Abstract
A tendency in cell biology is to divide and conquer. For example, decades of painstaking work have led to an understanding of endoplasmic reticulum (ER) and Golgi structure, dynamics, and transport. In parallel, cytoskeletal researchers have revealed a fantastic diversity of structure and cellular function in both actin and microtubules. Increasingly, these areas overlap, necessitating an understanding of both organelle and cytoskeletal biology. This review addressesconnections between the actin/microtubule cytoskeletons and organelles in animal cells, focusing on threetopics: ER structure/function, ER-to-Golgi transport; and Golgi structure/function. Making these connections has been challenging, due to 1) the small sizes and dynamic characteristics of some components, 2) the fact that organelle-specific cytoskeleton can easily be obscured by more abundant cytoskeletal structures, and 3) the difficulties in imaging membranes and cytoskeleton simultaneously, especially at the ultra-structural level. One major concept is that the cytoskeleton is frequently used to generate force for membrane movement, with two potential consequences: translocation of the organelle, or deformation of the organelle membrane. While initially discussing issues common to metazoan cells in general, we subsequently highlight specific features of neurons, since these highly polarized cells present unique challenges for organellar distribution and dynamics.
Keywords: myosin, kinesin, dynein, Arp2/3 complex, formins, ERGIC
Introduction
In some respects, cell biological research resembles a group of people putting together a very large jigsaw puzzle. The tried-and-true method for jigsaw puzzle solving is “divide and conquer”. For example, if the puzzle represents a horse in a pasture,one person might start with a bit of the horse's head, while another chips away at a leg (not necessarily knowing which leg) and a third might assemblesome pasture. As each expands their sphere, they eventually make connections. At that point, the process moves rather quickly, with un-anticipated connections.
Cell biology has, to some degree, cleared the first part of the process, reaching a point where connections are regularly made between previously separate fields. Sometimes, though, it is all too easy to stay close to the horse's head, avoiding linking up with the leg or the pasture. In this review, we consider connections between cytoskeleton and organelles (Figure 1), focusing on actin and microtubule involvement with ER and Golgi. We remain limited puzzle-solvers in several ways: ignoring other cytoskeletal elements such as intermediate filaments and septins; focusing on metazoans; and paying particular attention to one cell type, neurons.
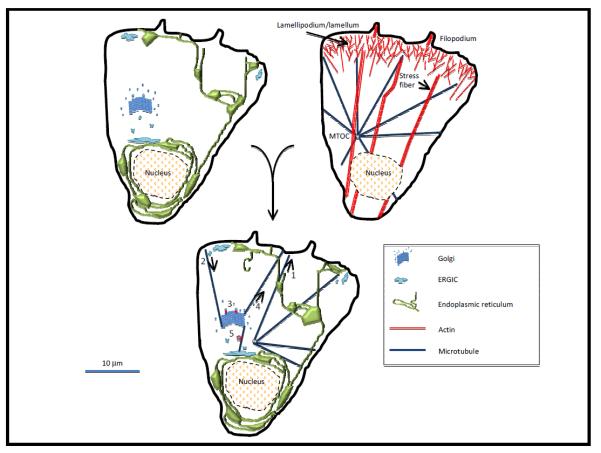
Figure 1. a cellular jigsaw puzzle of organelles and cytoskeleton.
Schematics of a “generic” metazoan cell. UPPER LEFT depicts three organelles: ER (green), Golgi (blue), and a mitochondrion (tan). ERGIC (both central and peripheral) is in light blue. UPPPER RIGHT depicts abundant cytoskeletal structures: lamellipodium/lamellum, filopodium and stress fibers for actin (red); and a microtubule array (blue) with its origin at the microtubule organizing center (MTOC). LOWER cell depicts known or postulated interactions between these organelles and cytoskeletal elements, including: 1) translocation of ER toward microtubule plus ends; 2) a set of microtubules originating at the cis-Golgi and being used for dynein-based transport from the ERGIC to the cis-Golgi; 3) actin involvement in membrane budding from the trans-Golgi network (TGN); 4) a set of microtubules originating at the TGN and being used for kinesin-based transport from the TGN to the periphery; 5) actin involvement in ER-to-Golgi movement of vesicles/tubules.
We come from the cytoskeletal field, and as such may commit some sins of omission in terms of background. For detailed background, please refer to the following publications: actin dynamics [1]; actin nucleation by Arp2/3 complex, formins, or other proteins[2]; myosin motors[3]; microtubules [4][5]; and microtubule motors [6, 7]. Here we briefly provide some salient features for actin and microtubules and largely direct readers to the above references for more details.
Actin filaments aretwo-stranded helical polymers of the 43 kDa actin monomer, and measure 7 nm in diameter. Filaments are polar, with “plus” and “minus” ends (which, for historical reasons are more often called “barbed” and “pointed” ends). In non-muscle cells, all known filament growth occurs at the barbed end. In addition, the barbed end tends to abut a membrane surface, with pointed end away from the membrane. Cytoplasmic actin concentration ranges from 50–200 μM in metazoan culture cells, with 50–70% polymerized at interphase ([8], our unpublished results).
A major control point in actin filament growth is nucleation of a new filament, and three classes of “nucleation factors” are known. Arp2/3 complex nucleates “branched filaments” (Figure 2). While there is only one Arp2/3 complex, functional diversity is provided by its activators such as WASP, N-WASP, Scar/WAVE proteins, WASH, and WHAMM, which are regulated by distinct mechanisms. A second class of actin nucleators are formin proteins, which remain at the barbed end after nucleation and subsequently control filament elongation. In fact, in some cases formins might serve chiefly as elongation factors for filaments nucleated by Arp2/3 complex or other proteins. There are multiple formins (15 in mammals, 6 in Drosophila and 7 in C. elegans), providing the potential for diverse cellular roles. Finally, COWs (compound WH2 domain proteins) represent a third class of nucleator and include proteins such as Spire and Cordon Bleu. COWs can also synergize with formin proteins in actin assembly [9].
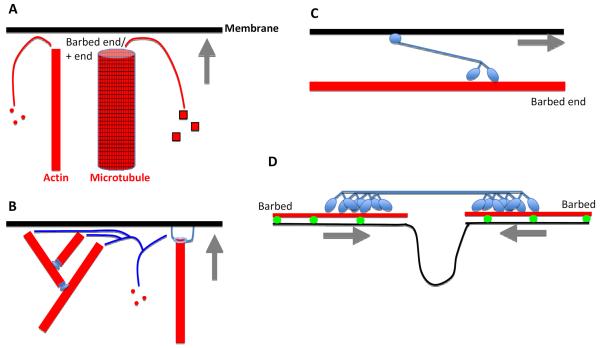
Figure 2. Mechanisms for cytoskeleton-based force generation on membranes.
Membrane in black, cytoskeletal element in red, cytoskeletal-interacting protein in blue. Gray arrows denote direction of membrane movement.
LEFT: Polymerization-based force generation. A) Membrane can be pushed by a polymerizing actin filament or microtubule by new polymer subunits adding to the + end (barbed end for actin), which abuts the membrane. B) Specific examples of actin polymerization-based force production. Arp2/3 complex (left) assembling a branched filament network (Arp2/3 complex is at the branches), or a formin protein (right) attached to the filament barbed end and to the membrane. Not shown are proteins containing multiple WH2 domains (Spire, Cordon Bleu), that can nucleate new actin filaments, possibly remaining at barbed ends in some cases.
RIGHT: motor-based force generation. C) Motor-based translocation along a filament. Dimeric myosin motor moving towards the actin filament barbed end using its motor head groups, and attached to a membrane through a tail motif. One myosin (myosin VI) moves toward the opposite end (the pointed end). Motors that translocate on microtubules include kinesins (toward plus end) and dyneins (toward minus ends). D) Motor-based contraction. A non-muscle myosin II mini-filament, in which the motor heads are bound to two different actin filaments, and moving towards their respective barbed ends. In the process, the myosin II causes constriction of the membrane attached to the actin filaments.
Actin filaments assemble in many places formany purposes. We count >20 known actin-based structures and there are certainly more remaining to be discovered[10]. While some actin-based structures such as stress fibers appear to be large and stable, the filaments in these structures are typically < 1 μm long and turn over on a time scale of minutes. Other cellular actin filaments are even shorter (< 200 nm) and shorter-lived, the clearest examples being the Arp2/3 complex-assembled “dendritic” networks at the leading edge of motile cells, around endosomes and in phagocytic cups. Structures assembled by formins or COWs are less well characterized in metazoan cells, but many are likely to be short and transient, such as those at mitochondrial fission sites [11]. We raise this point because short/transient actin filaments can be very difficult to identify by fluorescence or electron microscopy, and this has hindered elucidation of their roles in the secretory pathway (discussed in the Golgi section).
Microtubules arehollow tubes with 24 nm diameter: much larger than an actin filament. To illustrate the size difference between actin filaments and microtubules, two filaments could fit in the lumen of a microtubule. In most cellular circumstances, the tube is formed by 13 linear protofilaments that make lateral contacts. The building blocks of these protofilaments are hetero-dimers of α- and β-tubulin (both about 50 kDa). Like actin, microtubules are polar, with a plus- and a minus-end. Virtually all dynamics (growth, shrinkage) occur at the plus-end, which can disassemble very rapidly at times (catastrophe). In 3T3 cells, about 70% of the approximately 90 μM cytoplasmic tubulin dimer is polymerized at interphase (based on [12, 13]and our own calculations of cyotoplasmic volume). While tubulin represents about 4% of cellular protein in many cultured cells, it reaches 25% in brain [13], due partially to the high density of microtubules in axons and dendrites. Cellular microtubules tend to be longer than actin filaments and largely originate at one place, the microtubule organizing center (MTOC) or centrosome, with minus-ends remaining tightly embedded there. However, acentrosomal microtubules exist, the most relevant to this review being those originating from the Golgi[14, 15].
An important function of the cytoskeleton is to provide the force for membrane movement. This movement can be deformation (examples: yeast endocytosis,or leading edge extension during cell motility) or translocation from one point to another (example: GLUT4 vesicle translocation). Both actin and microtubules can generate force in two ways: by polymerization/depolymerization, or by serving as substrates for motor proteins (Figure 2). All myosin motor proteins for actin move in the barbed end direction, except myosin VI which is a pointed end motor. Myosin II is a special case, assembling into bi-polar filaments that can exert contractile force. For microtubules, dyneins are minus-end motors, while many kinesins are plus-end motors. However, there are kinesin motors that do not translocate along microtubules, and some of these are used for depolymerization or other functions. Of note, there are many members of the large myosin and kinesin families for which biochemical and/or cellular function is unknown. Lastly, while motor activity is often associated with membrane translocation and polymerization/depolymerization with deformation, motors certainly can affect deformation and polymerization/depolymerization can affect translocation.
While the importance of cytoskeleton to membrane movement has long been appreciated, new connections are now being made with organelles not commonly thought to be cytoskeleton-associated. In this review, a central theme is relating the function of actin and microtubules to either deformation or translocation of ER, Golgi, or transport intermediates.
Endoplasmic Reticulum
The ER mediates a wide variety of cellular processes such as: synthesis, modification, quality control, and transport of proteins; Ca2+ homeostasis; and lipid synthesis/distribution. First described for its “lace-like” reticular structure, this large organelle extends as a single membrane-bound entityto virtually all corners of the cell, and is composed of interconnecting sheets (also called cisternae) and tubules[16, 17]. Tubules are approximately 50 nm diameter in mammals, and sheets are flattened double-membrane structures with approximately 50 nm lumenal space. Some ER sheet can be fenestrated in yeast and mammalian culture cells[18–20], with the latter measuring about 75 nm in diameter [18]. Sheets and tubules can inter-convert in mammals[18, 21, 22].
ER tubules rely on several varieties of membrane-embedded proteins including reticulons, DP1, and REEPs[23] that stabilize the tightly curved membrane structure. New tubule branches can arise from the sides of existing tubules [24] and fuse with other tubules through the action of the dynamin family GTPase atlastin [25]. In sheets, several transmembrane proteins such as Climp63, p180 and kinectin are enriched in the flat region, while curvature-stabilizing proteins are excluded from these regions and instead enrich at the tightly curved sheet edges [26]. Suppression or deletion of curvature-stabilizing proteins causes reduction or abolition of tubules, whereas suppression of sheet-enriched proteins does not abolish sheets. These results might be explained by the fact that the tightly curved tubule is a high-energy structure compared to the flat sheet, which might be the default state. The regular spacing of membranes in the sheet does require stabilization, however, and interaction between the lumenal domains of Climp63 appears to serve this purpose in mammals[26]. Other specialized regions of ER include close connections with mitochondria, plasma membrane, Golgi, endosomes, and peroxisomes[27]. Mitochondria/ER contacts serve roles in exchange of molecules such as calcium and lipids, stress responses and mitochondrial fission, with some of these roles perhaps being interrelated.
The structural heterogeneity of ERclearly contributes toits functional compartmentalization, but it may be going too farto say there is absolute delineation of function to either sheets or tubules. For example, sheets are often equated with “rough ER” (containing translocon proteins and binding ribosomes), partially due to the abundance of sheet structure in “professional” protein secretors such as pancreatic beta cells or activated B lymphocytes. In addition, ribosome detachment might lead to an increase in tubules[18, 22] and Climp63 appears to immobilize translocon complexes[28], suggesting a reciprocal relationship between sheets and translation/translocation. However, ER tubules can also bind ribosomes in yeast [20] and contain translocon proteins in mammals [26], albeit at lower levels than in sheets. Further work is required to determine how much translation/translocation actually takes place on tubules. Stress response might also be enhanced in sheets, since cells appear to increase sheet:tubule ratio when under stress, perhaps to increase lumenal volume to handle increased unfolded protein levels[19, 20]. It is unclear what specific functions might be better conducted by tubules, but their transport on microtubule tips might suggest a role in Ca2+ dynamics (see below). Also, the association between ER and mitochondrial fission could be an ER tubule phenomenon from the examples shown [11, 29], although this has not been formally tested.
ER-Cytoskeletal interactions
Although ER can form a reticular network independently of cytoskeleton[30], ER distribution and sheet/tubule balance are influenced by microtubules in mammalian cells [31–35]. Microtubule-dependent ER movement comes in two flavors: sliding, involving motor-based transport along pre-existing stable acetylatedmicrotubules; and Tip Attachment Complex (TAC) mediated movement, whereby a plus end-attached ER tubule extends with the growing microtubule [34, 36] (Figure4). While motor-based transport represents the prevalent mechanism in the cells studied so far, more details are available for TAC mediated movement, although there are controversies as to mechanism and cellular function.
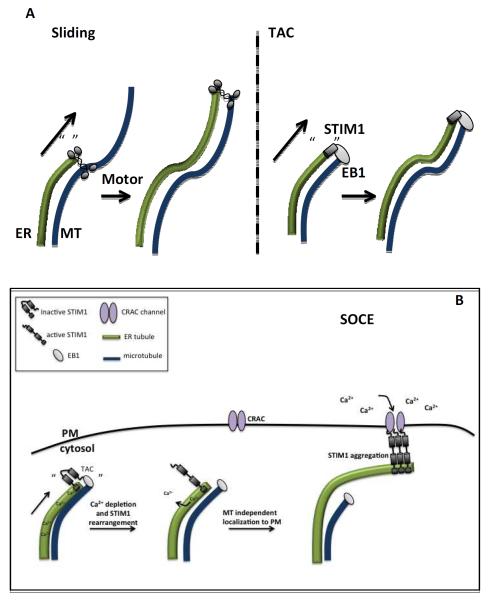
Figure 4. Microtubule-based ER motility.
A)Two mechanisms of microtubule-based ER translocation. “Sliding” refers to kinesin-mediated ER movement along an existing microtubule. “TAC” (or Tip Attachment Complex) refers to ER moving with the elongating plus end of a microtubule, through interaction between STIM1 on the ER and EB1 on the plus end.
B) TAC and ER calcium dynamics. In addition to binding EB1, STIM1 is an ER calcium sensor through its luminal EF hand motifs. Upon low ER calcium, STIM1 interacts with plasma membrane calcium channels (CRAC channels) to mediate Store Operated Calcium Entry (SOCE). One mechanistic model postulates that TAC is necessary to position STIM1-containing ER near appropriate sites for SOCE. Upon ER calcium depletion, STIM1 aggregates, loses association with the microtubule plus end, and engages the PM calcium channel.
In fact, there is the exciting possibility that TAC is directly involved in one of the major functions of ER, to act as a store of calcium ions that can be discharged upon appropriate stimulation. The ER lumen contains 10,000-fold higher Ca2+ than does cytosol[37], and must be replenished efficiently. One source of Ca2+ for ER is from the extracellular milieu, and ER tubules contact the plasma membrane (PM) for replenishment in a TAC-dependent manner in a process known as Store Operated Calcium Entry (SOCE) [38, 39]. The interacting proteins relevant for TAC motility are EB1, bound to the microtubule plus end, and STIM1, an ER transmembrane Ca2+ binding protein that serves as a lumenal Ca2+ sensor [36, 40, 41]. Upon depletion of ER Ca2+ stores, STIM1 aggregates and relocalizes to ER-PM junctions where it interacts with Ca2+ release activated calcium (CRAC) channels to promote influx of extracellular Ca2+ into the ER [40, 42, 43].
The precise role of TAC-based ER motility in SOCE is still debated and appears variable among cell types [40, 43–45]. Early theories postulated that, upon Ca2+ depletion, TAC is activated as an initial step in SOCE. However, STIM1 appears to release from EB1 upon its Ca2+-mediated aggregation[40]. A current model suggests that TAC-mediated ER movement is required prior to SOCE, to position STIM1 throughout the ER, with ER Ca2+ depletion causing microtubule-independent STIM1 translocation to the PM[45]. Interestingly, this process appears important in the pathogenesis of Clostridium difficile, which appears to hijack host cell ER through re-routing of STIM1-mediated TAC [46].
The molecules mediating and regulating sliding ER transport are less understoond. While kinesin-based plus-end transport to the cell periphery appears to occur[34, 36], the specific kinesin motor is not known. The sheet-enriched protein kinectin is known to bind kinesin [47]. While over-expression of kinectin's kinesin-binding domain perturbs ER dynamics[48], testing of kinectin-suppressed cells for kinesin-mediated ER dynamics would be more satisfying in ruling out indirect effects. There is clear evidence that ER can associate with dynein and move toward microtubule minus ends in Xenopus extracts [49, 50] and mammalian cells [51].
A number of other ER proteins have also been shown to bind microtubules, including Climp63, p180, and certain members of the REEP family (Figure5), although the exact role of microtubule binding is unclear in all cases. The short cytoplasmic N-terminal region of Climp63 binds microtubules directly [52], and Climp63-microtubule interaction appears to decrease the diffusion rate of the translocon complex[28]. A region of the extensive C-terminal cytoplasmic region of p180 binds and bundles microtubules, perhaps dependent on dimerization of this region[53], and over-expression results in increased microtubule acetylation. The function of p180 appears to be in the translation-independent localization of specific mRNAs to the ER membrane [54]. One member of the REEP protein family, REEP1, has been shown to bind microtubules directly through its C-terminal region, and sequence homology along with cellular experiments suggest that REEPs1-4, but not 5 and 6, bind microtubules[55]. Finally, a specific isoform of spastin, termed M1 spastin, is ER-associated through an N-terminal hairpin region that also confers atlastin binding [55]. Spastin contains a C-terminal hexameric AAA ATPase domain that has microtubule severing activity as well as a second microtubule interacting region between the hairpin and the AAA ATPase domain[56].
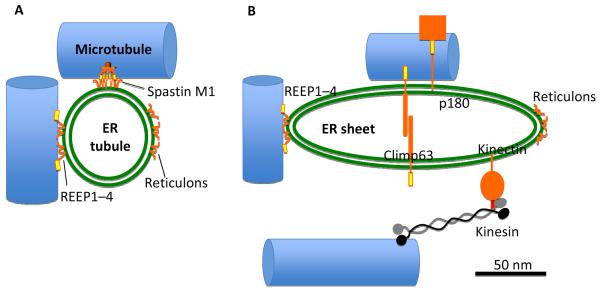
Figure 5. Known microtubule binding proteins on ER tubules and sheets.
Left – ER tubule. The mammalian ER tubule is shaped by proteins that stabilize tight membrane curvature. These proteins contain one or two hydrophobic hairpin segments that can embed into the cytoplasmic leaflet. The reticulons have no known microtubule binding capacity but some members the structurally similar REEPs (REEPs 1-4) have a C-terminal microtubule-interacting region. Spastin is a microtubule severing protein containing a hexameric AAA ATPase domain (the severing domain) and a separate microtubule binding region. In addition, spastin can be expressed with an N-terminal extension (called the spastin M1 variant) containing one hairpin sequence that can bind ER. Right – ER sheet. Curvature stabilizing proteins are excluded from the flat region of the sheet by mechanisms that are poorly understood. Among the sheet-enriched proteins are Climp63, p180 and kinectin. Climp63 has an extensive lumenal region that interacts homotypically and is thought to serve as the “spacer” that maintains a 50 nm lumenal width. P180 has an extensive cytoplasmic domain that contains a microtubule binding domain and may act in translation-independent localization of specific mRNAs to the ER membrane. Kinectin's cytoplasmic region interacts with a region near the C-terminus of kinesin. The diagrams are scaled to show accurate relative diameters of ER (50 nm for both tubule and sheet) and microtubule (24 nm) and length of kinesin. Climp63, p180 and kinectin are schematic, showing approximately the relative amounts within and without the ER lumen. Green double line – ER membrane, orange – proteins, yellow – microtubule binding region, red – kinesin binding region, blue – microtubule.
Mysteriously, microtubules appear to play an important role in the balance between ER sheets and tubules, since microtubule depolymerization causes an impressive accumulation of sheets within minutes of treatment[31, 35]. Given the many microtubule-associated ER proteins in both sheets (Climp63, p180, kinectin) and tubules (specific REEPs, spastin), the mechanism behind this transition is unclear. For example, it would be interesting to know how Climp63 phosphorylation, which apparently inhibits microtubule binding[57], influences sheet/tubule balance. Conversely, REEP1 and spastin M1 have curvature stabilizing domains that should drive tubule assembly, so how does microtubule binding modulate their localization? As a side note of no direct relevance here, it is interesting to the authors that both microtubules and ER tubules are tubes, of somewhat similar sizes. At times we idly wonder about the hidden world that might exist within the microtubule lumen.
In contrast to plants and yeast [58, 59], actindoes not appear to play a central role in ER movement and morphology in generic metazoan culture cells, although some evidence suggests actin and myosin act in retrograde ER transport[34, 60] and that a relationship exists between actin and ER through filamin proteins[61]. Interestingly, actin may play a role in the sheet-to-tubule transition through myosin 1c[62]. Also, actin functions in neuronal ER distribution into dendritic spines, as discussed below[63, 64]. A prenylated isoform of one formin, INF2, is tightly bound to ER but appears to play no clear role in ER dynamics[65]. Interestingly, however, ER-bound INF2 does play a role in mitochondrial fission[11], demonstrating the capacity of ER to influence other organelles.
Neurons- challenges for ER distribution and function
Given the enormous length and miniscule width of neurons (dendrite diameter 2–5 μm, axon diameter < 2 μm), it is a marvel that ER distributes as a continuous network from one end to the other, even entering dendritic spines [60, 66]. Super-resolution fluorescence microscopy and EM tomography show the intricacy of ER indendrites from hippocampal neurons both in culture and in situ, with multiple branched tubules often emanating off regions of higher complexity (which one might be tempted to call “sheets”) thatcorrelate with areas of high dendritic spine density[67]. This structural complexity results in functional compartmentalization in the ER, by reducing diffusion of a sub-set of newly synthesized plasma membraneproteins prior to ER exit [67]. Mechanisms for inducing this compartmentalization are not fully understood, but microtubules clearly play a rolethrough CLIMP63[67, 68].
Before discussing ER in neurons, we provide some general features of neuronal microtubules and actin (Figure 3). Both axons and dendrites are microtubule-rich, with microtubules uniformly oriented in axons (plus end-distal) and of mixed orientation in dendrites[69]. Axonal microtubules do not run the entire length but are staggered along the axon[70, 71]. Similar distribution is assumed in dendrites, but has not been observed directly to our knowledge. Many of these microtubules are acentrosomal (not emanating from the centrosome/MTOC), since the MTOC is in the cell body. Indeed, most microtubule nucleation in mature cultured hippocampal neurons is acentrosomal[72]. Although axons and dendrites contain much less actin than tubulin, actin does enrich at several places: axon initial segments, axonal termini and dendritic spines/filopodia. Interestingly, dendritic filopodia appear different from “normal” filopodia, in that their actin filaments are not uniformly oriented and they contain myosin II [73][74]. Finally, a recent super-resolution fluorescence study identified two additional low-abundance actin-based structures: longitudinally-running filaments in dendritic shafts, and regularly-spaced (190 nm periodicity) spectrin-actin structures forming bands around axonal shafts [75].
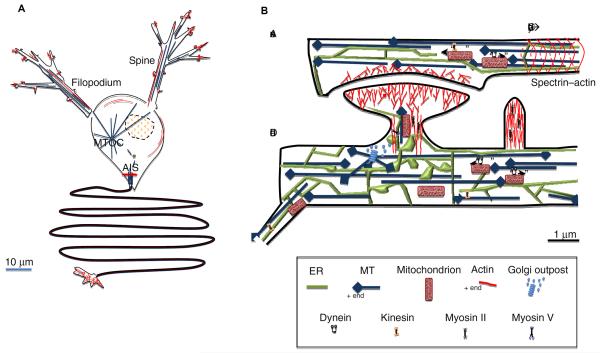
Figure 3. Cytoskeleton and organelles in neurons.
LEFT: Schematic of actin and microtubules in a “generic” neuron, with one axon and two dendrites emanating below and above the cell body, respectively. The narrow axon (> 2 μm diameter) is microtubule-rich, with actin filaments most abundant in the initial segment (AIS) and the synapse/distal terminus. The dendrite is also narrow (< 5 μm), but wider than the axon and tapers in width distally. As with axons, dendritic shafts contain abundant microtubules, with actin filaments enriching in dendritic filopodia and dendritic spines. Microtubules extend off of the MTOC, but are also produced by katanin/spastin-mediated severing in the cell body.
RIGHT: close-up of cytoskeleton and organelles in the vicinity of a synapse. Microtubules (blue) are of uniform orientation (plus ends marked with diamonds) in axons, and mixed orientation in dendrites. A recent study shows that there is a small number of actin filaments running longitudinally in dendrites, and a periodic spectrin-actin structure at 190 nm intervals in axons (only actin banding shown here). There are at least two populations of actin filaments in dendritic spines: an Arp2/3 complex-dependent branched filament network at the tip, and a set of anti-parallel filaments in the shaft. Myosin II also is present in the shaft, possibly making these structures contractile. A similar actin and myosin II arrangement exists in shafts of dendritic filopodia, but they are not bulbous at the tips. Both axons and dendrites contain ER. In dendrites, ER expands near dendritic spines, developing sheet-like structures and becoming highly branched. ER can move into the spine along actin filaments through myosin Va. Golgi is considered to be absent from axons and rare in dendrites, but small Golgi “outposts” are sometimes found near dendritic spines, and microtubules can originate at these outposts. Dynamic microtubules can enter the dendritic spine, but their origins are unknown.
Returning to ER, how does it get from the dendritic shaft into dendritic spines, where Ca2+ release from ER is crucial for synaptic plasticity and memory[76]? Microtubules can enter spines [77–80], but spines also contain actin filaments. A recent publication demonstrates an elegant mechanism whereby myosin Vaconducts short-range ER transport along actin filaments into the dendritic spine, with myosin Va localizing to the ER tip[81]. Thus, gross dendritic ER morphology might be microtubule-dependent but local ER import into spines might be actin-dependent. Given that ER enters these spines to release Ca2+, and STIM1 is involved in both Ca2+ sensing and ER transport, it will be interesting to see whether STIM1 plays any role in ER dynamics here. Of note, manipulation of actin filaments in hippocampal neurons affects Ca2+ release from ER [82].
As opposed to dendrites, axons are considered to be largely devoid of secretory machinery[83, 84](although this might not be true for some peripheral neurons[85]). Nevertheless, axons do contain extensive smooth ER that tracks all the way to axon termini[60, 83, 86]. In peripheral neurons, this axonal ER consists of a large number of branched tubules running parallel to the axonal axis, with some of these tubules in close proximity to the plasma membrane and occasional elaboration into sheet structures[86] (Blackstone and Terasaki personal communication). The importance of axonal microtubule-ER interactions in peripheral neurons is suggested by the disease hereditary spastic paraplegia (HSP), which affects axons in long peripheral neurons[55]. Two proteins mutated repeatedly in HSP are the microtubule binding proteins REEP1 and spastin. The exact functional connection between spastin and ER structure/function in axons is unclear, although M1 spastin is highly enriched in spinal chord neurons, those that are compromised in HSP[87], and is abundant at axonal branch points in cultured neurons[88]. One proposed neuronal role of spastin is to sever microtubules in the cell body for export to axons, which might affect ER distribution indirectly[89, 90]. However, spastin also co-localizes with REEP1 in axons[55] and spastin suppression causes neuronal defects even though katanin, another microtubule severer, is still present[88].
Golgi
Discovered over 100 years ago[91], the Golgi has become no less beautiful with age. The basic structural unit is the cisterna, a flattened membrane of roughly 20–40 nm thickness and 500 – 1000 nm in the other two dimensions[92]. Cisternae associate vertically to form a Golgi stack. In mammals, the number of cisternae per stack varies roughly between 4 and 11. The two outermost cisternae, the ER-facing cis-cisternum and the opposite-facing trans-Golgi network (TGN), are morphologically different from the more central (medial) cisternae, being more vesiculated (Figure 6). A more function-based nomenclature for Golgi cisternae has recently been proposed [93].
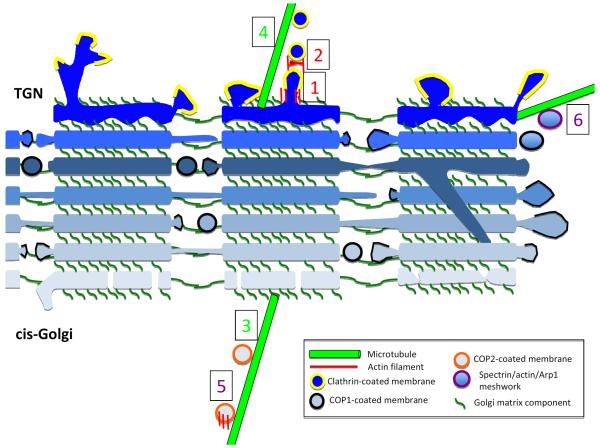
Figure 6. Possible roles for actin and microtubules in Golgi function.
Schematic cross-section of three laterally-connected mini-stacks from mammalian Golgi*, with individual cisternae in blue (lightest are cis- and darkest are TGN). Golgi matrix proteins (dark green) tether cisternae vertically within a mini-stack, and horizontally between mini-stacks. A sub-set of parallel cisternae establish lateral membrane connections, which are likely to be highly dynamic. Occasionally, inter-cisternae membrane connections might occur within a mini-stack (one depicted here). COP1-mediated vesicle transport (black banding around membrane) occurs from the edges of mini-stacks. COP2-coated vesicles arrive at cis-Golgi via ERGIC. At the TGN, exiting transport membranes are generally clathrin-coated (yellow banding). From our interpretation of the literature, we postulate six known or highly likely instances of cytoskeletal involvement in Golgi dynamics, some being actin-dependent (red numbers), some microtubule-dependent (green numbers) and some involving both actin and microtubules (purple numbers). (1) tubulation of TGN, involving actin, Arp2/3 complex and myosin 1B. (2) fission of transport vesicles from TGN, involving actin and myosin II. (3) transport of COP2 vesicles to cis-Golgi-attached minus ends of microtubules by dynein. (4) transport of clathrin vesicles away from TGN by kinesins. (5) transport of tubulovesicular membranes from ERGIC to cis-Golgi, involving actin, microtubules and WHAMM. (6) Spectrin coating of Golgi-derived membranes, possibly containing actin and/or Arp1, and interacting with microtubule motors in an as-yet un-defined manner.
Functionally, protein cargo arrive at cis-Golgi as vesicles or tubule-vesicular structures, undergo a series of modifications as they transit through the cisternae, then are packaged into clathrin-coated vesicles at the TGN for transport to multiple destinations (secretion, PM, organelles), which requires cargo sorting. The TGN also receives vesicles delivered from some of these regions. Golgi resident proteins are brought back to their proper region of the stack through COPI vesicle trafficking[93, 94].
The last two paragraphs paint a rather bucolic scene of harmony. In reality, Golgi structure/function is much more confusing and controversial [93–98]. Major questions for mammalian Golgi include: what is the mechanism of anterograde transport (through vesicles or through maturation/movement of an entire cisterna); how do large cargo (bigger than can be handled by 50 nm vesicles) get transported; and how are different rates of protein transport accommodated in the same Golgi[93, 99]?
In addition, metazoan Golgi structure is much more intricate than that depicted in most textbooks, with several well documented but often overlooked features (Figure 6). First, metazoan Golgi generally consists of a group of “mini-stacks” tethered in parallel to form a Golgi “ribbon”[92, 100], with a rough estimate of about 100 mini-stacks per Golgi ribbon in an interphase HeLa cell (A. Linstedt, personal communication). The mini-stacks are held together by lateral membrane fusion between a sub-set of adjacent cisternae, and these connections are likely to be highly dynamic[98]. We state “likely” because these connections are < 0.5 μm and are buried within the Golgi structure, thus difficult to image dynamically. Second, small pores or “fenestrae” exist in many cisternae, particularly in the connecting membranes between mini-stacks[92]. Third, there can periodically be tubular inter-cisternal connections within individual Golgi stacks[101, 102]. The precise significance of these structural features is unclear.
Another feature of metazoan Golgi is the matrix of proteins that coats each cisterna. This matrix is necessary for tethering transport vesicles as well as for maintenance of cisternal associations both longitudinally within mini-stacks and laterally between mini-stacks[103]. Made up primarily of peripheral membrane proteins in the GRASP and golgin families, the matrix is highly dynamic as these coiled-coil proteins associate and disassociate both with the Golgi and with each other, in part controlled by well-known kinase pathways[104]. The matrix is non-uniform, with some proteins such as GM130 associating towards the cis-face and others such as GCC185 associating with the TGN. Disruption of key matrix components causes Golgi fragmentation.
In a parallel universe, findings have accumulated suggesting that both actin and microtubules play important roles in Golgi structure and function. For the most part, however, clear mechanistic links have not been made. The study of Golgi/cytoskeleton interactions poses challenges for at least two reasons. First, the Golgi itself is relatively small, and its component mini-stacks are even smaller. Second, the Golgi is often found near the nucleus and the MTOC, and specific actin and microtubule interactions can easily be obscured by the abundance of both cytoskeletal elements in this region.
Before starting a more detailed discussion, we give a brief overview of Golgi/cytoskeleton connections. A general observation is that microtubule depolymerization causes Golgi fragmentation while actin depolymerization causes Golgi compaction[105, 106]. Microtubules are nucleated at the Golgi in a manner dependent on Golgi matrix proteins, both at the cis-Golgi and TGN, and maintain minus ends at these sites[14, 15]. On the cis-face, GM130 is thought to recruit γ-tubulin via an association with AKAP450. On the TGN-face, GCC185 association with CLASP proteins is thought to stabilize growing microtubule seeds. Dynein drives at least some vesicle transport towards Golgi, while kinesins takes vesicles away[6, 107]. Inhibition of dynein or its cargo adaptor dynactincauses Golgi fragmentation[108]. There is evidence for diverse Golgi-actin connections as well. Three actin polymerizing formin proteins have been linked to Golgi structure: FMNL1γ and INF2 promoting Golgi compaction, and mDia1 promoting Golgi fragmentation[109–111]. Other actin-binding proteins influencing Golgi structure are WHAMM, cortactin, and myosin 18A[112–115]. Arp2/3 complex associates with Golgi membranes[116], and plays a role in transport[117]. Cofilin is associated with the TGN, and plays a role in cargo sorting[118, 119]. There is also suggestion of a spectrin-actin meshwork at or close to the Golgi, with its integrity affecting Golgi function[108, 120, 121]. Finally, multiple myosins (in the I, II, VI and XVIII families) play roles in Golgi structure/function[113, 117, 122]. How might this all fit together? One topic we ignore at our peril is the Golgi structure/function of plants, which is better understood at many levels. As one example, most plant Golgi exists as individual mini-stacks that undergo rapid myosin-mediated transport along actin filaments[123].
What is “Golgi-associated actin”?
The evidence for actin association with Golgi is largely through immunofluorescence localization of actin-binding proteins[109, 117, 118, 122], and some have been shown to bind Golgi in cell-free assays[116]. Two issues should be raised here. First, these proteins often “enrich” at Golgi, but are also abundant in vesicular structures surrounding Golgi or elsewhere[115, 121]. Sometimes, the proteins shown to affect Golgi structure have little or no actual enrichment there[110, 111]. Second, the Golgi enrichment can be at one side of the stack, often towards the TGN[117, 122].
What is the evidence for actin filaments themselves on Golgi? Standard phalloidin staining does not conspicuously label Golgi in mammalian cells, but since the cytoplasm is often filament-rich, one may miss relatively small accumulations. Closer examination by fluorescence microscopy reveals actin “puncta” in the Golgi region[110, 117, 122]. These puncta are remarkably consistent in number (about 13/Golgi in U2OS cells), decrease upon suppression of the formin INF2, and often enrich with myosin 1b and Arp2/3 complex[110, 117]. Highly zoomed fluorescence images of these puncta suggest that they actually lie near the TGN membrane, not on it[117].
These actin puncta have not been obvious by EM, and actin is not mentioned in several cryo-EM tomography studies of Golgi[101, 102, 124]. However, there are major difficulties in EM imaging of both actin and membranes simultaneously, in that traditional fixation/contrast enhancement techniques for thin section EM are damaging to actin filaments[125, 126], whereas many techniques optimal for actin filaments either destroy membranes or provide little membrane contrast[127, 128]. Cryo-EM tomography techniques are extremely promising, but there are still cautionary notes. First, some cryo-EM techniques still include potentially actin-damaging treatments post-freezing[92, 101, 102]. Preservation of large structures such as stress fibers under these conditions provides imperfect evidence of actin integrity, since many of the organelle-interacting actin filaments are likely to be short and labile. Second, short actin filaments might be difficult to detect by cryo-EM tomography even if well preserved, due to their orientation relative to the electron beam or the to fact that they might be just plain short[129, 130]. As evidence for the difficulties in imaging short actin filaments by EM, there has been a 15-year debate on the morphology of actin filaments at the leading edge of motile cells [131], and in this case we actually knew filaments were there!
The association of a specific spectrin (βIII) with Golgi has been known for some time[132, 133], in addition to specific forms of spectrin-associated proteins ankyrin[134, 135] and protein 4.1[136]. Suppression of βIII spectrin disrupts Golgi structure[121]. However, the localization pattern of βIII spectrin is not confined to Golgi, and appears rich on vesicular structures elsewhere in the cytoplasm[121, 133]. This feature might suggest a role for spectrin more in transport to/from Golgi rather than directly in Golgi morphology.
Another feature of βIII spectrin might draw it closer to microtubules than to actin. The classical spectrin-actin network from erythrocytes consists of a short (14 subunit) actin filament at the intersection of much larger spectrin oligomers[137]. Interestingly, βIII spectrin associates with the actin-related protein, Arp1[138], which forms a short (8 subunit) filament as part of the dynactin complex which links the dynein motor to cargo[139]. One possibility is that a Golgi-linked βIII spectrin/dynactin meshwork could interact with dynein, possibly contributing to selective cargo transport.
What are “fragmented Golgi”?
What does the fragmented Golgi phenotype, induced by Golgi matrix disruption, microtubule depolymerization and other treatments[105, 109, 110],mean in terms of actual Golgi structure? For microtubule depolymerization, the fragments are mini Golgi stacks that localize near ERES and appear to be fully functional for transport[105]. Presumably, microtubule depolymerization disruptsinter-stack membrane connections within the ribbon, but it is unclear what the mechanism would be. It is also unclear how suppression of the formins FMNL1γ and INF2 might fragment Golgi, but this possibly could be related to the ability of formins to bind microtubules[140–142].
What is the dispersive force that drives fragmented Golgi to ERES? The fact that Golgi disperses upon microtubule depolymerization suggests that it is not a kinesin-driven process, although there could be stable microtubule tracks that remain. The fact that actin depolymerization[106] and suppression of three actin-associated proteins, mDia1, cortactin and myosin 18A[111, 113, 114], causes the opposite effect (compacted Golgi) does suggest that in some way actin contributes to Golgi fragmentation. There have been suggestions that myosin 18A activity on actin filaments might provide a tensile force to expand the Golgi[113], but an important caution is that no actual motor activity has been detected for myosin 18A[143, 144]. Interestingly, Golgi-associated Cdc42 has a negative effect on dynein-mediated Golgi transport and inhibits Golgi ribbon re-assembly after nocodazole wash-out in an actin-dependent manner[145]. This effect might be through Arp2/3-mediated actin polymerization[116, 146].
It is worth remembering that Golgi structure is highly sensitive to changes in membrane entry or removal[98]. Many of the effects mentioned above change import and/or export, thus could have indirect effects on Golgi structure[6, 108]. For example, cortactin suppression causes impressive changes in Golgi morphology, but acts on late endosome/lysosome dynamics[114].
Possible mechanisms for actin effects on Golgi
One interpretation of the current literature suggests that actin's roles are not on Golgi structure maintenance but on transport carrier production, either at the budding stage or in fission from the Golgi surface. The clearest picture is at the TGN, where Arp2/3 complex, myosin I, myosin II, and cofilin have been shown to play roles[117–119, 122, 147]. The combination of Arp2/3 complex and myosin 1 might mediate membrane deformation of the nascent transport carrier, which may be more of a tubule than a transport vesicle[117]. Subsequently, myosin II-based contractility might serve in fission of these tubules from TGN[122]. Cofilin's function might be to accelerate actin depolymerization during these processes, although a link between cofilin and the Ca2+ pump SPCA1 adds another level of complexity[118].
It must be noted that cargo sorting is of paramount importance at the TGN, with at least three types of export: to endosomes/lysosomes, to the PM, and an additional pathway for glycosylphophatidylinositol (GPI) linked proteins[148]. Many of the proteins noted above seem to affect only sub-sets of transported proteins. Possibly, one aspect of cargo sorting in this context may be transport carrier size and shape, a parameter that actin is well suited to influence through its ability to deform membranes.
Actin also appears to be involved in other phases of transport to/from Golgi. One example, discussed in a later section, is the role of the Arp2/3 complex activator WHAMM in ERGIC-to-Golgi transport, where the ability to tubulate membranes might be especially useful in transport of larger cargo[112]. Another role for actin might be in COP1-mediated retrograde transport. Arp2/3 complex, and its activators N-WASP and Cdc42, can associate with Golgi through the Arf1-stimulated COP1 coat[115, 116, 149].
We wonder about other possible functions for actin in Golgi structure/function, such as: dynamics of the tubular connections between mini-stacks in the Golgi ribbon, establishment of inter-cisternal connections within a stack, or dynamics/distribution of fenestrae within cisterna. These features can change with changes in secretory activity[98, 101, 102], and almost certainly vary greatly with cell type. A shift from generic culture cells to primary cell systems might be reveal possible connections more clearly.
Golgi in neurons – life at the outpost
Which brings us to neurons, where secretion poses particular issues. As mentioned earlier, small “Golgi outposts” are present in dendrites, in addition to the main Golgi ribbon in the cell body. It is unclear what role cytoskeleton might play in these specialized secretion systems, but the fact that dendritic Golgi outposts are microtubule nucleation sites[150] highlights the Golgi-microtubule link.
Mutations inβIII-spectrin can cause spinocerebellar ataxia type 5 (SCA5), a disease traceable in Abraham Lincoln's descendants and resulting in motor abnormalities through cerebellar degeneration[151]. βIII deletion in mice results in similar phenotypes, with altered organellar structure in Purkinje neuronsand mis-localization of several synaptic proteins[152, 153]. One SCA5 mutation appears to disrupt βIII/Arp1 interaction, and results in an excess βIII accumulation on Golgi[154]. These features might lend to a model in which βIII spectrin coordinates efficient post-Golgi cargo sorting through interactions with dynactin.
ER-Golgi Transport
ER-synthesized “cargo” proteins destined for secretion or delivery to the PM or other organelles get packaged into anterograde vesicles at specific ER exit sites (ERES) using the coatomer II (COPII) machinery[94, 155]. These vesicles ultimately get delivered to cis-Golgi. ER resident proteins that erroneously made this trip get returned to the ER by packaging into retrograde-transporting coatomer I (COPI) vesicles[94, 155].
In mammals, the distance between ERES and cis-Golgi is extremely heterogeneous, and can be large. Consider a HeLa cell, which contains many ERES scattered all over the cell, but one central Golgi[156]. Proteins packaged in all ERES must get to the Golgi, ranging between 1 μm to > 20 μm away. How does this occur efficiently?
One possible answer is motor-based transport along microtubules, starting at a structure called the ER-Golgi intermediate compartment (ERGIC, also called the VTC), which lies between ER and Golgi[94, 155]. The ERGIC is an irregular conglomeration of vesicles and tubules that can be flattened and highly branched, and debate exists as to whether this is a stable or transient structure. There is often a central ERGIC near the Golgi, but smaller peripheral ERGICs near peripheral ERES[157]. Dynein-based transport along microtubules plays a role in anterograde movement of vesicles between peripheral ERGIC and the central cis-Golgi[158], while several kinesins might be involved in Golgi-to-ERGIC retrograde transport[159].
The directionality of the motors implies that microtubule plus ends should be near ERGIC, while minus ends should be near cis-Golgi. This situation seems reasonable since the Golgi is near the MTOC in many mammalian cells. However, the microtubules nucleated from the cis-Golgi[15]might be direct routes for trafficking to/from ERGIC (Figure 6).
Does actin play a role in ER-to-Golgi transport? An intriguing possibility is presented by the protein WHAMM (WASP Homologue associated with Actin, Microtubules and Membranes). WHAMM localizes to ERGIC, cis-Golgi, and microtubule-associated ERGIC-derived structures that might be transport intermediates[112]. WHAMM suppression compromises anterograde ER-to-Golgi trafficking[112]. As its name suggests, WHAMM contains motifs that bind both microtubules and membranes directly[160]. WHAMM's association with actin is through its ability to activate Arp2/3 complex[112]. Thus, WHAMM might mediate an actin- and microtubule-dependent step between ERGIC and cis-Golgi. What this step might be is unclear, but WHAMM potently tubulates membranes, dependent on its activation of Arp2/3 complex[112]. Given that the ERGIC and ERGIC-derived membranes can be highly tubulated, WHAMM might contribute to this process.
ER-Golgi Transport in Neurons
Neuronal dendrites represent a fascinating variation of the typical secretory pathway. While the bulk of ERES and ERGIC and the main Golgi apparatus resides in the cell body, dendrites also contain significant ERES and ERGIC, in addition to small “Golgi outposts” which enrich at dendritic branch points [83, 84][161]. Protein secretion can take two routes from dendritic ERES: retrograde transport back to central Golgi; or to dendritic Golgi outposts. The fact that most cargo goes back to the central Golgi[84] and that only a sub-set of dendrites contains Golgi outposts[161] suggests that secretion through Golgi outposts might represent a pathway for specific cargo or situations[161]. The role of cytoskeleton in this specialized secretory pathway is unclear, but Golgi outposts are capable of nucleating microtubules[150].
Conclusions/Future Perspectives
We envision that mechanistic connections between cytoskeleton and ER/Golgi will be clarified significantly in the near future, in terms of the molecules involved, their roles (in translocation or deformation of membranes) and their interactions with known components of organelle structure/function. Particularly intriguing are connections between cytoskeleton and “membrane shaping” proteins such as reticulons/REEPs and Golgi matrix proteins, since these literally coat sections of their respective organelles. The development of super-resolution microscopy and a greater appreciation for EM preservation of both organelles and cytoskeleton will reveal ultra-structural relationships in a fundamentally new manner. We hope that this review helps, allowing new pieces to be inserted into thepuzzle. While some of our pieces might be inserted somewhat incorrectly, we hope their presence will allow others to replace them with better fitting ones.
Acknowledgements
We thank Peter Baas, Charlie Barlowe, Craig Blackstone, Kent Colosety, Peter Hollenbeck, Irena Kaverina, Adam Linstedt, Tim Mitchison, Jon Morrow, Dave Odde, James Putney, Trina Schroer, Andrew Staehelin, Graham Warren, and Gia Voeltz for valuable discussion. The fact that these gracious individuals helped us does not mean that they necessarily agree with all that is written here. We also thank our funding sources (NIH GM069818 and NIH GM160000 to HNH. NSF Pre-doctoral Fellowship to ALH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimers: this is an “artist's rendition” based on the available literature, and intended to provide food for thought. The membranous connecting regions between lateral cisternae could be very different from those depicted. The movement direction of the COP1 vesicles within Golgi is kept intentionally vague, due to uncertainty in the field. We make no attempt to depict cisternal maturation, though there is good evidence for this. From EM studies, the Golgi matrix is likely to be much more dense than depicted here. The matrix is also of heterogeneous protein composition, with some proteins (eg - GM130) at the cis-face and other proteins (eg -GCC185) at the TGN-face. Finally, we do not depict the fenestrae that have been documented in cisternae.
References
- 1.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartman MA, Spudich JA. The myosin superfamily at a glance. J Cell Sci. 2012;125:1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnham CP, Roll-Mecak A. The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton (Hoboken) 2012;69:442–463. doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etienne-Manneville S. Microtubules in Cell Migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 6.Caviston JP, Holzbaur EL. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Mallik R, Rai AK, Barak P, Rai A, Kunwar A. Teamwork in microtubule motors. Trends Cell Biol. 2013;23:575–582. doi: 10.1016/j.tcb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Heacock CS, Eidsvoog KE, Bamburg JR. The influence of contact-inhibited growth and of agents which alter cell morphology on the levels of G- and F-actin in cultured cells. Experimental cell research. 1984;153:402–412. doi: 10.1016/0014-4827(84)90609-8. [DOI] [PubMed] [Google Scholar]
- 9.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 11.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiller G, Weber K. Radioimmunoassay for tubulin: a quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhai Y, Borisy GG. Quantitative determination of the proportion of microtubule polymer present during the mitosis-interphase transition. J Cell Sci. 1994;107(Pt 4):881–890. doi: 10.1242/jcs.107.4.881. [DOI] [PubMed] [Google Scholar]
- 14.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. Embo J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter KR, Claude A, Fullam EF. A Study of Tissue Culture Cells by Electron Microscopy : Methods and Preliminary Observations. J Exp Med. 1945;81:233–246. doi: 10.1084/jem.81.3.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SH, Blackstone C. Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 2010;11:515–521. doi: 10.1038/embor.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol Biol Cell. 2012;23:2424–2432. doi: 10.1091/mbc.E10-12-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pendin D, McNew JA, Daga A. Balancing ER dynamics: shaping, bending, severing, and mending membranes. Curr Opin Cell Biol. 2011;23:435–442. doi: 10.1016/j.ceb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 24.English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 2013;15:169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011;147:1226–1231. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman JR, Voeltz GK. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 2011;21:709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikonov AV, Hauri HP, Lauring B, Kreibich G. Climp-63-mediated binding of microtubules to the ER affects the lateral mobility of translocon complexes. J Cell Sci. 2007;120:2248–2258. doi: 10.1242/jcs.008979. [DOI] [PubMed] [Google Scholar]
- 29.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreier L, Rapoport TA. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J Cell Biol. 2000;148:883–898. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terasaki M, Chen LB, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 33.Dabora SL, Sheetz MP. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cell extracts. Cell. 1988;54:27–35. doi: 10.1016/0092-8674(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 34.Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 35.Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 38.Terasaki M, Runft LL, Hand AR. Changes in organization of the endoplasmic reticulum during Xenopus oocyte maturation and activation. Mol Biol Cell. 2001;12:1103–1116. doi: 10.1091/mbc.12.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW, Jr., Hoogenraad CC, et al. STIM1 Is a MT-Plus-End-Tracking Protein Involved in Remodeling of the ER. Curr Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galan C, Dionisio N, Smani T, Salido GM, Rosado JA. The cytoskeleton plays a modulatory role in the association between STIM1 and the Ca2+ channel subunits Orai1 and TRPC1. Biochem Pharmacol. 2011;82:400–410. doi: 10.1016/j.bcp.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Redondo PC, Harper MT, Rosado JA, Sage SO. A role for cofilin in the activation of store-operated calcium entry by de novo conformational coupling in human platelets. Blood. 2006;107:973–979. doi: 10.1182/blood-2005-05-2015. [DOI] [PubMed] [Google Scholar]
- 45.Smyth JT, DeHaven WI, Bird GS, Putney JW., Jr. Role of the microtubule cytoskeleton in the function of the store-operated Ca2+ channel activator STIM1. J Cell Sci. 2007;120:3762–3771. doi: 10.1242/jcs.015735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwan C, Kruppke AS, Nolke T, Schumacher L, Koch-Nolte F, Kudryashev M, Stahlberg H, Aktories K. Clostridium difficile toxin CDT hijacks microtubule organization and reroutes vesicle traffic to increase pathogen adherence. Proc Natl Acad Sci U S A. 2014;111:2313–2318. doi: 10.1073/pnas.1311589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyoshima I, Yu H, Steuer ER, Sheetz MP. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Tee YH, Heng JK, Zhu Y, Hu X, Margadant F, Ballestrem C, Bershadsky A, Griffiths G, Yu H. Kinectin-mediated endoplasmic reticulum dynamics supports focal adhesion growth in the cellular lamella. J Cell Sci. 2010;123:3901–3912. doi: 10.1242/jcs.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allan VJ, Vale RD. Cell cycle control of microtubule-based membrane transport and tubule formation in vitro. J Cell Biol. 1991;113:347–359. doi: 10.1083/jcb.113.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Romano FB, Field CM, Mitchison TJ, Rapoport TA. Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J Cell Biol. 2013;203:801–814. doi: 10.1083/jcb.201308001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wozniak MJ, Bola B, Brownhill K, Yang YC, Levakova V, Allan VJ. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. Embo J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa-Goto K, Tanaka K, Ueno T, Kurata T, Sata T, Irie S. p180 is involved in the interaction between the endoplasmic reticulum and microtubules through a novel microtubule-binding and bundling domain. Mol Biol Cell. 2007;18:3741–3751. doi: 10.1091/mbc.E06-12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui XA, Zhang Y, Hong SJ, Palazzo AF. Identification of a region within the placental alkaline phosphatase mRNA that mediates p180-dependent targeting to the endoplasmic reticulum. J Biol Chem. 2013;288:29633–29641. doi: 10.1074/jbc.M113.482505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. The Journal of clinical investigation. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 57.Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griffing LR. Networking in the endoplasmic reticulum. Biochem Soc Trans. 2010;38:747–753. doi: 10.1042/BST0380747. [DOI] [PubMed] [Google Scholar]
- 60.Terasaki M, Reese TS. Interactions among endoplasmic reticulum, microtubules, and retrograde movements of the cell surface. Cell Motil Cytoskeleton. 1994;29:291–300. doi: 10.1002/cm.970290402. [DOI] [PubMed] [Google Scholar]
- 61.Lynch CD, Gauthier NC, Biais N, Lazar AM, Roca-Cusachs P, Yu CH, Sheetz MP. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol Biol Cell. 2011;22:1263–1273. doi: 10.1091/mbc.E10-08-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joensuu M, Belevich I, Ramo O, Nevzorov I, Vihinen H, Puhka M, Witkos TM, Lowe M, Vartiainen MK, Jokitalo E. ER sheet persistence is coupled to myosin 1c -regulated dynamic actin filament arrays. Mol Biol Cell. 2014 doi: 10.1091/mbc.E13-12-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabb JS, Molyneaux BJ, Cohen DL, Kuznetsov SA, Langford GM. Transport of ER vesicles on actin filaments in neurons by myosin V. J Cell Sci. 1998;111(Pt 21):3221–3234. doi: 10.1242/jcs.111.21.3221. [DOI] [PubMed] [Google Scholar]
- 64.Wagner W, Hammer JA., 3rd Myosin V and the endoplasmic reticulum: the connection grows. J Cell Biol. 2003;163:1193–1196. doi: 10.1083/jcb.200311077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farah CA, Liazoghli D, Perreault S, Desjardins M, Guimont A, Anton A, Lauzon M, Kreibich G, Paiement J, Leclerc N. Interaction of microtubule-associated protein-2 and p63: a new link between microtubules and rough endoplasmic reticulum membranes in neurons. J Biol Chem. 2005;280:9439–9449. doi: 10.1074/jbc.M412304200. [DOI] [PubMed] [Google Scholar]
- 69.Baas PW, Lin S. Hooks and comets: The story of microtubule polarity orientation in the neuron. Dev Neurobiol. 2011;71:403–418. doi: 10.1002/dneu.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bray D, Bunge MB. Serial analysis of microtubules in cultured rat sensory axons. J Neurocytol. 1981;10:589–605. doi: 10.1007/BF01262592. [DOI] [PubMed] [Google Scholar]
- 71.Yu W, Baas PW. Changes in microtubule number and length during axon differentiation. J Neurosci. 1994;14:2818–2829. doi: 10.1523/JNEUROSCI.14-05-02818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stiess M, Maghelli N, Kapitein LC, Gomis-Ruth S, Wilsch-Brauninger M, Hoogenraad CC, Tolic-Norrelykke IM, Bradke F. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 2010;327:704–707. doi: 10.1126/science.1182179. [DOI] [PubMed] [Google Scholar]
- 73.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holbro N, Grunditz A, Oertner TG. Differential distribution of endoplasmic reticulum controls metabotropic signaling and plasticity at hippocampal synapses. Proc Natl Acad Sci U S A. 2009;106:15055–15060. doi: 10.1073/pnas.0905110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 78.Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 79.Merriam EB, Millette M, Lumbard DC, Saengsawang W, Fothergill T, Hu X, Ferhat L, Dent EW. Synaptic regulation of microtubule dynamics in dendritic spines by calcium, f-actin, and drebrin. J Neurosci. 2013;33:16471–16482. doi: 10.1523/JNEUROSCI.0661-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shirao T, Gonzalez-Billault C. Actin filaments and microtubules in dendritic spines. J Neurochem. 2013;126:155–164. doi: 10.1111/jnc.12313. [DOI] [PubMed] [Google Scholar]
- 81.Wagner W, Brenowitz SD, Hammer JA., 3rd Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol. 2011;13:40–48. doi: 10.1038/ncb2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Mattson MP, Furukawa K. Endoplasmic reticulum calcium release is modulated by actin polymerization. J Neurochem. 2002;82:945–952. doi: 10.1046/j.1471-4159.2002.01059.x. [DOI] [PubMed] [Google Scholar]
- 83.Krijnse-Locker J, Parton RG, Fuller SD, Griffiths G, Dotti CG. The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol Biol Cell. 1995;6:1315–1332. doi: 10.1091/mbc.6.10.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Merianda TT, Lin AC, Lam JS, Vuppalanchi D, Willis DE, Karin N, Holt CE, Twiss JL. A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol Cell Neurosci. 2009;40:128–142. doi: 10.1016/j.mcn.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsukita S, Ishikawa H. Three-dimensional distribution of smooth endoplasmic reticulum in myelinated axons. Journal of electron microscopy. 1976;25:141–149. [PubMed] [Google Scholar]
- 87.Solowska JM, Morfini G, Falnikar A, Himes BT, Brady ST, Huang D, Baas PW. Quantitative and functional analyses of spastin in the nervous system: implications for hereditary spastic paraplegia. J Neurosci. 2008;28:2147–2157. doi: 10.1523/JNEUROSCI.3159-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu W, Qiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roll-Mecak A, Vale RD. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J Cell Biol. 2006;175:849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Errico A, Ballabio A, Rugarli EI. Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet. 2002;11:153–163. doi: 10.1093/hmg/11.2.153. [DOI] [PubMed] [Google Scholar]
- 91.Golgi C. Sulla struttura della cellula nervosa dei gangli spinali. Boll. Soc. Med. Chir. Pavia. 1898;13:60–70. [Google Scholar]
- 92.Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Day KJ, Staehelin LA, Glick BS. A three-stage model of Golgi structure and function. Histochem Cell Biol. 2013;140:239–249. doi: 10.1007/s00418-013-1128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klumperman J. Architecture of the mammalian Golgi. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer SR, Rabouille C, et al. Journeys through the Golgi--taking stock in a new era. J Cell Biol. 2009;187:449–453. doi: 10.1083/jcb.200909011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfeffer SR. How the Golgi works: a cisternal progenitor model. Proc Natl Acad Sci U S A. 2010;107:19614–19618. doi: 10.1073/pnas.1011016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rothman JE. The future of Golgi research. Mol Biol Cell. 2010;21:3776–3780. doi: 10.1091/mbc.E10-05-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sengupta D, Linstedt AD. Control of organelle size: the Golgi complex. Annu Rev Cell Dev Biol. 2011;27:57–77. doi: 10.1146/annurev-cellbio-100109-104003. [DOI] [PubMed] [Google Scholar]
- 99.Morriswood B, Warren G. Stalemate in the Golgi battle. Science. 2013;341:1465–1466. doi: 10.1126/science.1245656. [DOI] [PubMed] [Google Scholar]
- 100.Rambourg A, Clermont Y. Three-dimensional electron microscopy: structure of the Golgi apparatus. Eur J Cell Biol. 1990;51:189–200. [PubMed] [Google Scholar]
- 101.Marsh BJ, Volkmann N, McIntosh JR, Howell KE. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells. Proc Natl Acad Sci U S A. 2004;101:5565–5570. doi: 10.1073/pnas.0401242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Trucco A, Polishchuk RS, Martella O, Di Pentima A, Fusella A, Di Giandomenico D, San Pietro E, Beznoussenko GV, Polishchuk EV, Baldassarre M, et al. Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat Cell Biol. 2004;6:1071–1081. doi: 10.1038/ncb1180. [DOI] [PubMed] [Google Scholar]
- 103.Xiang Y, Wang Y. New components of the Golgi matrix. Cell Tissue Res. 2011;344:365–379. doi: 10.1007/s00441-011-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Truschel ST, Zhang M, Bachert C, Macbeth MR, Linstedt AD. Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphorylation. J Biol Chem. 2012;287:19870–19875. doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- 106.Valderrama F, Babia T, Ayala I, Kok JW, Renau-Piqueras J, Egea G. Actin microfilaments are essential for the cytological positioning and morphology of the Golgi complex. Eur J Cell Biol. 1998;76:9–17. doi: 10.1016/S0171-9335(98)80012-5. [DOI] [PubMed] [Google Scholar]
- 107.Allan V. Microtubule motors: moving forward on many fronts. F1000 Biol Rep. 2009;1:52. doi: 10.3410/B1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holleran EA, Holzbaur ELF. Speculating about spectrin: new insights into the Golgi-associated cytoskeleton. Trends Cell Biol. 1998;8:26–29. doi: 10.1016/s0962-8924(97)01195-1. [DOI] [PubMed] [Google Scholar]
- 109.Colon-Franco JM, Gomez TS, Billadeau DD. Dynamic remodeling of the actin cytoskeleton by FMNL1gamma is required for structural maintenance of the Golgi complex. J Cell Sci. 2011;124:3118–3126. doi: 10.1242/jcs.083725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramabhadran V, Korobova F, Rahme GJ, Higgs HN. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell. 2011;22:4822–4833. doi: 10.1091/mbc.E11-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zilberman Y, Alieva NO, Miserey-Lenkei S, Lichtenstein A, Kam Z, Sabanay H, Bershadsky A. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22:2900–2911. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. GOLPH3 Bridges Phosphatidylinositol-4-Phosphate and Actomyosin to Stretch and Shape the Golgi to Promote Budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirkbride KC, Hong NH, French CL, Clark ES, Jerome WG, Weaver AM. Regulation of late endosomal/lysosomal maturation and trafficking by cortactin affects Golgi morphology. Cytoskeleton (Hoboken) 2012;69:625–643. doi: 10.1002/cm.21051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- 116.Chen JL, Lacomis L, Erdjument-Bromage H, Tempst P, Stamnes M. Cytosol-derived proteins are sufficient for Arp2/3 recruitment and ARF/coatomer-dependent actin polymerization on Golgi membranes. Febs Letters. 2004;566:281–286. doi: 10.1016/j.febslet.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 117.Almeida CG, Yamada A, Tenza D, Louvard D, Raposo G, Coudrier E. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat Cell Biol. 2011;13:779–789. doi: 10.1038/ncb2262. [DOI] [PubMed] [Google Scholar]
- 118.von Blume J, Alleaume A-M, Cantero-Recasens G, Curwin A, Carreras-Sureda A, Zimmermann T, van Galen J, Wakana Y, Valverde MA, Malhotra V. ADF/Cofilin Regulates Secretory Cargo Sorting at the TGN via the Ca(2+) ATPase SPCA1. Dev Cell. 2011;20:652–662. doi: 10.1016/j.devcel.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 119.von Blume J, Duran JM, Forlanelli E, Alleaume AM, Egorov M, Polishchuk R, Molina H, Malhotra V. Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. Journal of Cell Biology. 2009;187:1055–1069. doi: 10.1083/jcb.200908040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.De Matteis MA, Morrow JS. Spectrin tethers and mesh in the biosynthetic pathway. J Cell Sci. 2000;113(Pt 13):2331–2343. doi: 10.1242/jcs.113.13.2331. [DOI] [PubMed] [Google Scholar]
- 121.Salcedo-Sicilia L, Granell S, Jovic M, Sicart A, Mato E, Johannes L, Balla T, Egea G. betaIII spectrin regulates the structural integrity and the secretory protein transport of the Golgi complex. J Biol Chem. 2013;288:2157–2166. doi: 10.1074/jbc.M112.406462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 123.Stamnes M. Regulating the actin cytoskeleton during vesicular transport. Curr Opin Cell Biol. 2002;14:428–433. doi: 10.1016/s0955-0674(02)00349-6. [DOI] [PubMed] [Google Scholar]
- 124.Marsh BJ, Mastronarde DN, Buttle KF, Howell KE, McIntosh JR. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc Natl Acad Sci U S A. 2001;98:2399–2406. doi: 10.1073/pnas.051631998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lehrer SS. Damage to actin filaments by glutaraldehyde: protection by tropomyosin. J Cell Biol. 1981;90:459–466. doi: 10.1083/jcb.90.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maupin P, Pollard TD. Improved preservation and staining of HeLa cell actin filaments, clathrin-coated membranes, and other cytoplasmic structures by tannic acid-glutaraldehyde-saponin fixation. J Cell Biol. 1983;96:51–62. doi: 10.1083/jcb.96.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Svitkina T, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actinmyosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 129.Hanein D. Tomography of actin cytoskeletal networks. Methods Enzymol. 2010;483:203–214. doi: 10.1016/S0076-6879(10)83010-1. [DOI] [PubMed] [Google Scholar]
- 130.Kudryashev M, Lepper S, Baumeister W, Cyrklaff M, Frischknecht F. Geometric constrains for detecting short actin filaments by cryogenic electron tomography. PMC Biophys. 3:6. doi: 10.1186/1757-5036-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ydenberg CA, Smith BA, Breitsprecher D, Gelles J, Goode BL. Cease-fire at the leading edge: new perspectives on actin filament branching, debranching, and cross-linking. Cytoskeleton (Hoboken) 2011;68:596–602. doi: 10.1002/cm.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beck KA, Buchanan JA, Malhotra V, Nelson WJ. Golgi spectrin: identification of an erythroid beta-spectrin homolog associated with the Golgi complex. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stankewich MC, Tse WT, Peters LL, Ch'ng Y, John KM, Stabach PR, Devarajan P, Morrow JS, Lux SE. A widely expressed betaIII spectrin associated with Golgi and cytoplasmic vesicles. Proc Natl Acad Sci U S A. 1998;95:14158–14163. doi: 10.1073/pnas.95.24.14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beck KA, Buchanan JA, Nelson WJ. Golgi membrane skeleton: Identification, localization and oligomerization of a 195 kDa ankyrin isoform associated with the Golgi complex. J Cell Sci. 1997;110:1239–1249. doi: 10.1242/jcs.110.10.1239. [DOI] [PubMed] [Google Scholar]
- 135.Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (Ank(G119)) in the kidney and muscle that binds beta I Sigma spectrin and associates with the Golgi apparatus. Journal of Cell Biology. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kang Q, Wang T, Zhang H, Mohandas N, An X. A Golgi-associated protein 4.1B variant is required for assimilation of proteins in the membrane. J Cell Sci. 2009;122:1091–1099. doi: 10.1242/jcs.039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bennett V, Gilligan DM. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annu Rev Cell Biol. 1993;9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- 138.Holleran EA, Ligon LA, Tokito M, Stankewich MC, Morrow JS, Holzbaur EL. beta III spectrin binds to the Arp1 subunit of dynactin. J Biol Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- 139.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 140.Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22:4575–4587. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Roth-Johnson EA, Vizcarra CL, Bois JS, Quinlan ME. Interaction between Microtubules and the Drosophila Formin Cappuccino and Its Effect on Actin Assembly. J Biol Chem. 2014;289:4395–4404. doi: 10.1074/jbc.M113.499921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guzik-Lendrum S, Heissler SM, Billington N, Takagi Y, Yang Y, Knight PJ, Homsher E, Sellers JR. Mammalian myosin-18A, a highly divergent myosin. J Biol Chem. 2013;288:9532–9548. doi: 10.1074/jbc.M112.441238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Taft MH, Behrmann E, Munske-Weidemann LC, Thiel C, Raunser S, Manstein DJ. Functional Characterization of Human Myosin-18A and its Interaction with F-actin and GOLPH3. J Biol Chem. 2013 doi: 10.1074/jbc.M113.497180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hehnly H, Xu W, Chen JL, Stamnes M. Cdc42 regulates microtubule-dependent Golgi positioning. Traffic. 2010;11:1067–1078. doi: 10.1111/j.1600-0854.2010.01082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dubois T, Paleotti O, Mironov AA, Fraisier V, Stradal TEB, De Matteis MA, Franco M, Chavrier P. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–U359. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- 147.Musch A, Cohen D, Rodriguez-Boulan E. Myosin II is involved in the production of constitutive transport vesicles from the TGN. J Cell Biol. 1997;138:291–306. doi: 10.1083/jcb.138.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Surma MA, Klose C, Simons K. Lipid-dependent protein sorting at the trans-Golgi network. Biochim Biophys Acta. 2012;1821:1059–1067. doi: 10.1016/j.bbalip.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 149.Chen JL, Fucini RV, Lacomis L, Erdjument-Bromage H, Tempst P, Stamnes M. Coatomer-bound Cdc42 regulates dynein recruitment to COPI vesicles. J Cell Biol. 2005;169:383–389. doi: 10.1083/jcb.200501157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ikeda Y, Dick KA, Weatherspoon MR, Gincel D, Armbrust KR, Dalton JC, Stevanin G, Durr A, Zuhlke C, Burk K, et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- 152.Perkins EM, Clarkson YL, Sabatier N, Longhurst DM, Millward CP, Jack J, Toraiwa J, Watanabe M, Rothstein JD, Lyndon AR, et al. Loss of beta-III spectrin leads to Purkinje cell dysfunction recapitulating the behavior and neuropathology of spinocerebellar ataxia type 5 in humans. J Neurosci. 2010;30:4857–4867. doi: 10.1523/JNEUROSCI.6065-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stankewich MC, Gwynn B, Ardito T, Ji L, Kim J, Robledo RF, Lux SE, Peters LL, Morrow JS. Targeted deletion of betaIII spectrin impairs synaptogenesis and generates ataxic and seizure phenotypes. Proc Natl Acad Sci U S A. 2010;107:6022–6027. doi: 10.1073/pnas.1001522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Clarkson YL, Gillespie T, Perkins EM, Lyndon AR, Jackson M. Beta-III spectrin mutation L253P associated with spinocerebellar ataxia type 5 interferes with binding to Arp1 and protein trafficking from the Golgi. Hum Mol Genet. 2010;19:3634–3641. doi: 10.1093/hmg/ddq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 159.Stephens DJ. Functional coupling of microtubules to membranes - implications for membrane structure and dynamics. J Cell Sci. 2012;125:2795–2804. doi: 10.1242/jcs.097675. [DOI] [PubMed] [Google Scholar]
- 160.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nature Reviews Molecular Cell Biology. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]