0.24 | 1.27E – 05 |
| hsa-miR-139-5p | 0.31 | 7.28E – 07 |
| hsa-miR-130b | 0.33 | 3.66E – 05 |
| hsa-miR-30c | 0.34 | 9.72E – 06 |
| hsa-miR-335 | 0.40 | 2.95E – 04 |
| hsa-miR-135a | 0.41 | 4.09E – 03 |
| hsa-miR-200c | 0.44 | 4.62E – 04 |
| hsa-miR-1290 | 0.47 | 2.93E – 03 |
| hsa-miR-30b | 0.49 | 2.34E – 03 |
| hsa-miR-96 | 0.55 | 5.45E – 03 |
| hsa-miR-26b | 0.56 | 2.10E – 03 |
| hsa-miR-30e | 0.56 | 2.91E – 03 |
| hsa-miR-101 | 0.60 | 9.90E – 03 |
| hsa-miR-148b | 0.62 | 8.21E – 03 |
| hsa-let-7f | 0.70 | 6.27E – 03 |
| hsa-miR-381 | 0.70 | 7.58E – 03 |
FIG. 1.
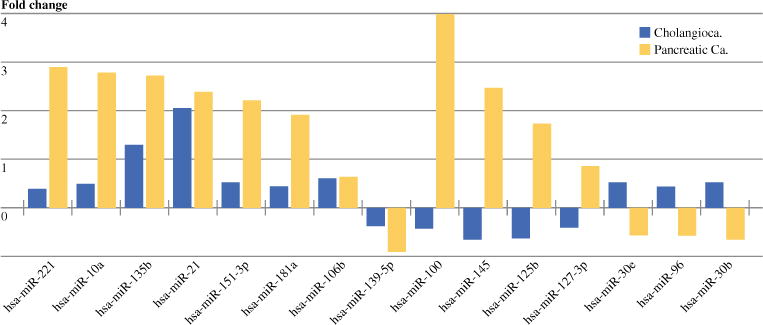
Comparison of the miR expression profiles established from cholangiocarcinoma and pancreatic adenocarcinoma versus their adjacent normal tissue
DISCUSSION
In practice, the differentiation between pancreatic cancer and cholangiocarcinoma is based solely on tumor location, as the two are virtually indistinguishable by standard histopathology. The distinction, however, is becoming more and more important as treatment algorithms diverge, particularly for advanced disease. As well, understanding the molecular differences and similarities between these two almost universally fatal diseases may prove important to understanding their pathophysiology. To our knowledge, this is the first study that attempts to identify such markers using microRNA expression profiles. We describe 15 potential miRs that are dysregulated in both tumor types, nearly half of which demonstrate opposite expression characteristics.
For this study, we used paraffin-embedded specimens for both tumor and matched benign tissue from patients with cholangiocarcinoma who had undergone surgical resection with curative intent. Given the inherent difficulty associated with RNA extraction from paraffin-embedded tissue and a limited number of tissue specimens available, only 31 pairs of cholangiocarcinoma samples had adequate RNA and appropriate controls for the final analysis. Additional RNA was available and adequate for analysis by the nanoString nCounter platform on nine pairs of pancreatic adenocarcinoma and matched benign tissue that had previously been analyzed using our fourth-generation microRNA microarray chip.19 These samples had similarly been obtained from paraffin-embedded tissue. While traditionally RNA quality has been too poor in such samples to allow meaningful conclusions to be made, miRNAs are uniquely resistant to such degradation owing mostly to their small size. Several studies have specifically addressed this issue and have shown that, although, RNA degradation is common, miRNAs are able to be accurately detected and quantified in FFPE tissue.24–26 Using the newer nCounter System from nanoString Technologies, we identified a profile of 41 significantly dysregulated miRs in cholangiocarcinoma relative to the adjacent normal bile duct epithelium. To date, an expression profile for microRNAs in cholangiocarcinoma has not been reported. Not surprisingly, miR-21 was the most overexpressed microRNA. More than any other, miR-21 has been shown in human and experimental models to play a role in cholangiocarcinoma carcinogenesis, likely through its targets PTEN and PDCD4.27–30 Using the same nanoString nCounter platform, 52 miRs were identified as being significantly dysregulated in pancreatic adenocarcinoma relative to adjacent normal tissue. Compared with our previous microRNA profile using our fourth-generation microarray chip, similar miRs were identified as differentially expressed, although fold changes varied (data not shown).19
Comparison of the two expression profiles identified 15 miRs that were significantly dysregulated in both cholangiocarcinoma and pancreatic cancers. Of these, eight miRs were similarly overexpressed or underexpressed in both, whereas the other seven miRs had inverse expression levels. While microRNA profiles have been reported for many tumor types, few studies have used these profiles to specifically differentiate cancers with similar clinical presentations. Schultz et al. used nonmicrodissected paraffin-embedded tissue (similar to our study) to compare microRNA profiles between pancreatic and ampullary adenocarcinoma.31 Similarly, Lubezky et al.32 used microRNA expression profiles to identify high-risk intraductal papillary mucinous neoplasms of the pancreas. While these studies along with ours use different profiling platforms, a recent comparison shows that expression profiles yield similar results across platforms in both fresh and paraffin-embedded tissue.33
This study reports the first microRNA expression profile for resected cholangiocarcinoma. We show that cholangiocarcinoma has a distinct microRNA profile from pancreatic adenocarcinoma. Our expression profiles may aid in further identifying key miRs, or their targets, responsible for the development of cholangiocarcinoma and pancreatic adenocarcinoma and may serve as targets for future therapies. However, given our small sample size, these findings will be strengthened by validation in a larger prospective dataset. The ability to distinguish between the two tumor types may guide physicians in treatment decisions and provide prognostic information to patients. Discrimination between these two tumor types could be possible with as few as seven miRs.
Acknowledgments
Research supported by the 2010 AACR-FNAB Fellows Grant for Translational Pancreatic Cancer Research Grant No. 10-30-14-COLL (ALC) and NCI CA13325-01 (MB).
Footnotes
CONFLICTS OF INTEREST None of the authors have any biomedical financial interests or potential conflicts of interest to disclose.
Presented at the 97th Annual Clinical Congress, American College of Surgeons, San Francisco, California 2011.
References
- 1.Francis H, Alpini G, DeMorrow S. Recent advances in the regulation of cholangiocarcinoma growth. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1–9. doi: 10.1152/ajpgi.00114.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatto M, Alvaro D. New insights on cholangiocarcinoma. World J Gastrointest Oncol. 2010;2:136–45. doi: 10.4251/wjgo.v2.i3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med. 1999;341:1368–78. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 4.Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology. 2005;128:1655–67. doi: 10.1053/j.gastro.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 5.Aljiffry M, Walsh MJ, Molinari M. Advances in diagnosis, treatment and palliation of cholangiocarcinoma: 1990–2009. World J Gastroenterol. 2009;15:4240–62. doi: 10.3748/wjg.15.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim JH, Park CK. Pathology of cholangiocarcinoma. Abdom Imaging. 2004;29:540–7. doi: 10.1007/s00261-004-0187-2. [DOI] [PubMed] [Google Scholar]
- 7.Farrow B, Berger DH, Rowley D. Tumor-derived pancreatic stellate cells promote pancreatic cancer cell invasion through release of thrombospondin-2. J Surg Res. 2009;156:155–60. doi: 10.1016/j.jss.2009.03.040. [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 9.Farley DR, Weaver AL, Nagorney DM. Natural history of unresected Cholangiocarcinoma: patient outcome after noncurative intervention. Mayo Clin Proc. 1995;70:425–9. doi: 10.4065/70.5.425. [DOI] [PubMed] [Google Scholar]
- 10.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366:1303–14. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 11.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–21. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. MicroRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med. 2005;11:712–4. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–4. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 15.Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–99. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067–74. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 17.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–6. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 19.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 20.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortina P, Surrey S. Digital mRNA profiling. Nat Biotechnol. 2008;26:293–4. doi: 10.1038/nbt0308-293. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 23.Gordon A, Glazko G, Qui X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Ann Appl Stat. 2007;1:179–90. [Google Scholar]
- 24.Weng L, Wu X, Gao H, Mu B, Li X, Wang JH, et al. MicroRNA profiling of clear cell renal cell carcinoma by whole-genome small RNA deep sequencing of paired frozen and formalin-fixed, paraffin-embedded tissue specimens. J Pathol. 2010;222:41–51. doi: 10.1002/path.2736. [DOI] [PubMed] [Google Scholar]
- 25.Ma Z, Lui WO, Fire A, Dadras SS. Profiling and discovery of novel miRNAs from formalin-fixed, paraffin-embedded melanoma and nodal specimens. J Mol Diagn. 2009;11:420–9. doi: 10.2353/jmoldx.2009.090041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng W, McElroy JP, Volinia S, Palatini J, Warner S, Ayers LW, et al. Comparison of MicroRNA deep sequencing of matched formalin-fixed paraffin-embedded and fresh frozen cancer tissues. PLoS One. 2013;8:e64393. doi: 10.1371/journal.pone.0064393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chusorn P, Namwat N, Loilome W, Techasen A, Pairojkul C, Khuntikeo N, et al. Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumour Biol. 2013;34:1579–88. doi: 10.1007/s13277-013-0688-0. [DOI] [PubMed] [Google Scholar]
- 28.Liu CZ, Liu W, Zheng Y, Su JM, Li JJ, Yu L, et al. PTEN and PDCD4 are bona fide targets of microRNA-21 in human cholangiocarcinoma. Chin Med Sci J. 2012;27:65–72. [PubMed] [Google Scholar]
- 29.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 30.He Q, Cai L, Shuai L, Li D, Wang C, Liu Y, et al. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases PTEN and PDCD4 by decreasing microRNA-21. Mol Carcinog. 2013;52:286–96. doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 31.Schultz NA, Werner J, Willenbrock H, Roslind A, Giese N, Horn T, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol. 2012;25:1609–22. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 32.Lubezky N, Loewenstein S, Ben-Haim M, Brazowski E, Marmor S, Pasmanik-Chor M, et al. MicroRNA expression signatures in intraductal papillary mucinous neoplasm of the pancreas. Surgery. 2013;153:663–72. doi: 10.1016/j.surg.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Kolbert CP, Feddersen RM, Rakhshan F, Grill DE, Simon G, Middha S, et al. Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS One. 2013;8:e52517. doi: 10.1371/journal.pone.0052517. [DOI] [PMC free article] [PubMed] [Google Scholar]


