Abstract
The enhanced neurotoxicity of the Parkinson’s disease-associated LRRK2 mutant, G2019S, than its wild-type counter-part has recently been reported. Overexpression of LRRK2 (G2019S) in cultured neural cells results in caspase-3-dependent apoptosis via a yet undefined signaling pathway. Elucidation of the mechanism underlying LRRK2 (G2019S) neurotoxicity may offer new insights into the pathogenesis of Parkinson’s disease. In this study, utilizing two-dimensional gel electrophoresis coupled with mass spectrometry, we have identified glutathione s-transferase P1 (GSTP1) as a selective target whose expression is negatively regulated at the transcriptional levels by LRRK2 (G2019S). Overexpression of LRRK2 (G2019S) in the human neuronal cell line SH-SY5Y markedly suppressed the expression of GSTP1 prior to any manifestation of cell death. This suppression of GSTP1 expression was due to LRRK2 (G2019S) – elicited production of peroxides and subsequent hyper-methylation on the promoter of GSTP1. Moreover, shRNA-mediated knockdown of endogenous GSTP1 expression exacerbated LRRK2 (G2019S) neurotoxicity, whereas overexpression of GSTP1 protected against LRRK2 (G2019S)-induced caspase-3 activation and neuronal apoptosis. In conclusion, the results suggest a previously undefined signaling mechanism underlying the neurotoxic effect of LRRK2 (G2019S), in which LRRK2 (G2019S) triggers oxidative stress in cells and, in turn, results in caspase-dependent apoptosis at least in part by suppressing the expression of GSTP1.
Keywords: LRRK2, G2019S, GSTP1, Apoptosis, Parkinson
2. Introduction
Parkinson disease (PD) is the second most common neurodegenerative disease behind Alzheimer’s disease. To date, there are over 1 million cases of PD in the United States and growing. Among them about 5% are accounted by mutations in PD related genes. Indeed, 13% of these cases can be accounted by over 20 specific mutations on leucine rich repeat kinase 2 (LRRK2)(also known as PARK8)(1–2) with G2019S as the most prevalent mutant of this gene (3–6). Unlike other PD-related genes, LRRK2 mutants such as G2019S induced clinical symptoms most similar to sporadic PD inclusive of their average age of clinical symptoms onset (3, 7–8). Therefore, understanding the physiological function of LRRK2 (wild-type and mutants) could provide the underlying cause/s of idiopathic PD. Since its identification, growing efforts has been invested in understanding the differential impact of LRRK2 wild-type and its mutants on cell viability and various cell death mechanisms.
To date, limited biochemical characterization of this mutant G2019S supports an elevated kinase activity as the cause of its toxicity (9–12) leading to an apoptotic mode of degeneration when over-expressed in dopaminergic cell line such as SHSY-5Y cells (11, 13). Despite the kinase activity dependent enhanced toxicity of G2019S as compared to its wild-type counterpart, no protein/s has been identified to account for its toxicity. From sequence and domain analysis, LRRK2 has been classified as a mixed-linage kinase (14) with the potential of utilizing stress-related signaling cascade to achieve its physiological function. Consistent to this notion, LRRK2 has been shown to regulate extracellular signal-regulated kinase (ERK) in response to oxidative stress (15). In addition, cell free in vitro studies also demonstrated the capacity for LRRK2 to activate various mitogen-activated protein kinase kinases (MAPKK), which in turn regulate p38 and JNK pathways (14, 16). So, it is likely that one of the physiological functions of LRRK2 is in maintaining the homeostasis of stress within the cells.
In this study, we used the proteomic approach comprising of two dimensional gel electrophoresis coupled to mass spectrometry to identify proteins that are responsive to the over-expression of LRRK2 wild-type and G2019S mutant respectively. From this approach, we have identified a transferase, GSTP1 (glutathione s-transferase P1), whose intracellular level appeared to be responsive to the expressions of LRRK2 wild-type and G2019S mutant in SHSY-5Y cells with a negative correlative relationship. In substantiation, G2019S over-expression also enhanced the methylation of GSTP1 promoter resulting in the depression of its expression. The lowering of GSTP1 level elicited a corresponding increase in cell death suggesting GSTP1 as a potential effector for G2019S toxicity in SHSY-5Y cells.
3. Material and Methods
3.1 Materials
The LRRK2 wild-type gene was kind gift from Dr. Matthew Farrer (Mayo Clinic, Jacksonville, FL, USA). All the chemicals are from Fisher Scientific (Fisher Scientific, Pittsburgh, PA, USA) unless otherwise stated. The lentivirus encoding GSTP1 shRNA is obtained commercially (Santa Cruz Biotech, Santa Cruz, CA).
3.2 Cell Culture
The human neuroblastoma SH-SY5Y cell line (ATCC: CRL-2266) was cultured in DMEM supplemented with 10% of Fetal Bovine Serum (Invitrogen, Carlsbad, CA, USA) at 37°C in 5% CO2 environment. The cells were sub-cultured when it reached 70–80% confluence to various configurations dictated by different experiments to ensure the cells were consistently in exponential phase.
3.3 DNA Transfection
The transfection efficiencies of plasmid encoding LRRK2 wild-type or G2019S genes were consistently lower than desired at 20% using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in SH-SY5Y cells. In order to perform cell count to determine percentage cell death, we have used co-transfection of plasmid encoding LRRK2 genes with FUEW plasmid encoding GFP. The plasmid ratio by wt: wt is 1:2 between PUEW and pcDNA3.1/LRRK2 wt/G2019S respectively. In experiments using 96 well plates, each well was seeded with 2×104 cells. In each well, 0.3 μg of total plasmid were used in the transfection process in accordance to manufacturer’s instruction. In brief, the spent media in each well was replaced with 100μl of GlutaMax media (Invitrogen, Carlsbad, CA, USA). Then, a ratio of 1:3 between DNA (μg): lipofectamine 2000 μl) was pre-mixed in 100 μl of GlutaMax medium (Invitrogen, Carlsbad, CA, USA) for 15 minutes before adding to each well. After 3 hours of incubation at 37°C/5% CO2, the transfection medium was replaced with normal growth medium and the cells were further incubate under the same conditions. Analytical assays were used after 24 – 48 hours.
3.4 Establishment of stable expression cell clones
The cDNA encoding human GSTP1 (tagged with HA at the c-terminal) was cloned into an expression vector pCDNA3.1 (Invitrogen, Carlsbad, CA, USA). The resulting plasmid was transfected into SH-SY5Y cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 hours, the tranfected cells were subcultured at the ratio of 1:3. Then, the cells were subjected to G418 selection at the final concentration 450 ng/ml 24 hours later. The transfected cells were continued to culture in the presence of G418 selection for 21 days to enable enrichment of cells expressing GSTP1. To confirm expression, transfected cells were harvested and the respective lysate probed for the presence of GSTP1 via Western blot analysis.
3.5 Sample preparation and Western Blot analysis
After treatments, cells destined for Western blot analysis were harvested and rinsed with cold phosphate buffered saline (PBS) three times consecutively before lysing with RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 1mM EDTA, 1 mM PMSF and protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA)). Afterwards, the lysates were subjected to 5 consecutive pulses of sonication, each lasting no more than 10 seconds while intermittently immersed in ice maintain the temperature of the lysates at 4°C. Then, the lysates were centrifuged at 15000xg for 10 minutes and the supernatant would contain all the soluble proteins. Finally, the protein concentrations of these samples were determined via the Bradford assay (Bio-Rad, Hercules, CA, USA).
Before each sample that was subjected to polyacrylamide gel electrophoresis (PAGE), they were normalized to the same amount of total protein. The samples were boiled for 3 minutes at 95oC before resolving by PAGE. Afterwards, the resolved protein profiles were transferred onto a PVDF membrane. Once the protein transfer was completed, the PVDF membrane was blocked in 5% low fat milk in 1x TBST solution for 1 hour. Then, the membrane was incubated overnight at 4°C in a 3% milk solution containing primary antibody diluted to the appropriate concentration. This was followed by one hour incubation in 3% milk solution containing the appropriate secondary antibody conjugated to Horse radish peroxidase (HRP). The protein of interest would be visualized by enhanced chemiluminescence (Pierce, Rockford, IL, USA).
3.6 Direct Immunofluorescence
Cells that are to be examined by immunofluorescence, they were seeded in 24 wells plates with each well containing SH-SY5Y cells. After treatments, they were fixed with 4% paraformaldeyde in 1xPBS and permeablized with 0.2% Triton x-100 for 30 minutes each. Afterwards, they were blocked with 1% BSA and 0.05% Tween-20 in 1x PBS for 1 hour at room temperature before incubated with the appropriate primary antibody overnight at 4°C. Then, they were washed three consecutive times with Wash Buffer (0.05% Tween 20 in 1x PBS) before incubated with an appropriate secondary antibody conjugated with Rodamine diluted at 1:300 (KPL Laboratories, Gaithersburg, MD, USA). These cells were then counter-stained with Hoechst 33342 (Sigma-Aldrich, St Louis, MO, USA) at the final concentration of 1 μg/ml after three consecutive washes with Wash Buffer. Finally, the immunostained cells were examined under the Leica fluorescence microscope model TCS SP5.
3.7 Cell Death Assessment
After co-transfection subsequent treatments, the percentage cell death is determined based on the ratio of the number of dying cells exhibiting apoptotic hallmarks such as nuclear condensation and fragmentation to the total cell population within the same visual field. Every sample reading was an average of three trials. In each trial, sufficient fields (typically 1–3) were captured for cell counting to ensure at least more than 100 GFP positive cells were present in the randomly chosen fields.
3.8 Two Dimensional gel electrophoresis and image analysis
To minimize noise from cells that do not express LRRK2 (wild-type or mutant), cells over-expressing these genes underwent flow cytometry (FACS caliber, BD) selection to enrich LRRK2 expressing cells. Then, the enriched cell populations were lysed in two-dimensional gel electrophoresis sample buffer (7M urea, 2M thiourea, 4% CHAPS, 65mM DTT, 0.2% IPG budder, 0.001% bromophenol blue and protease inhibitor cocktail (Sigma-Aldrich, St louis, MO, USA)) before centrifuging for 30 minutes at 15,000xg, 4°C. Then, the supernatant was removed as sample and its protein concentration ascertained via Bradford assay (Bio-Rad, Hercules, CA, USA). All the samples were normalized to the same amount of total protein (100μg) before subjected to two-dimensional gel electrophoresis. The first dimension electrophoresis (iso-electric focusing) was carried out in PROTEAN IEF Cell (Bio-Rad, Hercules, CA, USA). In brief, 100μg/100μl of each sample was applied to the 7cm IPG dry strips (pH range 3–10 Nonlinear). The loading volume was kept constant at 100μl. The rehydration and separation programs were processed as followed: 50V, 12 hours; 250V, 30 minutes; 1,000V, 1 hour; 4,000V, 3 hours and 4,000V until the total volt-hr reached 24,000. At the end of the iso-electric focusing step, the IPG strips were immediately equilibrated in Equilibration buffer I (0.375M Tris-HCl pH8.8, 6M urea, 30% glycerol, 2% SDS, 2% (w/v) DTT) for 15 minutes followed by another equilibration step in Equilibration buffer II (0.375M Tris-HCl pH 8.8, 6M urea, 30% glycerol, 2% SDS, 2.5% (w/v) iodoacetamide) for another 15 minutes. Then, the proteins in the IPG strips were further resolved by SDS poly-acrylamide gel electrophoresis (SDS PAGE) in the perpendicular direction. During SDS PAGE, the voltage was initially kept low (60V) until the dye front enters the resolving gel. Then, the voltage was increased to 100V until the dye front was at the end of the resolving gels. At the end of the electrophoresis, the gels were silver stained using the Bio-Rad silver stain kit (Bio-Rad, Hercules, CA, USA) in accordance to the manufacturer’s instruction. The stained gels were scanned with an Image Scanner (UMAX) and analyzed by the software PDQuest 7.3.1 to detect for differentially expressed protein spots.
3.9 In Gel Tryptic Digest
The protein spots that were considered significantly different in expression level were manually excised from the silver-stained gel before undergoing de-staining in the de-staining solution (15 mM potassium ferricyanide and 50 mM sodium thiosulfate (1:1 v/v)) fir 20 minutes at room temperature. After de-staining, the excised gels were washed twice with de-ionized water and shrunk by dehydration in ACN. The dried particles were rehydrated to 2μl of sodium bicarbonate in the presence of trypsin (Promega, Madison, WI, USA) at 4°C for 45 minutes in each well. Then, the samples were incubated at 37°C for 12 hours to ensure complete trypsin digestion. Afterwards, the peptide mixtures were extracted with 8ml of extraction solution (50% ACN/0.5% TFA) per well at 37°C for 1 hour before being dried under the protection of N2 gas.
3.10 MALDI-TOF MS and MS/MS analysis and database search
The peptides were spotted on the target plate after elution with 0.8 μl matrix solution (α-cyano-4-hydroxy-cinnamic acid (CHCA, Sigma, St. Louis, MO, USA) in 0.1% TFA, 50% ACN), and then air-dried and analyzed by 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems, Carlsbad, CA, USA) equipped with a 355 nm Nd:YAG laser. The proteins were identified by peptide mass fingerprinting (PMF) and tandem mass spectrometry (MS/MS) using the program MASCOT V2.1 (Matrix Science, London, UK) against NCBInr database with GPS explorer software (Applied Biosystems, Carlsbad, CA, USA). MASCOT protein scores (based on combined MS and MS/MS spectra) of greater than 61 were considered statistically significant (P<0.05).
3.11 Construction of Lentivirus encoding LRRK2 wild-type and G2019S mutant
To construct lentiviral vectors over-expressing truncated LRRK2 wild-type and G2019S mutant, these genes were inserted into the lentiviral transfer vector FSW under the control of the CMV promoter. The constructed transfer vectors were transformed into Stbl3 Escherichia coli and then isolated using the EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA). Large-scale production of the virus was performed using a protocol described previously (17). In brief, a plasmid mixture containing 435 μg of pCMV ΔR8.9 (packaging construct), 237 μg of pVSVG (envelope plasmid), and 675 μg of FSW (transfer vector) was suspended in 34.2 ml of CaCl2 (250 mM) and added volume for volume into 2× BES (2 N,N-bishydroxyethyl-2-aminoethane-sufonic acid) buffer, pH 6.95. The DNA-CaCl2 precipitate was added to human kidney 293 FT cells and allowed to incubate for 12 h before switching to fresh culture medium. The supernatant was collected 72 h after transfection, filtered through the 0.45 μm filter flask and centrifuged at 21,000 rpm for 2 h. Viruses were further purified by sucrose gradient ultracentrifuge. The pellet was suspended in 3 ml of PBS, loaded on the top of 2 ml of 20% sucrose solution, and centrifuged at 22,000 rpm for 2 h. The resulting pellet was resuspended in 200 μl of DMEM, aliquoted, and stored at −70°C. The titer of the vector stock was determined using ELISA
3.12 Real-time PCR
Total RNA was isolated from cells using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Valencia, CA), and 5 μg was used to synthesize the first strand of cDNA using random hexamer primers and the Superscript First-Strand Synthesis System for RT-PCR (invitrogen, Carlsbad, CA). PCR was performed on the Opticon 2 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using corresponding primers and SYBR gene PCR Master Mix (Applied Biosystems, Foster City, CA). The forward and reverse primers of GSTP1 were 5′-ATGACTATGTGAAGGCACTG-3′ and 5′-AGGTTCACGTACTCAGGGGA-3′, respectively. The cycle time (Ct) values of GSTP1 were first normalized with GAPDH of the same sample, and then the expression levels of GSTP1 mRNA under various experimental conditions were calculated and expressed as fold changes versus empty vector-transfection controls.
3.12 Methylation-specific PCR
The method was designed to distinguish methylated from unmethylated DNA in the promoter of genes of interests (18), based on that induced bisulfite modification in isolated DNA results in specific sequence alterations. In particular, in the bisulfite reaction, cytosines are converted to uracil, while methylated cytosines (5-methycytosine) are resistant to bisulfite reaction and remain as cytosines. To perform the assay, genomic DNA was extracted from cells using the DNeasy tissue isolation kit (Qiagen, Valencia, CA). Bisulfite modification was induced in DNA as described previously (18–19) with minor changes. In brief, DNA sample (3 μg) was denatured using 0.2 M NaOH for 10 min at 37°C, and then incubated for 20 hours at 50°C in the reaction solution (pH 5.0) containing 0.5 mM hydroquinone and 2.6 M sodium bisulfite. The post-reacted DNA was purified using DNA gel extraction kit (Qiagen, Valencia, CA) and eluted with TE buffer, treated with 0.3 M NaOH for 5 min at room temperature, and precipitated using ethanol. DNA was resuspended in sterilized water and freshly used for methylation-specific PCR. The PCR mixture contained dNTPs (200 μM of each), PCR primers (0.6 μM) (for detection of methylated DNA: forward 5′-TTCGGGGTGTAGCGGTCGTC-3′, reverse 5′-GCCCCAATACTAAATCACGACG-3′; for detection of un-methylated DNA: forward 5′-GATGTTTGGGGTGTAGTGGTTGTT-3′, reverse 5′ CCACCCCAATACTAAATCACAACA-3′), 200 ng of bisulfite-modified DNA, and 1.25 U of HotStar Taq polymerase (Qiagen, Valencia, CA). PCR was performed for 35 cycles under the following conditions: 30 seconds at 94 °C, 30 seconds at 55 °C, 30 seconds at 72 °C, and final extension at 72 °C for 8 min, which yielded products of methylated and unmethylated DNA at 91 bp and 97 bp, respectively. The amplification products were resolved on 8% polyacrylamide gels and stained with ethidium bromide.
3.13 Statistical analysis
Each experiment was performed at least three times. Values are expressed as mean ± S.E.M. Comparison of the results between two groups was analyzed by Student’s t-test. Statistical significance between multiple groups was performed using one-way analysis of variance (ANOVA). When ANOVA showed a significant difference, the post hoc Bonferroni/Dunn tests for multiple comparisons were performed. A value of p<0.05 was considered statistically significant.
4. Results
4.1 Cellular localization of truncated LRRK2 wild-type and G2019S
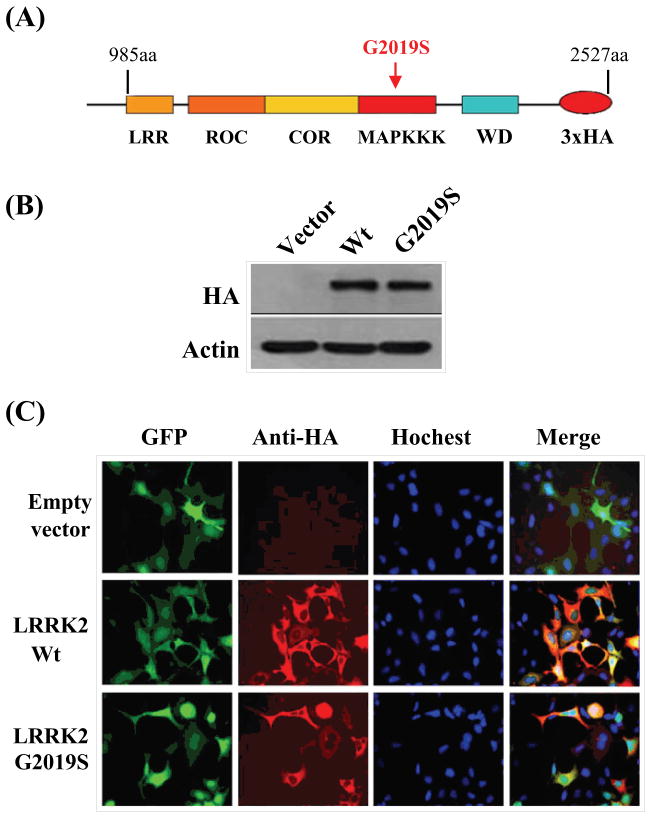
The cDNA encoding truncated wild-type LRRK2 or G2019S (985aa-2527aa) were fused with 3x-HA tag before cloning them respectively into the FUW plasmids suitable for eukaryotic expression (Fig. 1A). The truncation essentially occurred at the N-terminal where the ankryin repeats resided leaving the LRR, ROC, COR, the kinase (wild-type and mutant) and WD40 domains intact. Then each of the plasmid was transfected into SH-SY5Y cells to examine their localization profiles after 24 hours of expression. First, expressions of LRRK2 wild-type and G2019S were ascertained via Western blot analysis using antibody recognizing the HA tag (Fig. 1B). From immunocytochemistry, the cellular localization of LRRK2 wild-type and G2019S were mainly in the cytoplasm when over-expressed similar to that of GFP (merged image)(Fig. 1C).
Fig. 1.
Expression of LRRK2 mutants in neuronal SH-SY5Y cell line. (A) Schematic representation of LRRK2 wild-type and G2019S mutant used in subsequent studies; (B) Western blot analysis using anti-HA and anti-β-actin antibodies on protein extracts prepared from SH-SY5Y cells transiently transfected with plasmids encoding for the truncated versions of LRRK2 wild-type and G2019S mutant respectively; (C). Direct immunofluorescence using anti-HA antibody showing the cytoplasmic localization of LRRK2 wild-type and G2019S mutant when co-expressed with GFP in SH-SY5Y cell.
4.2 G2019S was more toxic than LRRK2 wild-type when over-expressed in SH-SY5Y cells
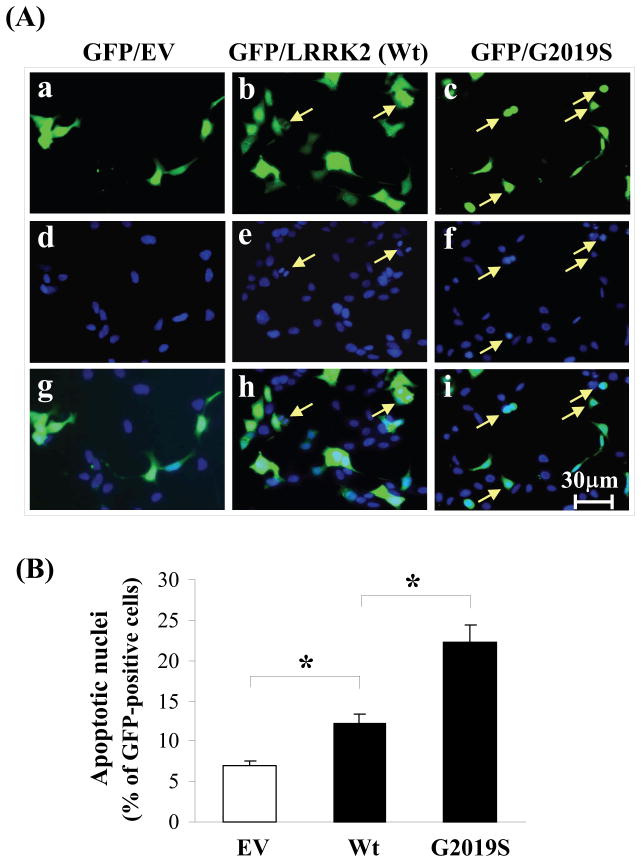
Despite the higher transfection efficiencies for the truncated wild-type LRRK2 and G2019S than full length counter-part (data not shown), the number of transfected cells were still too low for percentage cell death to be determined by cell count. Hence we adapted the strategy whereby each of the wild-type LRRK2 and G2019S was co-expressed with a plasmid encoding GFP used in other reported studies (1–2). As shown in Figure 2A, a higher number of cells exhibiting nuclear condensation and fragmentation (indicated by the arrows) for cells over-expressing G2019S than wild-type LRRK2. Moreover, cell count has quantified that SH-SY5Y cells over-expressing G2019S exhibited a 24% percentage cell death, 11% higher than those expressing wild-type LRRK2 with a 13% percentage cell death (Fig. 2B). This enhanced toxicity for G2019S observation is consistent with reported studies (11–12, 20–21). However, cells over-expressing wild-type LRRK2 also elicited a 6% higher in percentage cell death than those expressing GFP alone suggesting an innate toxicity in over-expressing wild-type LRRK2. In concert, the toxic characteristics of truncated wild-type LRRK2 and G2019S are identical with their full length counterpart.
Fig. 2.
G2019S mutant elicited higher percentage apoptosis than LRRK2 wild-type. (A). Immunofluorescence panels showing cells expressing GFP and staining with Hochest 33258 to indicate specific cell populations exhibiting condensed or fragmented neuclei (marked with arrow); (B) Quantification of apoptotic nuclei 48h after transfection. At least 100 GFP-positive cells from randomly chosen fields for each transfected constructs were counted, n=3. *p<0.01.
4.3 Expression of G2019S mutant decreased endogenous level of GSTP1
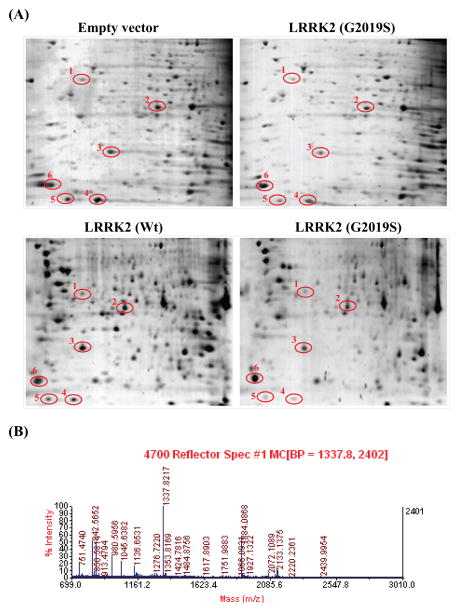
In the identification of proteins whose levels were affected by the expression of G2019S as compared to those expressing LRRK2 wild-type, we performed two- dimensional gel electrophoresis on cells expressing vector only, wild-type LRRK2 and G2019S mutant respectively for comparison. Due to the low transfection efficiencies for plasmids encoding G2019S and wild-type LRRK2, cell population expressing these LRRK2 proteins were enriched via sorting using flow cytometry. After the sorting process, the cell populations expressing LRRK2 (wild-type or mutant) increased from about 21% to 80%. In this way, the changes in the endogenous protein levels were magnified by nearly 4 fold. After comparing the differential protein profiles, spots of specific proteins whose expressions were significantly different were observed and showed (circled) in Figure 3A. Each of these spots was cut out from the resulting gel, subjected to tryptic digest and followed by MS analysis. Of all the proteins indentified, we have found that GSTP1 whose endogenous level was significant depressed by G2019S expression in SH-SY5Y cells.
Fig. 3.
Identification of proteins responsive to expression of wild-type LRRK2 and G2019S mutant respectively in SH-SY5Y cells. (A) Comparison of total proteins profiles from cells expressing vector alone, LRRK2 wild-type and G2019S mutant after subjected to two dimensional gel electrophoresis. Proteins levels responsive to LRRK2 (wild-type and mutant) expression are circled; (B) Typical mass spectrograph showing all the digested fragments of a single protein candidate (in this case, #4 spot, GSTP1).
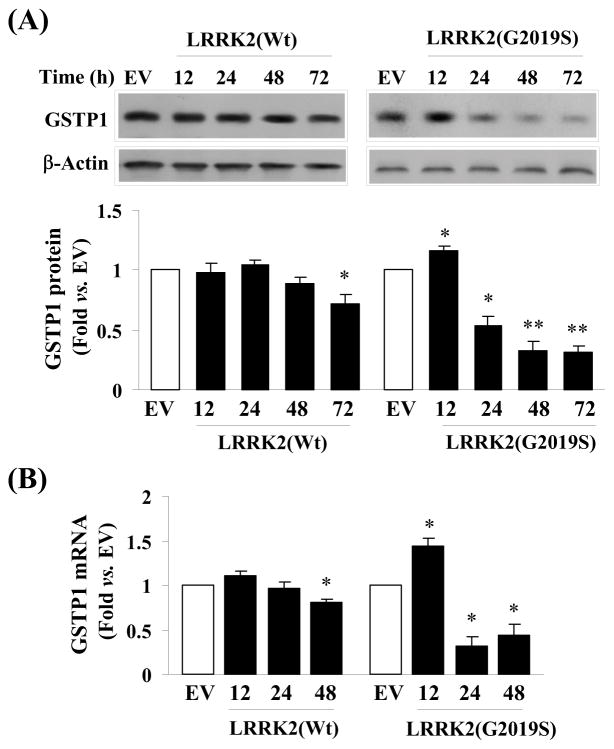
To verify this result, we charted the corresponding changes in the endogenous level of GSTP1 in cells expressing G2019S and LRRK2 wild-type over the duration of 72 hours. From Western blot analysis, we have observed a decreasing level of GSTP1 level in cells expressing G2019S beginning from 24 hours. A similar but less prominent drop in endogenous GSTP1 level was also seen in cells expressing wild-type LRRK2 (Fig. 4A). In other words, after 72 hours, a 20 and 70% drop in GSTP1 was observed in cells expressing LRRK2 wild-type and G2019S respectively. Moreover, these changes in protein levels of GSTP1 were mirrored by its mRNA level (Fig 4B) suggesting the decrease was regulated at the transcriptional level.
Fig. 4.
Expression of GSTP1 is different between cells over-expressing LRRK2 wild-type and G2019S mutant. (A) Comparative study showing cells expressing G2019S elicited a significant decrease in GSTP1 level than cells expressing LRRK2 wild-type visualized by Western blot analysis over duration of 72 hours. The changes in GSTP1 level were also quantified based on 3 experiments and averaged for ANOVA analysis. *p<0.05; **p<0.01 vs. EV; (B) Comparative study showing a corresponding significant decrease in GSTP1 mRNA in cells expressing G2019S over a duration of 48 hours than those expressing LRRK2 wild-type. Three independent studies were performed and averaged for ANOVA analysis. *p<0.05 vs. EV;
4.4 Over-expression of GSTP1 suppressed caspase-3 activation and cell death
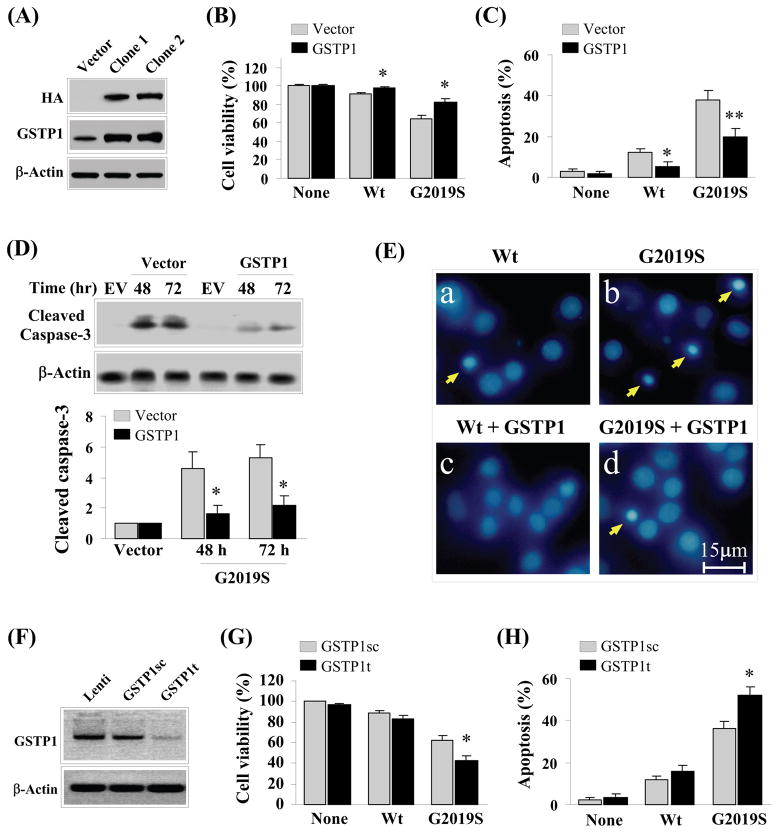
From literature, GSTP1 is a member of the glutathione transferase super-family that has been implicated in tumorigenesis and has the capacity to prevent cell death under cytotoxic insults (22–24). Under the same reasoning, it is probable that GSTP1 is functionally related to G2019S toxicity. To verify this notion, we generated two SH-SY5Y clones that constitutively over-expressed GSTP1-HA fusion protein. The exogenous expression of GSTP1 of these two clones were shown by Western blot analysis using anti-HA antibody (Fig. 6). In addition, we have also shown in the same figure that the overall endogenous levels of GSTP1 in these clones were at least 10 fold higher than control. Expression of LRRK2 wild-type and G2019S mutant in one of the clones with control showed a significant mitigation of G2019S toxicity as reflected by an increase in percentage cell viability (Fig 5B) and decrease in percentage apoptotic cells (Fig 5C). As expected, this mitigation of G2019S toxicity was also supported by a significant decrease in activated caspase-3 in cells with elevated GSTP1 within 72 hours of expressing G2019S visualized by Western blot analysis (Fig 5D) and Hoechst 33258 staining indicated by arrows showing nuclear condensation (Fig 5E). Quantitatively, the amount of cleaved caspase-3 was 2.5–4 folds lower than control (Fig. 5D) after these cells were expressing G2019S for 48 to 72 hours.
Fig. 6.
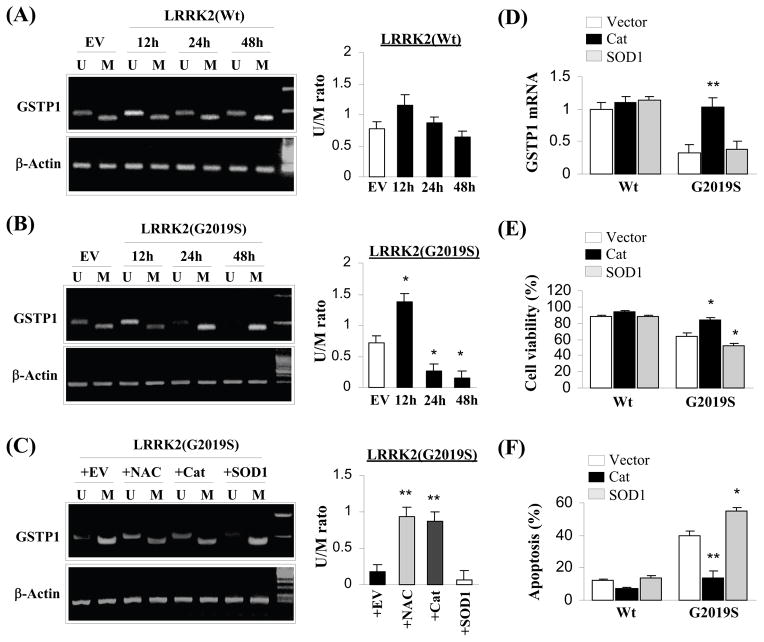
G2019S toxicity suppressed GSTP1 expression via hyper-methylation on its promoter regions. (A) Quantification of non-methylated (U) and methylated (M) promoter of GSTP1 in cells over-expressing LRRK2 wild-type by PCR visualized on an agarose gel over the duration of 48 hours. Three independent experiments were performed and the results were quantified and averaged for ANOVA analysis. No significant changes were observed; (B) Quantification of non-methylated (U) and methylated (M) promoter of GSTP1 in cells over-expressing G2019S mutant by PCR visualized on an agarose gel over the duration of 48 hours. Three independent experiments were performed and the results were quantified and averaged for ANOVA analysis. *p<0.05 vs. Empty vector (EV); (C) Quantification of non-methylated (U) and methylated (M) promoter of GSTP1 in four groups of cells over-expressing G2019S by PCR visualized on an agarose gel. Each of the four groups was pre-treated with over-expressing empty vector, N-acetyl cysteine, over-expressing catalase and over-expressing SOD1 respectively. Three independent experiments were performed and the results were quantified and averaged for ANOVA analysis. **p<0.05 vs. Empty vector (EV); (D) Cells over-expressing catalase but not SOD1 ameliorate the decrease in mRNA of GSTP1 due to G2019S toxicity. Three independent experiments were performed and averaged for ANOVA analysis. **p<0.05 vs. vector and SOD1; (E) Cells over-expressing catalase but not SOD1 attenuate cell death induced by G2019S toxicity. Three independent experiments were performed and averaged for ANOVA analysis. *p<0.05 vs. vector; (F) Cells over-expressing catalase but not SOD1 decrease the percentage of cell populations exhibiting condensed or fragmented nuclei due to G2019S toxicity. Three independent experiments were performed and averaged for ANOVA analysis. **p<0.01 vs vector, *p<0.05 vs. vector.
Fig. 5.
Alterations in GSTP1 level modulate G2019S toxicity induced cell death. (A) Western blot analysis showing the establishment of two clones with higher GSTP1 level by at least 10 fold; (B) Comparative study showing cells with higher GSTP1 exhibited higher tolerance towards G2019S toxicity. Average reading of three independent experiments was used for ANOVA analysis. *p<0.05 vs vector; (C) Comparative study indicating cells with higher GSTP1 exhibited significant lower percentage cell populations showing apoptotic characteristics. Average of three independent experiments was used for ANOVA analysis. *p<0.05; **p<0.01 vs. vector; (D) Western blot analysis visualizing cells with higher GSTP1 harbor lower caspase-3 activation over a duration of 72 hours induced by G2019S toxicity. In addition, the amount of cleaved caspase-3 was quantified and averaged over three independent experiments for ANOVA analysis. *p<0.05 vs vector; (E) Panels of Hoechst 33258 stained cells expressing LRRK2 wild-type and G2019S mutant respectively between cells with differential intracellular GSTP1 levels. Cells with higher GSTP1 showed lower number of cells with condensed or fragmented nuclei (indicated by arrows); (F) Knockdown of GSTP1 achieved by infection with lentivirus encoding GSTP1 knockdown sequence visualized by Western blot analysis. Random sequence control and lentivirus backbone were used as controls; (G) Cells with knockdown GSTP1 level exhibited lower tolerance towards G2019S toxicity. Average of three independent experiments was used for ANOVA analysis. *p<0.05 vs cells infected with lentivirus encoding random sequence control (GSTP1sc); (H) Cells with knockdown GSTP1 level exhibited higher percentage cell populations exhibiting apoptotic characteristics. Three independent experiments were performed and averaged to be used in ANOVA analysis. *p<0.05 vs. cells infected with lentivirus encoding random sequence control (GSTP1sc).
After establishing that increase in GSTP1 can ameliorate G2019S toxicity, we are interested in examining the impact of knockdown GSTP1 on G2019S toxicity. The knockdown sequence shRNA for GSTP1 as well as its sequence control packaged in lentiviral particle is available commercially. Applying these lentiviruses in SH-SY5Y cells followed by Western blot analysis showed a significant knockdown of GSTP1 can be achieved with GSTP1t (knockdown sequence) but not empty lentiviral particle and GSTP1sc (sequence control) (Fig 5F). Correspondingly, knockdown of GSTP1 by GSTP1t also elicited a decrease in percentage cell viability (Fig 5G) and an increase in percentage apoptotic cells (Fig 5H) as compared to GSTP1sc in cells expressing G2019S mutant.
4.5 G2019S down-regulate GSTP1 via promoter hyper-methylation
Typically, the expression of GSTP1 is dictated by the methylation state of its promoter (25–28). Adapting a method developed by Herman et al., 1996 to examine the methylation state of a DNA region, we determined the changes in the methylation state within the promoter of GSTP1 due to over-expression of LRRK2 wild-type and G2019S mutant and the results are shown in Fig 6A and 6B. Although the relative amount of methylation on GSTP1 promoter was somewhat increase with LRRK2 wild-type expression over time, the changes were not statistically significant. On the other hand, the same trend of changes in methylation on GSTP1 promoter was observed in cells expressing G2019S over the duration of 72 hours. In this instance, the sharp decrease was statistically significant. The increase in methylation of GSTP1 promoter corresponded well with the expected drop in the expression of this transferase (Fig 4A) implicating methylation of GSTP1 promoter depressed its expression.
In view of the function of GSTP1, a mark decrease in this enzyme is likely to result in an accumulation of peroxides, imparting oxidative stress to its host. Therefore, introduction of anti-oxidant such as N-acetyl-cysteine (NAC) or an increase in catalase to remove excess peroxides should ameliorate G2019S toxicity and probably reverse the suppression of GSTP1 expression. As speculated, introduction of NAC or catalase but not SOD1 decreased the methylation of GSTP1 promoter (Fig 6C). Concomitantly, the mRNA of GSTP1 was significantly increased due to an increase in catalase but not SOD1 (Fig 6D). In turn the physiologically impact due to the elevation in catalase can be quantified as an increase in percentage cell viability and decrease in percentage apoptosis of SH-SY5Y cells over-expressing G2019S mutant (Fig 6E and 6F).
5. Discussion
In this study, we have identified a member of the glutathione S-transferase super family called GSTP1 whose expression was suppressed due to the over-expression of G2019S resulting in the elevation of apoptotic cell death in SH-SY5Y cells. Exogenous alterations in the intracellular level of GSTP1 appear to modulate G2019S toxicity. In addition, over-expression of G2019S mediates a hyper-methylation of GSTP1 promoter resulting in the suppression its expression. On the whole, G2019S mutant regulates the downstream molecular GSTP1, which modulates its host towards G2019S toxicity.
GSTP1 has been characterized to be involved in the metabolism of a wide range of both endogenous and exogenous substrates (29–32). This enzyme, together with other members in the super family, formed the adaptive response to disturbance in the stress homeostasis by modulating the intracellular level of glutathione (GSH) (33–35). Of the six classes of GST (alpha, zeta, mu, pi, omega and theta), only GST pi has been shown to express in substantia nigrial neurons (31) and GSTP1 is a part of the GST pi sub-family. It has been reported that neuronal cells harboring mutated and inactivated GSTP1 was more susceptible to 1-methyl-1,4 phenyl 1,2,3,6 tetrahydropyridine (MPTP) (36–37). Moreover, members of the GSTP1 has also been reported to inhibit JNK signaling pathway via blocking c-Jun phosphorylation and hence apoptosis (38). In concert, these reports suggested that GSTP1 has the capacity to protect cellular demise from oxidative stress.
Consistent to this notion, elevating or knockdown of GSTP1 attenuates and acerbates cell death induced by G2019S, respectively, making it a potential downstream effector of G2019S toxicity. However, in the absence of any potential phosphorylation sites on GSTP1, it is unlikely that this protein can be a direct substrate of LRRK2 G2019S. Indeed, further examination has indicated the suppression of GSTP1 was due to the hyper-methylation of its promoter. This hyper-methylation phenomenon can be attenuated by the introduction of NAC and catalase. In other words, removing the accumulated oxidative stress within the cell can reverse the suppression of GSTP1. This also implies that the elevation in oxidative stress, probably accumulation of peroxides due to G2019S toxicity would cause the hyper-methylation phenomenon leading to the suppression of GSTP1. However, once GSTP1 suppression starts, it will further enhance the accumulation of peroxides leading to more hyper-methylation completing the viscous cycle to magnify G2019S toxicity.
From our results, it is clear that GSTP1 plays an important role in mediating cell death induced by G2019S toxicity. The capacity of G2019S toxicity to initiate a viscous cycle such as the suppression of GSTP1 expression and function demonstrates the importance of the temporal impact from G2019S toxicity. To date, it is yet uncertain whether there are other proteins whose expression is sensitive to G2019S toxicity exhibiting gradual mitigation of protective capacity such as GSTP1. Nevertheless, the time-dependent magnification of the impact due to the ever increasing suppression of GSTP1 expressing leading to neuronal demise may account for the late onset of Parkinsonian symptoms among the populations harboring LRRK2 mutations.
Acknowledgments
This project was supported by Special Research Funds from Chinese Ministry of Science & Technology to State Key laboratories (Y.G., M.J.Z., and J.C.). J.C. was supported in part by National Institutes of Health Grants NS062157 and NS059806. Y.G. was supported by the Chinese Natural Science Foundation Grants 30870794 and 30670642. We thank Pat Strickler for excellent secretary support.
References
- Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2–q13.1. Ann Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- Deng H, Le W, Guo Y, Hunter CB, Xie W, Jankovic J. Genetic and clinical identification of Parkinson’s disease patients with LRRK2 G2019S mutation. Ann Neurol. 2005;57:933–4. doi: 10.1002/ana.20510. [DOI] [PubMed] [Google Scholar]
- Goldwurm S, Di Fonzo A, Simons EJ, Rohe CF, Zini M, Canesi M, Tesei S, Zecchinelli A, Antonini A, Mariani C, Meucci N, Sacilotto G, Sironi F, Salani G, Ferreira J, Chien HF, Fabrizio E, Vanacore N, Dalla Libera A, Stocchi F, Diroma C, Lamberti P, Sampaio C, Meco G, Barbosa E, Bertoli-Avella AM, Breedveld GJ, Oostra BA, Pezzoli G, Bonifati V. The G6055A (G2019S) mutation in LRRK2 is frequent in both early and late onset Parkinson’s disease and originates from a common ancestor. J Med Genet. 2005;42:e65. doi: 10.1136/jmg.2005.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay DM, Zabetian CP, Factor SA, Nutt JG, Samii A, Griffith A, Bird TD, Kramer P, Higgins DS, Payami H. Parkinson’s disease and LRRK2: frequency of a common mutation in U.S. movement disorder clinics. Mov Disord. 2006;21:519–23. doi: 10.1002/mds.20751. [DOI] [PubMed] [Google Scholar]
- Khan NL, Jain S, Lynch JM, Pavese N, Abou-Sleiman P, Holton JL, Healy DG, Gilks WP, Sweeney MG, Ganguly M, Gibbons V, Gandhi S, Vaughan J, Eunson LH, Katzenschlager R, Gayton J, Lennox G, Revesz T, Nicholl D, Bhatia KP, Quinn N, Brooks D, Lees AJ, Davis MB, Piccini P, Singleton AB, Wood NW. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson’s disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128:2786–96. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- Goldwurm S, Zini M, Di Fonzo A, De Gaspari D, Siri C, Simons EJ, van Doeselaar M, Tesei S, Antonini A, Canesi M, Zecchinelli A, Mariani C, Meucci N, Sacilotto G, Cilia R, Isaias IU, Bonetti A, Sironi F, Ricca S, Oostra BA, Bonifati V, Pezzoli G. LRRK2 G2019S mutation and Parkinson’s disease: a clinical, neuropsychological and neuropsychiatric study in a large Italian sample. Parkinsonism Relat Disord. 2006;12:410–9. doi: 10.1016/j.parkreldis.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, Payami H, Nutt J, Maraganore DM, Czyzewski K, Styczynska M, Wszolek ZK, Farrer MJ, Toft M. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet. 2005;76:672–80. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowski JQ, Van Deerlin VM. Biochemical and pathological characterization of Lrrk2. Ann Neurol. 2006;59:315–22. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- Infante J, Rodriguez E, Combarros O, Mateo I, Fontalba A, Pascual J, Oterino A, Polo JM, Leno C, Berciano J. LRRK2 G2019S is a common mutation in Spanish patients with late-onset Parkinson’s disease. Neurosci Lett. 2006;395:224–6. doi: 10.1016/j.neulet.2005.10.083. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–7. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichling LJ, Riddle SM. Leucine-rich repeat kinase 2 mutants I2020T and G2019S exhibit altered kinase inhibitor sensitivity. Biochem Biophys Res Commun. 2009;384:255–8. doi: 10.1016/j.bbrc.2009.04.098. [DOI] [PubMed] [Google Scholar]
- Luzon-Toro B, Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson’s disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–9. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–68. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- Liou AK, Leak RK, Li L, Zigmond MJ. Wild-type LRRK2 but not its mutant attenuates stress-induced cell death via ERK pathway. Neurobiol Dis. 2008;32:116–24. doi: 10.1016/j.nbd.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LR, Toft M, Kvam SN, Farrer MJ, Aasly JO. MAPK-pathway activity, Lrrk2 G2019S, and Parkinson’s disease. J Neurosci Res. 2007;85:1288–94. doi: 10.1002/jnr.21240. [DOI] [PubMed] [Google Scholar]
- Stetler RA, Cao G, Gao Y, Zhang F, Wang S, Weng Z, Vosler P, Zhang L, Signore A, Graham SH, Chen J. Hsp27 protects against ischemic brain injury via attenuation of a novel stress-response cascade upstream of mitochondrial cell death signaling. J Neurosci. 2008;28:13038–55. doi: 10.1523/JNEUROSCI.4407-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- Anand VS, Braithwaite SP. LRRK2 in Parkinson’s disease: biochemical functions. FEBS J. 2009;276:6428–35. doi: 10.1111/j.1742-4658.2009.07341.x. [DOI] [PubMed] [Google Scholar]
- Heo HY, Park JM, Kim CH, Han BS, Kim KS, Seol W. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Exp Cell Res. 2010;316:649–56. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Dang DT, Chen F, Kohli M, Rago C, Cummins JM, Dang LH. Glutathione S-transferase pi1 promotes tumorigenicity in HCT116 human colon cancer cells. Cancer Res. 2005;65:9485–94. doi: 10.1158/0008-5472.CAN-05-1930. [DOI] [PubMed] [Google Scholar]
- Gate L, Majumdar RS, Lunk A, Tew KD. Influence of glutathione S-transferase pi and p53 expression on tumor frequency and spectrum in mice. Int J Cancer. 2005;113:29–35. doi: 10.1002/ijc.20540. [DOI] [PubMed] [Google Scholar]
- Ritchie KJ, Henderson CJ, Wang XJ, Vassieva O, Carrie D, Farmer PB, Gaskell M, Park K, Wolf CR. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Res. 2007;67:9248–57. doi: 10.1158/0008-5472.CAN-07-1764. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gomez P, Bello MJ, Arjona D, Lomas J, Alonso ME, De Campos JM, Vaquero J, Isla A, Gutierrez M, Rey JA. Promoter hypermethylation of multiple genes in astrocytic gliomas. Int J Oncol. 2003;22:601–8. [PubMed] [Google Scholar]
- Niu D, Zhang J, Ren Y, Feng H, Chen WN. HBx genotype D represses GSTP1 expression and increases the oxidative level and apoptosis in HepG2 cells. Mol Oncol. 2009;3:67–76. doi: 10.1016/j.molonc.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Qian ZR, Sano T, Asa SL, Yamada S, Kagawa N, Kudo E. Reduction of GSTP1 expression by DNA methylation correlates with clinicopathological features in pituitary adenomas. Mod Pathol. 2008;21:856–65. doi: 10.1038/modpathol.2008.60. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Chen Y, Ahsan H, Lunn RM, Chen SY, Lee PH, Chen CJ, Santella RM. Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma. Cancer Lett. 2005;221:135–43. doi: 10.1016/j.canlet.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Dourado DF, Fernandes PA, Ramos MJ. Mammalian cytosolic glutathione transferases. Curr Protein Pept Sci. 2008;9:325–37. doi: 10.2174/138920308785132677. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Henderson CJ, Wolf CR. Disruption of the glutathione transferase pi class genes. Methods Enzymol. 2005;401:116–35. doi: 10.1016/S0076-6879(05)01007-4. [DOI] [PubMed] [Google Scholar]
- Ye Z, Song H. Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur J Cancer. 2005;41:980–9. doi: 10.1016/j.ejca.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Asakura T, Sasagawa A, Takeuchi H, Shibata S, Marushima H, Mamori S, Ohkawa K. Conformational change in the active center region of GST P1-1, due to binding of a synthetic conjugate of DXR with GSH, enhanced JNK-mediated apoptosis. Apoptosis. 2007;12:1269–80. doi: 10.1007/s10495-007-0053-0. [DOI] [PubMed] [Google Scholar]
- Hegazy UM, Tars K, Hellman U, Mannervik B. Modulating catalytic activity by unnatural amino acid residues in a GSH-binding loop of GST P1-1. J Mol Biol. 2008;376:811–26. doi: 10.1016/j.jmb.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Kabler SL, Seidel A, Jacob J, Doehmer J, Morrow CS, Townsend AJ. Differential protection by human glutathione S-transferase P1 against cytotoxicity of benzo[a]pyrene, dibenzo[a,l]pyrene, or their dihydrodiol metabolites, in bi-transgenic cell lines that co-express rat versus human cytochrome P4501A1. Chem Biol Interact. 2009;179:240–6. doi: 10.1016/j.cbi.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbe LI, Di Iorio G, Markopoulou K, Athanassiadou A, Papapetropoulos S, Watts RL, Vance JM, Bonifati V, Williams TA, Spychala JR, Stenroos ES, Johnson WG. Glutathione S-transferase polymorphisms and onset age in alpha-synuclein A53T mutant Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:254–8. doi: 10.1002/ajmg.b.30450. [DOI] [PubMed] [Google Scholar]
- Smeyne M, Boyd J, Raviie Shepherd K, Jiao Y, Pond BB, Hatler M, Wolf R, Henderson C, Smeyne RJ. GSTpi expression mediates dopaminergic neuron sensitivity in experimental parkinsonism. Proc Natl Acad Sci U S A. 2007;104:1977–82. doi: 10.1073/pnas.0610978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Arifoglu P, Ronai Z, Tew KD. Glutathione S-transferase P1-1 (GSTP1-1) inhibits c-Jun N-terminal kinase (JNK1) signaling through interaction with the C terminus. J Biol Chem. 2001;276:20999–1003. doi: 10.1074/jbc.M101355200. [DOI] [PubMed] [Google Scholar]