Abstract
Mesenchymal stromal/stem cells (MSCs) are clinically useful for cell-based therapy, but concerns regarding their ability to replicate limit their human application. MSCs release extracellular vesicles (EVs) that mediate at least in part the paracrine effects of the parental cells. To understand the molecular basis of their biological properties, we characterized the RNA cargo of EVs from porcine adipose-tissue derived MSCs. Comprehensive characterization of mRNA and miRNA gene expression using high-throughput RNA sequencing (RNA-seq) revealed that EVs are selectively enriched for distinct classes of RNAs. For example, EVs preferentially express mRNA for transcription factors (e.g. MDFIC, POU3F1, NRIP1) and genes involved in angiogenesis (e.g. HGF, HES1, TCF4) and adipogenesis (e.g. CEBPA, KLF7). EVs also express Golgi apparatus genes (ARRB1, GOLGA4) and genes involved in TGF-β signaling. In contrast, mitochondrial, calcium signaling, and cytoskeleton genes are selectively excluded from EVs, possibly because these genes remain sequestered in organelles or intracellular compartments. RNA-seq generated reads for at least 386 annotated miRNAs, but only miR148a, miR532-5p, miR378, and let-7f were enriched in EVs compared to MSCs. Gene ontology analysis indicates that these miRNA target transcription factors and genes that participate in several cellular pathways, including angiogenesis, cellular transport, apoptosis, and proteolysis. Our data suggest that EVs transport gene regulatory information to modulate angiogenesis, adipogenesis, and other cell pathways in recipient cells. These observations may contribute to development of regenerative strategies using EVs to overcome potential complications of cell-based therapy.
Keywords: Mesenchymal stem cells, extracellular vesicles, microvesicles, exosomes, next generation sequencing (NGS), RNASeq, gene expression, miRNA
1. Introduction
Mesenchymal stromal/stem cells (MSCs) are undifferentiated non-embryonic stromal cells with multi-lineage potential reflecting their stem cell-like properties. Their ability to differentiate into a broad spectrum of mesenchymal cell lineages and their immunomodulatory properties offer therapeutic avenues for both tissue repair and regeneration1, 2. Importantly, MSCs can be isolated from a variety of tissues, including the stromal vascular fraction of subcutaneous adipose tissue, which is easily accessible and often abundantly available.
Considerable experimental evidence shows that delivery of MSCs can lead to structural and functional improvement of many organs and tissues3. In line with these observations, we have previously shown in porcine renovascular disease that adipose tissue-derived MSCs improved stenotic kidney function and structure after renal revascularization4, 5 and improved function in the non-revascularized stenotic-kidney6. Furthermore, several clinical studies have shown that MSCs are well tolerated and have an excellent safety record7, 8. Notwithstanding preclinical efficacy and safety in ongoing clinical trials, challenges remain in clinical applications as reports have documented that MSCs may promote tumor growth, malformation, or micro-infarctions9. Hence, safe and effective alternatives for their application are desired.
Recent data suggest that extracellular vesicles (EVs) released from MSCs mediate their paracrine effect by transferring proteins, lipids, and genetic material to target cells10, 11. Furthermore, experimental studies have shown that MSC-derived EVs emulate the effect of MSCs in various experimental models, stimulating cell proliferation and repair12, 13. Yet, safe and effective application of this therapy requires a thorough characterization of their molecular content.
Pigs are very effective disease models in biomedical research, particularly for translating findings to humans. The porcine model mimics several characteristics of human physiology, allowing deeper insight into clinically-relevant pathogenic mechanisms and developing regenerative strategies to ameliorate disease progression14. In this study, we addressed the molecular basis for the therapeutic potential of porcine MSC-derived EVs. We comprehensively characterized the mRNA and miRNA expression profile of EVs derived from porcine adipose tissue-MSCs using high-throughput RNA sequencing (RNA-seq) analysis. One key finding is that EVs from porcine MSCs are selectively enriched for distinct classes of mRNAs and miRNAs compared to the MSCs that produce them. Our results provide a molecular basis for understanding the therapeutic potential of EVs derived from MSCs.
2. Methods
2.1. MSC and EV characterization and culture
Autologous MSCs were collected from abdominal fat (5-10g) of 4 female domestic pigs. Adipose tissue was digested in collagenase-H for 45min, filtered, and cultured for 3 weeks in advanced MEM medium (Gibco/Invitrogen) supplemented with 5% platelet lysate (PLTmax, Mill Creek Life Sciences, Rochester, MN) in 37 degree/5% CO2. The 3rd passage was collected and kept in Gibco Cell Culture Freezing Medium (Life Technologies) at -80°C for in-vitro phenotype/function analysis. We avoided the use of any animal products (beyond porcine MSCs) in our cell culture procedures to approximate a clinical-grade tissue culture product. Cellular phenotype was examined in-vitro with immuno-fluorescent staining of porcine MSCs positive for CD90, CD44, and CD105, as previously described5, 15, 16 and consistent with our experience with human MSCs17, 18.
EVs were isolated from supernatants of 10×106 MSCs and cultured for 48h in advanced MEM medium without supplements. After centrifugation at 2000g, cell-free supernatants were ultra-centrifuged at 100,000g for 1h at 4°C, washed in serum-free medium containing HEPES 25mM and submitted to a second ultracentrifugation. EVs were collected and characterized based on the expression of both microvesicle (ß1-integrins, CD73, and CD40) and exosome (CD9 and CD81) markers using fluorescence activated cell sorting (FACS)19.
2.2. RNA sequencing & bioinformatic analysis
RNA sequencing and bioinformatic analysis was performed as previously described18. The following Annotation Sources were used to establish porcine libraries: UCSC Genome Browser assembly ID: susScr3; Sequencing/Assembly provider ID: Swine Genome Sequencing Consortium Sscrofa10.2; Assembly date: Aug. 2011; GenBank accession ID: GCA_000003025.4; NCBI Genome information: NCBI genome/84 (Sus scrofa); NCBI Assembly information: NCBI assembly/304498 (Swine Genome Sequencing Consortium Sscrofa10.2); BioProject information: NCBI Bioproject: 13421;Gene sets: NCBI & Ensembl.
Sequencing RNA libraries were prepared according to the manufacturer's protocol (TruSeq RNA Sample Prep Kit v2, Illumina). In brief, poly-A mRNA, purified from total RNA using oligo dT magnetic beads, was fragmented at 95°C for 8 minutes, eluted from the beads and primed for first strand cDNA synthesis. RNA fragments were copied into first strand cDNA using SuperScript III reverse transcriptase and random primers (Invitrogen), while second strand cDNA synthesis was done using DNA polymerase-I and RNase-H. A single AMPure XP bead (Agencourt) clean-up step purified the double-stranded cDNA. Then, cDNA ends were repaired and phosphorylated using Klenow, T4 polymerase, and T4 polynucleotide kinase followed by a single AMPure XP bead clean-up. Blunt-ended cDNAs were modified to include a single 3′ adenylate (A) residue using Klenow exo- (3′ to 5′ exo minus). Paired-end DNA adaptors (Illumina) with a single “T” base overhang at the 3′ end were immediately ligated to the ‘A tailed’ cDNA population. Unique indexes, included in the standard TruSeq Kits (12-Set A and 12-Set B) were incorporated at the adaptor ligation step for multiplex sample loading on the flow cells. The resulting constructs were purified by two consecutive AMPure XP bead clean-up steps. The adapter-modified DNA fragments were enriched by 12 cycles of PCR using primers included in the Illumina Sample Prep Kit. The concentration and size distribution of the libraries was determined on an Agilent Bioanalyzer DNA 1000 chip. A final quantification using Qubit fluorometry (Invitrogen) was performed to confirm sample concentrations.
Libraries were loaded onto flow cells at concentrations of 8-10pM to generate cluster densities of 700,000/mm2 following the standard protocol for the Illumina cBot and cBot Paired-end cluster kit version 3. Flow cells were sequenced as 51 × 2 paired end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HCS v2.0.12 data collection software. Base-calling was performed using Illumina's RTA version 1.17.21.3. The mRNA-Seq data were analyzed using the MAPRSeq v.1.2.1 system for RNA-sequencing data analysis (http://bioinformaticstools.mayo.edu/research/maprseq/), the Bioinformatics Core standard tool, which includes alignment with TopHat 2.0.620, 21 and gene counts with the featureCounts software22. The miRNA-Seq data were analyzed using CAP-miRSeq v1.123. Normalization and differential expression analysis were performed using edgeR 2.6.224.
2.3. mRNA expression analysis
Expression values for each gene were normalized to 1 million reads and corrected for gene length (reads per kilobasepair per million mapped reads, RPKM). Genes with RPKM>0.1, fold-change (EVs/MSCs) >1.4 and p values <0.05 (EVs vs. MSCs, Student's t-test) were classified as genes enriched in EVs18. Genes with RPKM>0.1 and fold-change (EVs/MSCs) <0.7 were considered excluded from EVs. Functional annotation clustering analysis was performed using DAVID6.7 database (http://david.abcc.ncifcrf.gov/)25, 26 to obtain a ranking of primary gene ontology categories for the up-regulated (enriched) and down-regulated (depleted) genes.
2.4. miRNA expression analysis
miRNA expression levels (normalized total reads) in EVs and MSCs, as well as the fold-change enrichment in EVs or MSCs were calculated. We used miRDB (Version 6.2) to predict target genes of miRNA with fold-change >1.4 and p-values <0.05 (Student's t-test), using a target prediction score ≥8027. Gene ontology analysis was performed using DAVID6.7.
3. Results
3.1. MSC and EV characterization in culture
Porcine MSCs from adipose tissue display a fibroblast-like, spindle-shaped morphology, expressed CD44, CD90, and CD105 markers, and trans-differentiated into osteocytes, chondrocytes, and adipocytes in-vitro (Figure 1A). Transmission and scanning electron microscopy demonstrated that cultured MSCs release substantial amounts of EVs (Figure 1B). Harvested EVs express primarily microvesicle (e.g. ß1-integrins, CD40, and CD73) and some exosome (e.g. CD9 and CD81) markers, as well as surface markers of their parent cells (CD44 and HLA Class 1) (Figure 1C).
Figure 1.
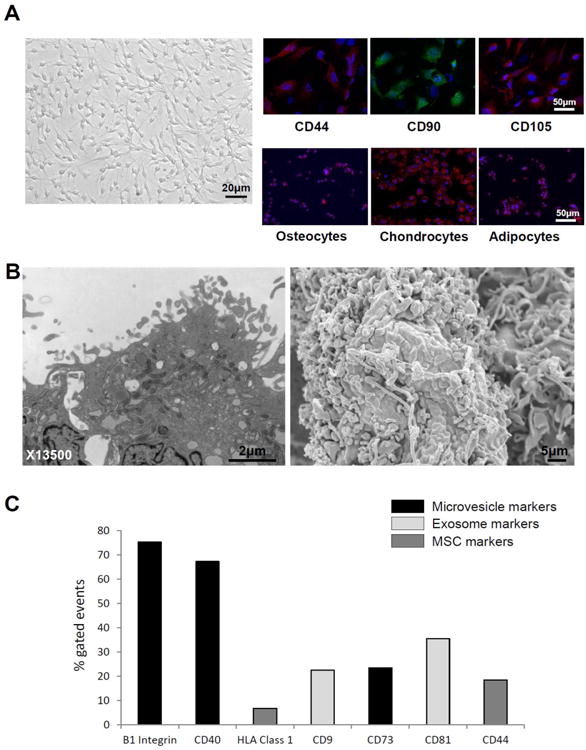
A: MSCs displayed a fibroblast-like, spindle-shaped morphology, expressed MSC markers, and transdifferentiated into osteocytes, chondrocytes, and adipocytes. B: Transmission (left) and scanning (right) electron microscopy demonstrated that cultured MSC release substantial amounts of EVs. C: EVs expressed microvesicle, exosome, and MSC markers.
3.2. mRNA expression
In both MSCs and EVs, the 100 most highly expressed genes account for 46% of all mapped transcripts (Figure 2A). These most abundant mRNAs (expressed at > 100 RPKM) encode mostly proteins involved in translation (e.g., ribosomal and extracellular matrix proteins) (Figure 2B). Of all annotated genes (n=17,628), mapping of RNA reads from cultured MSCs and their descendant EVs reveals that 30.2% (n=5,320) are expressed at levels >1 RPKM (Figure 2C). This proportion was similar between MSCs and EVs, yet EVs showed a higher number of extracellular matrix genes compared to their parent MSCs (Figure 2D). Only 3.5% of genes expressed at levels >1 RPKM changed by more than 4-fold in EVs compared to MSCs, while only a smaller subset showed >10-fold-change in mRNA expression (Figure 3A). Annotation analysis revealed that EVs preferentially contain mRNAs encoding for transcription factors (71.4%), Golgi apparatus components (21.4%), and proteins involved in TGF-β signaling (7.21%) (Figure 3B). In contrast, mRNAs for mitochondrial, calcium signaling, and cytoskeleton components were selectively depleted from EVs (Figure 4A-C).
Figure 2.
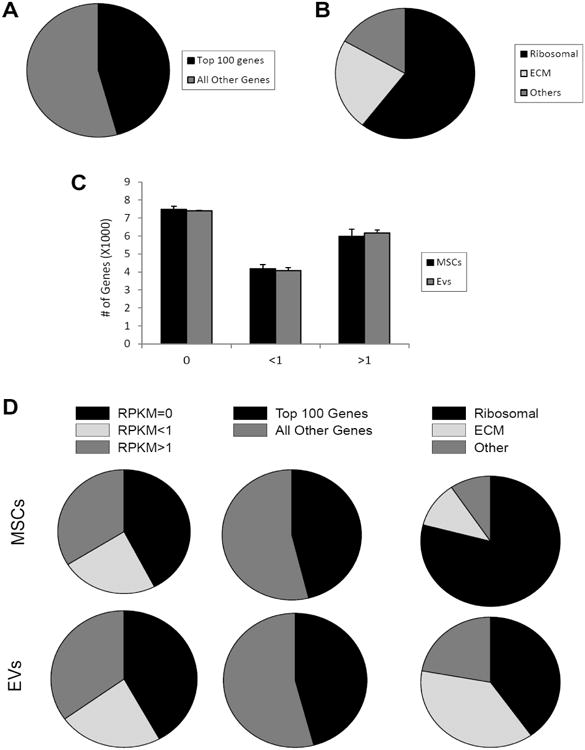
A: The 100 most highly expressed genes in MSCs and EVs account for 46% of all mapped transcripts. B: mRNAs expressed at >100 RPKM encode mostly proteins involved in translation. C: 30.2% of all annotated genes are expressed at levels >1 RPKM. D: Pie chart of data in panel C, expression (RPKM) of the 100 most abundant genes relative to other genes, and differential expression of genes in MSCs and EVs.
Figure 3.

Only 3.5% of genes expressed at levels greater than 1 RPKM changed by more than 4-fold when comparing MSCs and EVs, while a smaller subset showed a >10-fold-change in mRNA expression. B: mRNA RPKM (top) and fold-change (bottom) showing that EVs preferentially expressed transcription factors, Golgi apparatus genes, and genes involved in TGF-β signaling.
Figure 4.
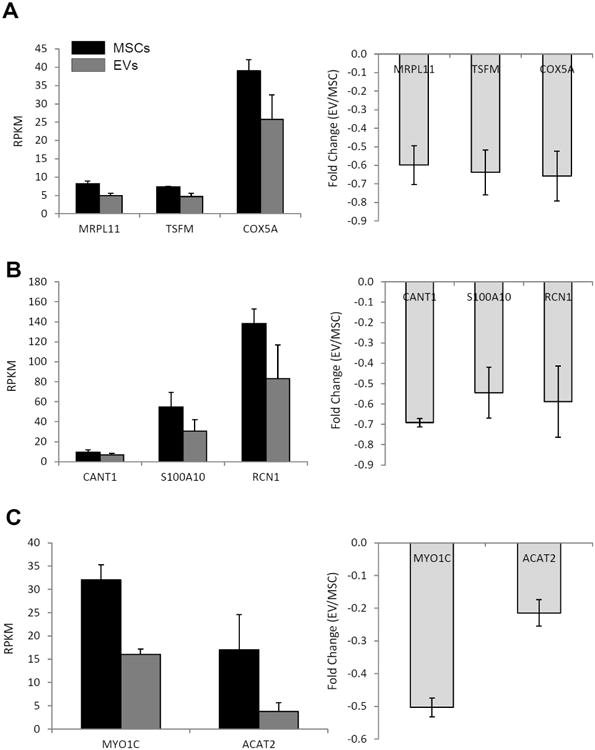
mRNA RPKM (left) and fold-change (right) showing that mitochondrial (A), calcium signaling (B), and cytoskeleton (C) genes were selectively depleted from EVs.
3.3. Selective enrichment of mRNAs enriched in EVs
The mRNAs for 40 transcription factors were selectively enriched in EVs (Table 1). This set of mRNAs encode positive or negative regulators of transcription, zinc-finger transcription factors, and proteins involved in related functions associated with alternative splicing, apoptosis, and chromosome organization. Fourteen genes (35%) were negative (e.g. MDM4, SKIL, HIVEP1, JARID2, LCOR, KDM6B, ZNF238, ZHX1) and 9 (22.5%) positive (e.g. MDFIC, POU3F1, NRIP1, REL) regulators of transcription. Seven (17.5%) zinc-finger transcription factors and 18 (45%) genes involved in alternative splicing (e.g. TMF1, BAZ2B, JMJD1C, MYNN, NFKBIZ, PEG3, KCNH6, RUNX1T1, SUFU) were also enriched in EVs. Finally, transcription factors involved in apoptosis (e.g. MDM4, IFT57, PEG3, PDCD4) and chromosome organization (e.g. FOXP3, JMJD1C, KDM6B) were present in higher levels in EVs compared to MSCs.
Table 1. Transcription factors enriched in extracellular vesicles.
| Official gene symbol | Gene name |
|---|---|
| CEBPA | CCAAT/enhancer binding protein (C/EBP)-alpha |
| KLF7 | Kruppel-like factor-7 |
| MXD1 | MAX dimerization protein-1 |
| MDM4 | Mdm4 p53 binding protein homolog |
| MDFIC | MyoD family inhibitor domain containing |
| POU3F1 | POU class 3 homeobox-1 |
| SKIL | SKI-like oncogene |
| TMF1 | TATA element modulatory factor-1 |
| BAZ2B | bromodomain adjacent to zinc-finger domain-2B |
| CREG1 | cellular repressor of E1A-stimulated genes 1 |
| FOXP3 | forkhead box P3 |
| HES1 | hairy and enhancer of split-1 |
| HIVEP1 | human immunodeficiency virus type-I enhancer binding protein-1 |
| RBAK | hypothetical LOC389458; RB-associated KRAB zinc-finger |
| IFT57 | intraflagellar transport-57 homolog |
| JMJD1C | jumonji domain containing-1C |
| JARID2 | jumonji, AT-rich interactive domain-2 |
| LCOR | ligand dependent nuclear receptor corepressor |
| KDM6B | lysine (K)-specific demethylase-6B |
| MYNN | myoneurin |
| NFKBIZ | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-zeta |
| NRIP1 | nuclear receptor interacting protein-1 |
| PEG3 | paternally expressed-3; PEG3 antisense RNA (non-protein coding); zinc finger, imprinted-2 |
| KCNH6 | potassium voltage-gated channel, subfamily-H (eag-related), member-6 |
| PDCD4 | programmed cell death-4 (neoplastic transformation inhibitor) |
| PROX2 | prospero homeobox-2 |
| RUNX1T1 | runt-related transcription factor-1; translocated to-1 (cyclin D-related) |
| SSBP2 | single-stranded DNA binding protein-2 |
| SNAPC1 | small nuclear RNA activating complex, polypeptide-1, 43kDa |
| SUFU | suppressor of fused homolog |
| TCF4 | transcription factor-4 |
| TRPS1 | trichorhinophalangeal syndrome-I |
| REL | v-rel reticuloendotheliosis viral oncogene homolog |
| ZBTB1 | Zinc-finger and BTB domain containing-1 |
| ZNF217 | Zinc-finger protein-217 |
| ZNF238 | Zinc-finger protein-238 |
| ZNF461 | Zinc-finger protein-461 |
| ZNF568 | Zinc-finger protein-568 |
| ZNF667 | Zinc-finger protein-667 |
| ZHX1 | Zinc-fingers and homeoboxes-1 |
EVs are also rich in transcription factors involved in pro-angiogenic pathways, including hepatocyte growth-factor (HGF, RPKM=13.0±3.5, fold-change=2.3±1.5), as well as hairy-and-enhancer of split (HES)1 and T-cell factor (TCF)4 (fold-change=1.6±0.2 and 1.8±0.3, respectively). Transcription factors involved in adipogenesis, like CCAAT/enhancer binding protein alpha (CEBPA, RPKM=1.8±0.8, fold-change=1.9±0.6) and Kruppel-like factor (KLF)7 (RPKM=3.5±0.4, fold-change=2.1±0.8) were also upregulated in these EVs compared to adipose tissue-derived MSCs.
Expression of Golgi apparatus genes was also upregulated in EVs compared to their parent MSCs. Arrestin beta (ARRB)1 and intraflagellar transport (IFT)57 were the most prominently up-regulated (RPKM=5.3±9.7 and 5.1±9.1, respectively), while Golgi autoantigen, golgin subfamily A (GOLGA)4 showed the highest fold-change (4.2±3.9). In addition, EVs express high levels of the TGF-β-related genes TGFB1, TGFB3, FURIN, and endoglin (ENG). EVs express robust levels of TGFB1 (RPKM=10.8±3.4, fold-change=2.7±0.5), and particularly FURIN (RPKM=46.2±12.8, fold-change=1.9±0.1). TGFB3 (RPKM=2.1±0.8, fold-change=1.7±0.3) and ENG (RPKM=0.6±0.2, fold-change=1.5±0.1) showed relatively lower enrichment in EVs.
3.4. Selective depletion of mRNAs in EVs
EVs do not contain appreciable levels of mitochondrial mRNAs, like mitochondrial ribosomal protein L11 (MRPL11, RPKM=4.9±0.7, fold-change=-0.6±0.1), translation elongation factor, mitochondrial (TSFM, RPKM=4.7±0.9, fold-change=-0.6±0.1), and cytochrome C oxidase subunit 5A (COX5A, RPKM=25.8±6.7, fold-change=-0.7±0.1), all encoded by the mitochondrial genome. In addition, EVs exhibit a selective depletion of mRNAs involved in calcium signaling, including S100 calcium binding protein A11 (S100A11, RPKM=30.5±11.7, fold-change=-0.5±0.1), calcium activated nucleotidase 1 (CANT1, RPKM=6.6±1.8, fold-change=-0.7±0.1), and reticulocalbin 1, EF-hand calcium binding domain (RCN1, RPKM=83.0±33.9, fold-change=-0.6±0.2). Furthermore, expression of cytoskeleton mRNA is at or below the level of detection, including actin, alpha 2, smooth muscle, aorta (ACTA2, RPKM=3.8±1.9, fold-change=-0.2±0.1) and myosin IC (MYO1C, RPKM=16.1±1.1, fold-change=-0.5±0.1).
3.5. Selective recruitment of several miRNAs in EVs
Analysis of miRNA species in either EVs or parent MSCs revealed that only 4 annotated miRNAs (1.2%) were enriched in EVs compared to MSCs (Figure 5A-B). We note that at present less than quarter of the known mammalian miRNAs has been annotated in the pig genome. Hence, the absolute number of enriched miRNA species is an under-estimate of the actual number of miRNAs expressed in MSCs that may be selectively recruited to EVs. We performed target gene prediction analysis of these four annotated miRNAs to understand the biological significance of their presence in MSC derived EVs. These miRNAs except let-7f are presented in order of their relative abundance in EVs.
Figure 5.
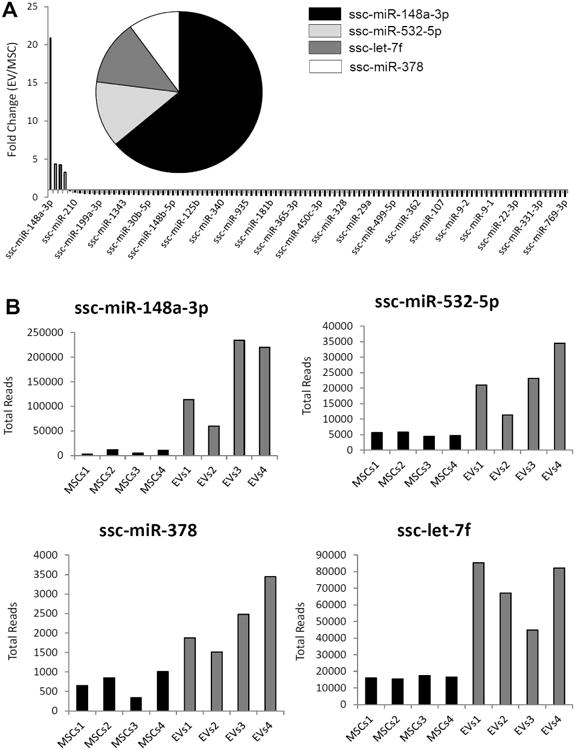
A: Only 4 miRs were enriched in EVs. B: miR148a, miR532-5p, miR378, and let-7f total reads in MSCs and EVs.
Let-7f is a member of a large family of let-7 related miRNAs. There are only 196 putative target genes for let-7f, but after filtering by target prediction score >80, only 16 genes remain. These genes have been associated to transcription (LIN28B, HMGA2, ADRB2) and membrane-associated proteins (GATM, IGDCC3, LRIG3).
miR148a-3p targets 371 genes, but only 56 showed target prediction score >80. Among them are transcription factors (EIF2C4, FMR1, TNRC6A) and genes that regulate apoptosis (ZFYVE26, MMD, GADD45A), proteolysis (ATP6AP2, RPS6KA5, USP33), angiogenesis (FLT1, QKI, MEOX2, S1PR1), and cellular transport (SOS2, YWHAB, ARFIP1).
miR532-5p modulates the expression of 367 genes, yet only 31 have a target prediction score >80. This set of genes incudes transcription factors (CPEB3, RAB11A, ATP13A3), mitochondrial genes (GABPA, MRPL18, SLC25A32), and genes involved in apoptosis (ZFP91, PAK7, CYCS) and membrane-associated proteins (FZD3, CHST6, UTP14A).
miR378 targets 481 genes, with 73 genes showing a target prediction score >80. These genes are related mainly to mitochondria (OPA3, MRP63, IVD), transcription (RRP7A, TRDMT1, PCBP2), cytoskeleton (TMOD2, GAS7, ENAH), calcium signaling (CACNG8, CCBE1, HPCAL4), apoptosis (DFFA, STAMBP, CTSB), and proteolysis (ABHD15, CMBL, GDPD1).
4. Discussion
This study examined the genetic content of EVs derived from MSCs isolated from porcine adipose tissue to understand the therapeutic efficacy of this clinically versatile source for tissue repair. We comprehensively determined the molecular characteristics of EVs using RNA-seq to interrogate mRNA and miRNA expression in EVs and their parent MSCs. Our data demonstrate that MSC-derived EVs contain a combination of mRNA and miRNA capable of regulating transcription of genetic information and modulating angiogenesis, adipogenesis, and other pathways in recipient cells. These observations could greatly contribute towards development of regenerative strategies using EVs to overcome the potential complications of cell-based therapy.
EVs are novel mediators of intercellular communication, which possess the ability to deliver complex biological messages among cells28. These vesicular structures, constantly released from all cells in response to several stimuli, travel in the extracellular space and can be internalized by target cells via membrane fusion or endocytosis, with subsequent release of their cargo29. Based on their origin and diameter EVs can be classified as exosomes (originating from multivesicular bodies, 10-100nm) or microvesicles (derived from the plasma membrane, 100-1000nm)29. EVs express a combination of microvesicle- and exosome-specific markers, such as ß1-integrins, CD73, and CD81, as well as surface markers of their parent cells. Importantly, EVs carry a broad range of functional mRNAs and miRNAs, which can be translated and induce phenotypic changes in the recipient cells30. Therefore, EVs may potentially serve as novel cell-free therapies in regenerative medicine.
Emerging evidence indicate that MSCs are avid producers of EVs10, 11, and that their delivery stimulates cell proliferation and repair12, 13. For example, MSC-derived EVs exert pro-survival effect on renal cells by inducing the expression of anti-apoptotic genes, and downregulating the expression of pro-apoptotic genes12. Similarly, MSC-derived EVs reduce infarct size in a pig model of ischaemia/reperfusion injury31. However, there is a need to elucidate the mechanisms responsible for their benefits to tailor regenerative therapies for patients with pathological conditions.
In this study, we characterized the mRNA and miRNA cargo of EVs harvested from adipose tissue-derived MSCs. We found that EVs express multiple transcription factors involved in related functions associated with alternative splicing, apoptosis, and chromosome organization. Among them are transcription factors that modulate the expression of genes involved in stem cell survival and function. For example, POU3F1 (TST-1, OCT6) is a transcription factor of the Pit-Oct-Unc (POU) family involved in developmental processes and stem cell functions32. Similarly, Jumonji, AT Rich Interactive Domain-2 (JARID2) is a transcriptional repressor that interacts with the Polycomb repressive complex-2 (PRC2), which plays an essential role in stem cell self-renewal33. In addition, EVs are enriched for mRNAs that regulate apoptosis via the p53 pathway, such as the p53-negative regulators MDM434 and paternally expressed-3 (PEG3)35. Taken together, the gene expression results show that EVs possess an important set of transcription factors, which might be able to reprogram target cells or otherwise modify their biological phenotype.
EVs are also rich in transcription factors involved in pro-angiogenic pathways, like HGF that stimulates proliferation and migration of endothelial and vascular smooth muscle cells36, 37. HES1 is a critical downstream effector of the Notch signaling pathway, and its activation regulates vascular remodeling and arterial fate of endothelial cells38. Importantly, activation of the Notch signaling pathway also increases human MSC survival39. Similarly, TCF4 is a key downstream effector of Wnt signaling, a canonical pathway that plays a central role in vascular development40 and in determining and maintaining the phenotype and functional properties of human stem cells41. Therefore, intercellular transmission of EVs containing HGF, HES1, and TCF4 may have both pro-angiogenic and pro-survival effects.
Also of note is the up-regulation of transcription factors involved in adipogenesis. CEBPA is a key transcriptional regulator of adipocyte differentiation, also involved in regulation of cell proliferation and differentiation in several organs42, 43. Likewise, KLF7 participates in regulation of cell functions in pre-adipocytes, mature adipocytes, pancreatic β-cells and skeletal muscle cells. KLF7 is also involved in the pathogenesis of type-2 diabetes44. These observations highlight the importance of EVs as key mediators for genetic transfer of metabolic information and that EVs have the ability to stimulate adipocyte differentiation of the recipient cells.
Apart from the transcription factors, EVs express robust levels of Golgi apparatus genes. GOLGA4, which showed the highest fold-change, is anchored to the trans-Golgi network membranes, interacts with microtubule proteins, and participates in delivery of transport vesicles to the cell periphery45. In contrast, ARRB1 and IFT57 are expressed on the outside of the Golgi cisternae and are mostly involved in post-Golgi vesicle-mediated transport46. In addition, EVs contain high levels of several mRNAs that encode for protein ligands within the TGFβ family that regulate proliferation, differentiation, extracellular matrix production and apoptosis. TGFB1 is a multifunctional protein secreted by a number of cell lineages, and the most prevalent isoform of TGF-β47. Previous studies have shown that TGFB1 increases mRNA expression of FURIN, its own converting enzyme48. Interestingly, FURIN showed the highest enrichment in EVs among all TGF-β-related genes. Thus, EVs from MSCs may control TGFβ signaling by direct delivery of mRNAs for TGFB1 and a key enzyme capable of regulating the bioavailability of TGFβ. In sum, our observations suggest that EVs have effects on diverse molecular functions of the target cells, such as molecular signaling, extracellular matrix remodeling, and intracellular transport.
Notably, our data show that mitochondrial, calcium signaling, and cytoskeleton mRNA are selectively depleted from MSC-derived EVs. The mitochondrial genome is located in the mitochondrial matrix and contains all polypeptides and molecules needed for intra-mitochondrial protein synthesis. Mitochondrial ribosomes translate mRNA synthesized from the mitochondrial genome in the matrix of the mitochondria49. Therefore, mitochondrial mRNA is enclosed within the double mitochondrial membrane (rather than free in the cytoplasm), which effectively would exclude its sequestration in EVs. From a technical perspective, the virtual absence of highly abundant mitochondrial mRNAs in EVs suggests that our EV preparations are a bona fide biological product rather than random membrane-encased cell remnants.
Our results show selective depletion in EVs of mRNAs involved in calcium signaling and the actin-cytoskeleton. Similar to organelle-specific seclusion of mRNAs that prevents their incorporation into EVs, sequestration by regionally-specialized ribosomes may clarify their relative absence from EV. For example, specific ribosomal sequestration of mRNAs involved in calcium signaling vesicles, which are linked to smooth endoplasmic-reticulum (ER)50, may account for their enrichment in EVs. While the smooth ER lacks ribosomes, ribosomes on the rough ER may selectively interact with mRNAs that function in calcium signaling thus effectively prevent their partitioning in EVs. Similarly, in several cell types, mRNAs for cytoskeletal proteins (e.g. β-actin) are located in peripheral regions of the cell in association with microtubules. Transport of mRNA into sites of actin polymerization serves to deliver cytoskeletal proteins over long distances51. Regional sequestration of mRNAs in actin-bound ribosomes may prevent packaging of actin mRNAs into EVs. Taken together, our observations suggest that mRNAs excluded from EVs are those sequestered in mitochondria with double membranes, in membrane-bound ribosomes attached to the endoplasmic reticulum, or in cytoskeleton-associated ribosomes.
Our results show that EVs contain several small noncoding RNAs and the production of these miRNAs is predicted to mediate post-transcriptional control of gene expression and as such modulate survival and metabolic activities of recipient cells. Interestingly, EVs contain only a small number of distinct miRNA types. The four annotated miRNAs characterized in our study are predicted to regulate expression of transcription factors, as well as genes that participate in cellular pathways like transport, apoptosis, angiogenesis and proteolysis.
Specifically, miR148a is known to modulates expression of transcription factors and genes involved in apoptosis, proteolysis, angiogenesis, and cellular transport52, 53, and may control MSC identity54. A second miRNA, miR532-5p, targets transcription factors, mitochondrial genes, and genes involved in apoptosis and membrane-associated proteins. Among these targets are the receptor for Wnt proteins frizzled-class-receptor-3 (FZD3), involved in MSCs differentiation into hepatocytes55 and the zinc-finger-protein-91 (ZFP91), which affect engraftment of hematopoietic stem cells56. Similarly, growth-arrest specific gene-7 (Gas7) and S/G2 cyclin, cyclin G2 (CCNG2), targets of mir378, regulate stem cell differentiation57, 58. Other targets of miR378 include genes related mainly to mitochondria, transcription, cytoskeleton, calcium signaling, apoptosis, and proteolysis, underscoring the versatility of EV-miRNA.
Finally, let-7f, also upregulated in EVs, targets genes associated to transcription and membrane-associated proteins with implications in cellular reprogramming and growth, like the upregulated lin-28 homolog-B (LIN28B)59, high-mobility-group AT-hook-2 (HMGA2)60, and the insulin-like growth factor-2 mRNA binding protein-1 (IGF2BP1)61. Collectively, our observations suggest that EVs are potential modulators of tissue repair by reprogramming target cells.
5. Conclusions
Our study used mRNA and miRNASeq analysis to identify gene expression signatures in EVs isolated from porcine adipose tissue-derived MSCs. We found that EVs preferentially are enriched for mRNAs encoding transcription factors (involved in angiogenic and adipogenic pathways), Golgi apparatus proteins, and proteins involved in TGF-β signaling, while proteins associated with mitochondrial, calcium signaling, and cytoskeletal functions were selectively depleted from EVs. Furthermore, we detected 4 miRNA that are highly enriched in EVs compared to MSCs. Among these were several miRNA that target transcription factors, as well as genes that participate in several cellular pathways, including angiogenesis, cellular transport, apoptosis, and proteolysis. Our data provide insight into the mechanistic underpinnings for the principal capabilities of EVs derived from MSCs with respect to their potential to modulate several pathways in the recipient cells, such as angiogenesis, adipogenesis, or extracellular matrix turnover. These observations provide a molecular framework for biomedical applications of clinical-grade MSC-derived EVs for tissue repair.
Acknowledgments
We thank the members of the Lerman and van Wijnen laboratories, as well as our Mayo Clinic colleagues Allan Dietz, Amir Lerman and Jennifer Westendorf for stimulating discussions and/or generous sharing of reagents and advice. We also appreciate the support of Jaime Davilla and Jean-Pierre Kocher for development of bioinformatic approaches for large animal models.
Funding: This study was supported by NIH grants numbers DK73608, DK100081, and HL121561 (to LOL), as well as AR049069 (to AvW).
Abbreviations
- MSCs
Mesenchymal stromal/stem cells
- EVs
Extracellular vesicles
- RNA-seq
RNA sequencing
- FACS
Fluorescence activated cell sorting
- RPKM
Reads per kilobasepair per million mapped reads
- HGF
Hepatocyte growth-factor
- HES
Hairy-and-enhancer of split
- TCF
T-cell factor
- CEBPA
CCAAT/enhancer binding protein alpha
- KLF
Kruppel-like factor
- ARRB
Arrestin beta
- IFT
Intraflagellar transport
- GOLGA
Golgi autoantigen, golgin subfamily A
- ENG
Endoglin
- MRPL11
Mitochondrial ribosomal protein L11
- TSFM
Translation elongation factor, mitochondrial
- COX5A
Cytochrome C oxidase subunit 5A
- S100A11
S100 calcium binding protein A11
- CANT1
Calcium activated nucleotidase 1
- RCN1
Reticulocalbin 1, EF-hand calcium binding domain
- ACTA2
Actin, alpha 2, smooth muscle, aorta
- MYO1C
Myosin IC
- POU
Pit-Oct-Unc
- JARID2
Jumonji, AT Rich Interactive Domain-2
- PRC2
Polycomb repressive complex-2
- PEG3
Paternally expressed-3
- FZD3
Frizzled-class-receptor-3
- ZFP91
Zinc-finger-protein-91
- Gas7
Growth-arrest specific gene-7
- CCNG2
S/G2 cyclin, cyclin G2
- LIN28B
Lin-28 homolog-B
- HMGA2
High-mobility-group AT-hook-2
- IGF2BP1
Insulin-like growth factor-2 mRNA binding protein-1
Footnotes
Disclosure: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Charbord P. Bone marrow mesenchymal stem cells: Historical overview and concepts. Hum Gene Ther. 2010;21:1045–1056. doi: 10.1089/hum.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 3.Lee HK, Lim SH, Chung IS, Park Y, Park MJ, Kim JY, Kim YG, Hong JT, Kim Y, Han SB. Preclinical efficacy and mechanisms of mesenchymal stem cells in animal models of autoimmune diseases. Immune Netw. 2014;14:81–88. doi: 10.4110/in.2014.14.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eirin A, Zhu XY, Krier JD, Tang H, Jordan KL, Grande JP, Lerman A, Textor SC, Lerman LO. Adipose tissue-derived mesenchymal stem cells improve revascularization outcomes to restore renal function in swine atherosclerotic renal artery stenosis. Stem cells (Dayton, Ohio) 2012;30:1030–1041. doi: 10.1002/stem.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebrahimi B, Eirin A, Li Z, Zhu XY, Zhang X, Lerman A, Textor SC, Lerman LO. Mesenchymal stem cells improve medullary inflammation and fibrosis after revascularization of swine atherosclerotic renal artery stenosis. PLoS One. 2013;8:e67474. doi: 10.1371/journal.pone.0067474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu XY, Urbieta-Caceres V, Krier JD, Textor SC, Lerman A, Lerman LO. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem cells (Dayton, Ohio) 2013;31:117–125. doi: 10.1002/stem.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathiasen AB, Haack-Sorensen M, Kastrup J. Mesenchymal stromal cells for cardiovascular repair: Current status and future challenges. Future Cardiol. 2009;5:605–617. doi: 10.2217/fca.09.42. [DOI] [PubMed] [Google Scholar]
- 8.Lalu MM, McIntyre L, Pugliese C, Fergusson D, Winston BW, Marshall JC, Granton J, Stewart DJ Canadian Critical Care Trials G. Safety of cell therapy with mesenchymal stromal cells (safecell): A systematic review and meta-analysis of clinical trials. PLoS One. 2012;7:e47559. doi: 10.1371/journal.pone.0047559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunter U, Rong S, Boor P, Eitner F, Muller-Newen G, Djuric Z, van Roeyen CR, Konieczny A, Ostendorf T, Villa L, Milovanceva-Popovska M, Kerjaschki D, Floege J. Mesenchymal stem cells prevent progressive experimental renal failure but maldifferentiate into glomerular adipocytes. Journal of the American Society of Nephrology : JASN. 2007;18:1754–1764. doi: 10.1681/ASN.2007010044. [DOI] [PubMed] [Google Scholar]
- 10.Yeo RWY, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, Lim SK. Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: A novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011;6:481–492. doi: 10.2217/rme.11.35. [DOI] [PubMed] [Google Scholar]
- 12.Bruno S, Grange C, Collino F, Deregibus MC, Cantaluppi V, Biancone L, Tetta C, Camussi G. Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS One. 2012;7:e33115. doi: 10.1371/journal.pone.0033115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera MB, Fonsato V, Gatti S, Deregibus MC, Sordi A, Cantarella D, Calogero R, Bussolati B, Tetta C, Camussi G. Human liver stem cell-derived microvesicles accelerate hepatic regeneration in hepatectomized rats. Journal of cellular and molecular medicine. 2010;14:1605–1618. doi: 10.1111/j.1582-4934.2009.00860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swindle MM, Makin A, Herron AJ, Clubb FJ, Jr, Frazier KS. Swine as models in biomedical research and toxicology testing. Vet Pathol. 2012;49:344–356. doi: 10.1177/0300985811402846. [DOI] [PubMed] [Google Scholar]
- 15.Eirin A, Zhu XY, Krier JD, Tang H, Jordan KL, Grande JP, Lerman A, Textor SC, Lerman LO. Adipose tissue-derived mesenchymal stem cells improve revascularization outcomes to restore renal function in swine atherosclerotic renal artery stenosis. Stem Cells. 2012;30:1030–1041. doi: 10.1002/stem.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu XY, Urbieta-Caceres V, Krier JD, Textor SC, Lerman A, Lerman LO. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem Cells. 2013;31:117–125. doi: 10.1002/stem.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudakovic A, Camilleri ET, Lewallen EA, McGee-Lawrence ME, Riester SM, Kakar S, Montecino M, Stein GS, Ryoo HM, Dietz AB, Westendorf JJ, van Wijnen AJ. Histone deacetylase inhibition destabilizes the multi-potent state of uncommitted adipose-derived mesenchymal stromal cells. J Cell Physiol. 2014 doi: 10.1002/jcp.24680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, Evans JM, Krych AJ, Smith J, Deyle DR, Stein JL, Stein GS, Im HJ, Cool SM, Westendorf JJ, Kakar S, Dietz AB, van Wijnen AJ. High-resolution molecular validation of self-renewal and spontaneous differentiation in adipose-tissue derived human mesenchymal stem cells cultured in human platelet lysate. J Cell Biochem. 2014 doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulsmans M, Holvoet P. Microrna-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013;100:7–18. doi: 10.1093/cvr/cvt161. [DOI] [PubMed] [Google Scholar]
- 20.Kalari KR, Nair AA, Bhavsar JD, O'Brien DR, Davila JI, Bockol MA, Nie J, Tang X, Baheti S, Doughty JB, Middha S, Sicotte H, Thompson AE, Asmann YW, Kocher JP. Map-rseq: Mayo analysis pipeline for rna sequencing. BMC Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y, Smyth GK, Shi W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Evans J, Bhagwate A, Middha S, Bockol M, Yan H, Kocher JP. Cap-mirseq: A comprehensive analysis pipeline for microrna sequencing data. BMC Genomics. 2014;15:423. doi: 10.1186/1471-2164-15-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X. Mirdb: A microrna target prediction and functional annotation database with a wiki interface. Rna. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrado C, Raimondo S, Chiesi A, Ciccia F, De Leo G, Alessandro R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int J Mol Sci. 2013;14:5338–5366. doi: 10.3390/ijms14035338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. Journal of proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 31.Timmers L, Lim SK, Arslan F, Armstrong JS, Hoefer IE, Doevendans PA, Piek JJ, El Oakley RM, Choo A, Lee CN, Pasterkamp G, de Kleijn DPV. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2007;1:129–137. doi: 10.1016/j.scr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Oatley JM, Oatley MJ, Kaucher AV, Avarbock MR, Brinster RL. The pou domain transcription factor pou3f1 is an important intrinsic regulator of gdnf-induced survival and self-renewal of mouse spermatogonial stem cells. Biol Reprod. 2010;82:1103–1111. doi: 10.1095/biolreprod.109.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M, Krogan N, Ren B, Song JS, Reiter JF. Polycomb-like 3 promotes polycomb repressive complex 2 binding to cpg islands and embryonic stem cell self-renewal. PLoS Genet. 2012;8:e1002576. doi: 10.1371/journal.pgen.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Lozano G. Molecular pathways: Targeting mdm2 and mdm4 in cancer therapy. Clin Cancer Res. 2013;19:34–41. doi: 10.1158/1078-0432.CCR-12-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broad KD, Curley JP, Keverne EB. Increased apoptosis during neonatal brain development underlies the adult behavioral deficits seen in mice lacking a functional paternally expressed gene 3 (peg3) Dev Neurobiol. 2009;69:314–325. doi: 10.1002/dneu.20702. [DOI] [PubMed] [Google Scholar]
- 36.Morishita R, Nakamura S, Hayashi S, Taniyama Y, Moriguchi A, Nagano T, Taiji M, Noguchi H, Takeshita S, Matsumoto K, Nakamura T, Higaki J, Ogihara T. Therapeutic angiogenesis induced by human recombinant hepatocyte growth factor in rabbit hind limb ischemia model as cytokine supplement therapy. Hypertension. 1999;33:1379–1384. doi: 10.1161/01.hyp.33.6.1379. [DOI] [PubMed] [Google Scholar]
- 37.Stewart N, Chade AR. Renoprotective effects of hepatocyte growth factor in the stenotic kidney. Am J Physiol Renal Physiol. 2013;304:F625–633. doi: 10.1152/ajprenal.00504.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitagawa M, Hojo M, Imayoshi I, Goto M, Ando M, Ohtsuka T, Kageyama R, Miyamoto S. Hes1 and hes5 regulate vascular remodeling and arterial specification of endothelial cells in brain vascular development. Mech Dev. 2013;130:458–466. doi: 10.1016/j.mod.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Moriyama H, Moriyama M, Isshi H, Ishihara S, Okura H, Ichinose A, Ozawa T, Matsuyama A, Hayakawa T. Role of notch signaling in the maintenance of human mesenchymal stem cells under hypoxic conditions. Stem Cells Dev. 2014 doi: 10.1089/scd.2013.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruotti N, Corrado A, Neve A, Cantatore FP. Systemic effects of wnt signaling. J Cell Physiol. 2013;228:1428–1432. doi: 10.1002/jcp.24326. [DOI] [PubMed] [Google Scholar]
- 41.Lu R, Qu Y, Ge J, Zhang L, Su Z, Pflugfelder SC, Li DQ. Transcription factor tcf4 maintains the properties of human corneal epithelial stem cells. Stem Cells. 2012;30:753–761. doi: 10.1002/stem.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of ccaat enhancer-binding protein beta by mapk and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birkenmeier EH, Gwynn B, Howard S, Jerry J, Gordon JI, Landschulz WH, McKnight SL. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding ccaat/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 44.Kawamura Y, Tanaka Y, Kawamori R, Maeda S. Overexpression of kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol. 2006;20:844–856. doi: 10.1210/me.2005-0138. [DOI] [PubMed] [Google Scholar]
- 45.Kakinuma T, Ichikawa H, Tsukada Y, Nakamura T, Toh BH. Interaction between p230 and macf1 is associated with transport of a glycosyl phosphatidyl inositol-anchored protein from the golgi to the cell periphery. Exp Cell Res. 2004;298:388–398. doi: 10.1016/j.yexcr.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 46.Gervais FG, Singaraja R, Xanthoudakis S, Gutekunst CA, Leavitt BR, Metzler M, Hackam AS, Tam J, Vaillancourt JP, Houtzager V, Rasper DM, Roy S, Hayden MR, Nicholson DW. Recruitment and activation of caspase-8 by the huntingtin-interacting protein hip-1 and a novel partner hippi. Nat Cell Biol. 2002;4:95–105. doi: 10.1038/ncb735. [DOI] [PubMed] [Google Scholar]
- 47.Kamato D, Burch ML, Piva TJ, Rezaei HB, Rostam MA, Xu S, Zheng W, Little PJ, Osman N. Transforming growth factor-beta signalling: Role and consequences of smad linker region phosphorylation. Cell Signal. 2013;25:2017–2024. doi: 10.1016/j.cellsig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Blanchette F, Day R, Dong W, Laprise MH, Dubois CM. Tgfbeta1 regulates gene expression of its own converting enzyme furin. The Journal of clinical investigation. 1997;99:1974–1983. doi: 10.1172/JCI119365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lightowlers RN, Rozanska A, Chrzanowska-Lightowlers ZM. Mitochondrial protein synthesis: Figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014 doi: 10.1016/j.febslet.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobie EA, Lederer WJ. Dynamic local changes in sarcoplasmic reticulum calcium: Physiological and pathophysiological roles. J Mol Cell Cardiol. 2012;52:304–311. doi: 10.1016/j.yjmcc.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill MA, Schedlich L, Gunning P. Serum-induced signal transduction determines the peripheral location of beta-actin mrna within the cell. The Journal of cell biology. 1994;126:1221–1229. doi: 10.1083/jcb.126.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng H, Lai M. Mir-148a promotes apoptosis by targeting bcl-2 in colorectal cancer. Cell Death Differ. 2011;18:1702–1710. doi: 10.1038/cdd.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J, Li Q, Xu Q, Liu L, Jiang B. Mir-148a inhibits angiogenesis by targeting erbb3. J Biomed Res. 2011;25:170–177. doi: 10.1016/S1674-8301(11)60022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giraud-Triboult K, Rochon-Beaucourt C, Nissan X, Champon B, Aubert S, Pietu G. Combined mrna and microrna profiling reveals that mir-148a and mir-20b control human mesenchymal stem cell phenotype via epas1. Physiological genomics. 2011;43:77–86. doi: 10.1152/physiolgenomics.00077.2010. [DOI] [PubMed] [Google Scholar]
- 55.Herencia C, Martinez-Moreno JM, Herrera C, Corrales F, Santiago-Mora R, Espejo I, Barco M, Almaden Y, de la Mata M, Rodriguez-Ariza A, Munoz-Castaneda JR. Nuclear translocation of beta-catenin during mesenchymal stem cells differentiation into hepatocytes is associated with a tumoral phenotype. PLoS One. 2012;7:e34656. doi: 10.1371/journal.pone.0034656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiem HP, Ironside C, Beard BC, Trobridge GD. A retroviral vector common integration site between leupaxin and zinc finger protein 91 (zfp91) observed in baboon hematopoietic repopulating cells. Experimental hematology. 2010;38:819–822. 822.e811–813. doi: 10.1016/j.exphem.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hung FC, Chang Y, Lin-Chao S, Chao CC. Gas7 mediates the differentiation of human bone marrow-derived mesenchymal stem cells into functional osteoblasts by enhancing runx2-dependent gene expression. J Orthop Res. 2011;29:1528–1535. doi: 10.1002/jor.21425. [DOI] [PubMed] [Google Scholar]
- 58.Baker SJ, Ma'ayan A, Lieu YK, John P, Reddy MVR, Chen EY, Duan Q, Snoeck HW, Reddy EP. B-myb is an essential regulator of hematopoietic stem cell and myeloid progenitor cell development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3122–3127. doi: 10.1073/pnas.1315464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, Stanger BZ, Lee JS, Rustgi AK. Lin28b promotes growth and tumorigenesis of the intestinal epithelium via let-7. Genes & development. 2013;27:2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, Shavlakadze T, Eash JK, Scaramozza A, Goddeeris MM, Kirsch DG, Campbell KP, Brack AS, Glass DJ. An hmga2-igf2bp2 axis regulates myoblast proliferation and myogenesis. Dev Cell. 2012;23:1176–1188. doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahaira LG, Katsara O, Pappou E, Iliopoulou E, Fortis S, Antsaklis A, Fotinopoulos P, Baxevanis CN, Papamichail M, Perez SA. Igf2bp1 expression in human mesenchymal stem cells significantly affects their proliferation and is under the epigenetic control of tet1/2 demethylases. Stem Cells Dev. 2014 doi: 10.1089/scd.2013.0604. [DOI] [PubMed] [Google Scholar]


