Abstract
Formins are cytoskeleton regulating proteins characterized by a common FH2 structural domain. As key players in the assembly of actin filaments, formins direct dynamic cytoskeletal processes that influence cell shape, movement and adhesion. The large number of formin genes, fifteen in the human, suggests distinct tasks and expression patterns for individual family members, in addition to overlapping functions. Several formins have been associated with invasive cell properties in experimental models, linking them to cancer biology. One example is FMNL1, which is considered to be a leukocyte formin and is known to be overexpressed in lymphomas. Studies on FMNL1 and many other formins have been hampered by a lack of research tools, especially antibodies suitable for staining paraffin-embedded formalin-fixed tissues. Here we characterize, using bioinformatics tools and a validated antibody, the expression pattern of FMNL1 in human tissues and study its subcellular distribution. Our results indicate that FMNL1 expression is not restricted to hematopoietic tissues and that neoexpression of FMNL1 can be seen in epithelial cancer.
Keywords: FMNL1, FRL1, formin, actin cytoskeleton, immunohistochemistry
Introduction
Formins are large multi-domain proteins that orchestrate a variety of cellular functions via the regulation of cytoskeletal dynamics. The conserved function of formins is polymerization of unbranched actin filaments. At the cellular level, they are involved in an abundance of processes, such as the maintenance of cell polarity, adhesion and migration. Formins are also implicated in morphogenesis, and have been linked to several human diseases (Faix and Grosse 2006).
Formins can be divided into subgroups based on phylogenetic analysis of the conserved formin homology 2 (FH2) domain that defines the family. The FH2 domain is essential for dimerization and subsequent actin filament assembly. The FH2 dimer stays attached to and moves along the actin filament when it is elongating, simultaneously capping it from severing molecules. N-terminal to the FH2 domain is the formin homology 1 (FH1) domain. The FH1 domain is essential for the physiological action of formins, as it recruits profilin-bound actin monomers to the growing actin filament.
Formin-like protein 1 (FMNL1, also known as FRL1) belongs to the group of diaphanous-related formins (DRFs). DRFs have additional autoregulatory domains: the GTPase binding domain (GBD) and a diaphanous autoregulatory domain (DAD). DRFs reside in an inactive conformation, and become activated by Rho GTPase binding to the GBD (Higgs 2005; Young and Copeland 2010). FMNL1 has two recognized Rho GTPase binding partners: Rac1 and RhoA (Yayoshi-Yamamoto et al. 2000; Gomez et al. 2007).
According to mRNA expression and western blot results, FMNL1 expression in human tissues is restricted to hematopoietic and lymphoid tissues such as thymus, spleen and bone marrow (Favaro et al. 2003; Han et al. 2009; Krainer et al. 2013). In addition, FMNL1 is overexpressed in non-Hodgkin lymphoma and in leukemic cell lines (Favaro et al. 2006; Schuster et al. 2007; Favaro et al. 2013). There are three FMNL1 isoforms, with differences at the C-terminal end (Fig. 1). Of these, isoform 3 (also known as FMNL1γ) has an intron retention, which may lead to its independence from autoregulation. Recently, many specific immunological functions have been attributed to FMNL1. In macrophages, FMNL1 participates in phagocytosis and migration and, in T-cells, FMNL1 is needed for cytotoxicity and maintenance of the Golgi complex (Yayoshi-Yamamoto et al. 2000; Seth et al. 2006; Gomez et al. 2007; Mersich et al. 2010; Colon-Franco et al. 2011). FMNL1 is also highly expressed in many epithelial cancer cell lines, which suggests it might be upregulated during malignant transformation (Colon-Franco et al. 2011). However, the expression and functional role of FMNL1 in human cancers is largely unknown. The main reason for this is the lack of antibodies that work in routine diagnostic material; i.e., in formalin-fixed paraffin-embedded (FFPE) tissues. This lack of antibodies for immunohistochemistry has been a major hurdle in investigating the possible alterations of FMNL1 expression in malignant disease.
Figure 1.
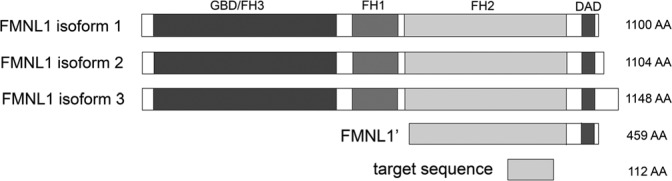
The domain structure of FMNL1 isoforms and the regions used for antibody production and transfections. FMNL1 isoforms 1 and 2 show only minor variations in their C-terminal region. The C-terminus of isoform 3 differs from the others because of an intron retention, which partly changes the DAD domain and endows constitutive activation of this isoform. The antigenic sequence for antibody production is within the common FH2 domain. The construct used for transfections harbors most of the FH2 domain and the DAD domain of isoform 1.
In this study, we describe and validate an antibody, which has been raised for immunohistochemical detection of the known FMNL1 isoforms, and describe the expression profile of FMNL1 in 28 human tissue types. Furthermore, we show that FMNL1 is overexpressed in basal type breast cancer with poor prognosis.
Materials & Methods
Cellular Studies
The human embryonic kidney cell line HEK 293T, the MDA-MB-231 breast cancer cell line and the IGROV1 ovarian cancer cell line were cultured in DMEM (Invitrogen, Carlsbad, CA). Melanoma cell lines WM164 and SK-Mel-28 were cultured in RPMI 1640 (Gibco-BRL; Paisely, UK) and MEM (Invitrogen), respectively. All media were supplemented with 10% fetal bovine serum (FBS), 5 mM ultraglutamine and 100 U/ml penicillin-streptomycin (Gibco-BRL).
The pCMV-sport6 vector containing the C-terminal half of human FMNL1 (BC021906.1) (abbreviated as FMNL1’, Fig. 1) was obtained from the Genome Biology Unit, Institute of Biotechnology, University of Helsinki, Finland. FMNL1’ was subcloned into the pEGFP-C1 vector (Clontech Laboratories; Mountain View, CA) for a GFP tag. Cultured cells were transfected with pEGFP-FMNL1’, with an empty pEGFP-C1 control vector (Clontech Laboratories) or pCMV-sport6-FMNL1’ using Lipofectamine 2000 (Invitrogen). The cells were used 48 hr after transfection.
For protein analyses, lysates were prepared and western blotting carried out as described (Gardberg et al. 2010). The primary rabbit anti-human FMNL1 antibody (HPA008129; Sigma-Aldrich Corporation; St Louis, MO) was used at a 1:1000 dilution. For simultaneous detection of FMNL2 and FMNL3, a primary mouse monoclonal anti-FMNL2 antibody was used (ab56963, Abcam; Cambridge, MA) at 1:1000 dilution. This antibody has been shown to cross-react with FMNL3 (Block et al. 2012).
To confirm the specificity of the antibodies, western blots were performed after FMNL2 and FMNL3 silencing in SK-Mel-28 cells using SMARTpool small interfering RNA (siRNA) (Dharmacon Research; Lafayette, CO). Non-targeting pooled siRNA was used as a control. Cells were transfected using DharmaFECT 4 transfection reagent (Dharmacon Research), according to the manufacturer’s instructions. FMNL2 or FMNL3 expression was silenced at 50 nM siRNA concentration. For double knockdown of FMNL2 and FMNL3, 25 nM of each siRNA was used. For subcellular imaging of FMNL1 or FMNL1’, cells were seeded 48 hr after the transfection on gelatine-coated coverslips and grown for additional 24 hr. Cells were fixed in 4% paraformaldehyde for 10 min at room temperature. The coverslips were washed and blocked with 3% bovine serum albumin (BSA), 5% dry milk, 0.5% Triton X-100 in PBS, and incubated for 1 hr with rabbit anti-FMNL1 (1:200; Sigma-Aldrich). The coverslips were subsequently incubated with Alexa Fluor 568 goat anti-rabbit IgG (Invitrogen), and filamentous actin was detected by Alexa Fluor 488 phalloidin (1:50; Invitrogen). The mounting media contained DAPI for staining nuclei (Vector Laboratories, Burlingame, CA). Images were taken with a Zeiss LSM780 confocal microscope (Carl Zeiss, Göttingen, Germany) or with an Olympus BX60 fluorescence microscope (Olympus Microscopes, Essex, UK).
In Silico Transcriptomics Analysis, Tissue Specimens and Immunohistochemistry
The GeneSapiens database was utilized to study the FMNL1 mRNA expression across all normal and neoplastic human tissues (Kilpinen et al. 2008). The samples included in this database were analyzed on the Affymetrix platform and, because of the unique normalization and data quality verifications, the gene expression profiles collected from different studies can be combined to generate an overview of the expression profile in human tissues.
Normal human tissues were collected, fixed and immunohistochemically stained, as described using a LabVision autostainer device (Gardberg et al. 2010). The collection of normal tissues for this study was approved by the Joint Committee on Ethics of the University of Turku and Turku University Hospital as well as written consent from the donors. Paraffin-embedded triple negative (basal type) breast cancer specimens were collected from the tissue archive of the Department of Pathology at Turku University Hospital with the approval of the Joint Committee on Ethics of the University of Turku and Turku University Hospital. According to Finnish legislation, the permission for research use of specimens collected for diagnostic purposes is granted by local institutional authorities when personal patient information is not included. The FMNL1 antibody (HPA008129, Sigma-Aldrich) was used at a 1:75 dilution. Individual tissues and cell types were evaluated by scoring the staining intensity using a grading from 0 (no staining) to 3 (strong staining). For double staining, the FMNL1 antibody (1:75) and a CD68 antibody (1:100, clone PG-M1 Dako, Glostrup, Denmark) were stained with a Benchmark XT autostainer device (Roche Tissue Diagnostics/Ventana Medical Systems, Tucson, AZ) by pretreatment with extended CC1, antibody incubations at 37C, a FMNL1 incubation time of 44 min, denaturation incubations at 95C for 4 min, a CD68 incubation time of 32 min followed by an Amplification Kit and counterstaining with Hematoxylin for 4 min and Bluing for 4 min. For detection, an UltraView Universal DAB Detection Kit and an UltraView Universal Alkaline Phosphatase Red Detection Kit were used (Roche Tissue Diagnostics/Ventana Medical Systems).
Results
Antibody Validation
The FMNL1 anti-human antibody was designed and produced by the Swedish Human Protein Atlas (HPA) on our initiative, in a project that placed emphasis on generating antibodies for immunohistochemistry. A sequence specific for FMNL1 (Uniprot O95466, amino acids 880-991) was chosen for recombinant protein production of Protein Epitope Signature Tags (PrESTs) and for immunization (Fig. 1). This sequence has only a 50% homology with the most closely related DRFs—FMNL2 and FMNL3. Rabbit polyclonal antibodies were affinity purified and initially validated by an antigen array of 384 different PrESTs. In this assay, the antibody only interacted with its own antigen.
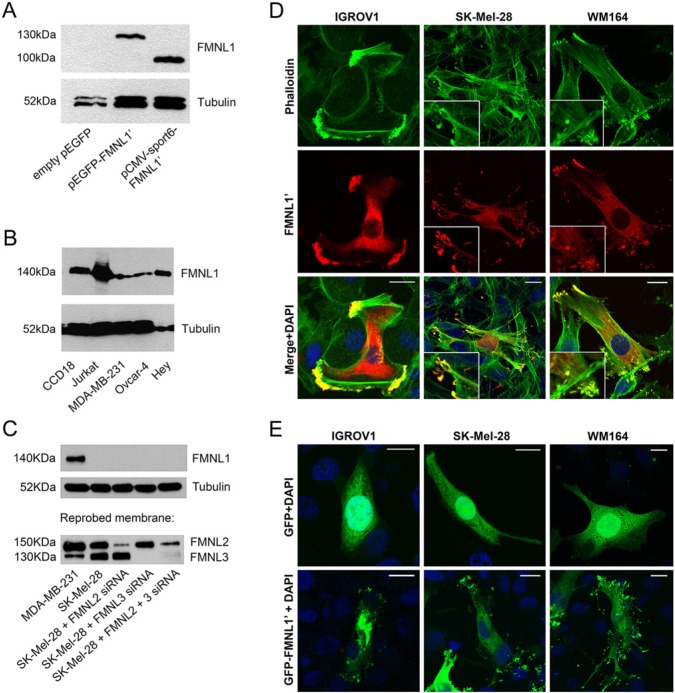
In western blotting, human embryonic kidney HEK 293T, ovarian adenocarcinoma IGROV1, and melanoma cell lines WM164, SK-Mel-28 and Bowes did not react with FMNL1 antibody (not shown). These cell lines were chosen for overexpression studies using the C-terminal half of FMNL1 isoform 1 (denoted FMNL1’) and the green fluorescent protein (GFP)-tagged FMNL1’. After transfection, cell lysates blotted with the FMNL1 antibody contained single bands of size 100 and 130 kDa, respectively, indicating that the antibody specifically detects FMNL1 (HEK 293T depicted in Fig. 2A).
Figure 2.
Detection of the FMNL1’ construct and endogenous FMNL1 by western blotting and fluorescence microscopy. (A) HEK 293T cells were transfected with pCMVsport6-FMNL1’, pEGFP-FMNL1’ or empty pEGFP expression constructs. In control cells transfected with empty pEGFP, no FMNL1 band was detected in western blotting. In transfected cells, a 130-kDa band corresponding to GFP-tagged FMNL1’, and a 100-kDa band corresponding to non-tagged FMNL1’ is visible. (B) Endogenous FMNL1 is detected as a single band of 140 kDa in several cell lines: myofibroblasts (CCD18), T-cell lymphoma (Jurkat), breast cancer (MDA-MB-231) and ovarian cancer (Ovcar-4 and Hey). (C) To confirm that the FMNL1 antibody does not cross-react with FMNL2 or FMNL3, cell lysates were first blotted with the FMNL1 antibody, and reprobed with an antibody that cross-reacts with both FMNL2 and FMNL3. In MDA-MB-231 cells, a single band corresponding to FMNL1 was visible in MDA-MB-231 cells but not in SK-Mel-28 melanoma cells. In both cell lines, bands corresponding to FMNL2 and FMNL3 were present, indicating that the FMNL1 antibody specifically detects FMNL1, not FMNL2 or FMNL3. The specificity of the FMNL2 and FMNL3 antibody was further confirmed by reduced reactivity after corresponding siRNA treatments. Tubulin was blotted as a loading control. (D) IGROV1 ovarian cancer cells and the WM164 and SK-Mel-28 melanoma cell lines, which do not express endogenous FMNL1, were transfected with an FMNL1’ construct and stained with green phalloidin to visualize actin filaments (upper panel) and FMNL1 (red, middle panel). In membrane protrusions, FMNL1’ co-localizes with bundled F-actin (merge in yellow, bottom panel). Higher magnification of membrane protrusions are depicted in insets. (E) IGROV1, WM164, and SK-Mel-28 cells were transfected with a construct expressing GFP-FMNL1’ or GFP only. GFP shows a diffuse cytoplasmic distribution (upper panel), whereas GFP-tagged FMNL1’ is partly located at the membrane protrusions. Scale bars, 20 μm. Nuclei were stained with DAPI (blue).
For further validation of the antibody, we performed western blotting on lysates from several cell lines. The predicted molecular mass for FMNL1 isoforms 1 and 2 is 122 kDa and for isoform 3 124 kDa. The antibody detected a single band slightly higher than expected (140 kDa) (which is in line with the band for FMNL1 reported in the literature) in the following human cell lines: CCD18 (myofibroblast), Jurkat (T-cell lymphoma), MDA-MB-231 (breast cancer), Ovcar-4 and Hey (both ovarian cancer) (Fig. 2B). Strong reactivity was seen in Jurkat, CCD18 and Hey cells.
To confirm that the FMNL1 antibody does not react with the two closely related formins, FMNL2 and FMNL3, we further performed western blotting with an antibody that detects FMNL2 and FMNL3 but not FMNL1 (Block et al. 2012). In SK-Mel-28 cells, no reactivity was seen with the FMNL1 antibody. Reprobing the same membrane with the FMNL2 and FMNL3 antibody detected bands corresponding to FMNL2 (150 kDa) and FMNL3 (130 kDa). These bands were clearly weaker or even undetectable after silencing with corresponding siRNAs (Fig. 2C). The western blotting was repeated in the inverse order: first using the FMNL2 and FMNL3 antibody and subsequently reprobing the membranes with the FMNL1 antibody. Identical results were obtained (data not shown). These findings show that the FMNL1 antibody does not cross-react with FMNL2 or FMNL3 and also demonstrate that the down-regulation of FMNL2 and/or FMNL3 does not lead to compensatory neoexpression of FMNL1, at least not in the timeframe of the siRNA experiments.
Next, we visualized FMNL1’ in transfected cell lines by immunofluorescence staining with the FMNL1 antibody, and stained actin filaments with fluorescent phalloidin. In cell lines IGROV1, SK-Mel-28 and WM164, the staining pattern was highly similar: the antibody detected FMNL1’ in the cytoplasm and at the cell membranes of transfected cells. Membrane staining was seen at protrusions at the short ends of elongated cells, where it co-localized with abundant actin filaments (Fig. 2D).
In order to confirm that the antibody detected FMNL1’ specifically, we furthermore visualized GFP-FMNL1’ in the corresponding transfected cell lines. GFP-FMNL1’ was present in transfected cells in the same locations as untagged FMNL1’ detected with FMNL1 antibody (Fig. 2E). Interestingly, the FMNL1’ constructs seemed to increase actin-enriched membrane protrusions in both ovarian cancer and melanoma cells. This is a novel finding, since others have shown that an overexpression of an FMNL1 construct containing FH1 and FH2 domains fails to induce filopodia or actin-rich membrane protrusions (Harris et al. 2010). Also, full length FMNL1 and FMNL2 constructs have been reported to remain cytoplasmic. Constitutively active FMNL1 constructs and FMNL1 isoform 3 target the cell membrane and induce membrane blebbing. This targeting requires N-myristoylation (Han et al. 2009). Our results indicate that there could be additional mechanisms intrinsic to the C-terminal FH2 and DAD domains that guide FMNL1 to the membrane, and further induce actin-rich membrane protrusions.
FMNL1 Expression in Human Tissues
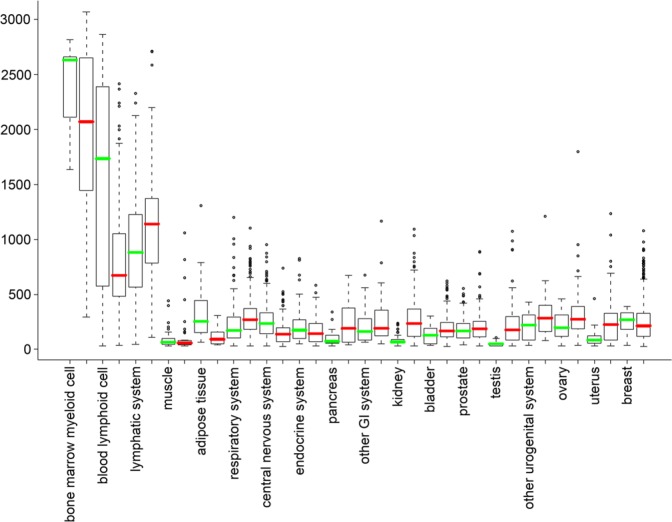
To evaluate the overall expression levels of FMNL1 mRNA across human tissues, we conducted an in silico survey on the GeneSapiens database (Kilpinen et al. 2008). We found that, in normal tissues, FMNL1 mRNA expression indeed is high in lymphocytes and hematopoietic tissues, whereas the expression is low or absent in most other tissues. Similarly, FMNL1 mRNA expression is high in hematopoietic malignancies, but low or absent in other cancer types. Interestingly, some cancer groups, such as breast cancer, had a group of high-expressing outliers, suggesting that a subpopulation among these cancers either have a marked lymphoid component or that the tumor cells express FMNL1 (Fig. 3; Kilpinen et al. 2008).
Figure 3.
FMNL1 mRNA expression profile in human normal and neoplastic tissues. The normalized expression values (y-axis) of FMNL1 across normal tissues and cancer types are presented as box-plots. The box extends from the first to the third quartile of the data and the median is indicated as a green line for normal tissue, and a red line for cancer. The whiskers extend to the extreme values unless there are outliers. The data observations that lie more than 1.5 × interquartile range (IQR) lower than the first quartile or 1.5 × IQR higher than the third quartile were considered as outliers and are indicated separately. The highest expression levels are seen in bone marrow myeloid cell and lymphatic system and their malignancies. In most tissues and cancers, FMNL1 expression is low or absent. Among cancer tissues, a substantial number of cases express FMNL1 more than the corresponding normal tissues.
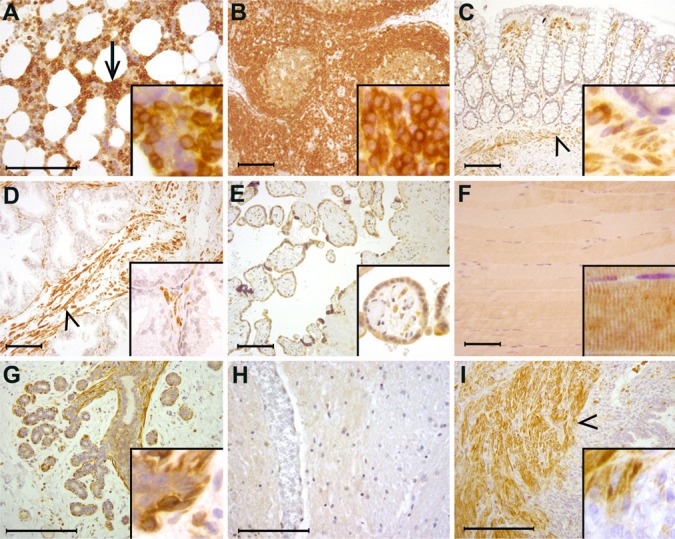
To investigate FMNL1 protein expression in human tissues, we immunohistochemically stained 28 different human tissues that were evaluated as morphologically normal in hematoxylin and eosin staining. The immunohistochemical staining pattern of FMNL1 was distinct. Moderate to strong expression was seen only in leukocytes and their bone marrow precursors, smooth muscle cells and myoepithelial cells in exocrine tissues. In bone marrow, FMNL1 expression was restricted to myeloid cells, whereas erythroid cells and megakaryocytes remained negative (Fig. 4A). In lymph nodes, mature lymphocytes stained strongly (Fig. 4B), as did lymphocytes in other tissues, such as the colon (Fig. 4C). Smooth muscle cells in all tissues stained strongly, including those in prostatic stromal smooth muscle (Fig. 4 D) and in all smooth muscle-walled blood vessels. Weak staining was seen in placental trophoblastic cells (Fig. 4E), skeletal muscle (Fig. 4F) and a few epithelial cell types; i.e., mammary epithelium (Fig. 4G), esophageal squamous epithelium and gastric epithelium. Examples of tissues that do not express FMNL1 are the central nervous system (Fig. 4H), peripheral nerve, pancreas, and endometrium (Fig. 4I). In all positively stained cells, FMNL1 staining was cytoplasmic. The results from the immunohistochemical analysis are summarized in Table 1.
Figure 4.
Distribution of FMNL1 in human tissues. (A) In the bone marrow, megakaryocytes and erythroid cells do not express FMNL1, whereas myeloid cells stain strongly (arrow). (B) In lymph nodes, mature lymphocytes stain strongly. (C) In the colon, epithelial cells do not express FMNL1, whereas stromal lymphocytes and smooth muscle show strong reactivity (arrowhead). (D) Prostatic epithelium does not express FMNL1, whereas stromal smooth muscle cells stain strongly (arrowhead). (E) In the placenta, trophoblastic cells express FMNL1. (F) Skeletal muscle cells stain weakly. (G) In the female breast, epithelial cells express little FMNL1. Myoepithelial cells, however, stain strongly (arrowhead). (H) The central nervous system is an example of tissues where parenchymal cells do not express FMNL1. Neither neurons nor glial cells express FMNL1. (I) In the uterus, endometrial epithelial cells and endometrial stromal cells do not express FMNL1. Small lymphocytes stain positively in the endometrial stroma. Myometrial (smooth muscle) cells stain strongly (arrowhead). Detailed staining is presented in insets. Scale bar, 200 μm.
Table 1.
FMNL1 Expression across Human Tissues and Cell Types.
| Tissue | Cell type/Localization | FMNL1 staining* |
|---|---|---|
| Adrenal gland | ||
| cortical cells | 0 | |
| medullary cells | 0 | |
| Bone marrow | ||
| erythropoietic cells | 0 | |
| myelopoietic cells | 3 | |
| megakaryocytes | 0 | |
| Breast | ||
| ductal epithelium | 1 | |
| lobular epithelium | 1 | |
| myoepithelium | 3 | |
| Brain | ||
| glial cells | 0 | |
| neurons | 0 | |
| Colon | ||
| epithelium | 0 | |
| stromal cells | 0 | |
| stromal lymphocytes | 3 | |
| Duodenum | ||
| epithelium | 0 | |
| stromal cells | 0 | |
| stromal lymphocytes | 3 | |
| smooth muscle cells | 3 | |
| Esophagus | ||
| squamous epithelium | 1a | |
| Heart | ||
| cardiomyocytes | 0 | |
| Kidney | ||
| mesangial cells | 0 | |
| endothelium | 0 | |
| arteriolar smooth muscle cells | 3 | |
| tubular epithelium | 0 | |
| stromal cells | 0 | |
| Liver | ||
| hepatocytes | 0 | |
| biliary epithelium | 0 | |
| Kupffer cells | 2 | |
| Lung | ||
| pneumocytes | 0 | |
| alveolar macrophages | 3 | |
| Lymph node | ||
| germinal center lymphocytes | 2 | |
| other lymphocytes | 3 | |
| Ovary | ||
| stroma | 0 | |
| granulosa cells | 0 | |
| follicles | 0 | |
| Pancreas | ||
| acinar cells | 0 | |
| ductal epithelium | 0 | |
| Langerhans islet cells | 0 | |
| Parathyroid gland | ||
| chief cells | 0 | |
| oxyphilic cells | 0 | |
| Parotid gland | ||
| acinar cells | 0 | |
| ductal cells | 0 | |
| Peripheral nerve | ||
| Schwann cells | 0 | |
| Placenta | ||
| trophoblast cells | 1 | |
| stromal cells | 0 | |
| endothelium | 0 | |
| Prostate | 0 | |
| epithelium | 0 | |
| smooth muscle cells | 3 | |
| Skeletal muscle | ||
| myocytes | 1 | |
| Skin | ||
| keratinocytes | 0 | |
| melanocytes | 0 | |
| sweat gland epithelium | 0 | |
| sebaceous gland cells | 0 | |
| smooth muscle cells | 3 | |
| Small intestine | ||
| epithelium | 0 | |
| stromal cells | 0 | |
| lymphocytes | 3 | |
| Spleen | ||
| white pulp cells | 3 | |
| red pulp cells | 3 | |
| Stomach | ||
| epithelium | 1 | |
| stromal cells | 0 | |
| smooth muscle cells | 3 | |
| Testis | spermatocytes | 0 |
| Leydig cells | 0 | |
| Sertoli cells | 0 | |
| Thyroid gland | ||
| follicular epithelium | 0 | |
| Urinary bladder | ||
| urothelial cells | 0 | |
| submucosal fibroblasts | 0 | |
| Uterus | ||
| endometrial epithelium | 0 | |
| endometrial stromal cells | 0 | |
| stromal lymphocytes | 3 | |
| myometrium smooth muscle cells | 3 | |
Staining intensity in tissue sections immunohistochemically stained with the anti-FMNL1 antibody. *0=no staining; 1=weak staining; 2=moderate staining; 3=strong staining; a =weaker staining towards epithelial surface.
FMNL1 Expression in Basal Type Breast Cancer
Breast cancer of the basal type is triple negative; i.e., it does not express estrogen receptor, progesterone receptor, or human epidermal growth receptor 2 (HER-2). The basal type breast cancer is a relatively rare subgroup, which has an aggressive clinical behavior.
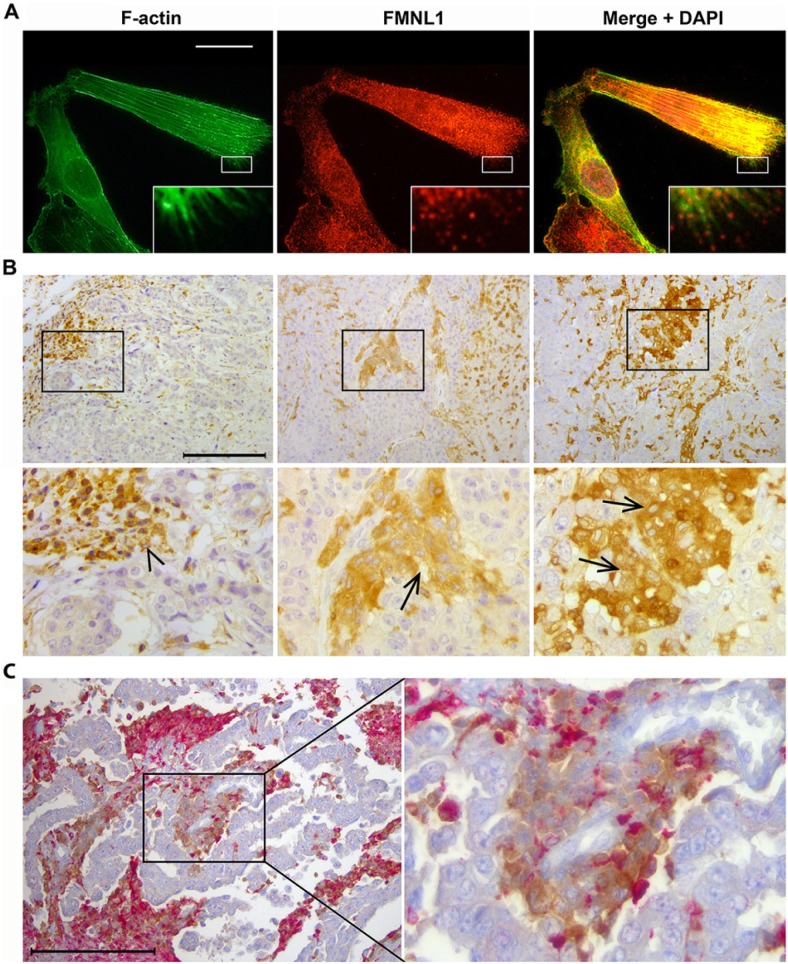
In our database search, we noted high FMNL1 mRNA expression in a set of outliers among the generally low FMNL1-expressing breast cancer tissue samples (Fig. 3). Additionally, among 63 human breast cancer cell lines included in this database, only one expressed high levels of FMNL1 mRNA. The FMNL1-expressing cell line was MDA-MB-231, a basal breast cancer cell line frequently used as a model for epithelial-to-mesenchymal transition (EMT) studies. Since high FMNL1 expression has previously been reported in breast cancers with a poor prognosis (Arjonen et al. 2011), we asked whether basal type breast cancers expressed FMNL1. To answer this question, we stained MDA-MB-231 cells and eight basal type breast cancer samples for FMNL1 expression. In immunofluorescence staining of MDA-MB-231 cells, FMNL1 was localized at cytoplasmic granules and at filopodial tips (Fig. 5A). In immunohistochemical staining of human basal breast cancers, all eight tumors contained FMNL1-positive cells. In three cases, FMNL1 expression was virtually restricted to inflammatory cells (lymphocytes and macrophages within the tumor) and present at the infiltrative margin (Fig. 5B, left micrograph). In five samples, however, FMNL1 staining was additionally seen in a subset of the malignant epithelial cells, mostly near the infiltrative margin of the otherwise negatively staining tumor (Fig. 5B, middle and right micrographs). Since macrophages within tumors can closely resemble neoplastic cells, we further double-stained the positive samples with anti-FMNL1 and the macrophage-specific anti-CD68. In these sections, CD68-negative and FMNL1-positive tumor cells were detected, often in close proximity with CD68- and FMNL1-positive macrophages. The presence of FMNL1 in epithelial cancer cell lines has been reported earlier, but this is, to our knowledge, the first time epithelial cancer cells in human cancer specimens have been shown to express FMNL1.
Figure 5.
FMNL1 staining in basal type breast cancer cells and tissues. (A) MDA-MB-231 breast cancer cells stained for F-actin and FMNL1. FMNL1 is present in lamellipodia, at filopodia tips (inset), and in the cytoplasm. Scale bar, 20 μm. (B) Among basal type breast cancers, FMNL1 expression is heterogeneous. In the left micrograph, cancer cells across the tumor are virtually negative, whereas strong staining is seen in lymphocytes and in other inflammatory cells present in the tumor (arrowhead). In the middle and right micrographs, most breast cancer cells express little FMNL1. However, subpopulations of cells near the tumor margin show strong reactivity (arrows). Scale bar, 200 μm. (C) FMNL1 (brown) and CD68 (red) double staining of basal breast cancers demonstrates that part of the FMNL1-positive cells are CD68-negative carcinoma cells and therefore distinct from macrophages. Scale bar, 200 μm.
Discussion
In this study, we characterized FMNL1 expression across human tissues using an antibody suitable for FFPE tissue analysis. The FMNL1 antibody detects all three FMNL1 isoforms that have been described. In normal tissues, we found that FMNL1 expression is restricted; it was rarely expressed in epithelia, and not present in neural parenchymal cells. Strong expression was seen in myelopoietic cells in the bone marrow and in lymphocytes of the lymph node, spleen and other tissues where myeloid cell descendants are present. These findings are largely in line with earlier studies describing the presence of FMNL1 mRNA or protein in different tissues (Favaro et al. 2003; Han et al. 2009; Krainer et al. 2013). A novel finding is that FMNL1 is strongly expressed in smooth muscle cells and myoepithelial cells. These cell types are present in most tissues; for instance, as layers of smooth muscle around arterioles and myoepithelial cells around ducts of secreting tissues. Previously, T-cells recognizing FMNL1-derived peptide have been shown to have antitumoral activity in hematological malignancies, making FMNL1 an attractive therapeutic target (Schuster et al. 2007).The discovery of FMNL1 as a smooth muscle formin, not only a leukocyte formin, makes such targeting challenging because of its wider than expected expression profile in normal tissues. Although FMNL1 has been attributed to several functional roles in hematopoietic cells, its roles in smooth muscle cells remain to be elucidated in future studies.
We have earlier characterized the expression of two other related formins in human tissues: formin-like protein 2 (FMNL2) and formin homology 2 domain containing 1 (FHOD1) (Gardberg et al. 2010; Gardberg et al. 2013). Of these, FMNL2 is widely expressed, predominantly in nervous and epithelial tissues, whereas FHOD1 is absent from both the nervous system and most epithelial tissues. FHOD1 is predominantly expressed in mesenchymally derived tissues, especially in the endothelium. The expression of FMNL1 is the most restricted, and clearly the highest of these three in hematological cells, smooth muscle and myoepithelium.
Through mRNA profiling, we found that FMNL1 expression was high in a breast cancer cell line of basal type and, whereas most breast cancers express little or no FMNL1, a small group of breast cancers expressed FMNL1. We hypothesized that this subpopulation might include basal type breast cancers. In immunohistochemical staining of basal type breast cancers, FMNL1 was indeed present in all samples. However, the cell types with positive staining were in many cases macrophages and lymphocytes, which are present in many forms of cancer, especially in carcinomas. We conclude that the results for FMNL1 mRNA levels in clinical cancer tests should be interpreted with some caution. As the presence of inflammatory cells can give rise to false positive results, immunohistochemical validation to histologically confirm which cell types are expressing FMNL1 is needed.
Interestingly, FMNL1 was expressed in neoplastic cells in over half of the breast cancers of the basal type. To our knowledge, this is the first report of FMNL1 protein expression in tumors other than hematological malignancies. The expression was seen near the infiltrating edge of the tumors, which suggests a role in invasion. In cultured basal type breast cancer cells, we noted FMNL1 staining at the filopodial tips. This finding extends the results of a recent study, in which macrophage filopodia induced by Borrelia burgdorferii were shown to contain FMNL1 (Naj et al. 2013). Filopodia are thought to be involved in directional migration and adhesion. Since FMNL1 regulates cell motility in macrophages (Yayoshi-Yamamoto et al. 2000), and also promotes proliferation and transendothelial migration in leukemia cells (Favaro et al. 2013), it is possible that basal breast cancer cells utilize similar mechanisms by upregulating FMNL1. The question whether FMNL1 is involved in basal breast cancer progression requires future study.
Acknowledgments
The authors are grateful to Sinikka Kollanus, Piia Mikkonen and Paula Merilahti for skilful technical assistance and to Jaakko Liippo for helping with the photography.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was financially supported by the Finnish Cancer Organisations, Medical Society of Finland, the Juselius foundation, the K. Albin Johansson foundation, the Perklén foundation, the National Graduate School of Clinical investigation and Turku University Hospital grants.
References
- Arjonen A, Kaukonen R, Ivaska J. (2011). Filopodia and adhesion in cancer cell motility. Cell Adh Migr 5:421-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, Geyer M, Stradal TE, Faix J, Rottner K. (2012). FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol 22:1005-1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Franco JM, Gomez TS, Billadeau DD. (2011). Dynamic remodeling of the actin cytoskeleton by FMNL1gamma is required for structural maintenance of the Golgi complex. J Cell Sci 124:3118-3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R. (2006). Staying in shape with formins. Dev Cell 10:693-706 [DOI] [PubMed] [Google Scholar]
- Favaro P, Traina F, Machado-Neto JA, Lazarini M, Lopes MR, Pereira JK, Costa FF, Infante E, Ridley AJ, Saad ST. (2013). FMNL1 promotes proliferation and migration of leukemia cells. J Leukoc Biol 94:503-512 [DOI] [PubMed] [Google Scholar]
- Favaro PM, de Souza Medina S, Traina F, Basseres DS, Costa FF, Saad ST. (2003). Human leukocyte formin: a novel protein expressed in lymphoid malignancies and associated with Akt. Biochem Biophys Res Commun 311:365-371 [DOI] [PubMed] [Google Scholar]
- Favaro PM, Traina F, Vassallo J, Brousset P, Delsol G, Costa FF, Saad ST. (2006). High expression of FMNL1 protein in T non-Hodgkin’s lymphomas. Leuk Res 30:735-738 [DOI] [PubMed] [Google Scholar]
- Gardberg M, Kaipio K, Lehtinen L, Mikkonen P, Heuser VD, Talvinen K, Iljin K, Kampf C, Uhlen M, Grenman R, Koivisto M, Carpen O. (2013). FHOD1, a Formin Upregulated in Epithelial-Mesenchymal Transition, Participates in Cancer Cell Migration and Invasion. PLoS One 8:e74923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardberg M, Talvinen K, Kaipio K, Iljin K, Kampf C, Uhlen M, Carpen O. (2010). Characterization of Diaphanous-related formin FMNL2 in human tissues. BMC Cell Biol 11:55-2121-11-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Kumar K, Medeiros RB, Shimizu Y, Leibson PJ, Billadeau DD. (2007). Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity 26:177-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Eppinger E, Schuster IG, Weigand LU, Liang X, Kremmer E, Peschel C, Krackhardt AM. (2009). Formin-like 1 (FMNL1) is regulated by N-terminal myristoylation and induces polarized membrane blebbing. J Biol Chem 284:33409-33417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Gauvin TJ, Heimsath EG, Higgs HN. (2010). Assembly of filopodia by the formin FRL2 (FMNL3). Cytoskeleton (Hoboken) 67:755-772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN. (2005). Formin proteins: a domain-based approach. Trends Biochem Sci 30:342-353 [DOI] [PubMed] [Google Scholar]
- Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H, Pisto T, Saarela M, Skotheim RI, Bjorkman M, Mpindi JP, Haapa-Paananen S, Vainio P, Edgren H, Wolf M, Astola J, Nees M, Hautaniemi S, Kallioniemi O. (2008). Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 9:R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer EC, Ouderkirk JL, Miller EW, Miller MR, Mersich AT, Blystone SD. (2013). The multiplicity of human formins: Expression patterns in cells and tissues. Cytoskeleton (Hoboken) 70:424-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersich AT, Miller MR, Chkourko H, Blystone SD. (2010). The formin FRL1 (FMNL1) is an essential component of macrophage podosomes. Cytoskeleton (Hoboken) 67:573-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj X, Hoffmann AK, Himmel M, Linder S. (2013). The formins FMNL1 and mDia1 regulate coiling phagocytosis of Borrelia burgdorferi by primary human macrophages. Infect Immun 81:1683-1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster IG, Busch DH, Eppinger E, Kremmer E, Milosevic S, Hennard C, Kuttler C, Ellwart JW, Frankenberger B, Nossner E, Salat C, Bogner C, Borkhardt A, Kolb HJ, Krackhardt AM. (2007). Allorestricted T cells with specificity for the FMNL1-derived peptide PP2 have potent antitumor activity against hematologic and other malignancies. Blood 110:2931-2939 [DOI] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK. (2006). Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol 174:701-713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayoshi-Yamamoto S, Taniuchi I, Watanabe T. (2000). FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol Cell Biol 20:6872-6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KG, Copeland JW. (2010). Formins in cell signaling. Biochim Biophys Acta 1803:183-190 [DOI] [PubMed] [Google Scholar]