Abstract
Development of sensitive and specific biomarkers, preferably those circulating in body fluids is critical for early diagnosis of cancer. This study performed profiling of microRNAs (miRNAs) in exocrine pancreatic secretions (pancreatic juice) by microarray analysis utilizing pancreatic juice from 6 pancreatic ductal adenocarcinoma (PDAC) patients and two pooled samples from 6 non-pancreatic, non-healthy (NPNH) as controls. Differentially circulating miRNAs were subsequently validated in 88 pancreatic juice samples from 50 PDAC, 19 chronic pancreatitis (CP) patients and 19 NPNH controls. A marked difference in the profiles of four circulating miRNAs (miR-205, miR-210, miR-492, and miR-1427) was observed in pancreatic juice collected from patients with PDAC and those without pancreatic disease. Elevated levels of the four miRNAs together predicted PDAC with a specificity of 88% and sensitivity of 87%. Inclusion of serum CA19-9 level increased the sensitivity to 91% and the specificity to 100%. Enrichment of the four miRNAs in pancreatic juice was associated with decreased OS, as was the combination of miR-205 and miR-210. Higher contents of miR-205 and miR-210 were also associated with lymph node metastasis. Elevated levels of circulating miR-205, miR-210, miR-492, and miR-1247 in pancreatic juice are, therefore, promising candidate biomarkers of disease and poor prognosis in patients with PDAC.
Keywords: miRNA, circulating, biomarker, pancreatic juice, pancreatic cancer.
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the United States, with a 5‑year survival rate of about 6% 1. When the tumor is detected at an early stage, surgical resection and adjuvant chemotherapy or radiotherapy can increase the 5-year survival rate to 15-40% 2. Thus, screening strategies for detection of early stage cancer or precursor lesions in individuals at highest risk, as well as pancreatic cancer biomarker assays for biological fluids, are urgently needed to reduce the high morbidity and mortality associated with this malignancy.
Exocrine pancreatic secretions, or pancreatic juice, collected from the second part of the duodenum after stimulation with intravenous synthetic human secretin using an upper endoscope 3-5 have been analyzed for molecular markers associated with pancreatic ductal adenocarcinoma (PDAC) 6, 7. Protein and gene expression profiles in the pancreatic juice of patients with PDAC have revealed novel protein markers and indicator genes that might be useful for PDAC diagnosis 8, 9. More recently, NGAL and MIC-1 in pancreatic juice were reported to be useful markers for diagnosing benign and malignant disease of the pancreas 10. However, since tumor-associated proteins may constitute only a minor fraction of the vast and dynamic range of proteins in blood plasma and other body fluids, proteomic-based strategies for identification of circulating cancer biomarkers have had limited success so far 11.
MicroRNAs (miRNAs) are a class of conserved small non-coding RNAs, which regulate gene expression via complementarity with the 3'-untranslated region of their target mRNAs, repressing translation and causing degradation of multiple target mRNAs 12. MicroRNAs have critical regulatory functions in cell development, proliferation, differentiation, apoptosis, and stress response. A number of studies in the past few years have established the regulatory roles of miRNAs in complex genetic networks underlying various cellular pathways 13. Aberrant expression of several miRNAs has been detected in PDAC and its precursor, pancreatic intraepithelial neoplasm 14. Varying levels of circulating miRNAs in body fluids have been associated with corresponding tumor-specific aberrant expression and proposed to represent novel biomarkers for detection and diagnosis of various cancers 15, 16. We have earlier demonstrated that miRNAs in the plasma of patients with pancreatic ductal adenocarcinoma (PDAC) may serve as novel blood-based biomarkers of disease 17. Serum and pancreatic cyst fluid levels of miR‑21, miR‑210 and miR-155 have been found elevated in patients with PDAC 18-21. Elevated serum level of miR-1290 has recently been reported to accurately distinguish patients with low stage pancreatic cancer from healthy and disease controls 22.
Altered levels of miRNAs in blood and body fluids can distinguish patients with cancer from healthy individuals 16, 17, 23. In view of a published report that relative expression levels of miR-21 and miR-155 in tissue and pancreatic juice from PDAC patients were significantly higher than those with chronic pancreatitis 24, it appears that unbiased profiling of circulating miRNAs in pancreatic juice may offer a viable approach for developing reliable biomarker assays for detecting PDAC. In this study, we performed microarray profiling of miRNAs in the pancreatic juice of patients with PDAC compared with those from individuals with chronic pancreatitis and non-pancreatic disease controls to investigate the utility of differentially circulating miRNAs as biomarkers for pancreatic cancer.
Materials and Methods
Patients
This prospective study was approved by the Mayo Clinic and M. D. Anderson Cancer Center Institutional Review Boards. Patients presenting at the Mayo Clinic with epigastric pain or discomfort warranting evaluation by endoscopic ultrasound-guided fine needle aspiration and later diagnosed with chronic pancreatitis (CP), pancreatic cancer or no pancreatic disease (non-pancreatic, non-healthy [NPNH] controls) were enrolled. Pancreatic juice and serum samples were collected. Sera were assayed for CA19-9 and the definitive final diagnosis was made. A multiphase, case-control study was designed to identify pancreatic juice miRNAs as potential markers for PDAC. In the initial biomarker identification stage, miRNA microarray profiling was performed on six pancreatic juice samples of PDAC patients and two pooled pancreatic juice samples from 6 NPNH controls to assay the circulating miRNA profiles. Validation of four differentially enriched miRNAs was then performed using qRT-PCR assay. All the pancreatic juice samples were separated randomly into training set (6 PDAC and 6 NPNH controls) and validation set (44 PDAC, 19 CP and13 NPNH controls) prior to analysis from a total of 88 patients enrolled in the study as described previously 10.
Collection of Secretin-Stimulated Exocrine Pancreatic Secretions (Pancreatic Juice)
All patients underwent upper endoscopic intubation after an overnight fast. Patients were moderately sedated with midazolam and meperidine before the procedure. Synthetic human secretin (ChiRhoStim; ChiRhoClin Inc, Burtonsville, MD) at a dose of 0.2 μg/kg (or a total dose of 16 μg) was administered intravenously for 1 minute immediately before endoscopic intubation. Gastric fluid was aspirated before intubation of the pylorus to minimize contamination. With the endoscope positioned in the second portion of the duodenum, opposite the papilla of Vater (without cannulation), a 2.3-mm plastic aspiration catheter (Hobbs Medical, Stafford Springs, CT) was passed through the biopsy channel of the endoscope until it was visible on the endoscopic monitor screen. Pancreatic juice exiting the papilla was then suctioned through the catheter for 10 minutes into a dry 20-mL tube attached to the proximal end of the catheter. On average, 5-8 mL of fluid was collected from each patient. The collected fluid was immediately aliquoted into 2-mL vials, snap-frozen in liquid nitrogen, and stored at -80°C until assays for miRNA microarrays and real-time polymerase chain reaction were performed. A pan-protease inhibitor cocktail pill (Roche Diagnostic Corporation, Indianapolis, IN) was added to the collected pancreatic juice to inhibit proteolytic degradation.
Determination of Final Diagnosis
Three gastroenterologists (Massimo Raimondo, Michael B. Wallace, and Timothy A. Woodward), who were blinded to the results of the miRNA assays, determined the final diagnosis of the patients on the basis of clinical history and laboratory, radiologic (computed tomography or magnetic resonance imaging), or endoscopic (endoscopic ultrasound or endoscopic retrograde cholangiopancreatographic) findings (chronic pancreatitis) and cytologic or histologic findings (for those with pancreatic ductal adenocarcinoma). Patients with no prior history of pancreatic disease and who had normal pancreatic test results (ultrasound, cholangiopancreatography, computed tomography, or magnetic resonance imaging) were deemed to have normal pancreata and thus considered to be non-pancreatic, non-healthy (NPNH) controls.
CA19-9 Measurement
Concentrations of CA19-9 in the pancreatic juice and serum of patients were measured, as described previously 10. Briefly, CA19-9 antigen concentration was determined using a solid-phase radioimmunoassay (Centocor, Malvern, PA), according to the manufacturer's instructions. Quantities of CA19-9 were expressed in arbitrary units (U/mL) with 1 unit of activity corresponding to approximately 0.8 ng of purified antigenic protein for CA19-9.
miRNA Extraction and Purification
Pancreatic juice was cleared through centrifugation at 1,300 × g at 4°C for 10 minutes and stored at -80°C. Total RNA was extracted and purified from all pancreatic juice samples. For the purpose, cleared pancreatic juice was mixed with Trizol LS (1:3 ratio; Life Technologies, Grand Island, NY), and after phase separation by centrifugation, the aqueous phase was extracted once with phenol/chloroform and added to ethanol before being applied directly to a mirVana miRNA column (Ambion, Austin, TX) according to the manufacturer's instructions. The concentration of RNA samples was quantified using NanoDrop 2000 (NanoDrop, Wilmington, DE). The total amount of RNA extracted from 500 µL of pancreatic juice ranged from 0.21 to 18.90 µg (mean, 1.34 µg).
miRNA Microarray Labeling, Hybridization, and Data Analysis
The G4470A Agilent human miRNA microarray (Agilent Technologies, Santa Clara, CA) was used to profile miRNA. RNA labeling and hybridization were performed using the Agilent miRNA labeling reagent and hybridization kit according to the manufacturer's protocol. Samples containing 100 ng of RNA were labeled with pCp-Cy3 using T4 RNA ligase (Ambion) and hybridized to microarrays. Microarrays were scanned using an Agilent microarray scanner, data were extracted, and array quality control was performed using Agilent Feature Extraction software. The AgiMicroRna package was used to process Agilent miRNA array data. In addition, hierarchical clustering was performed using Pearson correlation metrics with Ward's linkage.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Taqman miRNA assays (Applied Biosystems, Foster City, CA) were used to perform expression profiling of the miRNAs of interest. RNA was pretreated with DNase (Invitrogen). Ten ng of DNAse-treated RNA from each sample of pancreatic juice was used for each assay. Assays were performed in 15-μL reactions containing the reverse transcription mixture and Taqman primer mix. miRNA expression levels were quantified using the ViiA™ 7 Real-Time PCR System (Life Technologies). Relative expression of the mature miRNAs in terms of fold change in the test samples (PDAC or CP) compared with the non-pancreatic, non-healthy (NPNH) controls was calculated using the comparative CT (2-ΔΔCT) method, using RNU6B as the endogenous control to normalize the data, where CT (cycle threshold) is defined as the number of cycles required for the FAM signal to cross the threshold in real-time PCR. ΔCT was calculated by subtracting the CT values of RNU6B from the CT values of the miRNA of interest. ΔΔCT was then calculated by subtracting the mean ΔCT of NPNH controls from the ΔCT of test samples (PDAC or CP).
Locked Nucleic Acid In Situ Hybridization (LNA-ISH)
For analyzing miRNA expression formalin-fixed, paraffin-embedded tumor tissue slides were toasted, de-waxed in xylenes, and rehydrated through an ethanol dilution series. Tissue sections were digested with 15 μg/mL proteinase K and then treated with 3% hydrogen peroxide. Tissue sections were then subjected to prehybridization solution at 50°C for 30 minutes. The prehybridization solution was replaced with 50 μL of hybridization solution containing 10 pmol of the fluorescein isothiocyanate-labeled LNA probes for the miRNAs. Slides were blocked with 5% bovine serum albumin, incubated with anti-Fluorescein-HRP conjugated antibodies, and subjected to the CSA II Biotin-free Tyramide Signal Amplification System Kit (DAKO, Carpenteria, CA) as described previously 25.
Statistical Analysis
All clinicopathologic variables and miRNA expression levels in pancreatic juice were analyzed using SPSS v19 (SPSS Inc, Chicago, IL). Unpaired t tests were used to compare pancreatic juice miRNA expression between groups and p values <0.05 were considered statistically significant. The relationship between miRNA expression and overall survival (OS) was evaluated using Cox proportional hazards regression models. Hazard ratios and 95% confidence intervals (CIs) were calculated, with adjustments made for sex, ethnicity, and other clinical factors that were significant predictors of OS in multivariate Cox regression models. Receiver operating characteristic (ROC) curves were constructed and the area under the curve (AUC) was calculated to evaluate the ability of each individual miRNA and CA19-9 either individually or in combination to detect pancreatic cancer. OS curves were estimated using the Kaplan-Meier method. The median expression level of each miRNA was used as the cutoff point in Kaplan-Meier survival analysis.
Results
Patient Characteristics
Among the 88 patients enrolled in the study (training and validation sets), 19 were NPNH controls, 19 were diagnosed with CP, and 50 were diagnosed with PDAC. Patient demographic and clinicopathologic information is presented in Table 1. Among the 50 patients with PDAC, univariate analysis by SPSS showed that OS did not differ by age, sex, smoking and alcohol history, diabetes mellitus, CA19-9 findings, or lymph node involvement. The median OS duration of the 50 patients with PDAC was 7.90 months (95% CI, 0.417-41.09 months). However, the median follow-up time was 12.43 months for the patients who were still alive at the time of analysis. Well to moderate tumor differentiation was associated with increased OS duration (P=0.005).
Table 1.
Characteristics of the Study Population.
| Characteristic | NPNH (n = 19) | CP (n = 19) | PDAC (n = 50) | |||
|---|---|---|---|---|---|---|
| No. of patients | No. of death | MST (months) | P value | |||
| Age, years | 0.188 | |||||
| <60 | 9 | 8 | 12 | 7 | 10.73 | |
| 61-70 | 6 | 5 | 18 | 15 | 8.70 | |
| >70 | 4 | 6 | 20 | 13 | 4.37 | |
| Sex | 0.109 | |||||
| Male | 5 | 11 | 30 | 22 | 5.70 | |
| Female | 14 | 8 | 20 | 13 | 10.73 | |
| Ethnicity | 0.047 | |||||
| White | 16 | 17 | 48 | 34 | 8.00 | |
| Black | 2 | 1 | 2 | 1 | 2.07 | |
| Hispanic | 1 | 0 | 0 | 0 | ||
| Asian | 0 | 1 | 0 | 0 | ||
| Smoking historya | 0.347 | |||||
| Current | 2 | 7 | 9 | 6 | 5.63 | |
| Former | 6 | 6 | 15 | 8 | 8.70 | |
| Never | 11 | 6 | 25 | 21 | 5.20 | |
| History of alcohol usea | 0.696 | |||||
| Current | 9 | 9 | 24 | 14 | 6.77 | |
| Former | 5 | 5 | 8 | 7 | 8.70 | |
| Never | 5 | 5 | 17 | 14 | 10.27 | |
| Diabetes mellitusa | 0.451 | |||||
| Yes | 3 | 5 | 10 | 7 | 3.93 | |
| No | 16 | 14 | 39 | 28 | 8.70 | |
| CA19-9, U/mL | 0.404 | |||||
| <50 | 9 | 6 | 6 | 3 | 9.77 | |
| 51-500 | 1 | 1 | 4 | 2 | 2.07 | |
| 501-1500 | 3 | 4 | 5 | 4 | 2.33 | |
| >1500 | 6 | 8 | 35 | 26 | 7.90 | |
| Lymph node b | 0.290 | |||||
| 0 | 19 | 19 | 35 | 26 | 8.00 | |
| 1 | 0 | 0 | 13 | 9 | 6.77 | |
| Tumor differentiationa | 0.005 | |||||
| Well to moderate | - | - | 9 | 5 | 18.33 | |
| Poor or not available | - | - | 40 | 30 | 5.63 | |
aSmoking history, history of alcohol use, diabetes status and tumor differentiation were not available for 1 patient with PDAC. b lymph node was not available for 2 patient with PDAC.
Pancreatic Juice miRNA Profiles of Patients with PDAC
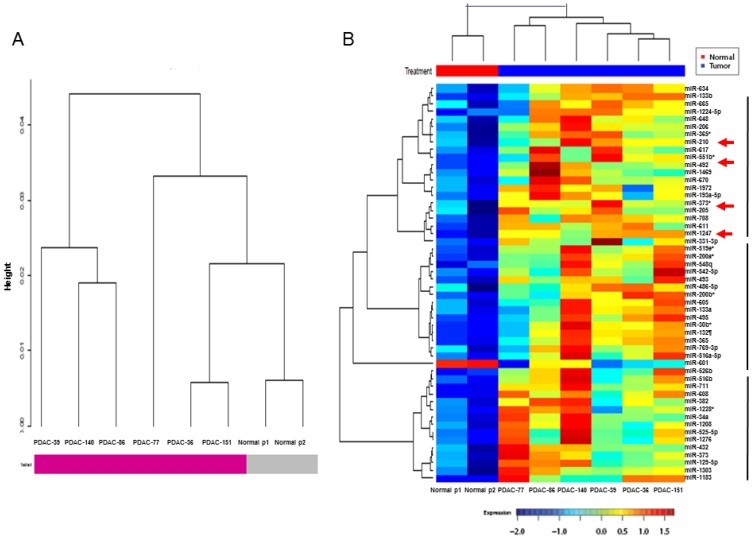
The pancreatic juice miRNA microarray profiles of 6 patients with PDAC were compared with those of pooled samples from 6 NPNH controls. Hierarchical clustering of miRNA expression using the Pearson correlation distance metric and Ward's linkage revealed clusters of patients with PDAC and NPNH controls after Robust Multi-Array Average (RMA) normalization (Fig. 1A). Levels of 49 miRNAs differed by at least 1.5-fold in the pancreatic juice of patients with PDAC compared with that of NPNH controls (P < 0.05; Additional File 1: Supplementary Table S1). To statistically analyze the similarity of pancreatic juice miRNA profiles among patients with PDAC, we generated a dendrogram of cancer-dependent miRNAs, in which PDAC pancreatic juice samples with similar miRNA profiles were grouped together (Fig. 1B). Multiple miRNAs with elevated profiles in the pancreatic juice, including miR-205 and miR-210, which are known to be overexpressed in PDAC tissue 26, 27, were found to segregate into distinct clusters in the dendrogram generated. We selected miR-492 and miR-1247, the top two miRNAs in the same cluster as miR-205 and miR-210, which have not been previously reported in PDAC, for validation as novel candidate biomarkers circulating at elevated levels in the pancreatic juice of patients with PDAC.
Figure 1.
Hierarchical clustering analysis of microRNA microarray expression profile. (A) Clusters of patients with pancreatic ductal adenocarcinoma (PDAC) and non-pancreatic, non-healthy controls (NPNH). (B) Hierarchical clustering of microRNA expression in PDAC pancreatic juice samples with similar miRNA profiles were grouped together, which showed that miR-492, miR-1247, miR-205 and miR-210 were in the same cluster.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis of Candidate miRNAs
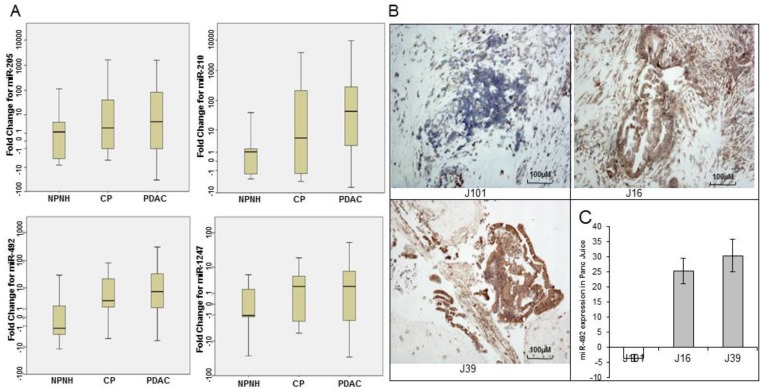
To validate the differential enrichment of four candidate miRNA biomarkers in pancreatic juice, identified on expression microarrays, we used qRT-PCR assay for 4 miRNAs (miR-205, miR-210, miR-492, and miR-1247) in individual pancreatic juice samples (training set). The relative abundance of four miRNAs, normalized to the level of RNU6B, significantly varied in pancreatic juice samples from patients with PDAC and NPNH. The levels of four miRNAs were further assayed in a larger cohort consisting of an independent set of pancreatic juice samples (validation set). The p values revealed that the levels of these miRNAs showed statistically significant differences among patients in these three groups (PDAC, CP and NPNH; miR-205, P = 0.04; miR-210, P = 1.90E-05; miR-492, P = 0.005; miR-1247, P = 0.04; Fig. 2A and Table 2). The mean fold changes of miRNAs and the p values reflected segregation of PDAC, CP, and NPNH samples in terms of varying miRNA levels, with the levels relatively high in PDAC, low in controls and intermediate in CP although the difference between PDAC and CP were not significant.
Figure 2.
Quantitative real-time polymerase chain reaction (qRT-PCR) and Locked nucleic acid in situ hybridization (LNA-ISH) analysis of expression levels of microRNAs. (A) qRT-PCR analysis of relative expression levels of 4 microRNAs (miR-205, miR-210, miR-492, and miR-1247) in pancreatic juice samples from patients with pancreatic ductal adenocarcinoma (PDAC), patients with chronic pancreatitis (CP), and non-pancreatic, non-healthy (NPNH) controls. The relative abundance of four miRNAs, normalized to the level of RNU6B, significantly varied in pancreatic juice samples from patients with PDAC, CP and NPNH. (B) LNA-ISH-IHC images showing miR-492 expression in 3 (J101, J16, J39) pancreatic ductal adenocarcinoma tissue samples (magnification ×100). (C) The graph shows qRT-PCR results for miR-492 in pancreatic juice samples from the same patients.
Table 2.
MicroRNA (miRNA) expression in samples of pancreatic juice from patients with pancreatic ductal adenocarcinoma (PDAC); Chronic pancreatitis (CP); Non-pancreatic, non-healthy controls (NPNH).
| miRNA expression | NPNH samples (n = 19), no. (%) | CP samples (n = 19), no. (%) | PDAC samples (n = 50), no. (%) | P value |
|---|---|---|---|---|
| miR-205 | 0.04 | |||
| Low | 14 (74) | 10 (53) | 20 (40) | |
| High | 5 (26) | 9 (47) | 30 (60) | |
| miR-210 | 1.90E-05 | |||
| Low | 18 (95) | 10 (53) | 16 (32) | |
| High | 1 (5) | 9 (47) | 34 (68) | |
| miR-492 | 0.005 | |||
| Low | 16 (84) | 10 (53) | 20 (40) | |
| High | 3 (16) | 9 (47) | 30 (60) | |
| miR-1247 | 0.04 | |||
| Low | 13 (76) | 9 (47) | 20 (41) | |
| High | 4 (24) | 10 (53) | 29 (59) | |
| miR-205/ miR-210 | 8.31E-04 | |||
| 0-1 | 19 (100) | 13 (68) | 26 (52) | |
| 2 | 0 | 6 (32) | 24 (48) |
The relative increase in the levels of the candidate miRNAs in pancreatic juice samples from patients with PDAC was significantly higher than in samples from NPNH controls (miR-205, P = 0.009; miR-210, P = 0.010; miR-492, P= 0.003; miR-1247, P= 0.041; Additional File 1: Supplementary Table S2). These results were in good agreement with microarray and training set results. Levels of the candidate miRNAs were higher in the CP samples than in the NPNH samples, although the difference was statistically significant for only miR-492. Levels of miR-205, miR-210, and miR-1247 did not differ between the CP samples and the NPNH samples. These miRNAs were significantly elevated in PDAC samples compared with NPNH samples but not compared with the CP samples.
Expression of miRNAs in PDAC Tumor Samples
To investigate whether varying levels of miRNAs detected in the pancreatic juice of patients with PDAC reflected differential expression of the miRNAs in pancreatic tumor tissues, we performed LNA-ISH with the miRNA probes on a limited set of 12 available tumor tissue samples. miRNAs detected by LNA-ISH in the tumor tissues (Fig. 2B) were consistent with those detected in the corresponding pancreatic juice samples (Fig. 2C).
To estimate miRNA-492 and miRNA-1247 differential expression in PDAC, qRT-PCR analysis of miRNA expression was also undertaken in 34 paired samples of PDAC tissue and adjacent normal tissue. Twenty PDAC tumor samples (59%) showed increased expression of miR-492, and only 13 samples (38%) showed increased expression of miR-1247 (data not shown).
Pancreatic Juice miRNA Profiles as Biomarkers of PDAC
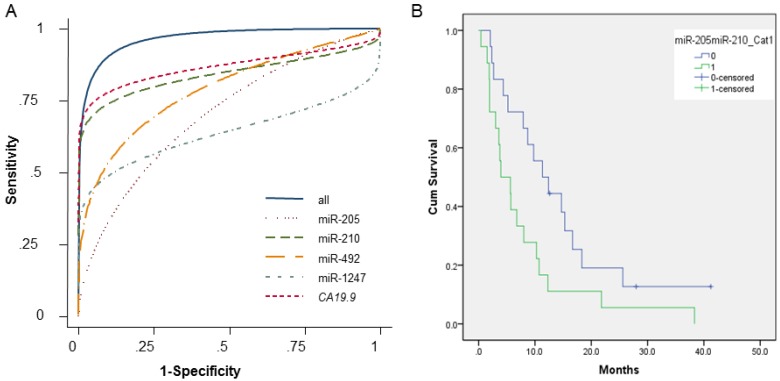
The ability of the 4 miRNAs to differentiate PDAC pancreatic juice samples from NPNH pancreatic juice samples was assessed using ROC curves, which indicated that the 4 miRNAs are candidate biomarker of PDAC (miR-205 AUC = 0.68; miR-210 AUC = 0.84; miR-492 AUC = 0.80; miR-1247 AUC = 0.63). For all 4 miRNAs combined, the AUC was 0.92, documenting that the altered levels of the 4 miRNAs in pancreatic juice can differentiate patients with PDAC from NPNH controls. When serum expression of CA19-9 was considered along with the 4 miRNAs, the AUC was 0.99, which was significantly improved compared with both CA19-9 alone (AUC = 0.89) and the combination of the 4 miRNAs without CA19-9 (Fig. 3A and Additional File 1: Supplementary Table S3).
Figure 3.
Receiver operating characteristic curves (ROC) and Kaplan-Meier plots showing association of pancreatic juice miRNA profiles in PDAC patients and their clinical outcome. (A) Receiver operating characteristic curves showing expression levels of individual microRNAs (miR-205, miR-210, miR-492, and miR-1247), CA19-9, and a combination. (B) Kaplan-Meier overall survival curves for patients with pancreatic ductal adenocarcinoma with high levels of both miR-205 and miR-210 at or above the median (shown in green); either miR-205 or miR-210, or neither upregulated in pancreatic juice samples (shown in blue). The curves were compared using univariate (log-rank) analysis.
ROC curves helped determine the sensitivities and specificities of the miRNAs at various cutoff values (Supplementary Table S3). Using the optimal cutoff points, sensitivity and specificity were 64% and 74%, respectively, for miR-205; 76% and 95% for miR-210; 73% and 82% for miR-492; and 53% and 88% for miR-1247. The sensitivity and specificity for the 4 miRNAs combined were 87% and 88%, respectively. Finally, the combination of the 4 miRNAs plus CA19-9 had a sensitivity of 91% and a specificity of 100%, which was significantly improved than CA 19-9 alone (77% sensitivity, 93% specificity).
Pancreatic Juice miRNAs as Biomarkers for PDAC Prognosis, Tumor Differentiation, and Lymph Node Metastasis
We performed Kaplan-Meier analyses to assess the relationship between pancreatic juice miRNA levels and clinical outcome, using the median expression level as a cutoff (at or above the median = high expression; below the median = low expression), in 36 PDAC patients for whom all the clinical data were available. Multivariate Cox regression models for individual miRNAs and combinations of miRNAs are shown in Table 3. Patients with low levels of miR-205 or miR-210 (individually) had longer OS durations than patients with high levels of these miRNAs, but the difference was not statistically significant (miR-205 log-rank P = 0.166; miR-210 log-rank P = 0.211). However, we found that elevated levels of all 4 miRNAs was associated with shorter OS (log-rank P = 0.050), as was high expression of both miR-205 and miR-210 together (log-rank P = 0.029). Patients with low levels of both miR-205 and miR-210 tended to have longer OS durations than did patients with high levels of both miR-205 and miR-210 (Fig. 3B).
Table 3.
Median overall survival time (MST) of pancreatic ductal adenocarcinoma (PDAC) patients with high and low levels of circulating miRNAs in pancreatic juice.
| miRNA expression | No. of patients | No. of deaths | MST, months | Log-rank P value | HR (95% CI)a | P value |
|---|---|---|---|---|---|---|
| miR-205 | ||||||
| Low b | 15 | 13 | 12.40 | 0.166 | 1.46(0.66-3.12) | 0.347 |
| High c | 21 | 20 | 5.63 | |||
| miR-210 | ||||||
| Low | 11 | 9 | 11.33 | 0.211 | 1.78(0.66-4.80) | 0.254 |
| High | 25 | 24 | 6.77 | |||
| miR-492 | ||||||
| Low | 17 | 15 | 7.90 | 0.678 | 0.82(0.36-1.83) | 0.620 |
| High | 19 | 18 | 8.70 | |||
| niR-1247 | ||||||
| Low | 14 | 14 | 6.77 | 0.586 | 1.47(0.67-3.23) | 0.334 |
| High | 21 | 18 | 7.90 | |||
|
miR-205/miR-210/ miR-492/miR-1247 |
||||||
| 0-3d | 32 | 29 | 8.70 | |||
| 4e | 4 | 4 | 3.77 | 0.050 | 2.87 (0.93-8.87) | 0.067 |
| miR-205/miR-210 | ||||||
| 0-1f | 18 | 15 | 11.33 | |||
| 2g | 18 | 18 | 3.93 | 0.029 | 2.14 (1.06-4.29) | 0.030 |
aHazard ratios (HR) derived from multivariate Cox regression models adjusted for sex, ethnicity, CA19-9 levels, and stage; bLow < the median expression level; cHigh ≥ the median expression level; d0-3: Patients with ≤ three miRNAs (miR-205, miR-210, miR-492 and miR-1247) upregulated in pancreatic juice; e4: Patients with four miRNAs upregulated in pancreatic juice; f0-1: Patients with either miR-205 or miR-210 upregulated in pancreatic juice; g2: Both miR-205 and miR-210 upregulated in pancreatic juice of the same patient.
Patients with low levels of both miR-205 and miR-210 appeared less likely to have lymph node metastasis than patients with high levels of both miR-205 and miR-210 Additional File 1: Supplementary Table S4). Patients with low levels of miR-205 also tended to have well to moderately differentiated tumors (P = 0.001) (Additional File 1: Supplementary Table S4).
Discussion
Elevated levels of microRNAs have been detected in precursor lesions of pancreatic cancer and were also associated with poorer survival of pancreatic cancer patients (26, 27. Our results indicate that miR-205, miR-210, miR-492, and miR-1247 in pancreatic juice are candidate biomarkers of PDAC, which perform even better when combined with serum CA19-9 levels. We also found that expression of these 4 miRNAs is associated with decreased OS in patients with PDAC. In addition, elevated amount of miR-205 and miR-210 was associated with lymph node metastasis, and expression of miR-205 was associated with poorly differentiated tumors, both indicators of worse prognosis. These findings suggest that pancreatic juice levels of miR-205, miR-210, miR-492, and miR-1247, as a combined signature can predict the clinical outcome of patients with PDAC.
Microarray analyses identified 49 miRNAs as differentially enriched in the pancreatic juice of patients with PDAC compared with controls; of these, miR-200a, miR-200b, and miR-30b have also been found to be upregulated in PDAC cells and tumor tissue 28. In addition, high expression of miR-205 was found in head and neck cancer cell lines 29, squamous cell carcinoma cell lines 30, and cervical cancer cell lines 31, as well as in clinical samples of metastatic head and neck squamous cell carcinoma 32, cervical cancer 31, non-small cell lung cancer 33, and endometrial cancer 34. Expression of miR-205 in clinical prostate cancer samples was associated with prostate cancer progression 35 and invasive bladder cancers 36. Up-regulation of miR-205 was also found to precede phenotypic changes in ductal cells in a conditional KRAS (G12D) mouse model of pancreatic cancer 27.
Aberrant high expression of miR-210 has been detected in many tumors 26, 37-42., Elevated miR-210 has been implicated in tumor progression 43 and is associated with negative clinical outcomes 26, 41, 42. Interestingly, increased plasma miR-210 levels were associated with lymph node metastases and trastuzumab resistance in breast cancer patients 44. These findings are to some extent consistent with our current observation that high miR-205 and miR-210 levels in pancreatic juice correlate with decreased OS, poorly differentiated tumors, and lymph node metastasis. These observations make a compelling case for miR-205 and miR-210 in pancreatic juice as informative biomarkers for PDAC.
A recent study has reported miR-1247 to be down-regulated in pancreatic cancer correlating with tumor grade and shorter overall survival of the patients 45. miR-492 has also been shown to potentially play an important role in the progression of malignant embryonal liver tumors 46. Although our study did not reveal significant differences in the levels of these miRNAs between CP and PDAC, it is plausible that this was due to relatively small sample size and thus lack of enough statistical power in the current study. However, in view of these miRNAs discriminating PDAC from NPNH controls with reasonably high sensitivity and specificity as well as showing significant correlation with decreased OS and lymph node metastasis, it is important that the miRNA signature be investigated in a larger patient population in the future. The current finding of the four miRNAs signature in pancreatic juice as a biomarker panel of pancreatic cancer suggest that these miRNAs are involved in critical cancer relevant genetic networks deregulated in PDAC.
Collection of pancreatic juice is a minimally invasive procedure which can easily detect circulating or secreted miRNAs as concentrations of miRNAs are higher than in blood (serum or plasma). This is important as circulating miRNAs in blood can potentially decrease over time whereas they can be still detectable in pancreatic juice. For detection of early PDAC, this procedure will be preferable as concentrations of miRNAs will be higher in pancreatic juice than in blood. Moreover, our technique can be used as part of routine diagnostic procedure to establish diagnosis/staging of PDAC and does not involve cannulation of pancreatic duct to collect pancreatic juice.
Finally, we recognize that the procedure is expensive if secretin is used and more invasive unlike detecting miRNAs in blood., The collection and storage procedures need to be standardized for a broader applicability of this technique. It is important to have stringent quality control standards for collection of pancreatic juice and pancreatic juice miRNAs expression analysis besides ensuring that sample size is adequate for selecting the most informative candidate biomarker miRNAs capable of discriminating patients with pancreatic cancer, pancreatitis versus no pancreatic disease.
In summary, our study demonstrates that miRNAs in exocrine pancreatic secretions are promising biomarkers of PDAC. Further comprehensive molecular and functional characterizations of miRNAs circulating at altered levels in pancreatic juice are warranted to elucidate the role of these miRNAs in pancreatic carcinogenesis and validate them in larger sample cohorts as biomarker panels for detection, prognosis as well as response to therapy for this highly lethal malignancy.
Supplementary Material
Supplementary Tables S1-S4.
Acknowledgments
The biomarker discovery work in Subrata Sen's laboratory is supported by a grant (U01 CA111302) from the Early Detection Research Network of the National Cancer Institute/National Institutes of Health, USA. The University of Texas MD Anderson Cancer Center is supported in part by a Cancer Center Support Grant (CA016672) from the National Institutes of Health.
Abbreviations
- AUC
area under the curve
- CA19-9
carbohydrate antigen 19-9
- CP
chronic pancreatitis
- LNA-ISH
locked nucleic acid in situ hybridization
- miRNA
microRNA
- NPNH
non-pancreatic, non-healthy
- OS
overall survival
- PDAC
pancreatic ductal adenocarcinoma
- qRT-PCR
quantitative real-time polymerase chain reaction
- ROC
receiver operating characteristic.
References
- 1.Jemal A, Siegel R, Xu J. et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Yeo CJ, Cameron JL, Lillemoe KD. et al. Pancreaticoduodenectomy for cancer of the head of the pancreas. 201 patients. Ann Surg. 1995;221(6):721–731. doi: 10.1097/00000658-199506000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohuchida K, Mizumoto K, Fujita H. et al. Sonic hedgehog is an early developmental marker of intraductal papillary mucinous neoplasms: clinical implications of mRNA levels in pancreatic juice. J Pathol. 2006;210(1):42–48. doi: 10.1002/path.2019. [DOI] [PubMed] [Google Scholar]
- 4.Noh KW, Pungpapong S, Wallace MB. et al. Do cytokine concentrations in pancreatic juice predict the presence of pancreatic diseases? Clin Gastroenterol Hepatol. 2006;4(6):782–789. doi: 10.1016/j.cgh.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Raimondo M, Imoto M, DiMagno EP. Rapid endoscopic secretin stimulation test and discrimination of chronic pancreatitis and pancreatic cancer from disease controls. Clin Gastroenterol Hepatol. 2003;1(5):397–403. doi: 10.1053/s1542-3565(03)00182-4. [DOI] [PubMed] [Google Scholar]
- 6.Ohuchida K, Mizumoto K, Yu J. et al. S100A6 is increased in a stepwise manner during pancreatic carcinogenesis: clinical value of expression analysis in 98 pancreatic juice samples. Cancer Epidemiol Biomarkers Prev. 2007;16(4):649–654. doi: 10.1158/1055-9965.EPI-06-0157. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima A, Murakami Y, Uemura K. et al. Usefulness of human telomerase reverse transcriptase in pancreatic juice as a biomarker of pancreatic malignancy. Pancreas. 2009;38(5):527–533. doi: 10.1097/MPA.0b013e3181a16d28. [DOI] [PubMed] [Google Scholar]
- 8.Tian M, Cui YZ, Song GH. et al. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer. 2008;8:241. doi: 10.1186/1471-2407-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira-Cunha M, Byers RJ, Siriwardena AK. Poly(A) RT-PCR measurement of diagnostic genes in pancreatic juice in pancreatic cancer. Br J Cancer. 2011;104(3):514–519. doi: 10.1038/sj.bjc.6606047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaur S, Baine MJ, Guha S. et al. Neutrophil gelatinase-associated lipocalin, macrophage inhibitory cytokine 1, and carbohydrate antigen 19-9 in pancreatic juice: pathobiologic implications in diagnosing benign and malignant disease of the pancreas. Pancreas. 2013;42(3):494–501. doi: 10.1097/MPA.0b013e31826a8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 12.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Sen S. MicroRNA functional network in pancreatic cancer: from biology to biomarkers of disease. J Biosci. 2011;36(3):481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- 14.Xue Y, Abou Tayoun AN. et al. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 2013;206(6):217–221. doi: 10.1016/j.cancergen.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Thosani N, Dasari CS, Bhutani MS. et al. Molecular pathogenesis of intraductal papillary mucinous neoplasms of the pancreas. Pancreas. 2010;39(8):1129–1133. doi: 10.1097/MPA.0b013e3181f66cdf. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Ba Y, Ma L. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Chen J, Chang P. et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho AS, Huang X, Cao H. et al. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Gao J, Du Y. et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 20.Ryu JK, Matthaei H, Dal Molin M. et al. Elevated microRNA miR-21 levels in pancreatic cyst fluid are predictive of mucinous precursor lesions of ductal adenocarcinoma. Pancreatology. 2011;11(3):343–350. doi: 10.1159/000329183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X, Du Y, Wang G. et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56(2):602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 22.Li A, Yu J, Kim H. et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19(13):3600–3610. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell PS, Parkin RK, Kroh EM. et al. Circulating microRNAs as stable blood-based markers for cancer detection. P Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadakari Y, Ohtsuka T, Ohuchida K. et al. MicroRNA expression analyses in preoperative pancreatic juice samples of pancreatic ductal adenocarcinoma. JOP. 2010;11(6):587–592. [PubMed] [Google Scholar]
- 25.Sempere LF, Preis M, Yezefski T. et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res. 2010;16(16):4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greither T, Grochola LF, Udelnow A. et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 27.du Rieu MC, Torrisani J, Selves J. et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56(4):603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 28.Kent OA, Mullendore M, Wentzel EA. et al. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol Ther. 2009;8(21):2013–2024. doi: 10.4161/cbt.8.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang JM, Lee EJ, Gusev Y. et al. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33(17):5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Ryan DG, Getsios S. et al. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. P Natl Acad Sci USA. 2008;105(49):19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Tang S, Le SY. et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PloS One. 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fletcher AM, Heaford AC, Trask DK. Detection of Metastatic Head and Neck Squamous Cell Carcinoma Using the Relative Expression of Tissue-Specific Mir-205. Transl Oncol. 2008;1(4):202–209. doi: 10.1593/tlo.08163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markou A, Tsaroucha EG, Kaklamanis L. et al. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54(10):1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 34.Karaayvaz M, Zhang C, Liang SR, Prognostic Significance of miR-205 in Endometrial Cancer. PloS One. 2012. 7(4) [DOI] [PMC free article] [PubMed]
- 35.Schaefer A, Jung M, Mollenkopf HJ. et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 36.Wszolek MF, Rieger-Christ KM, Kenney PA. et al. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol Oncol. 2011;29(6):794–801. doi: 10.1016/j.urolonc.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 37.Giannakakis A, Sandaltzopoulos R, Greshock J. et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7(2):255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porkka KP, Pfeiffer MJ, Waltering KK. et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 39.Lawrie CH, Gal S, Dunlop HM. et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 40.Iorio MV, Ferracin M, Liu CG. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 41.Camps C, Buffa FM, Colella S. et al. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 42.Gee HE, Camps C, Buffa FM. et al. hsa-mir-210 Is a Marker of Tumor Hypoxia and a Prognostic Factor in Head and Neck Cancer. Cancer. 2010;116(9):2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 43.Chan YC, Banerjee J, Choi SY. et al. miR-210: The Master Hypoxamir. Microcirculation. 2012;19(3):215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung EJ, Santarpia L, Kim J. et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118(10):2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi S, Lu Y, Qin Y. et al. miR-1247 is Correlated with Prognosis of Pancreatic Cancer and Inhibits Cell Proliferation by Targeting Neuropilins. Curr Mol Med. 2014;14:316–327. doi: 10.2174/1566524014666140228120014. [DOI] [PubMed] [Google Scholar]
- 46.von Frowein J, Pagel P, Kappler R. et al. MicroRNA-492 is Processed from the Keratin 19 Gene and Up-Regulated in Metastatic Hepatoblastoma. Hepatology. 2011;53(3):833–842. doi: 10.1002/hep.24125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1-S4.