Abstract
Photoexcitation with blue light of the flavin chromophore in BLUF photoreceptors induces a switch into a metastable signaling state that is characterized by a red-shifted absorption maximum. The red shift is due to a rearrangement in the hydrogen bond pattern around Gln63 located in the immediate proximity of the isoalloxazine ring system of the chromophore. There is a long-lasting controversy between two structural models, named Q63A and Q63J in the literature, on the local conformation of the residues Gln63 and Tyr21 in the dark state of the photoreceptor. As regards the mechanistic details of the light-activation mechanism, rotation of Gln63 is opposed by tautomerism in the Q63A and Q63J models, respectively. We provide a structure-based simulation of electrochromic shifts of the flavin chromophore in the wild type and in various site-directed mutants. The excellent overall agreement between experimental and computed data allows us to evaluate the two structural models. Compelling evidence is obtained that the Q63A model is incorrect, whereas the Q63J is fully consistent with the present computations. Finally, we confirm independently that a keto–enol tautomerization of the glutamine at position 63, which was proposed as molecular mechanism for the transition between the dark and the light-adapted state, explains the measured 10 to 15 nm red shift in flavin absorption between these two states of the protein. We believe that the accurateness of our results provides evidence that the BLUF photoreceptors absorption is fine-tuned through electrostatic interactions between the chromophore and the protein matrix, and finally that the simplicity of our theoretical model is advantageous as regards easy reproducibility and further extensions.
The biological photoreceptor proteins BLUF, for “sensors of blue-light using flavin adenine dinucleotide (FAD)”,1 are involved in numerous important physiological processes like phototaxis,2 photosynthetic gene regulation,3 virulence,4 and generation of bacterial biofilms;5 they are also employed as optogenetic tools in neuroscience, when coupled to specific enzymes.6,7
Two physiological states are reported for BLUF photoreceptors: a dark (inactive) state and a light-adapted (active) signaling state.3,8−13 The transformation from the dark to the light-adapted state occurs upon illumination with blue light on a 100 ps time scale14,15 and results in a 10 to 15 nm red shift16−27 of the absorption maximum of the photoreceptors. The signaling state decays back to the dark state on a much longer time scale ranging from seconds10,22,23 to almost 30 min,8−10,28 depending on the particular BLUF domain.
From a theoretical point of view, an observed absorption shift of 10 to 15 nm suggests a modulation of electrostatic interaction energies through conformational changes in the microenvironment of the conjugated π-system,29 the isoalloxazine ring system, of the chromophore (Figure S1 in the Supporting Information) and makes any covalent or redox modification of the chromophore, which would result in larger shifts, less likely.30,31 Therefore, it has been speculated since the discovery of the first BLUF domains that the spectroscopic shift is caused by a reshuffling of the hydrogen bond network in direct proximity to the isoalloxazine ring system.17,22,28,32,33
Hydrogen bond networks are a key element to describe accurately electrostatic interaction energies in proteinaeous systems.34−40 However, because light atoms diffract X-rays less than heavier ones, the hydrogen coordinates are usually not resolved in X-ray protein crystallography.41 The positions of those hydrogen atoms, which are involved in hydrogen bonds, might be deduced from proximal hydrogen bond donor and acceptor groups. But the procedure is further impeded as amide nitrogen and oxygen atoms have similar electron densities and are thereby often indistinguishable from their X-ray diffraction pattern at usual resolution.42 Missassignment of these atoms in the side-chains of asparagine and glutamine amino acids might lead to a prediction of incorrect local structures, including hydrogen bond networks.42
These shortcomings seemingly led to contradicting structural information on the atomic details of the BLUF photoreceptors in the dark state.29 The atomic details of the hydrogen bond networks of the photoreceptor are controversially described between two structural models designated in the literature as “tryptophan-in”44 (Trpin; Figure 1, left) and “tryptophan-out” (Trpout; Figure 1, right). The controversy originated from the opposing assignment of the side-chain amide nitrogen and the carbonyl oxygen of the central Gln63 with respect to the N5 atom and C4=O group of the isoalloxazine ring system of the chromophore (Figure 1).29
Figure 1.
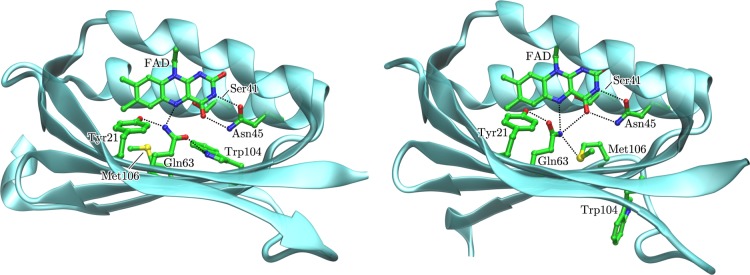
Ribbon diagrams of BLUF domain crystal structures enclosing the photoactive part as proposed by the Trpin model represented by the 1YRX crystal structure (AppA protein from Rhodobacter sphaeroides),43 coordinates set A (left), and the Trpout model represented by the 1X0P crystal structure (Tll0078 protein from Thermosynechococcus elongatus BP-1),21 coordinates set A (right).
Further differences between Trpin and Trpout concern the location of Trp104 and Met106 in both functional states (Figure 1) and an alternative light-activation mechanism, with distinct views on Gln63, where the Trpin model postulates a side-chain rotation43 and the Trpout, a keto–enol tautomerism;30,31,45 with31,46,47 or without30 side-chain rotation (Table S1 in the Supporting Information and Figure 2). Here and in the remainder of the publication, we follow the common nomenclature where Q63A and Q63J denote the rotameric conformations of Q63 proposed in Trpin (e.g., 1YRX crystal structure43) and in Trpout (e.g., 2IYG crystal structure44), respectively.
Figure 2.
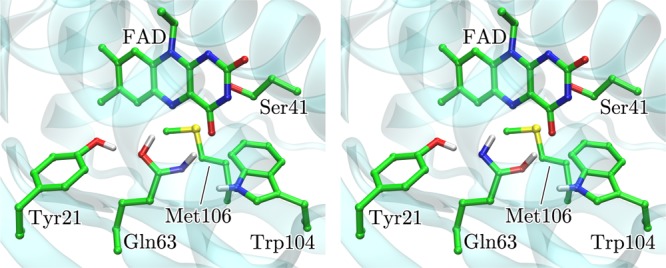
The two discussed forms of Gln63 as proposed by Sadeghian et al.30 (left) and Domratcheva et al.31 (right) for the light-adapted state.
The controversy on the molecular structures originated from opposing crystallographic models, and so far neither spectroscopic nor theoretical studies provided a unified view. In particular, recent theoretical investigations including structural refinements at different level of theory on the experimentally determined coordinates sets of AppA BLUF were inconclusive. A recent QM/MM study by Hsiao et al.48 with high-level QM theory suggested that the transition energy difference between the optimized 1YRX (Trpin, Q63A) and 2IYG (Trpout, Q63J) structures explains the measured absorption difference between the dark and light-adapted states. Hence, they concluded that Q63A and Q63J, respectively, represent the dark and light-adapted states structures. The view of Hsiao et al.48 was challenged in a recent QC study by Udvarhelyi and Domratcheva.47 Their optimized extended fragments of the photoactive part, treated as supermolecular clusters in the QC calculations, led to a strong deviation for the optimized Q63A and to good agreement for the optimized Q63J conformation with the observable electron density. According to Udvarhelyi and Domratcheva,47 the spectroscopic shift between the two functional states of the photoreceptor is explained by a tautomerization of Q63J, in constrast to the proposal of Hsiao et al.48 discussed above.
The seemingly contradicting results between QC47 and QM/MM48 studies motivated the present systematic structure-based analysis of measured electrochromic shifts of various BLUF photoreceptors, where Q63A versus Q63J and rotation versus tautomerization is analyzed including the orientation of the hydroxyl group of Tyr21. In summary, both structural models, the Trpin (Q63A)24,43,48−58 as well as the Trpout model (Q63J),20,21,25,30,31,44−47,59−63 are supported or rejected by several studies of the last years.
In the present study, we will resolve the controversy on the local conformation of the two functional states of the photoreceptors using a fairly simple physical model, which assumes that the fine-tuning of the optical properties of the chromophore by the surrounding protein matrix is accomplished by electrostatic interactions. This study is in the tradition of previous theoretical studies, showing electrostatic interactions to be fundamental in enzymatic reactions,64−67 electron transfer in redox-active complexes,68,69 and excitation energy transfer in photosynthetic light-harvesting complexes.70−75 Here, similar electrostatic calculations are applied to the class of photoreceptor proteins.
More specifically, we use the electronic transition of the isoalloxazine ring system of BLUF photoreceptors to monitor the local conformation of the critical amino acid residues in the active site, applying the charge-density coupling (CDC) method,71,72,74 a quantum chemical/electrostatic two-step approach that was successfully applied to reveal the energetics of optical pigment transitions in light harvesting complexes.75,76 This approach is in the spirit of Cohen et al.,77 who used the vibrational Stark effect caused by a series of mutants in the neighborhood of an active site of a protein to infer the local conformation of amino acid residues. Here we apply this idea to the electronic Stark effect of the chromophore in the active site. In our theoretical approach, quantum chemical calculations of the ground and first excited state of the chromophore in vacuo are combined with electrostatic calculations of the coupling of the respective charge densities with that of the whole protein. By comparing electrostatic interaction energies of the wild type and site-directed mutants (Figure 3), the shift in optical transition energy of the flavin chromophore between the wild type and the mutant will be calculated and compared with experimental data.2,3,16,17,21,24,33,45,55,78−80 The contradicting information from various coordinates sets of BLUF photoreceptors is evaluated and the hydrogen bond networks of its dark and light-adapted states are revealed.
Figure 3.
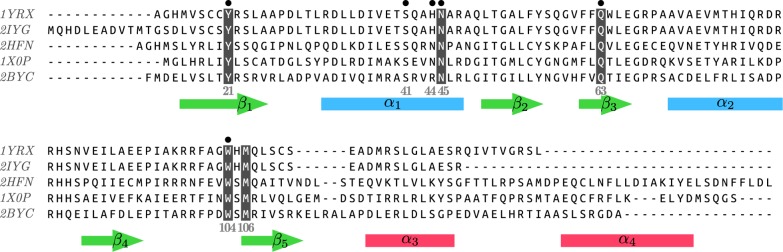
Multiple sequence alignments of BLUF domain-containing proteins and the secondary structural elements of Tll0078 (below). Highlighted residues are supposed to be involved in the photoreaction. Black dots indicate positions where measured absorption shifts induced by site-directed mutagenesis have been investigated in the present study.
The remaining of this paper is organized in the following way: we summarize the CDC method including its parametrization by quantum chemical calculations and explain how it can be used to calculate mutant-minus-wild type absorbance shifts. Next, the CDC method is applied to a variety of experimentally determined coordinates sets. By comparing the electrochromic shifts of various site-directed mutants, obtained with CDC, with the experimental values, we evaluate the different structural models, including those for the dark-to-light adapted state transition of BLUF photoreceptors.
Computational Details
Overview
The change in optical transition energy ΔE between the wildtype and a site directed mutant (mutant-minus-wild type) is obtained by applying the CDC method71,72,74 as
| 1 |
where QI(g) and QI are the atomic partial charges (APCs) of the Ith atom at position R⃗I of the chromophore in the ground and excited state, respectively; qj and qk are the ground state APCs of the jth atom of the mutated protein at position r⃗j and the kth atom of the protein in the wild type at position r⃗k, respectively. Equation 1 describes the Coulomb coupling of the difference in charge density between the excited and the ground state of the chromophore with the difference density between the wild type and the mutant of the protein. The effective dielectric constant εeff in eq 1 takes into account screening and local field effects of the Coulomb coupling in an effective way. In addition, εeff corrects for uncertainties in the absolute magnitudes of atomic partial charges QI(g/e) of the chromophore, obtained from quantum chemical calculations described below. Therefore, within certain limits, εeff is an adjustable parameter in the present calculations. We obtained quantitative agreement between calculated and measured ΔE values by setting εeff to 2.2. This value is in good agreement with earlier estimates on different chromophore–protein complexes.72,74
In order to apply eq 1, we need to know the APCs of the chromophore in the ground and first excited state and those of the protein in the wild type and the mutant, as well as the coordinates of all atoms of the chromophore–protein complex for the wild type and the mutant structures.
Quantum Chemical Calculations
The APCs of the wild type and mutant protein structures were taken from the molecular mechanics force field of the program CHARMM22,81,82 except for the keto–enol tautomer of Gln63, which does not exist in CHARMM22. The APCs of the keto–enol tautomer of Gln63 were obtained from a fit of the ab initio electrostatic potential (ESP) of the electronic ground state of a glycin-glutamin (keto–enol)-glycin tripeptide computed on the geometry optimized structure obtained with the Hartree–Fock method and a 6-31G* basis set, which is the way APCs of the amino acids are determined in the molecular mechanics force field of CHARMM22.81,82 The program Jaguar83 was used for this purpose. The numerical values of the APCs are given in the Table S6 in the Supporting Information. For the ribose part and the pyrophosphate of the chromophore, APCs were taked from the CHARMM27 all-hydrogen nucleic acid topology file.84 The APCs of the isoalloxazine ring system in the ground and excited state were obtained from a fit of the ESP of the respective charge densities of the two states obtained with density functional theory (DFT) for the ground state and with the time-dependent DFT (TD-DFT) in the Tamm–Dancoff approximation85 for the excited state, as described earlier.86,87 The calculations were performed on a fully optimized structure of methyl-isoalloxazine in vacuo. The geometry optimization was carried out with DFT using the B3LYP XC functional and a 6-31G* basis set with the program Jaguar.83 The quantum chemical calculations of the charge density of the ground and excited states were performed with Q-Chem.88
Several hybrid exchange-correlation (XC) functionals, which differ in the amount of exact exchange, were investigated. The charge densities for methyl-isoalloxazine with the Becke-Half-and-Half-LYP (BHHLYP) XC functional, which has been described in earlier work,86 yielded the closest match with the experimental difference dipole moment89,90 between the excited and the ground states of the flavin chromophore (Figure S3 in the Supporting Information). Therefore, the atomic partial charges (APCs) obtained with BHHLYP were used to calculate the electrochromic shifts. The numerical values of the APCs are given in Table S5 in the Supporting Information. We note that independent support for the BHHLYP XC functional comes from a comparison of the electronic structure of the isoalloxazine ring system in the ground and excited state, obtained with (TD)DFT and the high-level coupled cluster 2 wave function-based method.30
Electrostatic Computations
The heavy atom coordinates of the wild type were taken from the crystal structures. Hydrogen atom positions were generated and energetically optimized by molecular mechanics with CHARMM81 using the CHARMM22 force field. All heavy atom coordinates of the mutant proteins were kept as in the various crystal structures, except for the residue considered for mutation. For the mutated residue, we kept as much structural information as possible from the respective wild type residue. We inserted the mutations by manipulating the wild type protein coordinates files in the following way: for the Y21F, Y21I, and Y21W mutants, all possible atomic coordinates of Tyr21 were conserved to create the Phe21 mutant. Ile21 was equipped with all backbone atoms of Tyr21 plus the Cβ atom (in IUPAC recommended nomenclature91); the same holds for Trp21. For the N44A and N45A mutants, the heavy backbone atoms and the Cβ atom of the corresponding wild type residue were used to implement Ala44 and Ala45. For the S41A and Q63L mutants, the same procedure as for N44A and N45A was used. For Q63E, we maintained the entire set of coordinates of Gln63 for Glu63, but changed the side-chain nitrogen of Gln63 into the second carbonyl oxygen of Glu63. The subsequent minimization and the generation of a “more light-adapted state” are described below. The keto–enol tautomers of Gln63 were prepared as the Q63E mutant; that is, we maintained all heavy atoms of Gln63, but for the second tautomer we rotated the side-chain amide group by 180°.
To account for possible structural relaxation of the light-adapted state, we prepared a “more light-adapted state” of Q63E by rotating the end of the side-chain of the in silico mutant by ∼ –40°, which decreased the atomic distances between the NH group of Glu63 and the C4=O group of the isoalloxazine from ∼2.5 to ∼2.0 Å. In this way, the stronger hydrogen bond between residue 63 and the isoalloxazine ring system, which was inferred from infrared spectroscopy,17,32,92−95 was taken into account. The “more light-adapted state” of the keto–enol tautomer of Gln63 was created in the same way.
Results
General Overview
We estimated transition energy shifts ΔE (eq 1) between the wild type protein and nine in silico mutants for 25 coordinates sets contained in four different BLUF dark state crystal structures from the literature.21,43,44,96 Through this structure-based simulation and the comparison of the computed with the measured absorption shifts, we aim to exclude side-chain conformations which represent the artifacts of the crystallographic assignment.
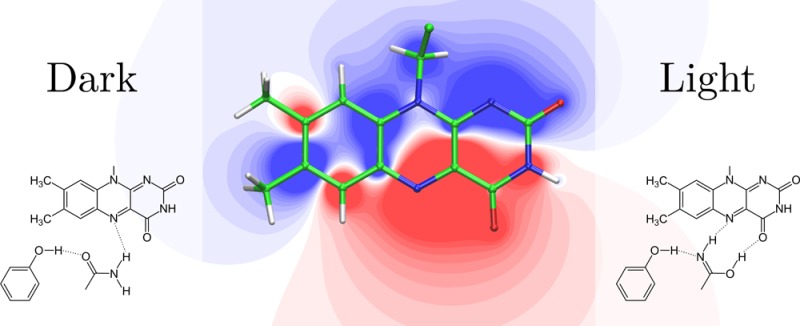
The mutations are located in different regions of the electrostatical difference potential between the ground and excited state of the isoalloxazine ring system, which determines the absorption shift upon mutation (Figure 4). The two-dimensional map of the difference in the plane of the chromophore with the projection of the mutated residues, shown in Figure 4, can be a useful tool to evaluate qualitatively the direction of the absorption shift. The general rule to derive the direction of the absorption shift (mutant-minus-wild type) is that the removal of an atom with a positive partial charge from the positive region of the difference potential (shown in blue in Figure 4) results in a red-shifted absorption maximum of the mutant, whereas the removal of a negative charge in the same region yields a blue-shifted absorption maximum. The inverse effect would be observed in the negative region of the difference potential (shown in red in Figure 4).
Figure 4.
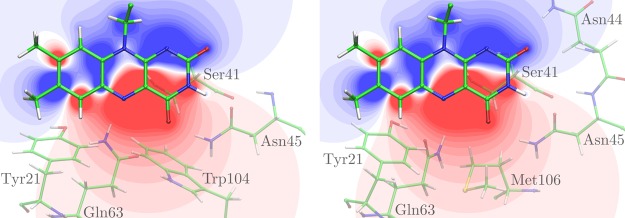
Electrostatic difference potential between ground and excited state of the isoalloxazine ring system of the flavin chromophore. The positive regions of the difference potential (excited-minus-ground state) are colored in shades of blue and the negative ones in shades of red. Relevant amino acids from the two different coordinates sets in Figure 1 representing the Q63A (Trpin, left) and the Q63J (Trpout, right) models are projected in the plane of the isoalloxazine ring system in translucent.
The selected mutations are located at six different positions along the protein sequence (Figure 3). Whereas for some of these residues, the side-chain conformation is identical in all crystal structures, for others it differs. One single side-chain orientation is proposed for asparagine 44 and 45. Computed absorption shifts for the N44A and N45A mutant reproduce accurately the experimental values (Table S2 in the Supporting Information).
Different side-chain conformations are discussed for Ser41 within the 25 coordinates sets, but the differences are not related to the controversy between the two structural models (Trpin and Trpout). Our structure-based computation of the absorption shift of S41A reveals that only one out of four proposed side-chain conformations explain the measured absorption shift (Table S2 in the Supporting Information). The distinguished side-chain conformation, which is seen, for instance, in all 10 coordinates sets of 1X0P, is the only conformation that is stabilized by a hydrogen bond (for further details, see Text S1 in the Supporting Information). Hence, there is independent support for this conformation.
Albeit different conformations being proposed for Trp104 in Trpin and Trpout, in both structural models the polar NH group of Trp104 lies in a region of weak to very weak difference potential of the flavin chromophore (Figure 4, left; Figure 1, right). Therefore, the W104F mutant shows only a very minor absorption shift in our calculations (Table S2 in the Supporting Information), in agreement with the experiment.
We now proceed to the discussion of the absorption shifts for the residues at positions 21 and 63, whose side-chain orientations differ systematically between the Q63A and Q63J structural models.
Calculation of the Electrochromic Shifts for the Y21F, Y21I, and Y21W Mutants
Regarding Tyr21, absorption shifts were measured for the site-directed mutations Y21F, Y21I, and Y21W.79,80 In summary, mutation of Tyr21 against a hydrophobic residue results in a minor to intermediate blue shift of the flavin absorption in the experiments (Table 1).
Table 1. Calculated Absorption Shifts in Units of Nanometer for the Site-Directed Mutants Y21F, Y21I, Y21W, Q63E (neutral, where the proton forms a permanent hydrogen bond to C4=O), and Q63L and the Enol Tautomerization of Gln63 Using the Structural Data from Different Crystal Structures That Contain Different Coordinates Sets (first column) Which Can Be Assigned to the Q63A and Q63J Structural Models (second column); Experimental Absorption Shifts Are Given in the Last Row (calculated absorption shifts in kilocalorie per mole are given in Table S7 in the Supporting Information).
| type | Y21F | Y21I | Y21W | Q63E | Q63Ea | Q63L | Q63Qenol a | Q63Qrotenol a | |
|---|---|---|---|---|---|---|---|---|---|
| 1YRX A | Q63A | +5.7 | +5.6 | +6.3 | +20.8 | +21.3 | +13.6 | n/a | n/a |
| 1YRX B | Q63A | +5.1 | +5.0 | +5.4 | +19.0 | +21.4 | +13.1 | n/a | n/a |
| 1YRX C | Q63A | +4.8 | +4.7 | +5.2 | +25.0 | +24.2 | +15.4 | n/a | n/a |
| 2HFN D | Q63A | +5.3 | +5.0 | +6.0 | +20.2 | +20.1 | +14.3 | n/a | n/a |
| 1X0P A | Q63J | –5.2 | –5.4 | –4.7 | +7.4 | +4.8 | –4.1 | +9.0 | +13.2 |
| 1X0P B | Q63J | –4.9 | –5.1 | –4.4 | +9.6 | +4.2 | –2.2 | +9.8 | +14.3 |
| 1X0P C | Q63J | –4.9 | –5.2 | –4.4 | +5.0 | +4.0 | –7.2 | +6.3 | +10.6 |
| 1X0P D | Q63J | –4.9 | –5.2 | –4.5 | +9.3 | +4.9 | –2.8 | +10.6 | +14.2 |
| 1X0P E | Q63J | –4.6 | –4.8 | –4.3 | +5.8 | +4.6 | –5.0 | +7.3 | +10.8 |
| 1X0P F | Q63J | –4.7 | –5.1 | –4.3 | +8.9 | +4.0 | –3.0 | +10.2 | +13.6 |
| 1X0P G | Q63J | –4.8 | –5.1 | –4.4 | +7.8 | +4.2 | –3.4 | +8.4 | +13.0 |
| 1X0P H | Q63J | –5.2 | –5.5 | –4.7 | +9.1 | +4.4 | –2.0 | +10.7 | +14.0 |
| 1X0P I | Q63J | –4.7 | –5.0 | –4.3 | +7.6 | +3.5 | –3.3 | +10.3 | +12.7 |
| 1X0P J | Q63J | –5.1 | –5.4 | –4.8 | +8.6 | +5.2 | –2.9 | +10.3 | +13.1 |
| 2HFN A | Q63J | –4.7 | –5.0 | –4.0 | +5.7 | +4.5 | –6.2 | +9.2 | +9.9 |
| 2HFN B | Q63J | –4.7 | –4.9 | –4.0 | +5.7 | +3.7 | –5.1 | +9.3 | +9.8 |
| 2HFN C | Q63J | –4.5 | –4.7 | –4.2 | +5.8 | +4.8 | –5.8 | +8.9 | +9.7 |
| 2HFN E | Q63J | –4.7 | –4.9 | –3.9 | +5.3 | +5.6 | –5.6 | +8.3 | +9.6 |
| 2HFN F | Q63J | –4.6 | –4.9 | –4.0 | +5.0 | +4.4 | –5.9 | +8.6 | +9.3 |
| 2HFN G | Q63J | –4.9 | –5.2 | –4.2 | +6.0 | +4.9 | –5.6 | +10.9 | +10.9 |
| 2HFN H | Q63J | –4.6 | –5.0 | –4.0 | +4.8 | +4.5 | –5.4 | +9.2 | +9.5 |
| 2HFN I | Q63J | –4.6 | –4.9 | –4.3 | +6.8 | +4.7 | –4.9 | +11.0 | +11.1 |
| 2HFN J | Q63J | –4.3 | –4.5 | –3.6 | +5.5 | +4.1 | –6.3 | +7.7 | +9.3 |
| 2IYG A | Q63J | –6.0 | –6.2 | –5.1 | +5.2 | +2.8 | –6.5 | +9.3 | +10.3 |
| 2IYG B | Q63J | –5.3 | –5.4 | –4.6 | +6.3 | +2.4 | –7.9 | +9.3 | +10.2 |
| measured value | –2/–4b | –4c | –4d | +3e | +3e | –6/–10f | +10 to +15g | +10 to +15g | |
The side-chain orientation of Gln63 has been manipulated in order to simulate the hydrogen bond distance between the side-chain amide and the C4=O of the isoalloxazine as related for the light-adapted state according to spectroscopic data (see text).
See refs (79 and 80) (the exact value was not given in ref (80), but from an overlay of the wild type and mutant absorption spectra, we could estimate the difference of the absorption maximum with an uncertainty of roughly 2 nm).
See ref (78).
See ref (33).
See ref (55).
In the Q63A conformation, the side-chain hydroxyl dipole of Tyr21 is oriented such that the hydrogen is close to the neutral region of the difference potential of the flavin, whereas the oxygen atom lies in the negative region of intermediate strength (Figure 4, left). Therefore, the mutation of Tyr21 against a nonpolar residue effectively corresponds to the elimination of a negative charge from the negative region of the difference potential (Figure 4, left), which should result in a red-shifted absorption maximum. The structure-based computation of absorption shifts for Tyr21 mutated against Phe, Ile, or Trp based on the Q63A conformation indeed all report a ∼5 nm red shift (Table 1), which is, however, in contradiction to the experimental results.33,78−81
In the Q63J conformation, the hydroxyl dipole is oriented differently (Figure 4, right): the polar hydrogen is now located in a more negative region of the difference potential than the oxygen atom. The hydrogen atom should, therefore, be the main side-chain contributor of Tyr21 to the absorption shift. In this case, the mutation effectively results in a removal of a positive charge from the negative region of the difference potential and should yield a blue-shifted absorption maximum. The computed shifts for Y21F, Y21I, and Y21W are indeed to the blue, and the values (−4 to −6 nm) are in good agreement with the experimental data33,70−80 (Table 1). Therefore, we conclude that the conformation of Tyr21 is as proposed by the Trpout model (Q63J).
Calculation of the Electrochromic Shifts for the Q63L and Q63E Mutants
According to the difference potential in the right part of Figure 4, the electrochromic shift induced by Gln63 in the Q63J model should be dominated by one hydrogen of the side-chain amide group, which is located in the strongly negative region of the difference potential of the chromophore. Consequently, the Q63L mutation is expected to yield a red-shifted absorption maximum. In the case of the Q63A model, a qualitative assessment of the shift is difficult since, besides the hydrogen, also the oppositely charged carbonyl oxygen of Gln63 is located in the strongly negative difference potential. Our electrostatic calculations assuming the Q63A conformation predict a red-shifted absorption for the Q63L mutant (Table 1), in contradiction to the measured values.24,45 In contrast, all computed absorption shifts using the structural templates proposing the Q63J configuration of Gln63 agree qualitatively, and in most cases quantitatively, with the experimental values (Table 1). Therefore, the computations of Q63L absorption shifts strongly support the Q63J structural model. A more thorough analyses of the absorption shift (Q63E, Q63L) reveals that the mutation induces a reorientation of the side-chain hydroxyl in the adjacent residue Tyr21 in the Q63A but not in the Q63J structural model. This reorientation of the Tyr21 hydroxyl group exerts a dominant effect on the red-shifted absorption shift.
As regards the Q63E mutation, biochemical experiments combined with NMR97 and ultrafast time-resolved infrared spectroscopy55 agree that the mutated residue locks the photoreceptor in a quasi-light-adapted state, where the glutamic acid is in its neutral (protonated) state, forming a permanent hydrogen bond with the C4=O group of the isoalloxazine ring system. The moderate experimental absorption shift of 3 nm to the red55 supports the idea of a neutral glutamic acid, which implies a shift of the 3 pKa value of the side-chain carboxyl group by more than 3 pKa units between aqueous solution and the protein.
For all the 25 coordinates sets, we computed absorption shifts assuming an anionic and a neutral glutamic acid. For the latter state, we generated two mutants with the side-chain either in hydrogen bond contact with the N5 atom or the C4=O group of the chromophore. The all-atom representation of the mutant was generated by imposing the position of all side-chain heavy atoms of Gln63 to the mutant residue, that is, the side-chain nitrogen becoming a carbonyl oxygen. Computed absorption shifts using Q63A structural templates fail to reproduce the experimental result55 independently of the protonation state of Glu63 (Table 1 for the neutral Glu63). Based on the structural information provided by the Q63J model, the computations assuming an anionic state of Glu63 deviate from the experimental results by a factor of ∼20 (Table S3 in the Supporting Information). If the absorption shift is computed with a neutral (protonated) Glu63, where the proton forms a permanent hydrogen bond with the N5 atom of the chromophore (Table S3 in the Supporting Information), the results are considerably improved, but good to excellent agreement is only obtained if the proton is assumed to be in hydrogen bond contact with the C4=O group of the chromophore (Table 1).
To take into account spectroscopic results, reporting a strong hydrogen bond between the side-chain hydroxyl of the neutral Glu63 and the C4=O group of the isoalloxazine ring system,17,32,92−95 we generated a “more light-adapted state” structure of the Q63E mutant as we decreased the hydrogen bond distance to ∼2.0 Å by rotating the side-chain of the mutant. Computed absorption shifts based on this structural refinement agree even better with the experimental result (Table 1).
In summary, we conclude: (i) the dark state configuration proposed for Tyr21 and Gln63 by the Q63J model and not by the Q63A model is the spectroscopically characterized conformation, and (ii) Glu63 in the Q63E mutant is indeed in its protonated (neutral) state and the proton is in permanent hydrogen bond contact with the C4=O group of the chromophore.
Summary: Trpin versus Trpout for the Dark State
The correlation plots in Figure 5 summarize the comparison between computed and measured absorption shifts for various mutants using coordinates sets representative for the Q63A (Figure 5, left) and Q63J (Figure 5, right) structural models. Compelling evidence is obtained that the Q63J structural model, which was originally proposed as part of the Trpout model, is very likely to be the spectroscopically characterized state.
Figure 5.
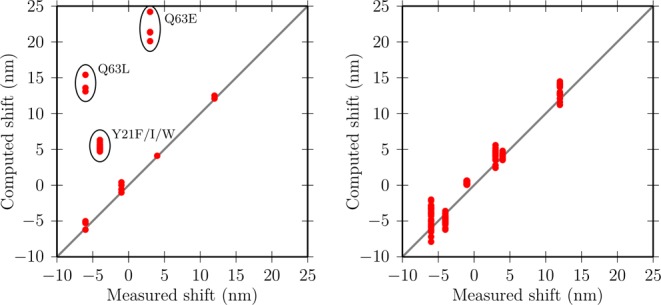
Correlation diagrams of the computed absorption shifts with the measured ones for representative coordinates sets of the Q63A (Trpin, left) and Q63J (Trpout, right) models. Encircled points highlight the measured data for Tyr21 and Gln63 which fail to be explained by the Q63A structural model.
Gln63 Keto–Enol Tautomerism as a Light-Activation Mechanism Explains the Absorption Shift
Within the framework set by the Q63J model, the local conformation of the light-adapted state in the vicinity of the conjugated π-system is mainly distinguished from the dark state by an enol tautomerization of Gln63, with31 or without30 flip of its side-chain (Table S1 in the Supporting Information). In the unrotated form, the hydroxyl group and the NH group of Gln63 form hydrogen bonds with the N5 atom and the C4=O group, respectively, of the isoalloxazine ring system (Figure 2, left). If the tautomerism involves rotation of the side-chain, the hydrogen bond partners are exchanged (Figure 2, right). In the following, we will denote the enol form of the Gln63 as Q63Qenol.
We evaluated if the two proposed Trpout light-adapted state structures explain the measured absorption shift of 10 to 15 nm to the red with respect to the dark state. In these calculations, the enol form of Gln63 formally takes over the role of a mutated residue. We generated the all-atom representation of the two enol forms (Q63Qenol and Q63Qrotenol) (Table 1) as for the Q63E mutant by imposing all side-chain heavy atoms of Gln63 on the enol form. This approach implies an exchange of the side-chain nitrogen with the carbonyl oxygen to generate the enol form of Gln63, involving rotation of the side-chain. Again, we also generated a “more light-adapted state” structure by decreasing the hydrogen bond distance of Gln63 with the C4=O group of the isoalloxazine ring system by rotation.
Computed absorption shifts, using the original coordinates of the crystal structures, agree with the measured value for both enol forms. Using the “more light-adapted state” structure with a decreased hydrogen bond distance, an improvement is observed for the rotated form (Table 1), but the discrepancies between the two forms are certainly to small to allow a final judgment.
Discussion
The present study provides an elaborate structure-based analysis of electrochromic shifts for BLUF photoreceptors. In total, we computed nine mutant-minus-wild type absorption shifts, simulated the spectroscopic shift between the two functional states, that is, the dark and light-adapted states of the photoreceptor and compared each result with the corresponding experimental value.
The overall accurateness of the presented data documents that the CDC method71,72,74 at the applied level of theory is appropriate to describe the electrochromic shift of the flavin photoreceptor.
The simplicity of the method, that neglects any change of the wave function of the flavin’s ground and excited states by the protein environment, shows that in essence the measured mutant-minus-wild type absorption shifts reflect the change in electrostatic interaction between the flavin chromophore and a particular residue and its mutant. The excellent agreement between computed and measured values strongly indicates that for BLUF photoreceptors in general the site-directed mutants have indeed only local effects; that is, neither the global conformation nor the overall hydrogen bond pattern is affected.80 The assumption is further supported by the moderate absorption shifts and the overall similarity between the absorption spectra of the wild type and mutant protein.80 Further support comes from the rigidity of the ferredoxin-like fold, which is adopted by all known BLUF proteins.20,25,29,43,44
An exception might be the N45A mutation, which renders the photoreceptor photoinactive,21 possibly because the loss of two hydrogen bonds (between Asn45 and the isoalloxazine ring system) causes a spatial rearrangement in the photoactive cleft. To account for such a dynamic behavior, the CDC method should be combined with MD simulations, but such calculations are beyond the scope of the present work. We note, however, that the present simple method, nevertheless, already yields the major contribution to the N45A absorption shift of the flavin chromophore.
As regards the ongoing debate on the molecular structures of BLUF photoreceptors our study provides independent support of the following issues: first, the Q63A conformation is rejected as a functional state; second, strong evidence is provided that Q63J represents the dominant functional dark state conformation of BLUF photoreceptors; and third, it is shown that the keto–enol tautomerism of Gln63 can account for the observed red shift of the flavin absorption spectrum between the dark and light-adapted states.
The rejection of the Q63A conformation is supported by the majority of structual studies on BLUF photoreceptors.20,21,25,44,59−61 Support is also granted by the work of Grinstead and co-workers49,50 who did not see the Q63A conformation in any of the 20 NMR structures they determined.50 This conclusion is also in accordance with a recent theoretical study of Udvarhelyi and Domratcheva,47 who showed that QC refined Q63A supermolecular clusters (extended fragments of the photoactive cleft) disagree with the experimentally observed electron density.47
In contrast, the Q63J conformation in our model explains all considered absorption shifts and in particular gives an excellent agreement between computed and measured values for the Tyr21 and Gln63 mutations. This result is also supported by the study of Udvarhelyi and Domratcheva47 who showed that the QC refined supermolecular clusters of the Q63J conformation represents a stable minimum and agrees with the observed electron density.
Furthermore, our absorption shifts calculated between the Q63J conformation and the keto–enol tautomer of Gln63 are in the range of 10 to 15 nm observed experimentally between the dark and light-adapted states and show that the transition from Q63A to Q63J does not account for the observed red shift of the light-adapted state. In accordance with Udvarhelyi and Domratcheva,47 we find that the transition energy difference between Q63J and Q63Qenol explains the 10 to 15 nm red shift observed between the dark and the light-adapted states. Hence, the present study provides independent support for the keto–enol tautomer of Gln63 in the light-adapted state of BLUF photoreceptors. This model was first evoked to interpret spectroscopic data of the photoreceptor domain45 and then confirmed by two independent theoretical studies30,31 A QM/MM approach revealed that the keto–enol tautomer is likely to evolve from the Q63J conformation30 and a QC study showed that this transition is consistent with the downshift of the vibrational frequency of the C4=O group.31 The latter is indicative for the formation of a strong hydrogen bond between residue 63 and the chromophore in the light-adapted state.
Our electrochromic shift computations do not allow us to distinguish between the possible Trp104 locations suggested in Trpin (Figure 1, left) and Trpout (Figure 1, right); that is, we cannot say if Trp104 is located inside the photoactive cleft or on the surface of the protein. Traditionally, Q63A and Q63J were assigned to Trpin and Trpout, respectively. But in the 20 NMR models of Grinstead et al.,50 Trp104 is reported inside the protein albeit the highly flexible Q63 did not converge to the Q63A structure. The Q63J structure in combination with the Trpin model would probably lead to a more flexible Q63 side chain, because the NH group of the indole ring of Trp104 would rather destabilize than stabilize the Q63J conformation. Consistent with this view, Udvarhelyi and Domratcheva47 computed flat energy barriers between different dark state conformations, where Q63J represents the most stable conformation. According to our results, a more flexible Q63 side chain rotating in the range of ±60° around the Q63J is possible. But this point requires a more systematic analysis, including a systematic dihedral scan of Gln63.
Conclusion
Two contradicting structural models, designated as Trpin and Trpout in the literature as the dark state structure of BLUF photoreceptor domains, were studied. The controversy is on the atomic structure of the photoactive side and in particular on the side-chain conformations of the two critical residues Tyr21 and Gln63, and hence not necessarily on the position of Trp104. Our structure-based simulations explain absorption shifts measured for a number of site-directed mutants, involving the critical residues, only if the Q63J rotamer of the Trpout model crystal structures are used. Striking deviations between simulations and experiments are obtained for the crystal structures containing the Q63A rotamer of the Trpin model. We, therefore, conclude that the Q63A rotamer is present in the dark state structure of BLUF photoreceptors characterized in spectroscopic experiments.
The advantage of the present investigation is its simplicity, which allows for a high throughput analysis in terms of structure and absorption shifts. To a major extent, our results independently support the findings of Udvarhelyi and Domratcheva.47
Besides solving a particular puzzle in the structure and function of BLUF photoreceptors, we believe that the structure-based computation of protein-induced electrochromic shifts of a photoactive dye is a powerful tool to determine local hydrogen bond patterns in photoreceptors and in other proteins with photoactive compounds.
Acknowledgments
Financial support by the Austrian Science Fund (FWF: P 24774-N27) is gratefully acknowledged.
Supporting Information Available
Additional discussion regarding calculation of the electrochromic shifts for the mutants N44A and N45A, detailed skeletal structure of the complete photoactive center according to the Trpout model, overlay of the 10 monomers of the 1X0P crystal structure, difference dipole moment of the methyl-isoalloxazine in dependence on the (TD)DFT functional used, skeletal structures of the different conformations of the photoactive part in the dark, transition and light-adapted states, calculated absorption shifts for the site-directed mutants S41A, N44A, N45A, and W104F, calculated absorption shifts for the site-directed mutant Q63E assuming different protonation states, calculated absorption shifts for the enol tautomerization of Gln63, computated atomic partial charges of the isoalloxazine ring system in the ground and excited state, chosen atomic partial charges for the enol form of Gln63, and calculated absorption shifts in units of kilocalorie per mole for the site-directed mutants Y21F, Y21I, Y21W, S41A, N44A, N45A, Q63E, Q63L, and W104F. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This paper published ASAP on September 5, 2014. Additional corrections were made and the revised version was reposted on September 8, 2014. Figure 5 was replaced and the revised version was reposted on September 15, 2014.
Supplementary Material
References
- Gomelsky M.; Klug G. BLUF: A Novel FAD-Binding Domain Involved in Sensory Transduction in Microorganisms. Trends Biochem. Sci. 2002, 27, 497–500. [DOI] [PubMed] [Google Scholar]
- Iseki M.; Matsunaga S.; Murakami A.; Ohno K.; Shiga K.; Yoshida K.; Sugai M.; Takahashi T.; Hori T.; Watanabe M. A Blue-Light-Activated Adenylyl Cyclase Mediates Photoavoidance in Euglena gracilis. Nature 2002, 415, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Masuda S.; Bauer C. E. AppA Is a Blue Light Photoreceptor That Antirepresses Photosynthesis Gene Expression in Rhodobacter sphaeroides. Cell 2002, 110, 613–623. [DOI] [PubMed] [Google Scholar]
- Mussi M. A.; Gaddy J. A.; Cabruja M.; Arivett B. a.; Viale A. M.; Rasia R.; Actis L. A. The Opportunistic Human Pathogen Acinetobacter baumannii Senses and Responds to Light. J. Bacteriol. 2010, 192, 6336–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust R.; Haigney A.; Lukacs A.; Gil A.; Hossain S.; Addison K.; Lai C.-T.; Towrie M.; Greetham G. M.; Clark I. P.; et al. Ultrafast Structural Dynamics of BlsA, a Photoreceptor from the Pathogenic Bacterium Acinetobacter baumannii. J. Phys. Chem. Lett. 2014, 5, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder-Lang S.; Schwärzel M.; Seifert R.; Strünker T.; Kateriya S.; Looser J.; Watanabe M.; Kaupp U. B.; Hegemann P.; Nagel G. Fast Manipulation of Cellular cAMP Level by Light in Vivo. Nat. Methods 2007, 4, 39–42. [DOI] [PubMed] [Google Scholar]
- Weissenberger S.; Schultheis C.; Liewald J. F.; Erbguth K.; Nagel G.; Gottschalk A. PACα − An Optogenetic Tool for in Vivo Manipulation of Cellular cAMP Levels, Neurotransmitter Release, and Behavior in Caenorhabditis elegans. J. Neurochem. 2011, 116, 616–625. [DOI] [PubMed] [Google Scholar]
- Gauden M.; Yeremenko S.; Laan W.; van Stokkum I. H. M.; Ihalainen J. A.; van Grondelle R.; Hellingwerf K. J.; Kennis J. T. M. Photocycle of the Flavin-Binding Photoreceptor AppA, a Bacterial Transcriptional Antirepressor of Photosynthesis Genes. Biochemistry 2005, 44, 3653–3662. [DOI] [PubMed] [Google Scholar]
- Dragnea V.; Waegele M.; Balascuta S.; Bauer C.; Bogdan D. Time-Resolved Spectroscopic Studies of the AppA Blue-Light Receptor BLUF Domain from Rhodobacter sphaeroides. Biochemistry 2005, 44, 15978–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirak P.; Penzkofer A.; Schiereis T.; Hegemann P.; Jung A.; Schlichting I. Absorption and Fluorescence Spectroscopic Characterization of BLUF Domain of AppA from Rhodobacter sphaeroides. Chem. Phys. 2005, 315, 142–154. [Google Scholar]
- Zirak P.; Penzkofer A.; Schiereis T.; Hegemann P.; Jung A.; Schlichting I. Photodynamics of the Small BLUF Protein BlrA from Rhodobacter sphaeroides. J. Photochem. Photobiol., B 2006, 83, 180–194. [DOI] [PubMed] [Google Scholar]
- Nakasone Y.; Ono T.-a.; Ishii A.; Masuda S.; Terazima M. Transient Dimerization and Conformational Change of a BLUF Protein: YcgF. J. Am. Chem. Soc. 2007, 129, 7028–7035. [DOI] [PubMed] [Google Scholar]
- Tanaka K.; Nakasone Y.; Okajima K.; Ikeuchi M.; Tokutomi S.; Terazima M. Time-Resolved Tracking of Interprotein Signal Transduction: Synechocystis PixD–PixE Complex As a Sensor of Light Intensity. J. Am. Chem. Soc. 2012, 134, 8336–8339. [DOI] [PubMed] [Google Scholar]
- Brust R.; Lukacs A.; Haigney A.; Addison K.; Gil A.; Towrie M.; Clark I. P.; Greetham G. M.; Tonge P. J.; Meech S. R. Proteins in Action: Femtosecond to Millisecond Structural Dynamics of a Photoactive Flavoprotein. J. Am. Chem. Soc. 2013, 135, 16168–16174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs A.; Brust R.; Haigney A.; Laptenok S. P.; Addison K.; Gil A.; Towrie M.; Greetham G. M.; Tonge P. J.; Meech S. R. BLUF Domain Function Does Not Require a Metastable Radical Intermediate State. J. Am. Chem. Soc. 2014, 136, 4605–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M.; Kaplan S. AppA, a Redox Regulator of Photosystem Formation in Rhodobacter sphaeroides 2.4.1, Is a Flavoprotein. J. Biol. Chem. 1998, 273, 35319–35325. [DOI] [PubMed] [Google Scholar]
- Masuda S.; Hasegawa K.; Ishii A.; Ono T.-A. Light-Induced Structural Changes in a Putative Blue-Light Receptor with a Novel FAD Binding Fold Sensor of Blue-Light Using FAD (BLUF); Slr1694 of Synechocystis sp. PCC6803. Biochemistry 2004, 43, 5304–5313. [DOI] [PubMed] [Google Scholar]
- Rajagopal S.; Key J. M.; Purcell E. B.; Boerema D. J.; Moffat K. Purification and Initial Characterization of a Putative Blue Light-Regulated Phosphodiesterase from Escherichia coli. Photochem. Photobiol. 2004, 80, 542–547. [DOI] [PubMed] [Google Scholar]
- Ito S.; Murakami A.; Sato K.; Nishina Y.; Shiga K.; Takahashi T.; Higashi S.; Iseki M.; Watanabe M. Photocycle Features of Heterologously Expressed and Assembled Eukaryotic Flavin-Binding BLUF Domains of Photoactivated Adenylyl Cyclase (PAC), a Blue-Light Receptor in Euglena gracilis. Photochem. Photobiol. Sci. 2005, 4, 762–769. [DOI] [PubMed] [Google Scholar]
- Jung A.; Domratcheva T.; Tarutina M.; Wu Q.; Ko W.-h.; Shoeman R. L.; Gomelsky M.; Gardner K. H.; Schlichting I. Structure of a Bacterial BLUF Photoreceptor: Insights into Blue Light-Mediated Signal Transduction. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 12350–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita A.; Okajima K.; Morimoto Y.; Ikeuchi M.; Miki K. Structure of a Cyanobacterial BLUF Protein, Tll0078, Containing a Novel FAD-Binding Blue Light Sensor Domain. J. Mol. Biol. 2005, 349, 1–9. [DOI] [PubMed] [Google Scholar]
- Hasegawa K.; Masuda S.; Ono T.-A. Spectroscopic Analysis of the Dark Relaxation Process of a Photocycle in a Sensor of Blue Light Using FAD (BLUF) Protein Slr1694 of the Cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 2005, 46, 136–146. [DOI] [PubMed] [Google Scholar]
- Fukushima Y.; Okajima K.; Shibata Y.; Ikeuchi M.; Itoh S. Primary Intermediate in the Photocycle of a Blue-Light Sensory BLUF FAD-Protein, Tll0078, of Thermosynechococcus elongatus BP-1. Biochemistry 2005, 44, 5149–5158. [DOI] [PubMed] [Google Scholar]
- Unno M.; Masuda S.; Ono T.-A.; Yamauchi S. Orientation of a Key Glutamine Residue in the BLUF Domain from AppA Revealed by Mutagenesis, Spectroscopy, and Quantum Chemical Calculations. J. Am. Chem. Soc. 2006, 128, 5638–5639. [DOI] [PubMed] [Google Scholar]
- Wu Q.; Gardner K. H. Structure and Insight into Blue Light-Induced Changes in the BlrP1 BLUF Domain. Biochemistry 2009, 48, 2620–2629. [DOI] [PubMed] [Google Scholar]
- Ryu M.-H.; Moskvin O. V.; Siltberg-Liberles J.; Gomelsky M. Natural and Engineered Photoactivated Nucleotidyl Cyclases for Optogenetic Applications. J. Biol. Chem. 2010, 285, 41501–41508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierl M.; Stumpf P.; Udwari D.; Gueta R.; Hagedorn R.; Losi A.; Gärtner W.; Petereit L.; Efetova M.; Schwarzel M.; et al. Light Modulation of Cellular cAMP by a Small Bacterial Photoactivated Adenylyl Cyclase, bPAC, of the Soil Bacterium Beggiatoa. J. Biol. Chem. 2011, 286, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauden M.; van Stokkum I. H. M.; Key J. M.; Lührs D. C.; van Grondelle R.; Hegemann P.; Kennis J. T. M. Hydrogen-Bond Switching Through a Radical Pair Mechanism in a Flavin-Binding Photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 10895–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S. Light Detection and Signal Transduction in the BLUF Photoreceptors. Plant Cell Physiol. 2013, 54, 171–179. [DOI] [PubMed] [Google Scholar]
- Sadeghian K.; Bocola M.; Schütz M. A Conclusive Mechanism of the Photoinduced Reaction Cascade in Blue Light Using Flavin Photoreceptors. J. Am. Chem. Soc. 2008, 130, 12501–12513. [DOI] [PubMed] [Google Scholar]
- Domratcheva T.; Grigorenko B. L.; Schlichting I.; Nemukhin A. V. Molecular Models Predict Light-Induced Glutamine Tautomerization in BLUF Photoreceptors. Biophys. J. 2008, 94, 3872–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M.; Sano R.; Masuda S.; Ono T.-A.; Yamauchi S. Light-Induced Structural Changes in the Active Site of the BLUF Domain in AppA by Raman Spectroscopy. J. Phys. Chem. B 2005, 109, 12620–1262. [DOI] [PubMed] [Google Scholar]
- Bonetti C.; Stierl M.; Mathes T.; van Stokkum I. H. M.; Mullen K. M.; Cohen-Stuart T. A.; van Grondelle R.; Hegemann P.; Kennis J. T. M. The Role of Key Amino Acids in the Photoactivation Pathway of the Synechocystis Slr1694 BLUF Domain. Biochemistry 2009, 48, 11458–11469. [DOI] [PubMed] [Google Scholar]
- Baker E. N.; Hubbard R. E. Hydrogen Bonding in Globular Proteins. Prog. Biophys. Mol. Biol. 1984, 44, 97–179. [DOI] [PubMed] [Google Scholar]
- Aaqvist J.; Warshel A. Simulation of Enzyme Reactions Using Valence Bond Force Fields and Other Hybrid Quantum/Classical Approaches. Chem. Rev. 1993, 93, 2523–2544. [Google Scholar]
- Honig B.; Nicholls A. Classical Electrostatics in Biology and Chemistry. Science 1995, 268, 1144–1149. [DOI] [PubMed] [Google Scholar]
- Jeffrey G. A.An Introduction to Hydrogen Bonding; Oxford University Press: New York, 1997. [Google Scholar]
- Warshel A.; Sharma P. K.; Kato M.; Xiang Y.; Liu H.; Olsson M. H. M. Electrostatic Basis for Enzyme Catalysis. Chem. Rev. 2006, 106, 3210–3235. [DOI] [PubMed] [Google Scholar]
- Sigala P. A.; Fafarman A. T.; Schwans J. P.; Fried S. D.; Fenn T. D.; Caaveiro J. M. M.; Pybus B.; Ringe D.; Petsko G. A.; Boxer S. G.; et al. Quantitative Dissection of Hydrogen Bond-Mediated Proton Transfer in the Ketosteroid Isomerase Active Site. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E2552–E2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikita H.; Saito K. Proton Transfer Reactions and Hydrogen-Bond Networks in Protein Environments. J. R. Soc. Interface 2014, 11, 20130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd M. F. C.; Palmer R. A.. Structure Determination by X-Ray Crystallography; Plenum Press: New York, 1977. [Google Scholar]
- Weichenberger C. X.; Sippl M. J. NQ-Flipper: Recognition and Correction of Erroneous Asparagine and Glutamine Side-Chain Rotamers in Protein Structures. Nucleic Acids Res. 2007, 35, W403–W406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.; Dragnea V.; Masuda S.; Ybe J.; Moffat K.; Bauer C. Structure of a Novel Photoreceptor, the BLUF Domain of AppA from Rhodobacter sphaeroides. Biochemistry 2005, 44, 7998–8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A.; Reinstein J.; Domratcheva T.; Shoeman R. L.; Schlichting I. Crystal Structures of the AppA BLUF Domain Photoreceptor Provide Insights into Blue Light-Mediated Signal Transduction. J. Mol. Biol. 2006, 362, 717–732. [DOI] [PubMed] [Google Scholar]
- Stelling A. L.; Ronayne K. L.; Nappa J.; Tonge P. J.; Meech S. R. Ultrafast Structural Dynamics in BLUF Domains: Transient Infrared Spectroscopy of AppA and Its Mutants. J. Am. Chem. Soc. 2007, 129, 15556–15564. [DOI] [PubMed] [Google Scholar]
- Khrenova M. G.; Nemukhin A. V.; Domratcheva T. Photoinduced Electron Transfer Facilitates Tautomerization of the Conserved Signaling Glutamine Side Chain in BLUF Protein Light Sensors. J. Phys. Chem. B 2013, 117, 2369–2377. [DOI] [PubMed] [Google Scholar]
- Udvarhelyi A.; Domratcheva T. Glutamine Rotamers in BLUF Photoreceptors: A Mechanistic Reappraisal. J. Phys. Chem. B 2013, 117, 2888–2897. [DOI] [PubMed] [Google Scholar]
- Hsiao Y.-W.; Götze J. P.; Thiel W. The Central Role of Gln63 for the Hydrogen Bonding Network and UV−Visible Spectrum of the AppA BLUF Domain. J. Phys. Chem. B 2012, 116, 8064–8073. [DOI] [PubMed] [Google Scholar]
- Grinstead J. S.; Hsu S.-T. D.; Laan W.; Bonvin A. M. J. J.; Hellingwerf K. J.; Boelens R.; Kaptein R. The Solution Structure of the AppA BLUF Domain: Insight into the Mechanism of Light-Induced Signaling. ChemBioChem 2006, 7, 187–193. [DOI] [PubMed] [Google Scholar]
- Grinstead J. S.; Avila-Perez M.; Hellingwerf K. J.; Boelens R.; Kaptein R. Light-Induced Flipping of a Conserved Glutamine Sidechain and Its Orientation in the AppA BLUF Domain. J. Am. Chem. Soc. 2006, 128, 15066–15067. [DOI] [PubMed] [Google Scholar]
- Gauden M.; Grinstead J. S.; Laan W.; van Stokkum I. H. M.; Avila-Perez M.; Toh K. C.; Boelens R.; Kaptein R.; van Grondelle R.; Hellingwerf K. J.; et al. On the Role of Aromatic Side Chains in the Photoactivation of BLUF Domains. Biochemistry 2007, 46, 7405–7415. [DOI] [PubMed] [Google Scholar]
- Ishikita H. Light-Induced Hydrogen Bonding Pattern and Driving Force of Electron Transfer in AppA BLUF Domain Photoreceptor. J. Biol. Chem. 2008, 283, 30618–30623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K. C.; van Stokkum I. H. M.; Hendriks J.; Alexandre M. T. A.; Arents J. C.; Avila Perez M.; van Grondelle R.; Hellingwerf K. J.; Kennis J. T. M. On the Signaling Mechanism and the Absence of Photoreversibility in the AppA BLUF Domain. Biophys. J. 2008, 95, 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M.; Kikuchi S.; Masuda S. Structural Refinement of a Key Tryptophan Residue in the BLUF Photoreceptor AppA by Ultraviolet Resonance Raman Spectroscopy. Biophys. J. 2010, 98, 1949–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs A.; Haigney A.; Brust R.; Zhao R.-K.; Stelling A. L.; Clark I. P.; Towrie M.; Greetham G. M.; Meech S. R.; Tonge P. J. Photoexcitation of the Blue Light Using FAD Photoreceptor AppA Results in Ultrafast Changes to the Protein Matrix. J. Am. Chem. Soc. 2011, 133, 16893–16900. [DOI] [PubMed] [Google Scholar]
- Rieff B.; Bauer S.; Mathias G.; Tavan P. DFT/MM Description of Flavin IR Spectra in BLUF Domains. J. Phys. Chem. B 2011, 115, 11239–11253. [DOI] [PubMed] [Google Scholar]
- Mathes T.; Zhu J.; van Stokkum I. H. M.; Groot M. L.; Hegemann P.; Kennis J. T. M. Hydrogen Bond Switching Among Flavin and Amino Acids Determines the Nature of Proton-Coupled Electron Transfer in BLUF Photoreceptors. J. Phys. Chem. Lett. 2012, 3, 203–208. [Google Scholar]
- Meier K.; Thiel W.; van Gunsteren W. F. On the Effect of a Variation of the Force Field, Spatial Boundary Condition and Size of the QM Region in QM/MM MD Simulations. J. Comput. Chem. 2012, 33, 363–378. [DOI] [PubMed] [Google Scholar]
- Obanayama K.; Kobayashi H.; Fukushima K.; Sakurai M. Structures of the Chromophore Binding Sites in BLUF Domains As Studied by Molecular Dynamics and Quantum Chemical Calculations. Photochem. Photobiol. 2008, 84, 1003–1010. [DOI] [PubMed] [Google Scholar]
- Wu Q.; Ko W.-H.; Gardner K. H. Structural Requirements for Key Residues and Auxiliary Portions of a BLUF Domain. Biochemistry 2008, 47, 10271–10280. [DOI] [PubMed] [Google Scholar]
- Barends T. R. M.; Hartmann E.; Griese J. J.; Beitlich T.; Kirienko N. V.; Ryjenkov D. A.; Reinstein J.; Shoeman R. L.; Gomelsky M.; Schlichting I. Structure and Mechanism of a Bacterial Light-Regulated Cyclic Nucleotide Phosphodiesterase. Nature 2009, 459, 1015–1018. [DOI] [PubMed] [Google Scholar]
- Sadeghian K.; Bocola M.; Schütz M. A QM/MM Study on the Fast Photocycle of Blue Light Using Flavin Photoreceptors in Their Light-Adapted/Active Form. Phys. Chem. Chem. Phys. 2010, 12, 8840–8846. [DOI] [PubMed] [Google Scholar]
- Khrenova M. G.; Nemukhin A. V.; Grigorenko B. L.; Krylov A. I.; Domratcheva T. M. Quantum Chemistry Calculations Provide Support to the Mechanism of the Light-Induced Structural Changes in the Flavin-Binding Photoreceptor Proteins. J. Chem. Theory Comput. 2010, 6, 2293–2302. [DOI] [PubMed] [Google Scholar]
- Getzoff E. D.; Cabelli D. E.; Fisher C. L.; Parge H. E.; Viezzoli M. S.; Banci L.; Hallewell R. A. Faster Superoxide Dismutase Mutants Designed by Enhancing Electrostatic Guidance. Nature 1992, 358, 347–351. [DOI] [PubMed] [Google Scholar]
- Lee L.-P.; Tidor B. Barstar Is Electrostatically Optimized for Tight Binding to Barnase. Nat. Struct. Mol. Biol. 2001, 8, 73–76. [DOI] [PubMed] [Google Scholar]
- Simonson T. Gaussian Fluctuations and Linear Response in an Electron Transfer Protein. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 6544–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk A. J.; Cao J.; Kamerlin S. C. L.; Warshel A. Catalysis by Dihydrofolate Reductase and Other Enzymes Arises from Electrostatic Preorganization, Not Conformational Motions. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 14115–14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein B.; Ullmann G. M.; Knapp E.-W. Energetics of Electron-Transfer and Protonation Reactions of the Quinones in the Photosynthetic Reaction Center of Rhodopseudomonas viridis. Biochemistry 1998, 37, 2488–2495. [DOI] [PubMed] [Google Scholar]
- Ishikita H.; Saenger W.; Biesiadka J.; Loll B.; Knapp E.-W. How Photosynthetic Reaction Centers Control Oxidation Power in Chlorophyll Pairs P680, P700, and P870. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müh F.; Madjet M. E.-A.; Adolphs J.; Abdurahman A.; Rabenstein B.; Ishikita H.; Knapp E.-W.; Renger T. α-Helices Direct Excitation Energy Flow in the Fenna-Matthews-Olson Protein. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 16862–16867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolphs J.; Müh F.; Madjet M. E.-A.; Renger T. Calculation of Pigment Transition Energies in the FMO Protein. Photosynth. Res. 2008, 95, 197–209. [DOI] [PubMed] [Google Scholar]
- Adolphs J.; Müh F.; Madjet M. E.-A.; Schmidt am Busch M.; Renger T. Structure-Based Calculations of Optical Spectra of Photosystem I Suggest an Asymmetric Light-Harvesting Process. J. Am. Chem. Soc. 2010, 132, 3331–3343. [DOI] [PubMed] [Google Scholar]
- Müh F.; Madjet M. E.-A.; Renger T. Structure-Based Identification of Energy Sinks in Plant Light-Harvesting Complex II. J. Phys. Chem. B 2010, 114, 13517–13535. [DOI] [PubMed] [Google Scholar]
- Schmidt am Busch M.; Müh F.; Madjet M. E.-A.; Renger T. The Eighth Bacteriochlorophyll Completes the Excitation Energy Funnel in the FMO Protein. J. Phys. Chem. Lett. 2011, 2, 93–98. [DOI] [PubMed] [Google Scholar]
- Renger T.; Madjet M. E.-A.; Schmidt am Busch M.; Adolphs J.; Müh F. Structure-Based Modeling of Energy Transfer in Photosynthesis. Photosynth. Res. 2013, 116, 367–388. [DOI] [PubMed] [Google Scholar]
- Renger T.; Müh F. Understanding Photosynthetic Light-Harvesting: A Bottom-up Theoretical Approach. Phys. Chem. Chem. Phys. 2013, 15, 3348–3371. [DOI] [PubMed] [Google Scholar]
- Cohen B. E.; McAnaney T. B.; Park E. S.; Jan Y. N.; Boxer S. G.; Jan L. Y. Probing Protein Electrostatics with a Synthetic Fluorescent Amino Acid. Science 2002, 296, 1700–1703. [DOI] [PubMed] [Google Scholar]
- Laan W.; van der Horst M. A.; van Stokkum I. H.; Hellingwerf K. J. Initial Characterization of the Primary Photochemistry of AppA, a Blue-Light-Using Flavin Adenine Dinucleotide-Domain Containing Transcriptional Antirepressor Protein from Rhodobacter Sphaeroides: A Key Role for Reversible Intramolecular Proton Transfer from the Flavin Adenine Dinucleotide Chromophore to a Conserved Tyrosine?. Photochem. Photobiol. 2003, 78, 290–297. [DOI] [PubMed] [Google Scholar]
- Kraft B. J.; Masuda S.; Kikuchi J.; Dragnea V.; Tollin G.; Zaleski J. M.; Bauer C. E. Spectroscopic and Mutational Analysis of the Blue-Light Photoreceptor AppA: A Novel Photocycle Involving Flavin Stacking with an Aromatic Amino Acid. Biochemistry 2003, 42, 6726–6734. [DOI] [PubMed] [Google Scholar]
- Okajima K.; Fukushima Y.; Suzuki H.; Kita A.; Ochiai Y.; Katayama M.; Shibata Y.; Kunio M.; Noguchi T.; Itoh S.; et al. Fate Determination of the Flavin Photoreceptions in the Cyanobacterial Blue Light Receptor TePixD (Tll0078). J. Mol. Biol. 2006, 363, 10–18. [DOI] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar]
- MacKerell A. D. Jr.; Bashford D.; Bellott M.; Dunbrack J. R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Bochevarov A. D.; Harder E.; Hughes T. F.; Greenwood J. R.; Braden D. A.; Philipp D. M.; Rinaldo D.; Halls M. D.; Zhang J.; Friesner R. A. Jaguar: A High-Performance Quantum Chemistry Software Program with Strengths in Life and Materials Sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar]
- MacKerell A. D.; Banavali N.; Foloppe N. Development and Current Status of the CHARMM Force Field for Nucleic Acids. Biopolymers 2000, 56, 257–265. [DOI] [PubMed] [Google Scholar]
- Hirata S.; Head-Gordon M. Time-Dependent Density Functional Theory Within the Tamm-Dancoff Approximation. Chem. Phys. Lett. 1999, 314, 291–299. [Google Scholar]
- Madjet M. E.-A.; Abdurahman A.; Renger T. Intermolecular Coulomb Couplings from Ab Initio Electrostatic Potentials: Application to Optical Transitions of Strongly Coupled Pigments in Photosynthetic Antennae and Reaction Centers. J. Phys. Chem. B 2006, 110, 17268–17281. [DOI] [PubMed] [Google Scholar]
- Madjet M. E.-A.; Müh F.; Renger T. Deciphering the Influence of Short-Range Electronic Couplings on Optical Properties of Molecular Dimers: Application to “Special Pairs” in Photosynthesis. J. Phys. Chem. B 2009, 113, 12603–12614. [DOI] [PubMed] [Google Scholar]
- Shao Y.; Molnar L. F.; Jung Y.; Kussmann J.; Ochsenfeld C.; Brown S. T.; Gilbert A. T. B.; Slipchenko L. V.; Levchenko S. V.; O’Neill D. P.; et al. Advances in Methods and Algorithms in a Modern Quantum Chemistry Program Package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [DOI] [PubMed] [Google Scholar]
- Stanley R. J.; Jang H. Electronic Structure Measurements of Oxidized Flavins and Flavin Complexes Using Stark-Effect Spectroscopy. J. Phys. Chem. A 1999, 103, 8976–8984. [Google Scholar]
- Stanley R. J.; Siddiqui M. S. A Stark Spectroscopic Study of N(3)-Methyl, N(10)-Isobutyl-7,8-Dimethylisoalloxazine in Nonpolar Low-Temperature Glasses: Experiment and Comparison with Calculations. J. Phys. Chem. A 2001, 105, 11001–11008. [Google Scholar]
- Markley J. L.; Bax A.; Arata Y.; Hilbers C. W.; Kaptein R.; Sykes B. D.; Wright P. E.; Wüthrich K. Recommendations for the Presentation of NMR Structures of Proteins and Nucleic Acids (IUPAC Recommendations 1998). Pure Appl. Chem. 1998, 70, 117–142. [DOI] [PubMed] [Google Scholar]
- Masuda S.; Hasegawa K.; Ono T.-A. Light-Induced Structural Changes of Apoprotein and Chromophore in the Sensor of Blue Light Using FAD (BLUF) Domain of AppA for a Signaling State. Biochemistry 2005, 44, 1215–1224. [DOI] [PubMed] [Google Scholar]
- Hasegawa K.; Masuda S.; Ono T.-A. Structural Intermediate in the Photocycle of a BLUF (Sensor of Blue Light Using FAD) Protein Slr1694 in a Cyanobacterium Synechocystis sp. PCC6803. Biochemistry 2004, 43, 14979–14986. [DOI] [PubMed] [Google Scholar]
- Bonetti C.; Mathes T.; van Stokkum I. H. M.; Mullen K. M.; Groot M.-L.; van Grondelle R.; Hegemann P.; Kennis J. T. M. Hydrogen Bond Switching Among Flavin and Amino Acid Side Chains in the BLUF Photoreceptor Observed by Ultrafast Infrared Spectroscopy. Biophys. J. 2008, 95, 4790–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T.; Watanabe A.; Iseki M.; Watanabe M.; Kandori H. Strong Donation of the Hydrogen Bond of Tyrosine During Photoactivation of the BLUF Domain. J. Phys. Chem. Lett. 2011, 2, 1015–1019. [Google Scholar]
- Yuan H.; Anderson S.; Masuda S.; Dragnea V.; Moffat K.; Bauer C. Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling. Biochemistry 2006, 45, 12687–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragnea V.; Arunkumar A. I.; Lee C. W.; Giedroc D. P.; Bauer C. E. A Q63E Rhodobacter sphaeroides AppA BLUF Domain Mutant Is Locked in a Pseudo-Light-Excited Signaling State. Biochemistry 2010, 49, 10682–10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


