Abstract
A gene for the Hfq protein is present in the majority of sequenced bacterial genomes. Its characteristic hexameric ring-like core structure is formed by the highly conserved N-terminal regions. In contrast, the C-terminal forms an extension, which varies in length, lacks homology, and is predicted to be unstructured. In Gram-negative bacteria, Hfq facilitates the pairing of sRNAs with their mRNA target and thus affects gene expression, either positively or negatively, and modulates sRNA degradation. In Gram-positive bacteria, its role is still poorly characterized. Numerous sRNAs have been detected in many Gram-positive bacteria, but it is not yet known whether these sRNAs act in association with Hfq. Compared with all other Hfqs, the C. difficile Hfq exhibits an unusual C-terminal sequence with 75% asparagine and glutamine residues, while the N-terminal core part is more conserved. To gain insight into the functionality of the C. difficile Hfq (Cd-Hfq) protein in processes regulated by sRNAs, we have tested the ability of Cd-Hfq to fulfill the functions of the E. coli Hfq (Ec-Hfq) by examining various functions associated with Hfq in both positive and negative controls of gene expression. We found that Cd-Hfq substitutes for most but not all of the tested functions of the Ec-Hfq protein. We also investigated the role of the C-terminal part of the Hfq proteins. We found that the C-terminal part of both Ec-Hfq and Cd-Hfq is not essential but contributes to some functions of both the E. coli and C. difficile chaperons.
Keywords: Hfq, Escherichia coli, Clostridium difficile, small noncoding RNAs, gene expression
INTRODUCTION
Hfq is a small protein (102 amino acid residues in Escherichia coli) encoded by the hfq gene. Hfq belongs to an ancient family of RNA-binding proteins that is implicated in RNA-mediated reactions in all three domains of life and plays a pivotal role in the control of gene expression (for a recent review, see Vogel and Luisi 2011). Eukaryotic and archaeal homologs of Hfq are, respectively, Sm-proteins and the closely related Sm-like (LSm) proteins. Sm proteins are arranged in a multimeric ring-like quaternary structure comprising seven different monomers. In contrast, the bacterial Hfq proteins form homohexameric rings. Hfq is a regulatory protein that has recently received much attention because of its crucial role in cellular processes controlled by small noncoding RNAs (sRNAs). Hfq facilitates the pairing of sRNAs with their target mRNAs, thereby affecting their expression either positively or negatively. Moreover, Hfq plays an important role in modulating mRNA degradation and RNA transcription (Folichon et al. 2003; Le Derout et al. 2010). Hfq is present in half of the bacterial sequenced genomes including many pathogens (Chao and Vogel 2010). Hfq was shown to act as a virulence factor in several bacterial pathogens (for review, see Hajnsdorf and Boni 2012), but it is not clear whether it can be considered as a general factor required for the virulence of all Hfq-containing pathogens.
The E. coli and Staphylococcus aureus Hfqs have been crystallized. The ring-like structure (the so-called core made up of two Sm motifs), is formed from the highly conserved N-terminal parts of Hfq molecule (aa 1–65 in E. coli). A number of amino acids in the E. coli protein have been identified as important for interaction with RNA. Three RNA-binding sites were defined by groups of residues and called the proximal (Q8, D9, F39, Y55, K56, F42), distal (Y25, I30), and the rim (R16, R19, and R17) RNA-binding surfaces (Mikulecky et al. 2004; Zhang et al. 2013). While the core of the various Hfq paralogs is rather conserved in sequence and structure (residues 7–66 in E. coli) (Fig. 1), in contrast, the C-terminal sequences have different lengths and lack homology in different species (Fig. 1). Hfq proteins of γ- and β-proteobacteria have an extended C terminus (up to 38, mainly hydrophilic, amino acids in E. coli and close relatives), whereas some Gram-positive bacteria, including Bacillus subtilis and Staphylococcus aureus, have Hfq proteins with short (<10 amino acid) C-terminal extensions (Sauter et al. 2003). The role of the C-terminal part has not been elucidated. In E. coli, it was shown that the hfq2::Ω mutation, which leaves 79 N-terminal amino acids intact out of 102 residues, does not affect its function and had no obvious phenotype (Tsui and Winkler 1994). The C-terminal domain of E. coli Hfq is dispensable for hexamer formation, a truncated form of E. coli Hfq deprived of its 19 C-terminal residues is fully able to bind to polyadenylated rpsO mRNA (Arluison et al. 2004) and sRNA DsrA (Sonnleitner et al. 2004). But more recent data show that the C-terminal domain is required for binding to long mRNAs like rpoS (Vecerek et al. 2008; Beich-Frandsen et al. 2011) and to activate GlmS expression by GlmY but not GlmZ (Salim et al. 2012). The C-terminal part possesses no identifiable motif, it is flexible and in the crystallographic structure extends laterally away from the hexameric core and exhibits features typical of intrinsically disordered proteins. Some intrinsically disordered domains have been implicated in facilitating intermolecular interactions (Babu et al. 2011). It has been suggested that a disordered C-terminal domain could participate in the interaction of Hfq with RNA molecules (Beich-Frandsen et al. 2011).
FIGURE 1.
Multiple sequence alignment of Hfq from different species. The ortholog cluster multiple alignment of the amino acids sequences of Hfq proteins of several Clostridia and various Gram-positive or Gram-negative model species was performed by using ClustalW on the website http://mbgd.genome.ad.jp. Proximal face residues important for the interaction with RNA are highlighted in gray, those important for distal face interaction and at the rim are double underlined and underlined, respectively, both in the Ec-Hfq (Mikulecky et al. 2004; Zhang et al. 2013) and Cd-Hfq. The numbering is based on Ec-Hfq. The secondary structure is indicated by boxes (α-helix) and arrows (β sheets). The Sm motifs are boxed with dotted lines. The core structured part of Ec-Hfq and Cd-Hfq are boxed with plain lines.
In Gram-negative bacteria, it is well-established that Hfq assists sRNAs in their regulatory function, while in Gram-positive bacteria, its role is still poorly characterized (Jousselin et al. 2009; Waters and Storz 2009; Chao and Vogel 2010). Although numerous sRNAs have been detected in many Gram-positive bacteria, it is not yet clear whether Hfq is implicated in sRNA network. In Listeria monocytogenes, Hfq has been shown to play a role in stress tolerance and virulence (Christiansen et al. 2004) and its inactivation does not generally affect sRNAs abundance, only three sRNAs coimmunoprecipitated with Hfq (Christiansen et al. 2006; Mandin et al. 2007; Toledo-Arana et al. 2009) with facilitation of LhrA binding to its targets (Nielsen et al. 2010, 2011). In S. aureus, Hfq is neither required for RNAIII function nor for sRNA stabilization (Boisset et al. 2007; Geissmann et al. 2009).
The role of Hfq homologs from several bacteria has been previously tested by complementation of hfq mutations in E. coli. These studies revealed that heterologous hfq genes may or may not reverse phenotypes associated with hfq deficiency in E. coli. As examples, Pseudomonas aeruginosa Hfq, which comprises only 82 amino acids, and the Moraxella catarrhalis hfq gene (210 amino acids) both functionally replace the E. coli protein (Sonnleitner et al. 2002; Attia et al. 2008), but the Hfq homologs of Synechocystis sp.PCC 6803 and Anabaena PCC 7120 (Boggild et al. 2009) are not able to replace E. coli Hfq while Staphylococcus aureus Hfq fails to substitute for Salmonella typhimurium Hfq (Rochat et al. 2012). However, the experimental conditions greatly varied in these studies regarding the expression system of the Hfq homologs as well as the functions examined.
An increasing amount of data shows that the Clostridia also contain sRNAs (Chen et al. 2011; Mraheil et al. 2011). For example, a great number and a large diversity of regulatory RNAs have been recently identified in the pathogenic clostridium Clostridium difficile (Soutourina et al. 2013). Moreover, a gene (CD1974), homologous to hfq, has been shown to be transcribed in this bacterium (I Verstraete, O Soutourina, pers. comm.). The C. difficile Hfq (Cd-Hfq) protein exhibits 46% identity (31 amino acids out of the first 66 amino acids) with Ec-Hfq. Most of the key residues that have been identified as important for interaction with RNA in the E. coli Hfq protein are present in Cd-Hfq: e.g., in the proximal binding surface (Q8, D9, F39, Y55, K56, but not F42 which is replaced by Y, also an aromatic residue), in the distal site (Y25, I30) and at the rim (R16, R19, but not R17, replaced by K and which retains the positive charge) (Mikulecky et al. 2004; Zhang et al. 2013). However, the Cd-Hfq C-terminal domain is very different; it is much shorter (16 aa) and includes an unusual stretch of seven asparagine residues, unique to this species (Fig. 1).
To investigate the functionality of the Cd-Hfq protein and its potential role in sRNA regulation of gene expression, we tested the ability of Cd-Hfq to complement various phenotypes associated with the E. coli hfq mutation. We have examined four specific functions of Hfq where the involvement of Ec-Hfq has previously been well-documented. We show that Cd-Hfq is as efficient as Ec-Hfq in the negative control of OppA expression and in the positive control of RpoS expression but with differential effects on the stability of the sRNAs involved. In addition, Cd-Hfq is as proficient as Ec-Hfq in controlling Ec-Hfq synthesis. Surprisingly, Cd-Hfq does not participate in the negative control of PtsG carried out by SgrS. In addition we show that deletion of the C-terminal extensions despite having different primary sequences similarly impact the regulatory function of the two proteins.
RESULTS
Cd-Hfq is expressed at similar levels as Ec-Hfq in E. coli
The Cd-hfq ORF was cloned in place of the Ec-hfq open reading frame of plasmid pTX381 (a low copy-number derivative of pACYC184) (Tsui et al. 1994). These two plasmids are hereafter designated as pCd-Hfq and pEc-Hfq, respectively. We also cloned the two core proteins without the C-terminal part: pEc-Hfq core and pCd-Hfq core, corresponding to the 65 and 68 N-terminal amino acids of the two proteins, respectively (Fig. 1).
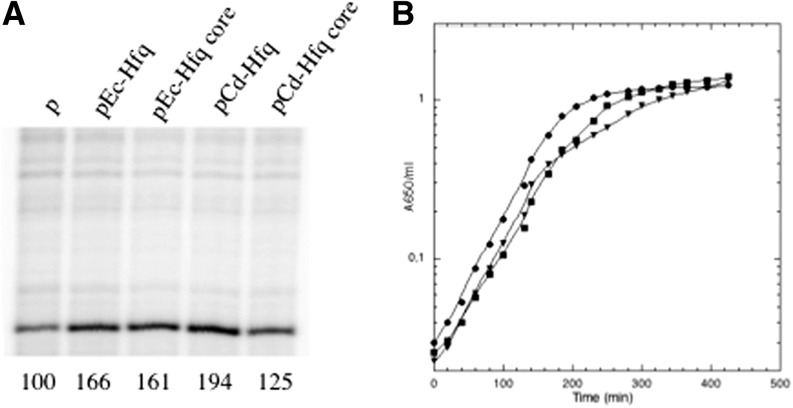
We first verified that the four Hfq proteins were expressed at comparable levels from the plasmids. All hfq variants are expressed from both P2hfq and P3hfq promoters present on the original pTX381 plasmid (Tsui et al. 1994, 1996, 1997). The relative expression levels of the different hfq variants was estimated by performing an abortive primer extension with an oligonucleotide that hybridizes to the 5′ UTR of the hfq gene present in all the constructs. This sequence is still present in the chromosomal hfq deletion and also in the hfq-lacZ chromosomal fusion in strain IBhfq95 Δhfq (Fig. 2A, lane p; Ziolkowska et al. 2006). The reverse transcript measured in the strain carrying the vector plasmid corresponds to these two transcripts (Fig. 2A). The presence of the plasmids expressing full-size and core Hfqs increases the amounts of the hfq UTR transcript less than twofold, showing that hfq is expressed from the plasmids at levels comparable to the physiological levels.
FIGURE 2.
Complementation by Hfq variants of growth phenotype and expression of Hfq. (A) Total RNA from strains IBhfq95 Δhfq transformed with the empty vector (p), pEc-Hfq, pEc-Hfq core, pCd-Hfq, or pCd-Hfq core were used as templates for reverse transcriptase primed with hfg primer, which hybridizes upstream of the hfq ORF, and carried out in abortive conditions. Quantification of the radioactivity (arbitrary units) was indicated below the gel. The transcript detected in the plasmid vector containing strain (IBhfq95 Δhfq/p) corresponds to the 5′ end of mRNA covering the hfq deletion on the chromosome and the 5′ end of the hfq-lacZ fusion present at lacZ (Ziolkowska et al. 2006); both should respond to Hfq autoregulation. Fainter bands detected on the same gel due to unspecific hybridization of the primer do not vary between samples, revealing that equivalent loading has been applied on the gel. (B) Wild-type strain carrying the empty vector (p) (▾) and the Δhfq mutant transformed with pEc-Hfq (●) and pCd-Hfq (▪) were grown in LB medium with appropriate antibiotics at 37°C. Turbidity of the cultures was monitored at 37°C.
An Hfq mutant exhibits several phenotypes compared to the wt strain. In LB liquid medium the growth of the mutant is diauxic with a decrease of growth rate and an entry into stationary phase at a lower OD than for the wt (Tsui et al. 1994). We first compared growth in LB medium and found that the hfq mutant transformed with pCd-Hfq exhibited a growth intermediary between the control strain (Δhfq/p) and the complemented strain (Δhfq/pEc-Hfq) (Fig. 2B), giving the first indication that the Cd-hfq gene is functional in E. coli. Removing the C-terminal tails had only a minimal effect on the complementation efficiency, suggesting that the C-terminal tail of Ec as well as Cd is probably dispensable in the tested conditions.
Cd-Hfq is functional for Hfq autoregulation in E. coli
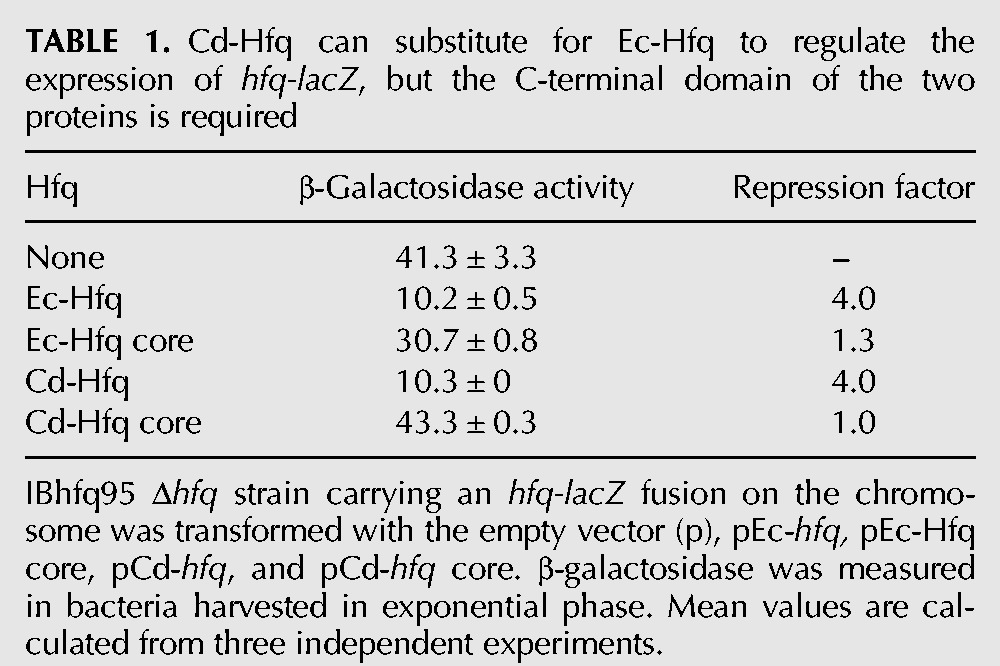
To further compare the Cd-Hfq and Ec-Hfq proteins, we examined their ability to negatively affect the expression of an hfq::lacZ fusion under the control of the P3hfq promoter. Hfq autoregulates its own expression fourfold by binding to its own mRNA, although the mechanism has not been completely elucidated (Vecerek et al. 2005; Ziolkowska et al. 2006). Table 1 shows that Ec-Hfq and Cd-Hfq exhibit the same repression factor (4.0) but that the plasmids expressing just the N-terminal cores of Ec-Hfq and Cd-Hfq were inactive in repression. This implies that the C-terminal unstructured region of the two proteins might be important for their interaction with the 5′ UTR of the hfq transcript or for the stability of the proteins.
TABLE 1.
Cd-Hfq can substitute for Ec-Hfq to regulate the expression of hfq-lacZ, but the C-terminal domain of the two proteins is required
Negative regulation of oppA expression by Cd-Hfq
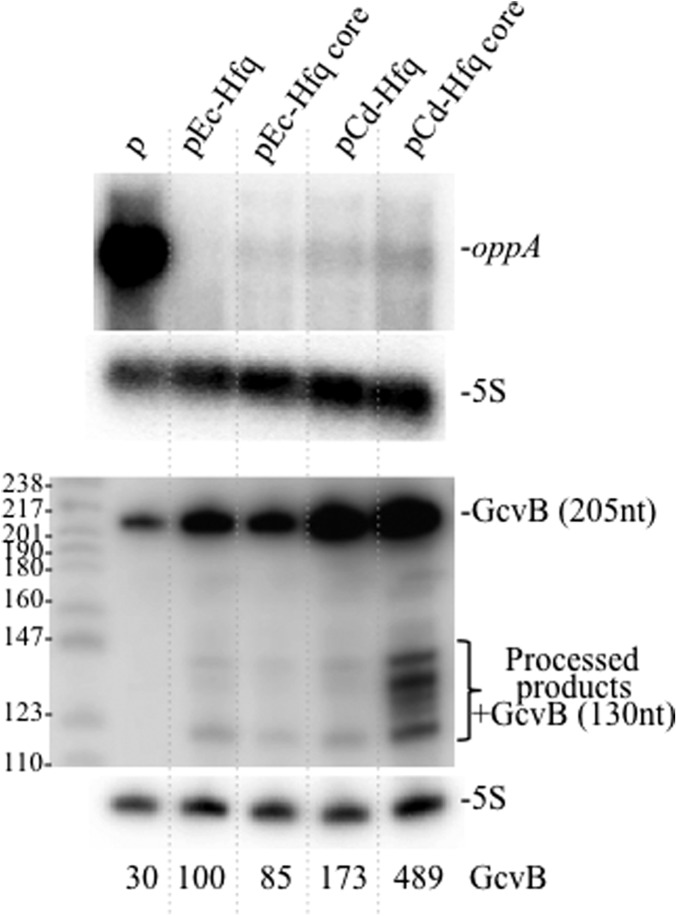
A 56-kDa protein identified as OppA, a periplasmic component of the oligopeptide transport system, was detected only in the hfq mutant (Ziolkowska et al. 2006). We took advantage of this easy assay to examine the effect of the various Hfq derivatives on oppA expression. Coomassie blue staining did not reveal any detectable band corresponding to OppA protein in the strains containing hfq genes expressed from the plasmid (data not shown). This result was confirmed by the absence of oppA mRNA transcript in the same strains indicating that all the Hfq proteins tested inhibit the synthesis of this polypeptide (Fig. 3). OppA repression is achieved by Hfq and GcvB, a conserved sRNA, which targets many ABC transporters of small peptides, amino acids, and also proteins involved in amino acid biosynthesis pathways (Sharma et al. 2007, 2011). GcvB is an Hfq-dependent sRNA, its transcription is independent on Hfq and it is unstable in the hfq mutant (Urban and Vogel 2007; Pulvermacher et al. 2009). We analyzed its level by Northern blot and found that the expression of Ec-Hfq increases its level threefold compared with the hfq mutant, and only a slightly reduced level was found in the strain with the deletion of the C terminus domain (Fig. 3). Surprisingly, GcvB levels were higher when the Cd-Hfq protein, with or without the C-terminal region, was expressed from the plasmid. Moreover, a 130-nt processed GcvB form, already observed by Urbanowski et al. (2000), and other degradation products were detected only when the Cd-Hfq core was present (Fig. 3). We conclude that Cd-Hfq is as efficient as the E. coli protein for the negative control of OppA expression and that the C-terminal domains of Ec-Hfq and Cd-Hfq are not necessary for this regulation. However, Cd-Hfq has a greater stabilizing effect on GcvB sRNA than Ec-Hfq with a role of the Cd-Hfq C-terminal domain.
FIGURE 3.
Cd-Hfq represses oppA expression. Northern blots showing the oppA transcript and GcvB sRNA detected in strain IBhfq95 Δhfq transformed with the empty vector (p), pEc-Hfq, pEc-Hfq core, pCd-Hfq, and pCd-Hfq core. Membranes were reprobed for 5S rRNA to normalize the RNA content. Relative SgrS levels after normalization are indicated below the Northern blot.
Cd-Hfq activates rpoS expression
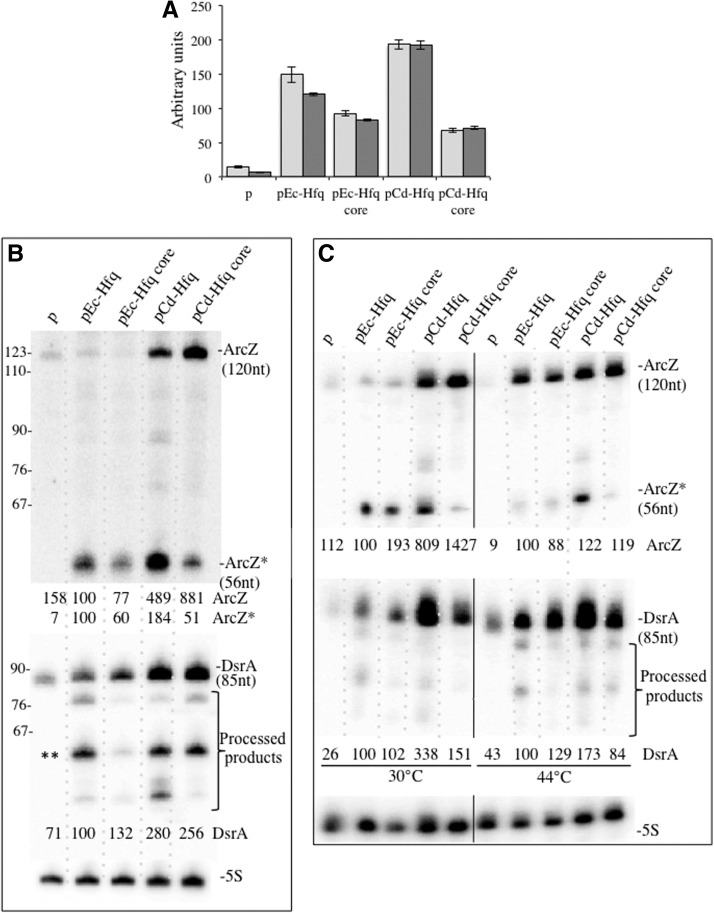
An increase in RpoS expression occurs under the conditions of nutrient deprivation, stress or at the entry into the stationary phase. This accumulation is regulated at multiple levels. Three sRNAs DsrA (Majdalani et al. 1998), RprA (Majdalani et al. 2001), and ArcZ (Mandin and Gottesman 2010) activate the translation of RpoS while OxyS represses it (Zhang et al. 1998), and all of these regulatory processes are Hfq-dependent. Therefore, rpoS appears to be an ideal target to examine the regulatory function of the various Hfq constructs. We used a translational rpoS-lacZ chromosomal fusion (Ziolkowska et al. 2006) and determined its expression in exponential growing bacteria in the presence of the various hfq-expressing plasmids (compared with the plasmid without an Hfq insert). Figure 4A shows that both Ec-Hfq and Cd-Hfq activate the expression of the fusion protein to comparable levels. Deletion of the C-terminal coding sequence of Ec-Hfq and Cd-Hfq partly reduces expression of the fusion, confirming that it is required for optimal regulation of rpoS expression (Vecerek et al. 2008; Beich-Frandsen et al. 2011). This latter result indicates that the sequence of the C-terminal extension is not crucial by itself.
FIGURE 4.
Complementation by Hfq variants of rpoS expression. (A) RO91-Δhfq (light-gray) and RO91-Δhfq arcZ mutant (dark-gray) were transformed with pACYC184 derivative plasmid expressing either wild-type or C-terminally truncated Hfq proteins from E. coli and C. difficile giving pEc-Hfq, pEc-Hfq core, pCd-Hfq, and pCd-Hfq core, respectively. The empty vector was used as a control (p). β-galactosidase activity from the rpoS-lacZ chromosomal fusion was measured in exponentially growing cells at 37°C (OD650 of 0.4). Each value is the mean of three independent experiments, with standard deviations not exceeding 15% of magnitude. (B) Northern blots showing ArcZ, ArcZ*, and DsrA in RO91-Δhfq transformed with the same plasmids (as in Fig. 3). (**) A 61 -nt long DsrA processed fragment identified in Repoila and Gottesman (2001). (C) Northern blots showing ArcZ, ArcZ*, and DsrA in IBPC 928 transformed with the same plasmids, grown at 30°C, and shifted for 15 min to 44°C to inactivate thermosensitive RNase E. Relative amounts of ArcZ and DsrA normalized to 5S RNA are indicated.
The various sRNAs that control the expression of RpoS are expressed under different stress conditions, notably in entrance to stationary phase or nutrient stress. We first concentrated on examining ArcZ since it is expressed in rich medium in exponential growth corresponding to our growth conditions and contributes to rpoS expression (Mandin and Gottesman 2010). Hfq coimmunoprecipitates with the ArcZ sRNA (Wassarman et al. 2001; Zhang et al. 2003) and ArcZ is required for RpoS activity (Papenfort et al. 2009). ArcZ (previously named as RyhA/SraH) is transcribed as a 121-nt RNA, which is subsequently processed to generate a smaller, stable RNA that consists of the last 56 nt of the original transcript (nt 66–121 of the full-length transcript, hereafter designated as ArcZ*) (Argaman et al. 2001; Papenfort et al. 2009; Mandin and Gottesman 2010), which base pairs within the rpoS mRNA leader (Mandin and Gottesman 2010). ArcZ* sRNA is abundant, while the full-length transcript is hardly detected in E. coli and S. typhimurium and the 5′ part is not detectable (Papenfort et al. 2009). Neither the full-length transcript nor the 5′ processed region were detected in S. typhimurium hfq mutant (Papenfort et al. 2009). Figure 4B shows that the full-length ArcZ sRNA is hardly detectable in the hfq mutant and in strains complemented by Ec-Hfq but accumulated in the Cd-Hfq complemented strains. Moreover, both the total amount of the two transcripts and their relative abundance varied depending on the origin of the Hfq protein. Since Hfq deficiency does not impair ArcZ transcription (Papenfort et al. 2009), our results suggest that the origin of the Hfq protein differently affected ArcZ stability and/or its processing into ArcZ*. ArcZ* levels are high in strains growing in early exponential phase complemented with either Cd-Hfq or Ec-Hfq. In both cases, higher levels of ArcZ* were detected in the presence of the full-length Hfq compared with the truncated form. Interestingly, the full-length sRNA was more abundant in the presence of Cd-Hfq and Cd-Hfq core. RNase E has been proposed to process the ArcZ sRNA (Papenfort et al. 2009). One interpretation of our results is that Cd-Hfq might be less efficient than Ec-Hfq to recruit RNase E. According to this hypothesis, thermal inactivation of RNase E increases the level of ArcZ and decreases that of ArcZ* in the presence of Ec-Hfq whereas there is no effect on the levels of both ArcZ and ArcZ* in the presence of Cd-Hfq (Fig. 4C). However, we cannot exclude that Cd-Hfq strongly interacts with ArcZ in the cell, thus decreasing its turnover. We conclude that Cd-Hfq participates in both RpoS synthesis and ArcZ sRNA stabilization, which are required for the activation of RpoS expression. In Ec as in Cd, the C-term domain has a minor role but could maximize the effects by favoring contact with rpoS mRNA as previously reported (Vecerek et al. 2008; Beich-Frandsen et al. 2011). Inactivation of ArcZ has only a faint impact on the expression of rpoS in the presence of the different Hfq variants (Fig. 4A). This is not surprising since rpoS transcription is very low in our experimental conditions and therefore its expression may be controlled by regulators that are functionally redundant. For this reason we then examined DsrA sRNA, which also contributes to rpoS expression in LB medium at 37°C (Mandin and Gottesman 2010) and is unstable in the hfq mutant when expressed from the chromosome (Sledjeski et al. 2001). DsrA is more abundant in Ec-Hfq complemented strains than in the mutant but as ArcZ, it is even more abundant in Cd-Hfq complemented strains (Fig. 4B). Both RNase E and RNase III have been shown to cleave DsrA and DsrA-rpoS duplex, respectively (Moll et al. 2003; Resch et al. 2008). The number of processed forms we detected is higher than that previously reported and their relative abundance greatly varies making it difficult to establish a link between these processing and the Hfq variant acting in the bacteria. As for GcvB and ArcZ, thermal inactivation of RNase E has no effect on the levels of these processing products while it stabilizes DsrA in Ec-Hfq but not Cd-Hfq containing cells (Fig. 4C), reinforcing our assumption that RNase E cannot interact with Cd-Hfq.
Cd-Hfq does not trigger ptsG control by SgrS
The addition of α-methylglucoside (αMG) to wild-type cells rapidly generates phosphosugar stress, resulting in induction of the SgrS sRNA (Vanderpool and Gottesman 2004), which in turn leads to destabilization of ptsG mRNA. PtsG mRNA degradation is dependent upon RNase E and Hfq in an RNase E-Hfq-SgrS ribonucleoprotein complex (Morita et al. 2003, 2004, 2006). As a consequence, an hfq mutant is unable to initiate the phosphostress response and does not grow on plates containing αMG (Fig. 5A). (Vanderpool 2007). The pCd-Hfq protein as well as the Ec-Hfq and Ec-Hfq core proteins were able to restore growth on αMG containing plates but not the Cd-Hfq core carrying strain (Fig. 5A). This indicates that Cd-Hfq deprived of its C-terminal domain cannot fully replace Ec-Hfq, and cannot properly regulate ptsG expression.
FIGURE 5.
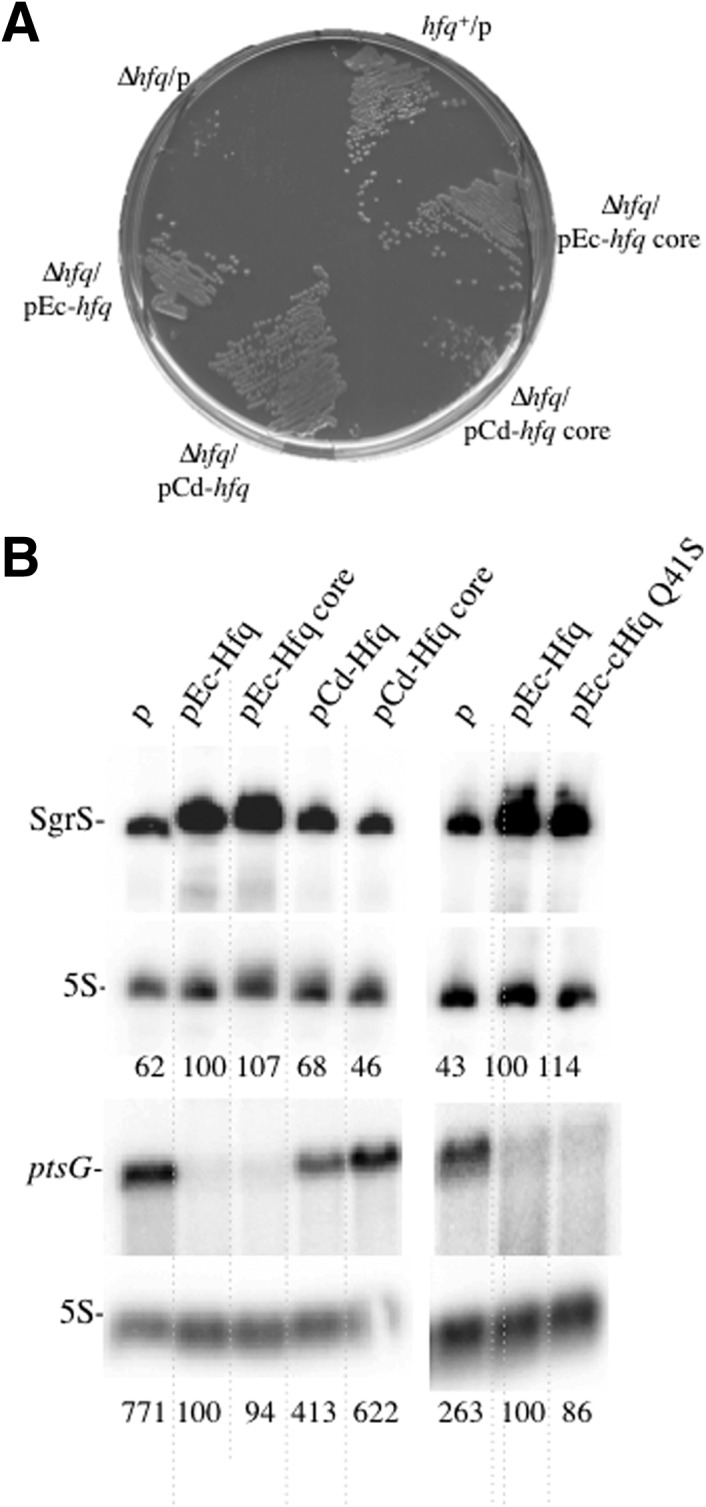
Cd-Hfq cannot substitute for Ec-Hfq in the negative control of ptsG expression. (A) RO91-Δhfq bacteria transformed with the various plasmids were grown at 37°C on LB plates containing appropriate antibiotics and 0.2% α-methylglucoside. (B) Northern blots showing SgrS and ptsG transcripts detected in strains grown in LB after addition of 1% α-MG for 20 min. The strain was also transformed with pEc-HfqQ41S.
However, strains containing Ec-Hfq and Ec-Hfq core grew more rapidly in liquid LB medium containing αMG (doubling time of 33 and 35 min, respectively), than strains with Cd-Hfq and Cd-Hfq core (generation time of 68 and 77 min, respectively) (data not shown). The addition of αMG for 15 min in liquid LB medium caused an increased synthesis of SgrS sRNA in the Ec-Hfq complemented strains compared with the control and pCd-Hfq-containing strains (Fig. 5B). Interestingly, ptsG mRNA degradation, which takes place in the strain complemented with Ec-Hfq as previously observed (Morita et al. 2005) and with Ec-Hfq core, is strongly reduced or absent in strains containing the Cd-Hfq proteins (Fig. 5B). In addition, we detected in all cases an inverse correlation between ptsG mRNA and SgrS levels. It has previously been postulated that the folded secondary structure of SgrS sRNA has to be modified by Hfq before it can anneal with the ptsG mRNA (Maki et al. 2010). The lower SgrS levels detected in strains containing Cd-Hfq constructs might mean that the C. difficile proteins inefficiently interact with SgrS sRNA in vivo and as a consequence do not provide the formation of the SgrS-Hfq-RNase E ribonucleic complex required for ptsG degradation in the presence of αMG. Our assumption on the inability of Cd-Hfq in binding to RNase E would strengthen this effect. In addition, our results indicate that for the E. coli Hfq, the C-terminal domain is not required for interaction with SgrS and/or ptsG mRNA. E. coli Hfq core protein is equally efficient as the full-length Hfq at controlling the expression of ptsG by SgrS and at promoting the degradation of the ptsG messenger carried out by RNase E.
Comparison of Ec-Hfq and Cd-Hfq RNA-binding capacity
The RNA-binding capacity of purified His-tagged Ec-Hfq and Cd-Hfq was evaluated in parallel by mobility-shift assay using SgrS, ArcZ, ArcZ*, and GcvB sRNAs and also the 5′-hfq UTR. Determination of the binding constants for half saturation (K1/2) shows that Cd-Hfq binds all RNAs but with a 13- to 240-fold higher K1/2 than Ec-Hfq (Table 2). This may be related to a difference in protein stability and/or hexamer formation of the two purified proteins. The two proteins were purified in parallel with similar yields but unlike the Ec-Hfq protein which, immediately after purification, was 50% recalcitrant to denaturation on an SDS gel, there was little hexamer (15%) detected with Cd-Hfq, (Fig. 6). There seemed to be no direct correlation between sRNA abundance in vivo and in vitro binding affinities for Cd-Hfq.
TABLE 2.
Relative interaction of Ec-Hfq and Cd-Hfq with sRNAs and 5′-UTR hfq
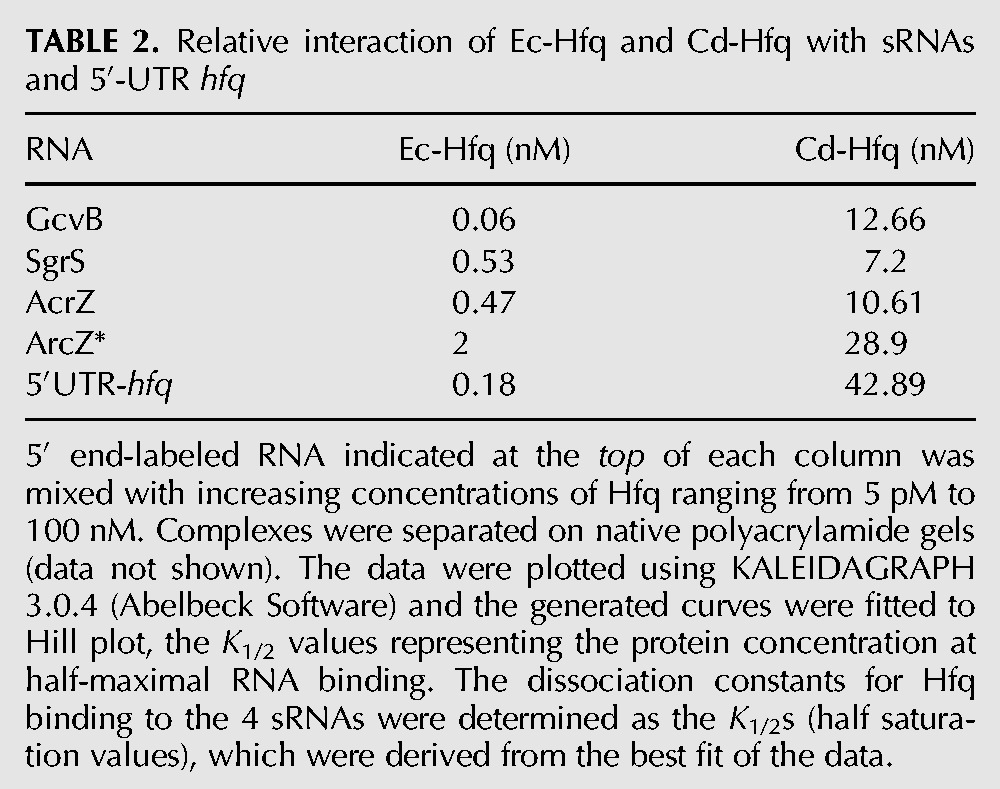
FIGURE 6.
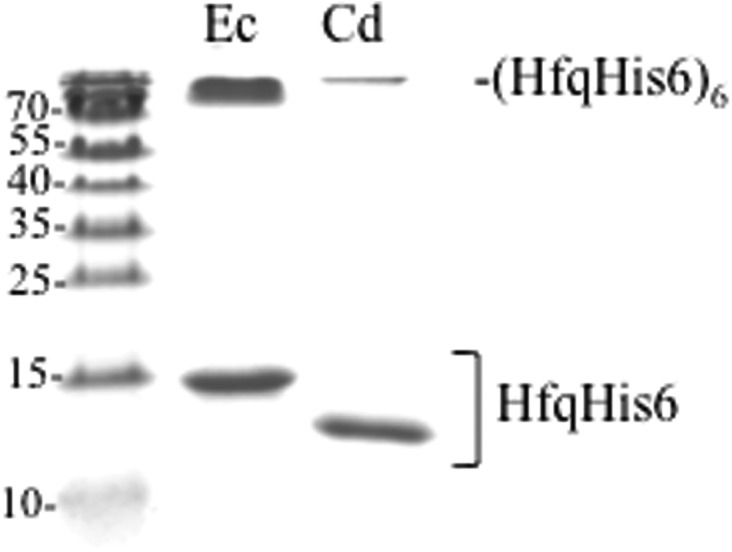
The Cd-Hfq hexamer is less stable than the EC-Hfq hexamer. Coomassie staining of an SDS–polyacrylamide gel showing Ec-Hfq and Cd-Hfq proteins after purification.
The ortholog cluster multiple alignment of the Hfq proteins of several Clostridia and various Gram-positive or Gram-negative model species (http://mbgd.genome.ad.jp) shows that there is an extra amino acid at the Sm1–Sm2 junction only in Cd-Hfq (NTV in Ec-Hfq and DNRQ in Cd-Hfq) (Fig. 1). The model of Cd-Hfq protein based on the known structure of Ec-Hfq (Protein Data Bank entry 1HK9) (Swiss-Model at http://expasy.org) shows a different orientation of the loop that could propagate along the β3 and β4 sheets and thus alter the interaction of the monomers in the hexameric form of Hfq (data not shown) and affect the stability of the Cd-Hfq hexamer.
DISCUSSION
Expression of Hfq is tightly regulated at both transcriptional and post-transcriptional levels (Tsui and Winkler 1994; Vecerek et al. 2005; Ziolkowska et al. 2006), and its availability is a limiting factor in sRNA-mediated silencing and activation of gene expression (Wagner 2013). To avoid any artifact due to overproduction, sudden induction of Hfq synthesis or formation of heterohexamers, we chose to express the Ec- and Cd-hfq genes in the hfq mutant at physiological levels under the control of the native E.coli hfq promoters and translation initiation sites. This is in contrast to previous studies where Hfq was synthesized from inducible promoters (Vecerek et al. 2008; Boggild et al. 2009; Salim et al. 2012).
We demonstrate that the Hfq protein of C. difficile can substitute for E. coli Hfq in the positive control of RpoS, the negative control of OppA expression, and in hfq autoregulation showing that it does act as an RNA chaperone, at least in E. coli for these functions. However, it is not functionally identical to Ec-Hfq, since Cd-Hfq is unable to carry out the negative control of ptsG expression. A possible explanation for this difference is discussed below.
Mutagenesis analysis has identified different conserved amino acids of Hfq implicated in regulation of mRNAs by sRNAs. These amino acids have defined three regions in the Hfq structure, two important for the interaction with sRNAs, the proximal face and lateral surface, while the proximal face is thought to contact the mRNA (Mikulecky et al. 2004; Ziolkowska et al. 2006; Sauer et al. 2012; Zhang et al. 2013). As described above, the Hfq protein of C. difficile retains most but not all the critical residues found to be essential for the function of the E. coli protein. Furthermore, the C-terminal domains are very different. By comparing both the full size and C-terminal deleted versions of Ec-Hfq and Cd-Hfq, we have also investigated whether the C-terminal region has a role in Hfq's RNA chaperon function.
We examined the role of Hfq in various sRNA-dependent gene regulations in E. coli, which implicate different binding surfaces of the protein. ArcZ regulation of rpoS expression requires Q8, F39, F42, K56, H57 on the proximal face, Y25 and I30 on the distal face, and R16 at the rim (Zhang et al. 2013). Among these essential residues only F42 is missing in Cd-Hfq but replaced by Y, another aromatic amino acid. We found that RpoS expression is fully activated in Cd-Hfq containing strains, indicating that Cd-Hfq actively cooperates with the sRNAs in charge of regulating RpoS expression. Our previous results indicated that different sites of Hfq were involved in the control of RpoS and OppA, since mutation of Ec-Hfq Valine 43 to Arginine abolished regulation of RpoS expression but did not impair OppA control (Ziolkowska et al. 2006). In Cd-Hfq V43 is replaced by an Isoleucine and in the case of Ec-Hfq a Cystein at this position is fully compatible with RpoS repression (Ziolkowska et al. 2006).
While Cd-Hfq is as efficient as Ec-Hfq in oppA repression and rpoS activation of gene expression, it has a greater impact on GcvB, DsrA, and ArcZ sRNAs stability. These three sRNAs are more abundant in Cd-Hfq than in Ec-Hfq containing cells. GcvB coimmunoprecipitates with Hfq and is unstable in its absence, but less than other sRNAs (Zhang et al. 2003, 2013; Pulvermacher et al. 2009; Busi et al. 2010). We show here that GcvB sRNA accumulates to higher levels in the Cd-Hfq containing cells than in the presence of Ec-Hfq. This indicates that Cd-Hfq has a higher protective effect against GcvB degradation than Ec-Hfq. ArcZ is one out of four sRNAs that contributes to rpoS translation (Mandin and Gottesman 2010). This sRNA is processed, probably by RNase E (Papenfort et al. 2009), to ArcZ*, which is the active form of the sRNA. Surprisingly, full-length ArcZ which is normally undetected in hfq+ cells, accumulated to high levels in the presence of Cd-Hfq and the processed form ArcZ*, and was also more abundant especially with the full-size Cd-Hfq. Thus, GcvB, ArcZ, and DsrA are inefficiently processed in the presence of Cd-Hfq. GcvB, DsrA, and ArcZ* are probably degraded by RNase E when associated with Ec-Hfq as a consequence of their mRNA base-pairing activity, as previously described (Massé et al. 2003; Morita et al. 2005). As RNase E does not exist in C. difficile, it is conceivable that Cd-Hfq does not allow recruitment of RNase E in E. coli, thus explaining the enhanced stability of certain mRNAs and small RNAs (Monot et al. 2011). In agreement, inactivation of thermosensitive RNase E that stabilizes ArcZ and DsrA in the presence of Ec-Hfq has no impact on their abundance when Cd-Hfq is substituted for Ec-Hfq (Fig. 4C), suggesting that Cd-Hfq may not be proficient in RNase E binding. In the case of oppA where the mRNA is as unstable with Cd-Hfq as with Ec-Hfq, degradation of the mRNA would be independent of the formation of the Hfq-sRNA-RNase ribonucleoprotein complex and most probably result from translational inhibition and accessibility of the messenger to the endoribonucleases. Alternatively, both GcvB and ArcZ may interact more tightly with Cd-Hfq than with Ec-Hfq, conferring a higher protection against ribonucleases.
A role for the C-terminal domain of Hfq
We show that both Ec- and Cd-Hfq proteins are able to negatively control oppA expression in the absence of the nonstructured C-terminal part of the proteins. This indicates that the two core proteins are stable and functional enough in vivo to regulate the expression of certain targets. Similarly Ec-Hfq and Ec-Hfq core are equally efficient in the control of ptsG expression. On the other hand, for other targets the C-terminal region seems to play a role with different graduations: the C-terminal domain of both Ec and Cd Hfq increases the activation of RpoS expression and is required for the autoregulation of Hfq. Those differences could be due to different affinities with the mRNAs. Despite the fact that the C-terminal regions are of different lengths and sequences, they confer similar functions that are not exhibited by the core regions alone. It could be the intrinsically disordered character of this part of the proteins that is required for the interaction with the rpoS messenger for both Hfqs.
The special case for SgrS
Two mechanisms have been proposed to account for sRNA-induced mRNA decay. In the first pathway, interruption of translation frees the messenger of ribosomes, allowing RNase E to access and cleave the mRNA (Morita et al. 2006). In the second pathway, proposed for degradation of ptsG and sodB mRNAs, formation of a sRNA-Hfq-RNase E ribonucleoproteic complex stimulates RNase E cleavage of the target mRNA (Morita et al. 2005) even at distance from the interaction site (Prevost et al. 2011). SgrS interaction with ptsG mRNA inhibits its translation promoting the rapid degradation of the SgrS–ptsG complex (Ikeda et al. 2011). The rapid turnover of ptsG mRNA requires the C-terminal scaffold region of RNase E, which interacts with Hfq as well as with Hfq bound to small RNAs (Morita et al. 2005). In addition, RNase E and Hfq bind similar sites on the RNA (Folichon et al. 2003) and base pairing of the sRNA and mRNA may allow Hfq displacement from the sRNA and access of RNase E to the duplex (Massé et al. 2003). We show here that Ec-Hfq carried out repression and degradation of ptsG transcript with no role of the C-terminal domain, indicating that the latter is not required for interacting with RNase E. Inhibition of ptsG translation and its degradation are not observed in Cd-Hfq-containing cells because in this case SgrS sRNA is not stabilized by Cd-Hfq. This situation is unique compared with the two other sRNAs we have examined here, ArcZ and GcvB, and also GlmZ (data not shown), because all of them are more stabilized by Cd-Hfq than by Ec-Hfq. SgrS sRNA was the first dual-function Hfq-dependent, sRNA regulator identified. It encodes a peptide, SgrT (Wadler and Vanderpool 2007), which is capable of inhibiting αMG uptake through a mechanism that is independent of the base-pairing function. If translation of SgrT is efficient in the presence of Cd-Hfq even when ptsG mRNA is not degraded, this may explain why Cd-Hfq-containing cells can survive in the presence of α-MG (Fig. 5A).
Similarities and differences of Ec-Hfq and Cd-Hfq
As discussed above, Ec-Hfq residues are important for ArcZ function, Q8, F39 (proximal face), Y25, and I30 (distal face) (Fig. 1; Zhang et al. 2013) are also present in Cd-Hfq, except that I30 is substituted by a Valine, which does not impact on ArcZ function in E. coli.
The proximal face of Hfq (Q8, D9, F39, F42, Y55, K56, H57) is important for SgrS function. It binds the terminator poly(U) tail. This interaction is crucial for PtsG silencing since a SgrS variant with a short poly(U) tail was inactive (Otaka et al. 2011). Q41 and F42 participate in the direct recognition of uracil (Sauer and Weichenrieder 2011). They are replaced by S41 and Y42 in Cd-Hfq. However, replacement of Q41 by S41 in Ec-Hfq neither stabilizes ptsG transcript nor destabilizes SgrS (Fig. 5). So, loss of one of the two U-contacting amino acids is not sufficient to prevent SgrS stabilization.
It has been recently demonstrated that conserved arginines on the rim of E. coli Hfq protein (RRER motif) are required for its chaperon activity. An interesting correlation has been emphasized between the number of positively charged amino acids (arginines or lysines) at the rim and the function of Hfq from Gram-positive bacteria in sRNA regulation (Panja et al. 2013). Indeed, among the Hfq proteins studied in Gram-positive bacteria, L. monocytogenes Hfq carries the RKEK rim motif and stabilizes at least one sRNA–mRNA complex (Nielsen et al. 2010, 2011), while B. subtilis Hfq having a RKEN motif associated with numerous RNAs was not required for sRNA regulation (Heidrich et al. 2007; Gaballa et al. 2008; Dambach et al. 2013). The function of S. aureus Hfq carrying the KANQ rim motif remains unclear (Bohn et al. 2007). Interestingly, the Cd-Hfq carries the most conserved RKER motif with just one amino acid substitution as compared with Ec-Hfq (Fig. 1), and thus maintains a similar surface charge with two arginines on the rim as the E. coli protein. It is interesting to correlate this conserved positive charge with the similar RNA chaperone functions of this Hfq homolog that we have demonstrated in this work.
On the other hand, there are some differences between the two Hfq proteins that may account for the reduced stability of the hexameric Cd-Hfq chaperone detected in our preparations. First, the presence of an extra amino acid at the junction of the Sm1 and Sm2 motifs in Cd-Hfq as compared with Ec-Hfq may modify the interactions of monomers in the hexameric form (Fig. 1). Second, the sequence of the C-terminal domain may also have an impact on the hexamer organization in agreement with our previous data, suggesting that in Ec-Hfq residues 84–102 protect the interface between monomers and possibly contribute to the thermodynamic stabilization of the hexameric Hfq structure (Arluison et al. 2004). Comparative studies of in vitro properties of these two proteins will be useful to compare the structure and the function of these RNA chaperons.
Perspectives
While the function of Hfq in Gram-negative bacteria is now well-documented, its role in Gram-positive bacteria is still under debate. Hfq was shown to act as a virulence factor in several bacterial pathogens but it is not at all clear how it acts or whether it cooperates with sRNAs to control physiological functions. In Gram-positive bacteria containing Hfq such as B. subtilis, S. aureus, and L. monocytogenes, the vast majority of the sRNA–mRNA duplexes characterized in vivo do not require Hfq (for review, see Brennan and Link 2007; Toledo-Arana et al. 2007; Repoila and Darfeuille 2009; Hajnsdorf and Boni 2012)). Here we show that Cd-Hfq is functional in sRNA-mediated regulation in E. coli. However, its role in C. difficile remains to be determined. Numerous potential trans-riboregulators have been identified in C. difficile by global deep sequencing (Soutourina et al. 2013). Indeed, among the 250 potential regulatory RNAs detected experimentally (with expression of 35 sRNAs confirmed by gene-specific experimental approaches), about 100 sRNAs are located in intergenic regions and might represent trans-RNA regulators. These identified RNAs could require Hfq for their regulatory action as observed in many other bacterial systems (Vogel and Luisi 2011). The growth phase-dependent expression of some of the identified sRNAs and the conservation of the majority of them within C. difficile strains strongly suggest that they have a functional role and could impact the physiopathology of this pathogen. Together with the present work showing that Cd-Hfq protein can function as an RNA chaperon, these data emphasize the potential importance of RNA-based regulatory mechanisms in C. difficile. Further studies will be required to assess the function of these new sRNA regulators as well as to identify their targets, mechanisms of action, and to determine the role of Hfq protein in related regulatory processes.
MATERIALS AND METHODS
Bacterial strains and plasmids
The strains and plasmids used in this study are listed in Supplemental Table S1 and the primers in Supplemental Table S2. New strains were constructed by P1 transduction.
The plasmids pCd-Hfq, pCd-Hfq core, pEc-Hfq core, and pEc-HfqQ41S used to complement the deletion of the hfq gene in E. coli were constructed by a two-step PCR procedure from the original pTX381 plasmid containing the miaA′-hfq-hflX′ region (Tsui et al. 1994). The first step was made of individual PCRs. The second step combined the former individual PCRs. The product of this PCR is then cleaved by restriction enzymes and recloned in the same sites of pACYC184.
- pCd-Hfq
The first step is made of three individual PCRs. The first PCR amplifies the 5′ UTR of the E. coli hfq gene (primers JC411 out and JC400 with pTX381 as the template). The second PCR allows the amplification of the Cd-Hfq ORF from the ATG to the stop codon with primers JC401 and JC414 and C. difficile chromosomal DNA as the matrix. The third PCR amplifies a sequence corresponding to 30 nt of the 3′ part of Cd Hfq ORF together with sequence of the plasmid pCA24N (Kitagawa et al. 2005) with primers JC413 and JC412out and pCA24N as the template. The second step is a PCR combining the three former PCRs with primers JC411out and JC412out.
- pCd-Hfq core construction
The first step is made of two individual PCRs. The first PCR is performed using primers JC411out and JC416 with pCd-Hfq as the template, which allows the amplification of the Cd hfq gene to the position corresponding to G68 codon with two stop codons. The second PCR is performed with primers JC415 and JC412out and pCd-Hfq DNA as the matrix. The second step combined these two PCR products with primers JC411out and JC412out.
- pEc-Hfq core
The first step is made of two individual PCRs. The first PCR is performed with primers JC411out and JC418 with pTX381 as the template. The second PCR is performed with primers JC417 and JC412out and pCA24N DNA as the matrix. We then used these two PCR products with primers JC411out and JC412out.
For these three constructions the PCR products were then cleaved by BamHI and cloned into the corresponding site of pACYC184 plasmid.
- pEc-HfqQ41S
The first two PCRs allow the amplification of Ec-Hfq in two parts with the pairs of oligos JC425–JC427 and JC426–JC428 and pTX381 as template, with JC425 and 426 harboring the Q41S mutation. The two PCR products were used with primers JC427 and JC428 in a second set of amplification. The resulting product was cleaved with HindIII and XmaI and cloned into the corresponding sites of pTX381.
- pCd-HfqHis6
The C. difficile hfq gene was amplified by PCR from C. difficile strain 630 DNA using oligonucleotides JC407 and JC408. The 280-bp amplified product was cloned into the PCRII blunt vector (Invitrogen), giving pTOPO-Cd-HfqHis6. The 263-bp NcoI–XhoI fragment of pTOPO-Cd-HfqHis6 was then cloned between corresponding sites of the pET28a expression vector downstream from the T7 promoter. The resulting plasmid pET28a-Cd-HfqHis6 allows expression in E. coli BL21 λDE3▵hfq of the C. difficile hfq protein bearing a C-terminal His-tag (Cd-HfqHis6).
β-Galactosidase assay
▵hfq derivative of RO91 containing the rpoS::lacZ chromosomal fusion (Lange and Hengge-Aronis 1994) and ▵hfq derivative of IBhfq95 containing the hfq::lacZ chromosomal fusion (Supplemental Table S1) were transformed with the pACYC184 series of hfq-expressing plasmids described above. Cells were grown at 37°C in Luria-Bertani (LB) medium with chloramphenicol. Cells were harvested in exponential phase, disrupted by sonication, and β-galactosidase activities were measured in clarified cell extracts. Specific β-galactosidase activity was expressed as nmol ONPG (o-nitrophenyl-β-D-galactopyranoside) hydrolyzed/min/mg of total soluble cell proteins (Ziolkowska et al. 2006). Protein concentrations in lysates were measured by Bradford assay (Bio-Rad). Means values are calculated from three independent experiments.
Protein purification and band-shift assay
Ec-HfqHis6 and Cd-HfqHis6 were overexpressed and purified in parallel as previously described (Ziolkowska et al. 2006). Templates for the synthesis of the different sRNAs were obtained by PCR amplification using the primers described in Supplemental Table S2. Band-shift assays were performed as in Folichon et al. (2005) with Hfq concentration expressed on the basis of the hexamer form. The radioactivity signal above the level of the free RNA was counted as retarded RNA on the gel. Data were analyzed and fitted to the Hill equation using Kaleidagraph and the K1/2 (half saturation values) were derived from the fit of the data.
RNA preparation, analysis, and labeling
Total RNA was prepared from bacteria grown to an A650= 0.35–0.4 in LB medium using the hot-phenol procedure described in Braun et al. (1996). Ten micrograms of total RNA were separated either on 1% agarose formaldehyde gel or 6% polyacrylamide gels for mRNA and sRNA analysis, respectively, and analyzed by Northern blotting (Hajnsdorf et al. 1994; Hajnsdorf and Régnier 1999). Templates for the synthesis of ArcZ, GcvB, SgrS, and oppA and ptsG RNA probes were obtained by PCR amplification using a oligonucleotide containing the T7 promoter and a RNA-specific oligonucleotide (indicated with prefix m) described in Supplemental Table S2. RNAs were synthesized by T7 RNA polymerase with [α-32P]UTP yielding uniformly labeled RNAs (Hajnsdorf and Régnier 2000). Membranes were also probed for 5S rRNA with a 5′-labeled oligonucleotide (Supplemental Table S2). RNA levels were quantified by a PhosphorImager. hfq RNA levels were determined by abortive reverse transcription using 5′-labeled hfg primer (Supplemental Table S2) as described in Hajnsdorf et al. (1995) except that reverse transcription was carried out with 0.25 mM dATP, 0.25 mM dGTP, 0.25 mM dTTP, and 0.25mM ddCTP. RNA levels were quantified using a PhosphorImager.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Centre National de la Recherche Scientifique (UPR9073; now FRE3630), University Paris-Diderot, Agence Nationale de la Recherche (asSUPYCO, ANR-12-BSV6-0007-03), and the “Initiative d'Excellence” program from the French State (Grant “DYNAMO,” ANR-11-LABX-0011). We thank P. Mandin for providing strains. We are indebted to Evelyn Sauer for comments and suggestions and O. Soutourina, I. Verstraete, and J. Plumbridge for discussions and critical reading of the manuscript. J.M. Desfontaines contributed to this work as an undergraduate student.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.043372.113.
REFERENCES
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, Hajnsdorf E, Regnier P 2004. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur J Biochem 271: 1258–1265 [DOI] [PubMed] [Google Scholar]
- Attia AS, Sedillo JL, Wang W, Liu W, Brautigam CA, Winkler W, Hansen EJ 2008. Moraxella catarrhalis expresses an unusual Hfq protein. Infect Immun 76: 2520–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, Gsponer J 2011. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol 21: 432–440 [DOI] [PubMed] [Google Scholar]
- Beich-Frandsen M, Vecerek B, Konarev PV, Sjoblom B, Kloiber K, Hammerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, et al. 2011. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res 39: 4900–4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggild A, Overgaard M, Valentin-Hansen P, Brodersen DE 2009. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J 276: 3904–3915 [DOI] [PubMed] [Google Scholar]
- Bohn C, Rigoulay C, Bouloc P 2007. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer A-C, Benito Y, Jacquier A, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F, Hajnsdorf E, Régnier P 1996. Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3′ hairpin. Mol Microbiol 19: 997–1005 [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM 2007. Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Busi F, Le Derout J, Cerciat M, Regnier P, Hajnsdorf E 2010. Is the secondary putative RNA-RNA interaction site relevant to GcvB mediated regulation of oppA mRNA in Escherichia coli? Biochimie 92: 1458–1461 [DOI] [PubMed] [Google Scholar]
- Chao Y, Vogel J 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13: 24–33 [DOI] [PubMed] [Google Scholar]
- Chen Y, Indurthi DC, Jones SW, Papoutsakis ET 2011. Small RNAs in the genus Clostridium. mBio 2: e00340–e00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol 186: 3355–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambach M, Irnov I, Winkler WC 2013. Association of RNAs with Bacillus subtilis Hfq. PLoS One 8: e55156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E 2003. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res 31: 7302–7310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M, Allemand F, Regnier P, Hajnsdorf E 2005. Stimulation of poly(A) synthesis by E. coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J 272: 454–463 [DOI] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci 105: 11927–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, et al. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37: 7239–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Boni IV 2012. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94: 1544–1553 [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E, Régnier P 1999. E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J Mol Biol 286: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E, Régnier P 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A)polymerase I. Proc Natl Acad Sci 97: 1501–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P 1994. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams-rnb-pnp mutant. EMBO J 13: 3368–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Braun F, Haugel-Nielsen J, Regnier P 1995. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci 92: 3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich N, Moll I, Brantl S 2007. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res 35: 4331–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Yagi M, Morita T, Aiba H 2011. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol 79: 419–432 [DOI] [PubMed] [Google Scholar]
- Jousselin A, Metzinger L, Felden B 2009. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol 17: 399–405 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12: 291–299 [DOI] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev 8: 1600–1612 [DOI] [PubMed] [Google Scholar]
- Le Derout J, Boni IV, Regnier P, Hajnsdorf E 2010. Hfq affects mRNA levels independently of degradation. BMC Mol Biol 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S 2001. Regulation of rpoS by a novel small RNA: the characterization of RprA. Mol Microbiol 39: 1382–1394 [DOI] [PubMed] [Google Scholar]
- Maki K, Morita T, Otaka H, Aiba H 2010. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Mol Microbiol 76: 782–792 [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29: 3094–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35: 962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11: 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9: 1308–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monot M, Boursaux-Eude C, Thibonnier M, Vallenet D, Moszer I, Medigue C, Martin-Verstraete I, Dupuy B 2011. Reannotation of the genome sequence of Clostridium difficile strain 630. J Med Microbiol 60: 1193–1199 [DOI] [PubMed] [Google Scholar]
- Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem 278: 15608–15614 [DOI] [PubMed] [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, Aiba H 2004. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol 54: 1063–1075 [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci 103: 4858–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, et al. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39: 4235–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH 2010. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res 38: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JS, Larsen MH, Lillebaek EM, Bergholz TM, Christiansen MH, Boor KJ, Wiedmann M, Kallipolitis BH 2011. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One 6: e19019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, Aiba H 2011. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci 108: 13059–13064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja S, Schu DJ, Woodson SA 2013. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res 41: 7536–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J 2009. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158 [DOI] [PubMed] [Google Scholar]
- Prevost K, Desnoyers G, Jacques JF, Lavoie F, Masse E 2011. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV 2009. Role of the Escherichia coli Hfq protein in GcvB regulation of oppA and dppA mRNAs. Microbiology 155: 115–123 [DOI] [PubMed] [Google Scholar]
- Repoila F, Darfeuille F 2009. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell 101: 117–131 [DOI] [PubMed] [Google Scholar]
- Repoila F, Gottesman S 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J Bacteriol 183: 4012–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A, Afonyushkin T, Lombo TB, McDowall KJ, Blasi U, Kaberdin VR 2008. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 14: 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Bouloc P, Yang Q, Bossi L, Figueroa-Bossi N 2012. Lack of interchangeability of Hfq-like proteins. Biochimie 94: 1554–1559 [DOI] [PubMed] [Google Scholar]
- Salim NN, Faner MA, Philip JA, Feig AL 2012. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res 40: 8021–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Weichenrieder O 2011. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci 108: 13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Schmidt S, Weichenrieder O 2012. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci 109: 9396–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D 2003. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res 31: 4091–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J 2007. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 21: 2804–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J 2011. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81: 1144–1165 [DOI] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, Zhang A 2001. HFq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol 183: 1997–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Moll I, Bläsi U 2002. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology 148: 883–891 [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Napetschnig J, Afonyushkin T, Ecker K, Vecerek B, Moll I, Kaberdin VR, Blasi U 2004. Functional effects of variants of the RNA chaperone Hfq. Biochem Biophys Res Commun 323: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppee JY, et al. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9: e1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Repoila F, Cossart P 2007. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol 10: 182–188 [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- Tsui HC, Winkler ME 1994. Transcriptional patterns of the mutL-miaA superoperon of Escherichia coli K-12 suggest a model for posttranscriptional regulation. Biochimie 76: 1168–1177 [DOI] [PubMed] [Google Scholar]
- Tsui H-CT, Leung H-CE, Winkler ME 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 13: 35–49 [DOI] [PubMed] [Google Scholar]
- Tsui H-CT, Feng G, Winkler ME 1996. Transription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K12 from clustered Eσ32-specific promoters during heat-shock. J Bacteriol 178: 5719–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H-CT, Feng G, Winkler ME 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol 179: 7476–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35: 1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV 2000. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol 37: 856–868 [DOI] [PubMed] [Google Scholar]
- Vanderpool CK 2007. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol 10: 146–151 [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54: 1076–1089 [DOI] [PubMed] [Google Scholar]
- Vecerek B, Moll I, Blasi U 2005. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA 11: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Blasi U 2008. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res 36: 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci 104: 20454–20459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG 2013. Cycling of RNAs on Hfq. RNA Biol 10: 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15: 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G 1998. The oxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-1) protein. EMBO J 17: 6061–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S 2013. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425: 3678–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska K, Derreumaux P, Folichon M, Pellegrini O, Regnier P, Boni IV, Hajnsdorf E 2006. Hfq variant with altered RNA binding functions. Nucleic Acids Res 34: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.