Abstract
Imbalanced splicing of premessenger RNA is typical of tumorous malignancies, and the regulatory mechanisms involved in several tumorigenesis-associated splicing events are identified. Elevated expression of serine-arginine protein kinase 1 (SRPK1) may participate in the pathway responsible for the dysregulation of splicing events in malignant tumor cells. In this study, we observed a correlation between the cytoplasmic accumulation of RNA-binding motif protein 4 (RBM4) and up-regulated SRPK1 in breast cancer cells. The production of the IR-B and MCL-1S transcripts was induced separately by the overexpression of RBM4 and SRPK1 gene silencing. Overexpressed RBM4 simultaneously bound to the CU-rich elements within the MCL-1 exon2 and the downstream intron, which subsequently facilitated the exclusion of the regulated exon. Breast cancer cells are deprived of apoptotic resistance through the RBM4-mediated up-regulation of the IR-B and MCL-1S transcripts. These findings suggest that the splicing events regulated by the SRPK1-RMB4 network may contribute to tumorigenesis through altered sensitivity to apoptotic signals in breast cancer cells.
Keywords: alternative splicing, breast cancer, MCL-1, RNA binding motif protein 4
INTRODUCTION
Alternative splicing constitutes a molecular mechanism participating in various processes of tumorigenesis. The alternatively spliced transcripts of Bcl-2-like 1 (BCL2L1) and myeloid cell leukemia 1 (MCL-1) genes encode either the anti-apoptotic or pro-apoptotic isoforms that manipulate the therapeutic response of cancer cells (Moore et al. 2010; Gautrey and Tyson-Capper 2012). The inclusion of cyclin D intron 4 leads to the production of the cyclin D1b isoform which acts as an oncogene in prostate cancer cells (Burd et al. 2006; Olshavsky et al. 2010). An increase in the short S6K1 variant facilitates the transformation of breast epithelial cells, whereas the presence of the long S6K1 isoform mediates the opposite effect by inhibiting Ras-induced transformation and tumor growth (Ben-Hur et al. 2013). The loss of the survivin 2B isoform enhances the cell proliferation of breast cancer cells (Végran et al. 2011). These results demonstrate the functional influence of imbalanced splicing events on tumorigenic processes.
MCL-1 is a member of the Bcl-2 family, the elevated level of which was first reported in differentiating myeloid leukemia cells (Kozopas et al. 1993). The full-length transcript of the MCL-1 gene encodes the MCL-1L isoform which contains the Bcl-2 homology (BH) domains 1, 2, and 3. The associations between MCL-1L and other Bcl-2 family members maintain cell viability through an anti-apoptotic mechanism (Kozopas et al. 1993). An exon 2-skipping variant, MCL-1S, encodes the BH3 domain-only protein which was identified in the human placenta (Bae et al. 2000; Bingle et al. 2000). Dimerization of the MCL-1L and MCL-1S isoforms acts as a pro-apoptotic factor by antagonizing the effect of the MCL-1L-mediated inhibition of apoptosis (Bae et al. 2000). The elevated expression of the SRSF1, SRSF3, and SRSF5 genes has been demonstrated to exert an oncogenic effect through increases in the level of the MCL-1L isoform, which results in the anti-apoptotic response of MCF-7 and MDA-MB-231 breast cancer cells to cytotoxic agents (Gautrey and Tyson-Capper 2012). However, the detailed mechanism underlying the splicing profile of MCL-1 pre-mRNA in malignant tissues is unclear.
Although the molecular mechanism responsible for tumor-related alternative splicing events is not comprehensively understood, changes in the expression level of splicing factors correlate with alternative splicing events in malignant tumor cells (Cohen-Eliav et al. 2013; Iborra et al. 2013). Serine-arginine (SR) proteins comprise a family of splicing factors that exert influence on the utilization of the splice site by interacting with the cis-elements of candidate pre-mRNAs and trans-splicing factors (Wang et al. 2005). SR protein kinase 1 (SRPK1) induces the phosphorylation of numerous SR proteins, including SRp30, 40, 75, and SRSF1 (previously designated as ASF) (Manley and Krainer 2010; Zhou and Fu 2013). The Akt-SRPK1 pathway drives the translocation of phosphorylated SRSF1 from the cytoplasm to the nucleus, which substantially affects its influence on the splicing pattern of E1A pre-mRNA (Ngo et al. 2005; Zhou et al. 2012; Wang et al. 2014). SRPK1 is anchored in the cytoplasm by interacting with chaperones but translocates into the nucleus under osmotic stress and EGF treatment (Zhong et al. 2009; Zhou et al. 2012). The presence of nuclear SRPK1 induces the cytoplasmic accumulation of SR proteins and subsequently alters downstream splicing events (Aubol et al. 2013). Knocking down SRPK1 expression by using small interfering RNA (siRNA) results in the up-regulation of pro-apoptotic proteins, which subsequently sensitizes breast, colorectal, and pancreas tumor cells to apoptosis (Hayes et al. 2006, 2007). Thus, the expression level of SRPK1 may constitute a molecular switch in manipulating the tumorigenic process.
In this study, the relatively high levels of SRPK1 and RBM4 proteins were widely observed in human breast cancer tissues and breast cancer cells compared to noncancerous tissues and cells. The influence of SRPK1 reduction on the subcellular distribution of endogenous RBM4 was evaluated in MCF-7 cells. The effect of SRPK1 knockdown and RBM4 overexpression on breast cancer-associated splicing events, such as the insulin receptor (IR) and MCL-1 splice variants, and related cellular processes were also examined. Moreover, the RBM4-mediated mechanism involved in regulating the splicing profile of MCL-1 pre-mRNA was identified by adopting multiple approaches. The results indicate that the SRPK1-RBM4 network regulates splicing events that subsequently manipulate the apoptotic resistance of breast cancer cells toward a cytotoxic agent.
RESULTS
Expression of RBM4 and SRPK1 in human breast cancer tissues and cell lines
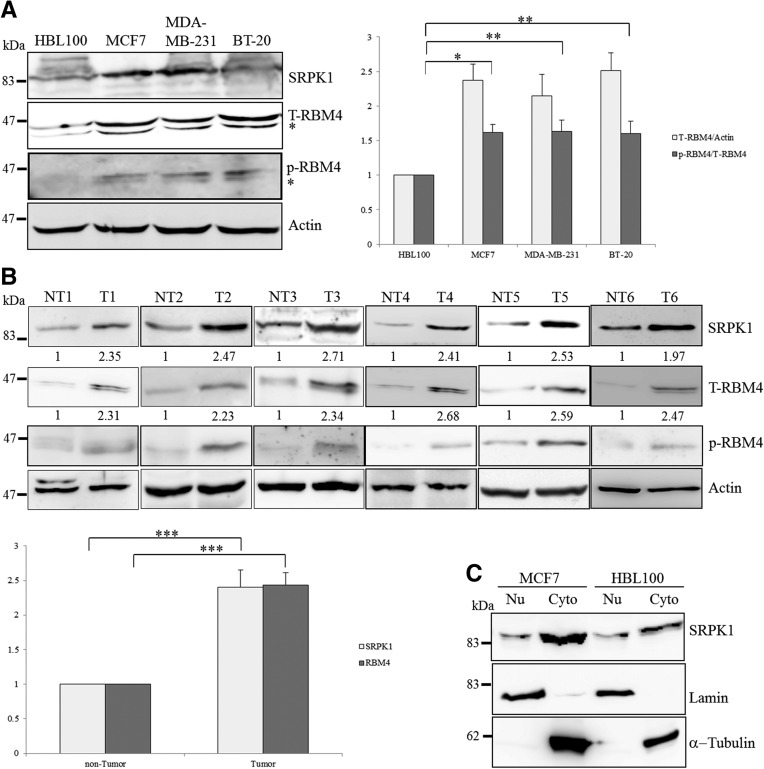
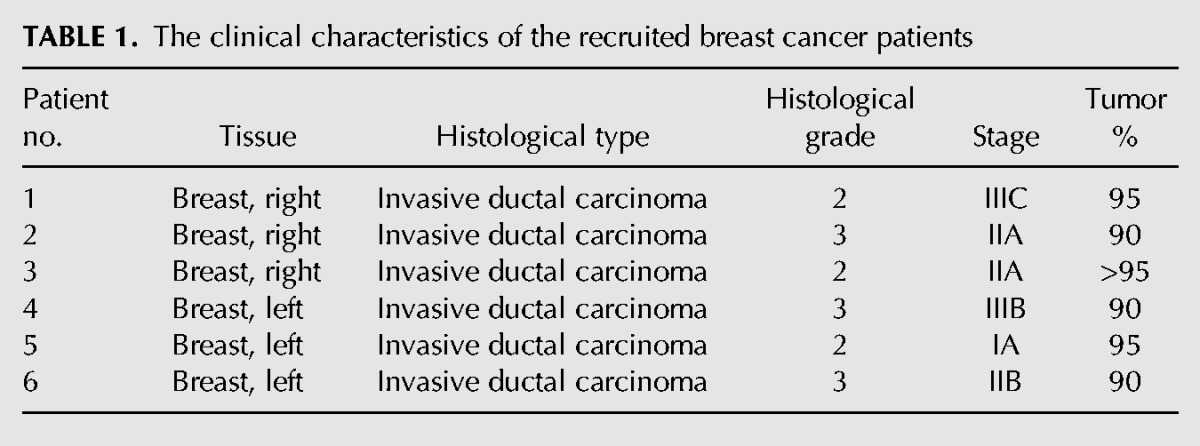
The up-regulation of SRPK1 has been widely reported in colorectal, pancreatic, and breast cancer (Hayes et al. 2007). In addition to SRPK1, the immunoblotting analysis also revealed relatively high levels of RBM4 expression in various types of breast cancer cells, compared with that in the noncancerous breast epithelial cells, HBL100 (Fig. 1A). The use of the anti-phospho Ser309 antibody indicated the elevated phosphorylation of RBM4 in breast cancer cell lines (Fig. 1A, p-RBM4). Higher levels of the SRPK1, total RBM4, and phosphorylated RBM4 proteins were also observed in the total extract prepared from the arbitrary breast cancer tissues, compared with those in the adjacent noncancerous tissues (n = 6) (Fig. 1B). The clinical characteristics of the breast cancer patients from whom the tissue samples were collected are listed in Table 1. Although the nuclear translocation of SRPK1 can be triggered by osmotic stress and EGF treatment (Zhong et al. 2009; Zhou et al. 2012), the majority of SRPK1 protein was enriched in the cytoplasm of MCF-7 cells, and similar results were observed in HBL100 cells (Fig. 1C). The subcellular fractionation analysis consistently showed the relatively high level of total SRPK1 in the MCF-7 cells compared to the HBL100 cells.
FIGURE 1.
Relatively high level of SRPK1 and RBM4 proteins in breast cancer tissues and cell lines. (A) The cell lysates isolated from cultured cells (MCF-7 and HBL100) and commercially produced cell lysates (MDA-MB-231 and BT-20; Santa Cruz Biotechnology) were analyzed by using an immunoblotting assay with specific primary antibodies as indicated. The bar graph shows the relative level of total RBM4 to actin (light gray) and phosphorylated RBM4 to total RBM4 (dark gray) in three independent experiments. (*) P < 0.05, (**) P < 0.01. (B) The protein extracts were prepared from human breast cancer tissues (T) and the adjacent noncancerous tissues (NT; n = 6) and analyzed by using immunoblotting analysis with specific primary antibodies. The bar graph presents the relative level of SRPK1 and total RBM4 in breast cancer tissues. (***) P < 0.005. (C) The subcellular fractions of HBL100 and MCF-7 cells were extracted and analyzed using an immunoblotting assay with primary antibodies against the endogenous SRPK1, lamin, and α-tubulin proteins.
TABLE 1.
The clinical characteristics of the recruited breast cancer patients
Elevated SRPK1 induces the cytoplasmic localization of RBM4 in MCF7 cells
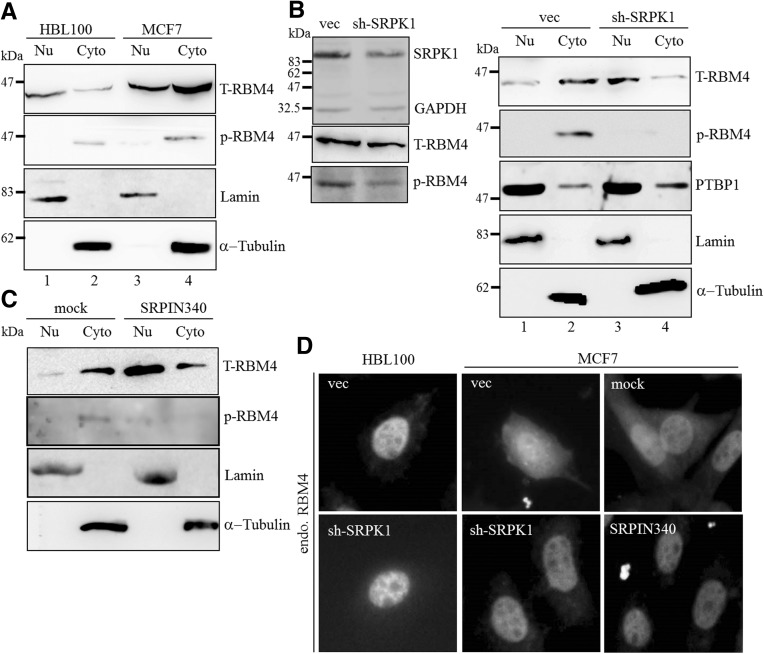
SRPK1 was demonstrated to phosphorylate SRSF1 at the N-terminal part of the RS domain by binding to its docking motif, which subsequently induces the cytoplasmic accumulation of SRSF1 (Ngo et al. 2005). SRPK1-mediated phosphorylation and translocation of SRSF1 abolishes its effect on the selection of the proximal 5′ splice site in the E1A minigene (Zhou et al. 2012). The SRPK1-mediated phosphorylation of RBM4 at Ser309 causes the cytoplasmic accumulation of RBM4 in cells subjected to oxidative stress (Lin et al. 2007). Therefore, we suspected that the increase in SRPK1 expression influences the cellular localization of RBM4 in breast cancer cells. The subcellular fractionation analysis showed that more than half of RBM4 was localized in the nuclear fraction of HBL100 cells (Fig. 2A, lane 1), whereas most RBM4 proteins were present in the cytoplasmic fraction from the MCF-7 cells (Fig. 2A, lane 4). Higher levels of phosphorylated RBM4 were present in the cytoplasmic fraction of the MCF-7 cells, compared to that observed in the HBL100 cells (Fig. 2A, p-RBM4). The siRNA-interference of SRPK1 expression substantially reduced the phosphorylation of RBM4 with a concomitant decline in endogenous SRPK1 (Fig. 2B, left row). The subcellular fractionation indicated that SRPK1 silencing restored the nuclear distribution of RBM4 (Fig. 2B, right row, lane 3), whereas the cellular distribution of polypyrimidine tract-binding protein 1 (PTBP1) was not affected by siRNA-mediated SRPK1 knockdown. Phosphorylated RBM4 was only detected in the cytoplasm of control cells transfected with the empty pSilencer1.0 U6 plasmid (Fig. 2B, p-RBM4). Serine-309 of RBM4 has been proven to be the major site for SRPK1-mediated phosphorylation both in vivo and in vitro (Lin et al. 2007). The insignificant effect of SRPK1 overexpression on the subcellular distribution of FLAG-tagged S309A was consistent with such findings (Supplemental Fig. S1). Treatment with the SRPK1 inhibitor SRPIN340 restored the nuclear localization of RBM4 (Fig. 2C, T-RBM4) and reduced the level of phosphorylated RBM4 in the MCF-7 cells. The indirect immunostaining assay consistently indicated the level of RBM4 protein in the nuclear fraction of the majority of the MCF-7 cells treated with SRPIN340 or transfected with the sh-SRPK1 expression plasmid (Fig. 2D, sh-SRPK1 and SRPIN340), compared to those transfected with the empty pSilencer1.0 U6 plasmid and the mock-treated MCF-7 cells. The silencing of SRPK1 expression exerted no substantial effect on the nuclear localization of endogenous RBM4 in the HBL100 cells. These results demonstrated the correlation between the elevated SRPK1 expression and the Ser309-phosphorylation-dependent cytoplasmic accumulation of RBM4 in MCF-7 cells.
FIGURE 2.
SRPK1 modulates the cellular localization of RBM4 in MCF7 cells. (A) Subcellular fractions were prepared from HBL100 and MCF-7 cells and analyzed using an immunoblotting assay with primary antibodies against RBM4, phosphorylated RBM4, lamin, and α-tubulin. (B) MCF-7 cells were transfected with the shSRPK1 expression plasmid or the empty shRNA expression vector for 24 h. The whole-cell lysate (left) and subcellular fractions (right) were analyzed using an immunoblotting assay with the indicated primary antibodies. (C) Subcellular fractions prepared from mock- and SRPIN340-treated MCF7 cells (10 µM, 48 h) were analyzed by using an immunoblotting assay with the primary antibodies as described for the previous panels. (D) MCF-7 and HBL100 cells were transiently transfected with the shSRPK1 expression plasmid or the empty shRNA vector or treated with SRPIN340 and analyzed by using indirect immunofluorescence with an anti-RBM4 antibody.
Splicing of IR and MCL-1 pre-mRNA is modulated by the SRPK1-RBM4 network in breast cancer cells
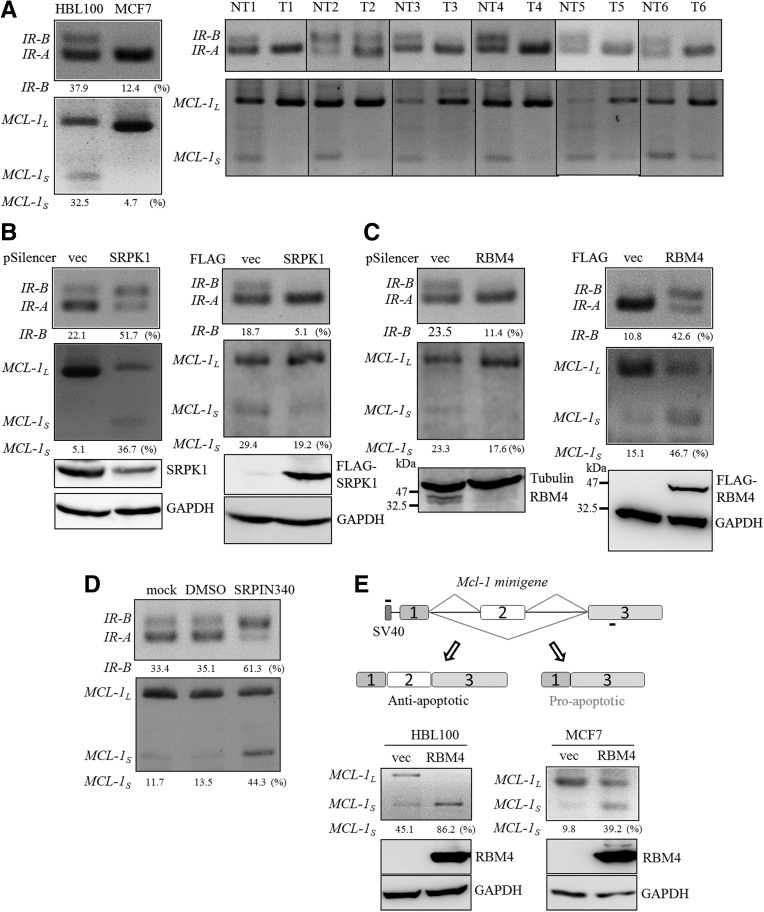
The cytoplasmic accumulation of splicing factors reduces their relative abundance in the nucleus, which subsequently interferes with the regulation of alternative splicing (Licatalosi and Darnell 2010). We investigated whether the cytoplasmic accumulation of RBM4 correlated with the splicing event in breast cancer cells. Exon 11-excluding IR-A and exon 11-including IR-B isoforms originate from the alternatively spliced transcripts of the IR gene, which has been characterized as the candidate target of RBM4 (Lin and Tarn 2011; Lin et al. 2014). The relatively high level of the anti-apoptotic IR-A transcript was examined in the origins of various malignancies (Kalli et al. 2002; Kalla Singh et al. 2011; Chettouh et al. 2013). The RT-PCR analysis showed that the predominant expression of the IR-A isoform was noted in MCF-7 breast cancer cells compared to those in HBL100 cells (Fig. 3A, left row). The level of the MCL-1L transcript was also higher in the MCF-7 cells (Fig. 3A, left row), which is consistent with the results reported in other malignancies (Palve and Teni 2012; Gao et al. 2013). The relatively high level of IR-A and MCL-1L was also observed in the breast cancer tissue samples, but not in the adjacent noncancerous tissues (Fig. 3A, right row). The silencing of SRPK1 expression led to an increase in the IR-B and MCL-1S transcript in MCF-7 cells, as was shown in the RBM4-overexpressing cells (Fig. 3B, left; Fig. 3C, right). In contrast, the overexpression of SRPK1 and the knockdown of RBM4 conversely induced the expression of IR-A and MCL-1L in MCF-7 cells (Fig. 3B, right; Fig. 3C, left). Treatment with SRPIN340 substantially increased the production of IR-B and MCL-1S transcript (Fig. 3D) as observed in the SRPK1-knockdown MCF-7 cells. To investigate the mechanism involved in regulating the splicing profile of MCL-1 pre-mRNA, an in vivo splicing assay was performed using the pCH-MCL-1 minigene plasmid (Fig. 3E, top row). HBL100 and MCF-7 cells were cotransfected with the pCH-MCL-1 minigene reporter and the RBM4 expression plasmid. Using a 5′ primer targeted to the SV40 promoter, an increase in the exogenous MCL-1S transcript was observed in the HBL100 cells and MCF-7 cells overexpressing RBM4 (Fig. 3E, bottom panel). These results clearly showed that an increase in RBM4 enhanced the production of the IR-B and MCL-1S transcript in various types of cell.
FIGURE 3.
The SRPK1-RBM4 network modulates the splicing pattern of the IR and MCL-1 pre-mRNAs. (A) Total RNA was extracted from HBL100 and MCF-7 cells (left panel) and from human breast cancer tissues (T) and the adjacent noncancerous tissues (NT; right panel). The RT-PCR analysis was conducted with sequence-specific primers designed to demonstrate the splicing pattern of the IR and MCL-1 genes. The PCR products were analyzed using electrophoresis on a 2% agarose gel. (B) MCF-7 cells were transfected with the empty expression vector, the shSRPK1 expression plasmid (left), or the SRPK1 expression plasmid (right) for 24 h. An RT-PCR was used to examine the splicing patterns of the IR and MCL-1 genes as described for the previous panel. The cell lysates were analyzed using an immunoblotting assay with anti-SRPK1, anti-FLAG, and anti-GAPDH antibodies. (C) Total RNA and whole-cell lysates were prepared from MCF-7 cells transfected with the empty expression plasmid, the shRBM4 expression plasmid (left), or the RBM4 expression plasmid (right). The splicing profile of IR and MCL-1 was examined using an RT-PCR as described for panel A. (D) The total RNA was isolated from mock-, vehicle-, or SRPIN340-treated MCF7 cells, and the splicing profile of IR and MCL-1 was examined by using an RT-PCR analysis as described for panel A. The PCR products were analyzed using electrophoresis on a 2% agarose gel. (E) The diagram presents the MCL-1 minigene (top panel) in the plasmid that was cotransfected with the empty plasmid or the RBM4 expression plasmid into HBL100 and MCF-7 cells. An RT-PCR was conducted using a primer set complementary to the SV40 promoter and MCL-1 exon 3. The PCR products were analyzed using electrophoresis on a 2% agarose gel. The cell lysates were subjected to an immunoblotting assay by using the anti-FLAG and anti-GAPDH antibodies.
Phosphorylation reduces the effect of RBM4 on pre-mRNA splicing
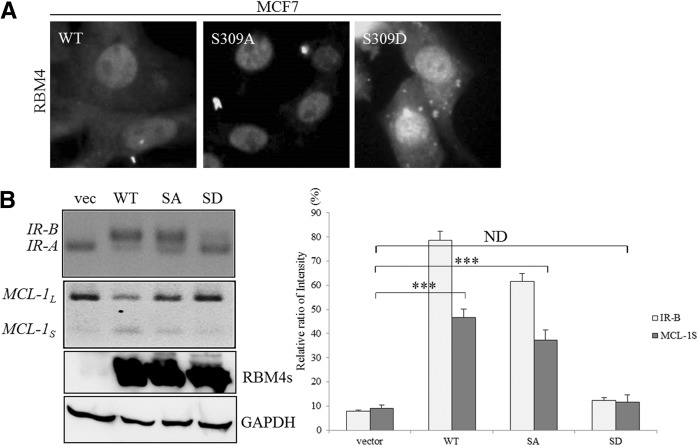
We investigated whether the phosphorylation of RBM4 also affected the splicing pattern of IR and MCL-1 transcripts in a manner similar to that caused by the cytoplasmic accumulation of RBM4. To evaluate this possibility, the impact of Ser309 phosphorylation on the cellular localization of RBM4 in MCF-7 cells was first examined. The MCF-7 cells were transfected with plasmids expressing the nonphosphorylatable S309A or the phosphomimetic S309D mutant RBM4 proteins. The results of an indirect immunostaining assay showed that the S309A mutant was less inclined to be distributed in the cytoplasm, whereas the S309D mutant had a high tendency for cytoplasmic localization compared to the wild-type RBM4 (Fig. 4A). The RT-PCR analysis indicated that the wild-type RBM4 and the S309A mutant exerted statistically similar effects regarding the levels of the IR-B and MCL-1S transcripts (Fig. 4B, WT and SA), whereas the S309D mutant had no substantial effect on the splicing patterns of the IR and MCL-1 transcripts in MCF-7 cells (Fig. 4B, SD). The influence of overexpressing RBM4 proteins on the splicing profile of IR and MCL-1 pre-mRNA was clearly dependent on its phosphorylation and biological activity since the expression levels were equal (Fig. 4B, bottom panel). These results indicated that the phosphorylation-mediated subcellular localization of RBM4 affected its influence on the splicing patterns of the IR and MCL-1 transcripts.
FIGURE 4.
The phosphorylation status of RBM4 modulates its cellular localization and effect on alternative splicing events. MCF-7 cells were transiently transfected with an expression vector encoding wild-type RBM4, the S309A mutant (nonphosphorylatable; SA), or the S309D mutant (phosphomimetic; SD) for 24 h. (A) Localization of overexpressing RBM4 proteins was examined using indirect immunostaining with an anti-FLAG antibody. (B) Total RNA was isolated from the transfected MCF-7 cells and analyzed using RT-PCR, as described in Figure 3A. The cell lysates were subjected to an immunoblotting assay by using anti-FLAG and anti-GAPDH antibodies. The bar graph presents the relative ratio of IR-B and MCL-1S in the transfected cells in three independent experiments. (***) P < 0.005.
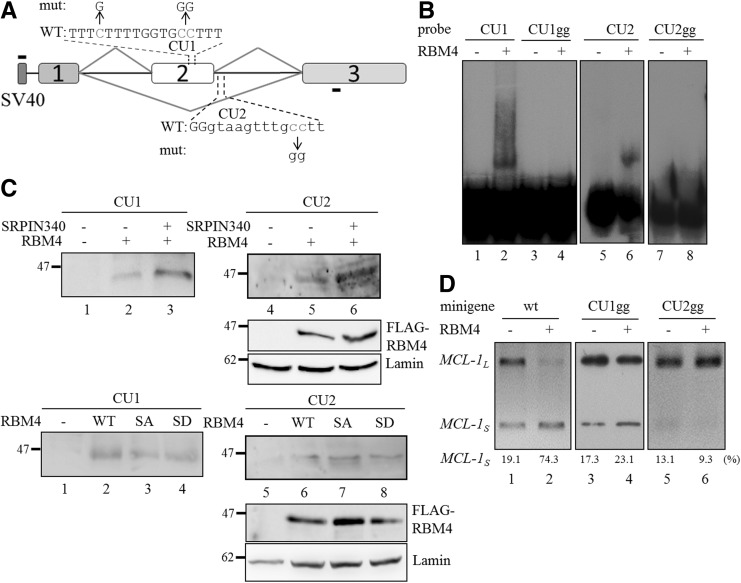
Utilization of MCL-1 exon 2 is regulated by RBM4 through binding to CU-rich elements
The overexpression of RBM4 has been demonstrated to enhance the inclusion of α-tropomyosin exon 9a by binding to the intronic CU-rich element markedly downstream from the regulated exon (Lin and Tarn 2005). However, the simultaneous binding of RBM4 to the intronic and exonic CU elements mediates the skipping of alternatively spliced exons in other candidate genes (Lin and Tarn 2011). CU-rich elements are also present in MCL-1 exon 2 of and the downstream intron adjacent to the 5′ splice site (Fig. 5A), and we, therefore, investigated the interaction between RBM4 and these elements. The DIG-labeled probes originated from the CU-rich elements in MCL-1 exon 2 and intron 2 (the latter contains the CU-rich element) (Fig. 5A) were incubated with the mock eluate or the recombinant His-tagged RBM4 protein, and the mixtures were analyzed using the RNA electrophoretic mobility shift assay (REMSA). The DIG-labeled probes derived from the exonic and intronic elements formed ribonucleoprotein (RNP) complexes with the recombinant RBM4 protein (Fig. 5B, lanes 2 and 6), whereas the cytosine-to-guanine nucleotide substitutions in the CU-rich element (Fig. 5A, top panel) substantially reduced RNP complex formation (Fig. 5B, CU1gg and CU2gg, lanes 4 and 8). The REMSA assay indicated that the interactions between RBM4 and the CU1 and 2 elements were sequence-specific. To investigate the impact of phosphorylation status on the binding of RBM4 to the CU-rich elements, the DIG-labeled probes were incubated with the nuclear extract of the mock-transfected MCF-7 cells or that of cells expressing FLAG-tagged RBM4 that were treated with SRPIN340 or vehicle control. Treatment of SRPIN340 increased the level of RBM4 in the nuclear extract, which subsequently enhanced RNP complex formation (Fig. 5C, top panel, lanes 3 and 6). Nevertheless, no obvious difference was noted in the association of RNP complexes when the DIG-labeled probes were incubated with the nuclear extract of cells expressing the wild-type RBM4 protein, the S309A RBM4 mutant, or the S309D RBM4 mutant (Fig. 5C, bottom panel). These results suggested that the phosphorylation status of RBM4 had no impact on its binding to the CU-rich elements. The in vivo splicing assay indicated that the cytosine-to-guanine nucleotide substitutions in the CU1 and 2 elements abolished the repressive effect of RBM4 on Mcl-1 exon 2 inclusion, resulting in a relatively high level of the MCL-1L mRNA originated from the mutant minigene (Fig. 5D, lanes 4 and 6). These results indicated that the direct binding of RBM4 to the exonic and intronic CU-rich elements subsequently enhanced the production of MCL-1S transcript.
FIGURE 5.
RBM4 mediates the exclusion of MCL-1 exon 2 by binding directly to the exonic and intronic CU-rich elements. (A) The diagram presents the sequence of the exonic and downstream intronic CU-rich elements of MCL-1 exon 2. The mutant reporters contained cytosine-to-guanine nucleotide substitutions in the CU1 and CU2 elements. (B) The mock eluate from Ni2+-nitrilotriacetic acid-agarose resin or 1 µg of recombinant His-tagged RBM4 protein was incubated with 10 nM DIG-labeled probes. The mixtures were fractionated in 8% native acrylamide gel and transferred to the nylon membrane. The membrane was probed using an HRP-conjugated anti-DIG Fab fragment. (C) MCF-7 cells were mock-transfected or transfected with the FLAG-RBM4 expression plasmid, followed by vehicle or SRPIN340 treatment. The nuclear extracts prepared from the various transfected cells were incubated with the DIG-labeled CU1 and CU2 probes, followed by UV crosslinking. The reactions were analyzed using an immunoblotting assay with the anti-FLAG M2 antibody. (D) The wild-type MCL-1 minigene and the derived mutants (CU1gg and CU2gg) were cotransfected with the empty vector or FLAG-RBM4 expression vector into MCF-7 cells. The PCR product of spliced transcript of each minigene was analyzed using electrophoresis on a 2% agarose gel.
The SRPK1-RBM4 network modulates the apoptotic resistance of breast cancer cells
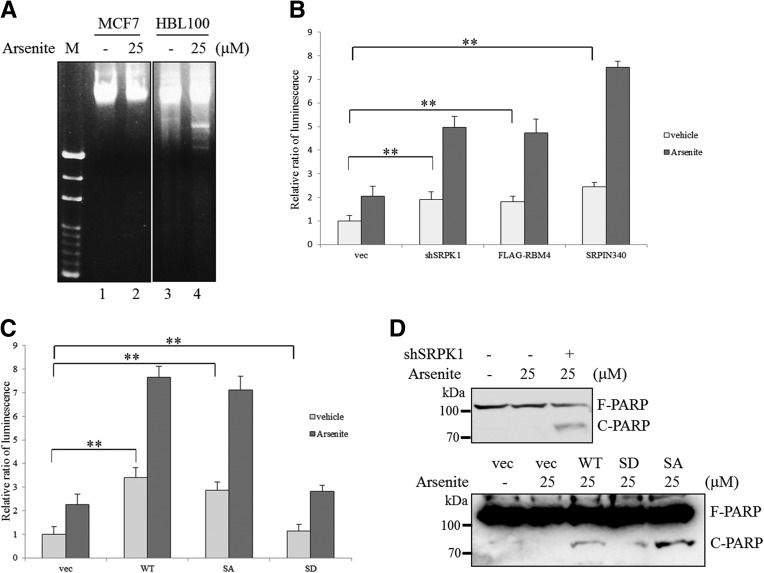
Relatively high levels of the IR-A and MCL-1L isoforms have been proven to exert anti-apoptotic effects in various cancer cells (Kalli et al. 2002; Gautrey and Tyson-Capper 2012; Palve and Teni 2012; Chettouh et al. 2013). We evaluated whether RBM4 affected the apoptotic sensitivity of breast cancer cells to cytotoxic agent through the modulation of IR and MCL-1 alternative splicing. The fragmented DNA is a well-characterized marker for apoptotic cells. The DNA fragmentation analysis indicated that MCF-7 cells exhibited cytotoxic resistance to 25 μM sodium arsenite for up to 8 h (Fig. 6A, lane 2), whereas HBL100 cells underwent apoptosis under the same condition (Fig. 6A, lane 4). The caspase 3 activity assay was performed to evaluate whether the apoptotic feature of MCF-7 cells was affected by SRPK1 knockdown and RBM4 overexpression. A relatively small increase in caspase 3 activity was observed in the SRPK1-silencing, RBM4-overexpressing, and SRPIN340-treated MCF-7 cells (Fig. 6B, light gray). In contrast, in the cells treated with SRPIN340, the knockdown of SRPK1 expression and the simultaneous overexpression of RBM4 caused a synergistic increase in the arsenite-induced apoptosis of MCF-7 cells, compared with that observed using sodium arsenite treatment alone (Fig. 6B, dark gray). The presence of the nonphosphorylatable S309A mutant, but not the phosphomimetic S309D mutant, exerted a similar effect as wild-type RBM4 on sensitizing the MCF-7 to the cytotoxic agent (Fig. 6C). It is well known that the active caspase cascade cleaves the DNA repair enzyme poly-ADP-ribose polymerase (PARP), resulting in its inactivation in apoptotic cells (Laha et al. 2014). The shRNA-mediated knockdown of SRPK1 expression increased the cleavage of PARP in the MCF-7 cells treated with sodium arsenite, relative to that observed in the cells transfected with the control plasmid (Fig. 6D, top panel). A substantial increase in the level of cleaved PARP was also observed in the MCF-7 cells overexpressing RBM4 following sodium arsenite treatment, whereas such an increase was not observed in the MCF-7 cells overexpressing the S309D RBM4 mutant (Fig. 6C, bottom panel, WT and SD). The overexpression of the S309A RBM4 mutant induced a relatively high level of PARP cleavage following sodium arsenite treatment (Fig. 6C, bottom panel, SA). Nevertheless, these results suggested that the SRPK1-RBM4 network modulated the sensitivity of breast cancer cells toward pro-apoptotic agents.
FIGURE 6.
The SRPK1-RBM4 network sensitizes MCF-7 cells to sodium arsenite-induced apoptosis. (A) HBL100 and MCF-7 cells were treated with 25 µM sodium arsenite or vehicle control for 8 h. The chromosomal DNA was extracted and subjected to electrophoresis on a 1.5% agarose gel, followed by ethidium bromide staining. (B) MCF-7 cells were transfected with the empty expression vector, the shSRPK1 expression plasmid, the FLAG-RBM4 expression plasmid, or pretreated with SRPIN340 before exposure to DMSO or 25 µM sodium arsenite for 8 h. The caspase 3 activity was quantified by adding a caspase 3-specific substrate in three independent experiments. (**) P < 0.01. (C) MCF7 cells were transfected with the expression vectors encoding FLAG-RBM4s and then exposed to DMSO or 25 µM sodium arsenite for 8 h. The caspase 3 activity in three independent experiments was quantified as in B. (**) P < 0.01. (D) MCF7 cells were transfected with the shSRPK1 expression plasmid (top) or the FLAG-RBM4 expression plasmid (bottom), followed by exposure to 25 µM sodium arsenite for 8 h. Whole-cell lysates were subjected to SDS-PAGE and analyzed using immunoblotting with anti-PARP antibody.
DISCUSSION
Alternative splicing events have been reported in diverse malignancies (Chen and Weiss 2014), and numerous splicing events have been proven to be critically regulated in tumorigenic cells. An increase in the intracellular level of SRPK1, a major kinase of SR proteins, has been demonstrated to be critical in the regulation of the alternative splicing profile of malignant cells (Gout et al. 2012; Odunsi et al. 2012; Zhou et al. 2013). Mutations of Wilms’ tumor suppressor gene (WT1) lead to the up-regulation of SRPK1, which diminishes the SRSF1-mediated up-regulation of VEGF165b (Amin et al. 2011). The elevated ratio of VEGF165 to VEGF165b correlates closely with the angiogenesis which facilitates the tumor growth (Merdzhanova et al. 2010). Elevated SRPK1 modulates the influence of SRSF2 on caspase-8 mRNA splicing and subsequently strengthens the therapeutic resistance of cancer cells to cisplatin treatment (Edmond et al. 2011). In vitro studies have shown that siRNA-mediated knockdown of SRPK1 expression sensitizes the malignant cells to chemotherapeutic treatment (Hayes et al. 2007). However, the reduced level of SRPK1 also contributes to retinoblastoma and tumorigenesis of male germ cells (Krishnakumar et al. 2008). Nevertheless, these results indicate that altered SRPK1 levels in either course are closely correlated with the progression and immortality of cancer cells.
The isoform shift of the IR gene is considered a tumorigenic event in various malignancies, including breast cancer (Kalli et al. 2002; Kalla Singh et al. 2011; Chettouh et al. 2013). Conventional approaches have revealed a close correlation between a high IR-A:IR-B ratio and malignant features of breast cancer, such as high proliferation and poor prognosis (Costa et al. 2008; Mester and Redeuilh 2008; Brierley et al. 2010). However, the comprehensive molecular mechanism involved in the isoform shift of IR transcript remains unclear. The overexpression of SRSF3 has been proven to antagonize the effect of CUGBP1-mediated exclusion of IR exon 11 in hepatic cancer cells (Sen et al. 2009). The aberrant expression of hnRNPA1 and SRSF1 also affects the utilization of IR exon 11 in myotonic dystrophy (Talukdar et al. 2011). The results of our previous study showed that elevated intracellular levels of RBM4 antagonize the repressive effect of PTBP1 on the inclusion of IR exon 11 during myogenesis (Lin and Tarn 2011). In this study, the relatively high expression of SRPK1 mediated the cytoplasmic accumulation of RBM4 but not that of PTBP1 (Fig. 2B, right panel). The reduced ratio of RBM4:PTBP1 in the nuclei of MCF-7 cells subsequently diminished the influence of RBM4 on the inclusion of IR exon 11 in alternatively spliced IR variants.
The splicing pattern of the MCL-1 gene has been reported to correlate with the sensitivity of a wide variety of tumor cells to cytotoxic agents (Palve and Teni 2012; Gao et al. 2013; Pennarun et al. 2013). Recent studies have demonstrated a link between the up-regulation of the MCL-1L transcript and SR proteins, such as SRSF1 and SRSF5 (Gautrey and Tyson-Capper 2012), which act as proto-oncogenes in various types of cancer (Das et al. 2012; Breig and Baklouti 2013; Fregoso et al. 2013). An increase in SRSF1 has been demonstrated to facilitate the translational efficiency and stability of the MCL-1L isoform in breast cells (Gautrey and Tyson-Capper 2012). However, a modest impact of SRSF1 silencing was noted on the apoptotic sensitivity of breast cancer cells toward cytotoxic agents (Gautrey and Tyson-Capper 2012). These results imply that an uncharacterized mechanism might also modulate the action of MCL-1 on apoptosis in malignant cells. In this study, the results of an in vivo splicing assay showed that the overexpression of RBM4 markedly increased the level of the MCL-1S transcript originating from the endogenous MCL-1 gene and exogenous MCL-1 minigene reporter in both mammary epithelium cells and breast cancer cells (Fig. 3C,E). The elevated caspase 3 activity and an increase in the cleaved PARP product indicated that the elevation of RBM4-favored MCL-1S modulated the apoptotic resistance of breast cancer cells toward cytotoxic agents (Fig. 6B–D). The depletion and inhibition of SRPK1 exerted a substantial effect on the induction of apoptosis in sodium arsenite-treated MCF-7 cells, which implies that another SRPK1-modulated mechanism might also modulate the apoptotic resistance of breast cancer cells.
Previous studies have investigated the interaction between RBM4 and the CU-rich elements within its candidate genes (Lin and Tarn 2005, 2011). RBM4 protein was first observed to enhance the inclusion of exon 9a in an α-TM splicing variant by binding to the CU-rich element within the downstream intron (Lin and Tarn 2005). RBM4 has also been proven to mediate the exclusion of exon 11 in a PTBP1 splicing variant by binding to CU-rich elements located within and adjacent to exon 11(Lin and Tarn 2011), which might interfere with the association between the spliceosome and the 5′ splice site. In this study, the binding of RBM4 to CU-rich elements within and downstream from MCL-1 exon 2 correlated with the exclusion of MCL-1 exon 2 (Fig. 5B,D), which is similar to the exclusion of PTBP1 exon 11. Our results and those of previous studies collectively indicate that the binding of RBM4 to CU-rich sequences in exons and adjacent introns might activate the exclusion of alternatively spliced exons in other RBM4-specific target genes. However, the association of RBM4 protein and other cis-elements was observed by using CLIP analysis (Uniacke et al. 2012; Wang et al. 2012). Future investigations are warranted to determine whether RBM4 modulates alternative splicing events through these molecular mechanisms.
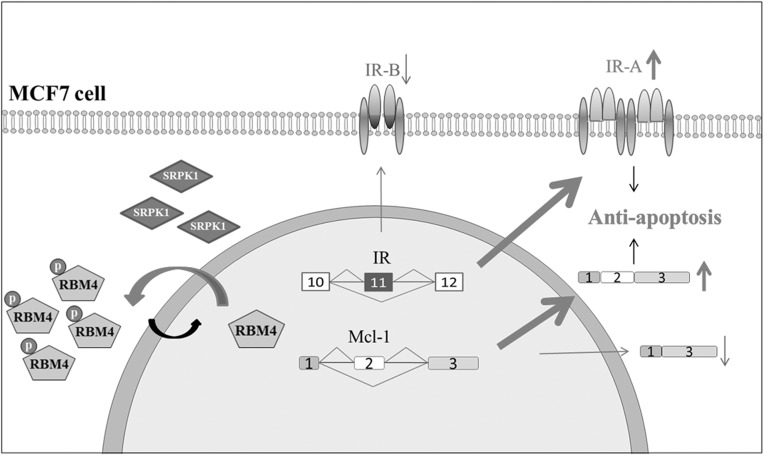
In conclusion, our results indicated that the elevation in SRPK1 protein induced the cytoplasmic accumulation of RBM4, which abolished the effect of RBM4 on the expression of pro-apoptotic IR-B and MCL-1S transcripts (Fig. 7). Our results constituted a molecular mechanism that the simultaneous binding of RBM4 to the CU-rich elements within and adjacent to MCL-1 exon 2 modulated its selection. We propose that the action of the SRPK-RBM4 network on the modulation of apoptotic sensitivity may support the therapeutic potential of SRPK1 and the associated splicing factor in breast cancer.
FIGURE 7.
SRPK1-RBM4 network constitutes an apoptotic circuit by regulating alternative splicing events in breast cancer cells. Elevated SRPK1 induces the cytoplasmic accumulation of phosphorylated RBM4, which abolishes its pro-apoptotic effect by inducing the IR-B and MCL-1S transcripts. The depletion of endogenous SRPK1 restores the pro-apoptotic influence of RBM4 by enhancing the expression of IR-B and MCL-1S isoforms in MCF-7 cells.
MATERIALS AND METHODS
Cell culture and patient samples
Human breast tumor samples (n = 10) were requested as anonymous specimens from the TMU Joint Biobank. The application was reviewed and approved by the Institutional Review Board of the Taipei Medical University (approval number: 201308023). All of the recommendations of the Declaration of Helsinki for biomedical research involving human subjects were followed. The HBL100 cells and MCF-7 human breast carcinoma cell line were kind gifts from Dr. C. H. Lee (Taipei Medical University, Taipei, Taiwan). The MCF-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM), and the HBL100 cells were maintained in DMEM/Ham's F12 nutrient mixture supplemented with 10% fetal bovine serum, 600 mg/mL glutamine, 100 units/mL penicillin, and 100 mg/mL streptomycin (Invitrogen).
Plasmid construction and transfection
Complementary DNAs encoding human SRPK1 were amplified by polymerase chain reaction using a cDNA library prepared from the human fetal brain as the template and then inserted into the pcDNA3.1 FLAG vector (Invitrogen). The vector expressing a SRPK1-targeting small hairpin RNA was constructed as follows. The short hairpin primers were designed around the potential sequence, namely sh1-SRPK1 (5′-GTGCAGCAGAAATTAATT-3′), which corresponds to nucleotides 1171–1189 of the SRPK1 transcript (GI: 47419935), and the sequence was annealed and then ligated into the PstI site of the pSilencer1.0 U6 vector (Ambion). The MCL-1 minigene reporter was constructed by inserting a complete human MCL-1 genomic fragment containing exons 1, 2, and 3 and intra-exon introns. The genomic fragments were amplified by PCR using the genomic DNA prepared from MRC5 fibroblasts as the template and then inserted into the HindIII/SacI sites to replace the β-galactosidase gene of pCH110 (Amersham Pharmacia). Mutant pCH-MCL-1 vectors containing changed nucleotides were constructed using the QuikChange site-directed mutagenesis system (Stratagene).
Subcellular fractionation
MCF-7 and HBL100 cells were collected and resuspended in 500 μL of RSB-100 buffer containing 40 mg/mL digitonin (Calbiochem). After incubation on ice for 5 min, the cells were centrifuged at 2000g for 8 min. The supernatant was collected as the cytosolic fraction, and the pellet was resuspended in 500 μL of RSB-100 containing 0.5% Triton X-100 and incubated on ice. After centrifugation, the supernatant was collected as the nuclear fraction.
Immunoblotting assay
The immunoblot analysis was conducted using an enhanced chemiluminescence (ECL) system (Millipore), and the images were analyzed with the LAS-4000 imaging system (Fujifilm). The primary antibodies used in this study included polyclonal anti-RBM4 and anti-pS309, which probed the total and phosphorylated RBM4, monoclonal anti-PTB (EMD Millipore), monoclonal anti-SRPK1 (BD Transduction Laboratories), monoclonal anti-FLAG M2 (Sigma-Aldrich), polyclonal anti-GAPDH (MDBio), polyclonal anti-lamin A/C (Santa Cruz Biotechnology), monoclonal anti-actin (Chemicon Millipore), and monoclonal anti-α Tubulin (EMD Millipore).
In vivo splicing assay
MCF-7 and HBL100 cells were grown to 50%–60% confluency in six-well plates and transfected with 2 μg of the effector expression vector alone or cotransfected with 1 μg of the minigene using PolyJet according to the manufacturer's instructions (SignaGen Laboratories). Total RNA from the cultured cells was collected using Trizol (Invitrogen). For the reverse-transcription PCR analysis, 5 μg of RNA were reverse-transcribed with SuperScriptase III (Invitrogen) in a 10-μL reaction. The PCR analysis was performed using specific sets of primers (see Table 2), and the products were resolved using agarose gel electrophoresis.
TABLE 2.
List of PCR primer sets
Indirect immunostaining
The indirect immunofluorescence was measured using a previously described procedure (Lin et al. 2007). A polyclonal anti-RBM4 antibody or monoclonal anti-FLAG M1 antibody (10 ng/mL; Sigma) was used as the primary antibody.
DNA fragmentation assay
The transfected cells were grown in the presence of sodium arsenite in 6-cm dishes for 8 h. The cells were washed twice with phosphate-buffered saline (PBS) and lysed in 300 μL of lysis buffer (20 mM Tris-HCl, pH 7.4, 5 mM EDTA, 150 mM NaCl, and 0.5% Triton X-100). After incubation on ice for 30 min, the cells were centrifuged at 12,000 rpm for 15 min. RNase A (200 μg/mL) was poured into the supernatant, and the mixture was incubated at 37°C for 1 h. After the addition of proteinase K (200 μg/mL), the mixture was incubated at 37°C for 1 h. The fragmented DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1 v/v/v) and precipitated with 95% ethanol. An equal amount of DNA was resolved in a 2% agarose gel and then stained with EtBr.
Apoptosis assay
Caspase 3 activity was measured using the Caspase-Glo 3/7Assay kit (Promega). Ten thousand cells were seeded in each well of 96-well plates 24 h prior to the assay. One hundred microliters of reaction reagent were added to each well followed by gentle shaking. The luminescence intensity relative to the caspase activity was measured at excitation/emission wavelengths of 485/535 nm using a Synergy HT multi-mode microplate reader (BioTek).
RNA-protein interaction
The DNA oligonucleotide containing the SP6 RNA promoter was synthesized in vitro. The sequence of the CU1 probe was 5′-tttcttttggtgcctttgtggctaaacacttgaagacca, and the sequence of the CU2 probe was 5′-gtaagtttgccttaaggatgaaaggggccttggagtg. The RNA probe was transcribed in vitro using SP6 RNA polymerase and then subjected to 5′-end DIG-labeling (Roche) according to the manufacturer's protocol. The DNA template was removed by treatment with DNase I (Promega) at 37°C for 15 min. The RNA probe was purified through a Sephadex G-25 column (Pharmacia Biotech) and quantified by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Tech). The 10-μL nuclear extract prepared from the empty-vector and FLAG-RBM4-overexpressing cells was incubated with the DIG-labeled probe (5 ng) in a 20-μL mixture at 30°C for 30 min. The reaction mixtures were irradiated on ice under 254-nm light for 10 min, and the unbound probes were digested with 1 μg of RNase A (Sigma) at 30°C for 20 min. The samples were subjected to 12% SDS-PAGE and then electron-transferred to a nitrocellulose membrane. The membranes were probed with anti-FLAG M2 antibody. An immunoblot analysis was conducted as previously described.
RNA electrophoretic mobility shift assay
Recombinant His-tagged proteins were prepared as described previously (Lin and Tarn 2011). RNA probes used were MCL-1 exon 2 (CU1, 39 nt) and the downstream intron (CU2, 39 nt). Exonic and intronic elements were in vitro-transcribed and used as probes. For RNA-protein interaction, 1 μg of recombinant protein was incubated with 10 nM of DIG-labeled probe in a 20-μL reaction containing 10 mM HEPES, pH 7.9, 50 μ MEDTA, 10% glycerol, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 μg/mL bovine serum albumin, and 12.5 ng/mL tRNA for 15 min at room temperature. The reactions were analyzed by electrophoresis on an 8% nondenaturing polyacrylamide gel in TBE buffer (45 mM Tris-HCl, 45 mM boric acid, and 1 mM EDTA, pH 8.0). The binding complexes were transferred to a nylon membrane (Hybond N, Amersham Bioscience) that was irradiated under 254-nm light for 60 sec. The immunoblotting was conducted by incubating the membranes with HRP-conjugated anti-DIG Fab fragments (Roche).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Chia-Hwa Lee (Taipei Medical University, Taiwan) for kindly providing the HBL100 and MCF-7 cells. This work was supported by grant NSC 102-2320-B-038-045 from the National Science Council of Taiwan.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045583.114.
REFERENCES
- Amin EM, Oltean S, Hua J, Gammons MV, Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, et al. 2011. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell 20: 768–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD 2013. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J Mol Biol 425: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J, Leo C, Hsu S, Hsueh A 2000. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem 275: 25255–25261 [DOI] [PubMed] [Google Scholar]
- Ben-Hur V, Denichenko P, Siegfried Z, Maimon A, Krainer A, Davidson B, Karni R 2013. S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep 3: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle C, Craig R, Swales B, Singleton V, Zhou P, Whyte MK 2000. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem 275: 22136–22146 [DOI] [PubMed] [Google Scholar]
- Breig O, Baklouti F 2013. Proteasome-mediated proteolysis of SRSF5 splicing factor intriguingly co-occurs with SRSF5 mRNA upregulation during late erythroid differentiation. PLoS One 8: e59137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley GV, Macaulay SL, Forbes BE, Wallace JC, Cosgrove LJ, Macaulay VM 2010. Silencing of the insulin receptor isoform A favors formation of type 1 insulin-like growth factor receptor (IGF-IR) homodimers and enhances ligand-induced IGF-IR activation and viability of human colon carcinoma cells. Endocrinology 151: 1418–1427 [DOI] [PubMed] [Google Scholar]
- Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, Lu S, Fenoglio-Preiser CM, Li J, Knudsen ES, et al. 2006. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci 103: 2190–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Weiss WA 2014. Alternative splicing in cancer: implications for biology and therapy. Oncogene 10.1038/onc.2013.570 [DOI] [PubMed] [Google Scholar]
- Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Clapéron A, Chrétien Y, Rey C, Scatton O, Soubrane O, Conti F, et al. 2013. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res 73: 3974–3986 [DOI] [PubMed] [Google Scholar]
- Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Ørntoft TF, Mu D, Karni R 2013. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol 229: 630–639 [DOI] [PubMed] [Google Scholar]
- Costa V, Foti D, Paonessa F, Chiefari E, Palaia L, Brunetti G, Gulletta E, Fusco A, Brunetti A 2008. The insulin receptor: a new anticancer target for peroxisome proliferator-activated receptor-γ (PPARγ) and thiazolidinedione-PPARγ agonists. Endocr Relat Cancer 15: 325–335 [DOI] [PubMed] [Google Scholar]
- Das S, Anczuków O, Akerman M, Krainer AR 2012. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep 1: 110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, Gazzeri S, Eymin B 2011. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J 30: 510–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregoso OI, Das S, Akerman M, Krainer AR 2013. Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol Cell 50: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Li L, Wu M, Liu M, Xie X, Guo J, Tang H, Xie X 2013. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS One 8: e65138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautrey HL, Tyson-Capper AJ 2012. Regulation of MCL-1 by SRSF1 and SRSF5 in cancer cells. PLoS One 7: e51497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout S, Brambilla E, Boudria A, Drissi R, Lantuejoul S, Gazzeri S, Eymin B 2012. Abnormal expression of the pre-mRNA splicing regulators SRSF1, SRSF2, SRPK1 and SRPK2 in non-small cell lung carcinoma. PLoS One 7: e46539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Beck AM, Miller LJ 2006. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res 66: 3819–3827 [DOI] [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Miller LJ 2007. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res 67: 2072–2080 [DOI] [PubMed] [Google Scholar]
- Iborra S, Hirschfeld M, Jaeger M, Zur Hausen A, Braicu I, Sehouli J, Gitsch G, Stickeler E 2013. Alterations in expression pattern of splicing factors in epithelial ovarian cancer and its clinical impact. Int J Gynecol Cancer 23: 990–996 [DOI] [PubMed] [Google Scholar]
- Kalla Singh S, Brito C, Tan QW, De León M, De León D 2011. Differential expression and signaling activation of insulin receptor isoforms A and B: a link between breast cancer and diabetes. Growth Factors 29: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalli KR, Falowo OI, Bale LK, Zschunke MA, Roche PC, Conover CA 2002. Functional insulin receptors on human epithelial ovarian carcinoma cells: implications for IGF-II mitogenic signaling. Endocrinology 143: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW 1993. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci 90: 3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, Mohan A, Kandalam M, Ramkumar HL, Venkatesan N, Das RR 2008. SRPK1: a cisplatin sensitive protein expressed in retinoblastoma. Pediatr Blood Cancer 50: 402–406 [DOI] [PubMed] [Google Scholar]
- Laha D, Pramanik A, Maity J, Mukherjee A, Pramanik P, Laskar A, Karmakar P 2014. Interplay between autophagy and apoptosis mediated by copper oxide nanoparticles in human breast cancer cells MCF7. Biochim Biophys Acta 1840: 1–9 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Tarn WY 2005. Exon selection in α-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol Cell Biol 25: 10111–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Tarn WY 2011. RBM4 down-regulates PTB and antagonizes its activity in muscle cell-specific alternative splicing. J Cell Biol 193: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Hsu M, Tarn WY 2007. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci 104: 2235–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Tarn WY, Hsieh WK 2014. Emerging role for RNA binding motif protein 4 in the development of brown adipocytes. Biochim Biophys Acta 1843: 769–779 [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24: 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdzhanova G, Gout S, Keramidas M, Edmond V, Coll JL, Brambilla C, Brambilla E, Gazzeri S, Eymin B 2010. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene 29: 5392–5403 [DOI] [PubMed] [Google Scholar]
- Mester J, Redeuilh G 2008. Proliferation of breast cancer cells: regulation, mediators, targets for therapy. Anticancer Agents Med Chem 8: 872–885 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G 2005. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell 20: 77–89 [DOI] [PubMed] [Google Scholar]
- Odunsi K, Mhawech-Fauceglia P, Andrews C, Beck A, Amuwo O, Lele S, Black JD, Huang RY 2012. Elevated expression of the serine-arginine protein kinase 1 gene in ovarian cancer and its role in Cisplatin cytotoxicity in vitro. PLoS One 7: e51030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshavsky NA, Comstock CE, Schiewer MJ, Augello MA, Hyslop T, Sette C, Zhang J, Parysek LM, Knudsen KE 2010. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res 70: 3975–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palve VC, Teni TR 2012. Association of anti-apoptotic MCL-1L isoform expression with radioresistance of oral squamous carcinoma cells. Radiat Oncol 7: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun B, Kleibeuker JH, Boersma-van Ek W, Kruyt FA, Hollema H, de Vries EG, de Jong S 2013. Targeting FLIP and MCL-1 using a combination of aspirin and sorafenib sensitizes colon cancer cells to TRAIL. J Pathol 229: 410–421 [DOI] [PubMed] [Google Scholar]
- Sen S, Talukdar I, Webster NJ 2009. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol 29: 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar I, Sen S, Urbano R, Thompson J, Yates JR III, Webster NJ 2011. hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PLoS One 6: e27869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S 2012. An oxygen-regulated switch in the protein synthesis machinery. Nature 486: 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Végran F, Boidot R, Bonnetain F, Cadouot M, Chevrier S, Lizard-Nacol S 2011. Apoptosis gene signature of Survivin and its splice variant expression in breast carcinoma. Endocr Relat Cancer 18: 783–792 [DOI] [PubMed] [Google Scholar]
- Wang J, Smith PJ, Krainer AR, Zhang MQ 2005. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res 33: 5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ma M, Xiao X, Wang Z 2012. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol 19: 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhou Z, Hu A, Ponte de Albuquerque C, Zhou Y, Hong L, Sierecki E, Ajiro M, Kruhlak M, Harris C, et al. 2014. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol Cell 54: 378–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD 2009. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev 23: 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Fu XD 2013. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122: 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. 2012. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell 47: 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Li Y, Deng Q, Wang H, Wang Y, Cai B, Han ZG 2013. SRPK1 contributes to malignancy of hepatocellular carcinoma through a possible mechanism involving PI3K/Akt. Mol Cell Biochem 379: 191–199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.